Abstract
Signal transducer and activator of transcription 3 (STAT3) is involved in cell proliferation, differentiation, and cell survival during immune responses, hematopoiesis, neurogenesis, and other biological processes. STAT3 activity is regulated by a variety of mechanisms, including phosphorylation and nuclear translocation. To clarify the molecular mechanisms underlying the regulation of STAT3 activity, we performed yeast two-hybrid screening. We identified ARL3 (ADP-ribosylation factor-like 3) as a novel STAT3-binding partner. ARL3 recognizes the DNA-binding domain as well as the C-terminal region of STAT3 in vivo, and their binding was the strongest when both proteins were activated. Importantly, small interfering RNA-mediated reduction of endogenous ARL3 expression decreased IL-6-induced tyrosine phosphorylation, nuclear accumulation, and transcriptional activity of STAT3. These results indicate that ARL3 interacts with STAT3 and regulates the transcriptional activation of STAT3 by influencing its nuclear accumulation of STAT3.
Keywords: gene transcription, nuclear translocation, protein phosphorylation, signal transduction, STAT3
Introduction
Upon stimulation with cytokines and growth factors, signal transducer and activator of transcription 3 (STAT3) is selectively activated and phosphorylated at various phosphorylation sites, particularly at Tyr-705 and Ser-727 (1–3). The activated STAT3 translocates into the nucleus and binds to specific promoter sequences to exert transcriptional regulation on various genes (1–3). Thus, STAT3 is a key molecule in the regulation of a variety of cellular processes, including proliferation, survival, and migration (4, 5). A number of experimental models have revealed that STAT3 is essential for embryo development and stem cell pluripotency (6–8). Furthermore, it is an important modulator of inflammatory and malignant diseases (7, 9). Indeed, constitutive or dysregulated expression of STAT3 has been found in cancer cells and oncogene-transfected cells (4, 5, 9).
The expression and function of STAT3 are tightly regulated by multiple molecular mechanisms (10, 11). For example, cytoplasmic tyrosine phosphatases, such as Src homology 2-containing phosphatase 1 (SHP1), SHP2, and protein-tyrosine phosphatase 1B (PTP1B), remove STAT3 phosphorylation (10, 11). In addition, the expression and activity of STAT3 are regulated at both pre- and post-transcription, and STAT3 functions are also dependent on its intracellular localization (12). We have identified several STAT3-interacting molecules, such as death domain-associated protein (DAXX) (13), zipper-interacting protein kinase (ZIPK) (14), Krüppel-associated box-associated protein 1 (KAP1) (15), PDZ and LIM domain 2 (PDLIM2) (16), Y14 (17, 18), and signal-transducing adaptor protein-2 (STAP-2) (19, 20). DAXX negatively and ZIPK positively regulate STAT3-mediated transactivation (13, 14). Within the nucleus, KAP1 regulates the phosphorylation and transactivation of STAT3 by interacting with HDAC3 (15). PDLIM2, a nuclear E3 ligase for STAT3, terminates STAT3-mediated signaling (16). Y14 influences tyrosine phosphorylation and positively regulates cytokine-induced STAT3 transactivation (17, 18). STAP-2 interacts with and enhances STAT3 activity (19, 20).
We performed yeast two-hybrid screening with the C-terminal region of STAT3 as bait. As a result, we identified ADP-ribosylation factor (Arf)-like 3 (ARL3) as a new binding partner of STAT3. The ARF2 family, which is characterized by the capacity to bind guanosine triphosphate binding, is structurally grouped as ARFs, ARLs, and SAR (secretion-associated and Ras-related) (21). Members of this family are activated by GDP to GTP exchange, and their activity is terminated upon the hydrolysis of GTP. The ARF family shares regulation of the budding and formation of vesicles in the endocytic and exocytic pathways (21). However, ARLs are likely to have specific functions and biochemical properties, which are divergent from ARFs. Indeed, mice deficient for the Arl3 gene exhibit abnormal development of renal, hepatic, and pancreatic epithelial tubule structures, which are seen in polycystic kidney diseases (22). Although the biological actions of ARFs are thought to come from their specific interactions with a large number of effectors, such as coat complexes, adaptor proteins, and lipid-modifying enzymes, much less information is available for ARLs. Here, we present new evidence that ARL3 plays a critical role in the phosphorylation and nuclear retention of STAT3.
Experimental Procedures
Reagents and Antibodies
Recombinant human LIF was purchased from INTERGEN (Purchase, NY). Human recombinant IL-6 was a kind gift from Ajinomoto (Tokyo, Japan). Recombinant human G-CSF was kindly provided by Chugai Pharmaceutical Co. (Tokyo, Japan). Expression vectors for FLAG-tagged STAT1–6 and STAT3-C were kindly provided by J. N. Ihle (St. Jude Children's Research Hospital, Memphis, TN), J. F. Bromberg (Rockefeller University, New York), and N. Yokosawa (Sapporo Medical School, Sapporo, Japan). Epitope-tagged STAT3 and its mutants were previously described (15). Expression vectors for STAT3-F, STAT3-D, and STAT3-LUC were kindly provided by Dr. T. Hirano (Osaka University Medical School, Osaka, Japan) (15, 23). Expression vectors for binder of ADP-ribosylation factor-like 2 (BART) was described previously (24). Myc-tagged ARL3 and its mutants were generated by PCR and sequenced (primer sequences are available upon request). Anti-Myc, anti-GST, anti-STAT3, and anti-ARL3 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-FLAG and anti-HA antibodies were from Sigma. Anti-phospho-STAT3 Tyr-705 (pSTAT3 Tyr-705) and anti-phospho-STAT3 Ser-727 (pSTAT3 Ser-727) were purchased from Cell Signaling Technologies (Beverly, MA).
Yeast Two-hybrid Screen
Gal4-STAT3 was constructed by fusing the coding sequence for the C-terminal region (amino acids 483–748) of STAT3 in-frame to the Gal4 DNA-binding domain in the pGBKT7 vector (Clontech). Saccharomyces cerevisiae AH109 cells were transformed with pGal4-STAT3 and then mated with Y187 cells containing a pretransformed mouse 11-day embryo MATCHMAKER cDNA library (Clontech), and ∼2.6 × 106 colonies were screened as described previously (14). Plasmid DNAs derived from positive clones were extracted from the yeast and sequenced.
Cell Culture, Transfection, siRNA, Quantitative Real Time PCR (qPCR)
A human cervix carcinoma cell line (HeLa) and human embryonic kidney carcinoma cell line (293T) were maintained in DMEM containing 10% FCS. An interleukin (IL)-3-dependent murine pro-B cell line, BaF-G133, was maintained in RPMI 1640 medium supplemented with 10% FCS with 10% of WEHI-3B conditioned medium as a source of IL-3 (25, 26). ARL3-knockdown HeLa cell lines (cell lines 2–4 and 2–10) were established by transduction with pGPU6/GFP/Neo vector (Shanghai GenePharm, Shanghai, China) bearing short hairpin RNA (shRNA) targeting ARL3 (5′-GCAGCTTGCATCTGAAGACAT-3′) and then selected with G418 (1 mg/ml; Sigma) (27). Similarly, control shRNA (nonsilencing, 5′-TTCTCCGAACGTGTCACGT-3′)-transfected HeLa cell line (shCont) was also established. The 293T cells were transfected using a standard calcium precipitation protocol (28). siRNAs targeting ARL3 used in this study were as follows: si-human (hu) ARL3#1, 5′-GGGUCAGGAACUAGCGGAATT-3′; huARL3#2, 5′- CACCUACACAGGUUUCAATT-3′; si-mouse (mu) ARL3, 5′-GCAAGAAUGUCAACGCAAATT-3′. Control siRNA was obtained from Qiagen (nonsilencing; catalog 1022076). HeLa cells were plated on 24-well plates at 2–3 × 104 cells/well and incubated with an siRNA/Lipofectamine 2000 (Invitrogen) mixture at 37 °C for 8 h, followed by the addition of fresh medium containing 10% FCS. At 48 h after siRNA treatment, cells were collected and analyzed for Western blotting or qPCR. BaF-G133 cells were transfected using a Nucleofector (Amaxa Biosystems, Cologne, Germany). Cells were transfected with 200 pmol of siRNA in Nucleofector solution V using program X-001. Immediately following transfection, medium was added to the BaF-G133 cells, which were then plated in 6-well tissue culture plates and incubated overnight. Cells were harvested, and total RNAs were prepared by using TRI Reagent (Molecular Research Center, Cincinnati, OH). First-strand cDNA was synthesized from 1 μg of total RNA with ReverTra Ace (TOYOBO, Osaka, Japan). qPCR analysis of mRNA transcripts was carried out using a combination of a KAPA SYBR FAST qPCR master mix (KAPA Biosystems, Woburn, MA) with an Mx3005P real time PCR system (Stratagene, Santa Clara, CA). Primers used for qPCR were as follows: G3PDH, 5′-GAAATCCCATCACCATCTTCCAGG-3′ (sense), and 5′-CAGTAGAGGCAGGGATGATGTTC-3′ (antisense); huARL1, 5′-TGGGATTTAGGAGGACAGACAAGTAT-3′ (sense), and 5′-GGCCTGTTCCATGTCCTGTTTATT-3′ (antisense); huARL2, 5′-ACGCTGGGCTTCAACATCAA-3′ (sense), and 5′-CGGATGGCGTTAGAGGACAGT-3′ (antisense); huARL3, 5′-GGACAGAGGAAAATCAGACCATACT-3′ (sense), 5′-GTCGCGGATGGTATGCAGGT-3′ (antisense); huSOCS3, 5′-TCACCCACAGCAAGTTTCCCGC-3′ (sense), and 5′-GTTGACGGTCTTCCGACAGAGATGC-3′ (antisense); muArl1, 5′-CAGTTGTGACCGAGATCGAA-3′ (sense), and 5′-ATTCCATTGCCTCATCAAGG-3′ (antisense); muArl2, 5′-GGAGAAGACGTGGACACCAT-3′ (sense), and 5′-AGTAGGCTCTGCAGCTCTCG-3′ (antisense); muArl3, 5′-GACGGGTCAGGAACTAACGG-3′ (sense), and 5′-CAGTTCATGCCATCCTGGAC-3′ (antisense); muSOCS3, 5′-TTTTCTTTGCCACCCACGGA-3′ (sense), and 5′-CAGGAACTCCCGAATGGGTC-3′ (antisense); muBcl2l1, 5′-CCTTGGATCCAGGAGAACGG-3′ (sense), and 5′-CGACTGAAGAGTGAGCCCAG-3′ (antisense); Bcl2l11, 5′-CCGGAGATACGGATTGCACA-3′ (sense), and 5′-GCCTTCTCCATACCAGACGG-3′ (antisense); muCdkn1a, 5′-GCAGATCCACAGCGATATCCA-3′ (sense), and 5′- GACAACGGCACACTTTGCTC-3′ (antisense); and muCdkn1b, 5′-CAAACTCTGAGGACCGGCAT-3′ (sense), and 5′-CTTAATTCGGAGCTGTTTACGTCT-3′ (antisense).
Dual-Luciferase Assay
Luciferase activities of HeLa cells transfected with STAT-LUC, in which the α2-macroglobulin promoter drives expression of a luciferase (LUC) reporter gene, were measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer's instructions.
Immunoprecipitation, Immunoblotting, Pulldown, and Affinity Precipitation Assay
Immunoprecipitation and Western blotting were performed as described previously (17). The cells were harvested and lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 0.15 m NaCl, 1% Nonidet P-40, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 10 mg/ml each of aprotinin, pepstatin, and leupeptin). The immunoprecipitates from cell lysates were resolved on SDS-PAGE and transferred to an Immobilon filter (Millipore; Bedford, MA), which was then immunoblotted with antibodies. Pulldown assay using glutathione S-transferase (GST) fusion proteins was performed as described previously (29). Active GTP-bound forms of GTPases were detected by affinity precipitation assays, as described previously (29). GST fusion proteins of BART (GST-BART) were bacterially expressed and purified on glutathione-Sepharose. The 293T cells were stimulated with LIF (100 ng/ml) for the indicated periods. Cells were lysed with the above lysis buffer and incubated with 10 μg of purified GST or GST-BART and glutathione-Sepharose beads. The beads were washed three times with lysis buffer. The precipitates were analyzed by immunoblotting with antibodies.
GTPγS Treatment
The total cell lysates were treated with GTPγS (100 μm) (Sigma) in the presence of 1 mm EDTA at 30 °C for 15 min, terminating the reaction by addition of MgCl2 (65 mm), as described previously (30).
Assay for GTPase Cycle
HeLa cells in a 6-well plate were transfected with Myc-tagged ARL3 using Lipofectamine 2000. The cells were stimulated with LIF (100 ng/ml) or IL-6 (10 ng/ml) at 48 h post-transfection and then lysed in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, and 1 mm EDTA for 10 min on ice. After centrifugation, an aliquot of the supernatant was transferred to a well of a 96-well plate, and then the activities of GTPase cycle were measured using GTPase-Glo (Promega) according to the manufacturer's instructions.
Wound Healing Assay
Wound healing assay was performed as described previously (31). siRNA-transfected HeLa cells were cultured in 24-well tissue culture plates as a confluent monolayer, and an artificial wound was created by scraping with a yellow pipette tip. Cells migrating into open space were monitored microscopically, and the width of the wound was measured immediately and after 30 h at 10 randomly chosen points per well.
Cell Proliferation Assay
The numbers of viable BaF-G133 cells after the indicated treatments were measured using a WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) assay (cell counting kit-8; Wako Pure Chemicals) (26). Briefly, 10 μl of WST-8 solution was added to the cells in each well and incubated for 2 h. The absorbances were measured at a test wavelength of 450 nm and a reference wavelength of 595 nm using a microplate reader (Bio-Rad).
Indirect Immunofluorescence Microscopy
HeLa cells (5 × 104) seeded on a glass plate were fixed with 4% paraformaldehyde and reacted with respective antibodies. The cells were then reacted with FITC-conjugated anti-rabbit IgG or rhodamine-conjugated anti-mouse IgG (Chemicon) and observed under a confocal laser fluorescent microscope (32). Images were obtained by using a Zeiss LSM 510 laser scanning microscope with an Apochromat ×63/1.4 oil immersion objective and ×4 zoom. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Wako, Osaka, Japan). Approximately 100 cells were classified according to FITC signals in the cytoplasm or nucleus. Results are representative of three independent experiments, in which 100 cells were counted.
Statistical Methods
The significance of differences between group means was determined by Student's t test.
Results
Molecular Interactions between STAT3 and ARL3
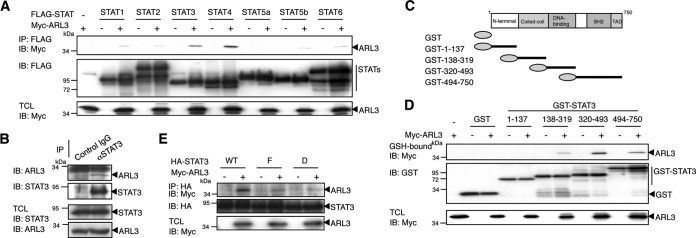
We performed a yeast two-hybrid screen of a mouse embryo cDNA library using the C-terminal region of STAT3 (amino acids 483–748) as bait. We screened about 2.6 × 106 transformants and identified several positive clones. Sequence analysis revealed that one of them encoded the entire ARL3 protein (amino acids 1–182). We first examined whether ARL3 binds to STAT3 and/or other STATs in mammalian cells. 293T cells were transfected with a series of FLAG-tagged STAT expression vectors together with Myc-tagged ARL3. Western blot analysis of anti-FLAG antibody immunoprecipitates revealed that ARL3 interacted strongly with STAT3 and STAT4 and weakly with STAT1, STAT2, STAT5a, STAT5b, and STAT6 (Fig. 1A). ARL3 is a ubiquitously expressed GTP-binding protein in the ARF family (33). Although STAT3 and STAT4 are more closely related to each other than to any other STATs, STAT4 expression is restricted to the testis, thymus, and spleen, whereas STAT3 is ubiquitously expressed (34). These results suggested that the STAT3-ARL3 interaction may have a wider range of biological functions. Therefore, we focused on the functional association between ARL3 and STAT3. To confirm that endogenous ARL3 interacts with STAT3 in vivo, co-immunoprecipitation experiments were performed using cell extracts obtained from 293T cells, in which both proteins could be detected by specific antibodies. The immunoprecipitate with anti-STAT3 antibody contained endogenous ARL3 protein, indicating that the binding of ARL3 to STAT3 occurs at physiological expression levels (Fig. 1B).
FIGURE 1.
ARL3 physically interacts with STAT3 in vivo. A, 293T cells (5 × 106) were transfected with Myc-tagged ARL3 (5 μg) and/or FLAG-tagged STAT1–6 (10 μg). At 48 h after transfection, the cells were lysed and immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with anti-FLAG or anti-Myc antibodies. An aliquot of total cell lysates (TCL) (1%) was blotted with anti-Myc antibody. B, HeLa cells (2 × 107) were lysed and immunoprecipitated with control or anti-STAT3 antibody and then immunoblotted with anti-STAT3 or anti-ARL3 antibodies. TCL (1%) was blotted with anti-ARL3 and anti-STAT3 antibodies. C, domain structure of STAT3 and its GST-fused mutant fragments are shown schematically. D, 293T cells (1 × 107) were transfected with GST, GST-STAT3(1–137), GST-STAT3(138–319), GST-STAT3(320–493), and GST-STAT3(494–750) (10 μg), together with or without Myc-tagged ARL3 (10 μg). At 48 h after transfection, the cells were lysed and pulled down with glutathione-Sepharose beads, followed by immunoblotting with anti-Myc or anti-GST antibodies. TCL (1%) was blotted with anti-Myc antibody. E, 293T cells (1 × 107) were transfected with HA-STAT3 WT or STAT3-F or STAT3-D (10 μg) with or without Myc-ARL3 (10 μg). At 48 h after transfection, the cells were lysed and immunoprecipitated with anti-HA and then blotted with anti-HA or anti-Myc antibodies. TCL (1%) was blotted with anti-HA or anti-Myc antibodies.
To delineate the regions of STAT3 involved in the ARL3-STAT3 interaction, various deletion constructs of GST-fused STAT3 (Fig. 1C) were subjected to pulldown assays in 293T cells. As shown in Fig. 1D, the DNA-binding domain (amino acids 320–493) of STAT3 interacted most strongly with ARL3, and weak associations of ARL3 with the coiled-coil (amino acids 138–319) and the C-terminal (amino acids 494–750) regions of STAT3 were also detected.
We further confirmed the involvement of the STAT3 activation state in these interactions. Expression vectors encoding Myc-tagged ARL3 and HA-tagged STAT3 WT (wild type), STAT3-F, and STAT3-D mutants were transiently transfected into 293T cells. STAT3-F has a phenylalanine substitution for the tyrosine residue at 705 and fails to be phosphorylated at Tyr-705. STAT3-D has glutamic acid substitutions for alanines at 434 and 435 and acts as a dominant-negative form of STAT3 because of the mutated DNA-binding domain (23). The transfected 293T cells were lysed and subjected to immunoprecipitation with anti-HA antibody. The immunoprecipitates were then used in Western blot analysis with an anti-Myc antibody. As shown in Fig. 1E, STAT3-F and -D showed much less binding potential to ARL3 than STAT3 WT, suggesting that activation state and an intact DNA-binding domain of STAT3 are required for full interaction between ARL3 and STAT3.
LIF Activates ARL3 and the Active Form of ARL3 Functionally Interacts with STAT3
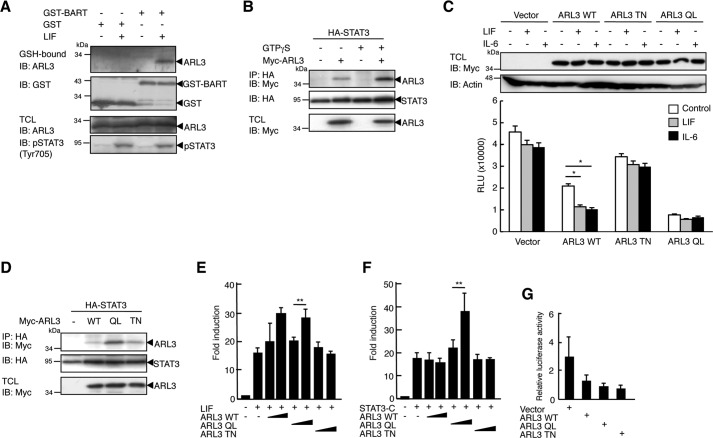
The ARL family of GTPases is known to be activated by GDP to GTP exchange. BART interacts with GTP-bound, but not GDP-bound, ARL3 (35, 36). We investigated whether LIF stimulation generates the GTP-bound form of ARL3 using GST-BART. As shown in Fig. 2A, GST-BART-bound ARL3 was detected in LIF-stimulated, but not unstimulated, 293T cell lysates. Thus, LIF stimulation activates ARL3 and generates a GTP-bound form of ARL3. We then tested whether the activation state of ARL3 is required for the physical interaction between ARL3 and STAT3. To produce GTP-bound ARL3, we employed the non-hydrolyzable GTP analog GTPγS. As shown in Fig. 2B, STAT3 bound much more strongly to ARL3 in the GTPγS-treated 293T cell lysates. To further examine this issue, we generated ARL3 mutants Q71L (QL) and T31N (TN) (the numbers refer to the amino acids in ARL3), corresponding to the classical Ras mutations Q61L and T17N (37), respectively. Oncogenic Ras Q61L is a dominant activating mutation because of the decreased ability of Ras GTPase-activating proteins to stimulate the hydrolysis of bound GTP, resulting in increased levels of GTP-bound Ras in cells. Ras T17N acts in a dominant-negative manner by sequestering the Ras exchange factor Sos, thereby blocking activation of endogenous Ras. We also assessed the activities of GTPases, GTPase-activating proteins and guanine nucleotide-exchange factors, which are components of the GTPase cycle in these ARL3 mutants, using a GTPase-Glo assay. This assay assesses the activities of GTPases by detecting the amount of GTP remaining after GTP hydrolysis in a GTPase reaction. In this system, the remaining GTP is converted into ATP and then detected as a luminescent signal. Therefore, activation of GTPase activity is observed by reduction of luminescence. These activities in cell lysates of ARL3 WT-transfected HeLa cells were enhanced compared with those in cell lysates of empty vector-transfected HeLa cells (Fig. 2C). Importantly, LIF or IL-6 treatment of ARL3 WT-transfected HeLa cells resulted in a significant enhancement of these activities. Enhancement of these activities was not observed in cell lysates of ARL3 TN-transfected HeLa cells, whereas further enhanced activities were observed in cell lysates of ARL3 QL-transfected HeLa cells when compared with those in cell lysates of ARL3 WT-transfected HeLa cells. However, a significant effect was not observed in these cells by LIF or IL-6 treatment, and further investigation will be required to clarify this issue. As shown in Fig. 2D, a weak interaction of STAT3 with ARL3 WT and TN was observed, whereas STAT3 interacted strongly with ARL3 QL in 293T cells. Therefore, LIF stimulation induces GTP-bound ARL3, which shows strong binding to STAT3.
FIGURE 2.
LIF stimulation activates ALR3 and enhances ARL3-STAT3 interaction. A, 293T cells (1 × 107) were treated with LIF (100 ng) for 30 min. Cells were lysed and mixed with GST or GST-BART protein (10 μg). The mixture was then pulled down with glutathione-Sepharose beads, followed by immunoblotting (IB) with anti-ARL3 or anti-GST antibodies. An aliquot of TCL (1%) was blotted with anti-ARL3 antibody or anti-pSTAT3 antibody. B, HeLa cells (1 × 107) were transfected with HA-STAT3 WT (10 μg) with or without Myc-ARL3 (10 μg). At 48 h after transfection, the cells were lysed and mixed with GTPγS. The immunoprecipitate (IP) with anti-Myc was then blotted with anti-HA or anti-Myc antibodies. TCL (1%) was blotted with anti-HA or anti-Myc antibody. C, HeLa cells in a 6-well plate were transfected with empty vector, Myc-tagged ARL3 WT, ARL3 QL, or TN (2 μg). The cells were stimulated with LIF (100 ng/ml) for 30 min or IL-6 (10 ng/ml) for 60 min. The activities of the GTPase cycle were calculated from luciferase activity of the lysate. *, p < 0.05. D, 293T cells (1 × 107) were transfected with HA-STAT3 WT (WT) with or without Myc-ARL3 WT, QL, or TN (10 μg). 48 h after transfection, the cells were lysed and immunoprecipitated with anti-Myc, followed by immunoblotting with anti-HA or anti-Myc antibodies. TCL (1%) was blotted with anti-Myc antibody. E, 293T cells in a 24-well plate were transfected with STAT3-LUC (100 ng) with or without ARL3 WT, ARL3 QL or ALR3 TN (100 ng). At 36 h after transfection, cells were treated with LIF (100 ng/ml) for an additional 12 h. The cells were harvested and assayed for luciferase activity. The results are indicated as fold induction of luciferase activity from triplicate experiments, and the error bars represent S.D. F, 293T cells in a 24-well plate were transfected with STAT3-LUC (100 ng) with or without STAT3-C and ARL3 WT, ARL3 QL, or ALR3 TN (100 ng). At 48 h after transfection, cells were harvested and assayed for luciferase activity as described above. **, p < 0.01. G, 293T cells in a 24-well plate were transfected with STAT3-LUC (100 ng) with empty vector, ARL3 WT, ARL3 QL, or ALR3 TN (100 ng). Relative luciferase activity represents the ratio of luciferase to β-galactosidase activity. Shown is a representative experiment, which was repeated at least three times with similar results.
ARL3 Enhances the Transcriptional Activity of STAT3 after IL-6 Stimulation
We examined whether activation of ARL3 influences STAT3-mediated transactivation. As shown in Fig. 2E, ectopic expression of ARL3 WT and QL, but not TN, enhanced LIF-induced STAT3-LUC activation in 293T cells, indicating that ARL3 WT and QL enhance LIF-induced STAT3 transactivation. To further examine a direct activation of STAT3 by ARL3, we used a constitutively active form of STAT3, STAT3-C. Ectopic expression of ARL3 QL, but not WT or TN, enhanced STAT3-LUC activation by STAT3-C, even in the absence of LIF stimulation (Fig. 2F). These results indicate that activated ARL3 positively regulates STAT3-mediated transactivation through a direct interaction. We also tested the effects of these mutants on STAT3-LUC activation without LIF stimulation. ARL3 WT, QL, or TN did not show any significant effect on STAT3-LUC activation without stimulation (Fig. 2G).
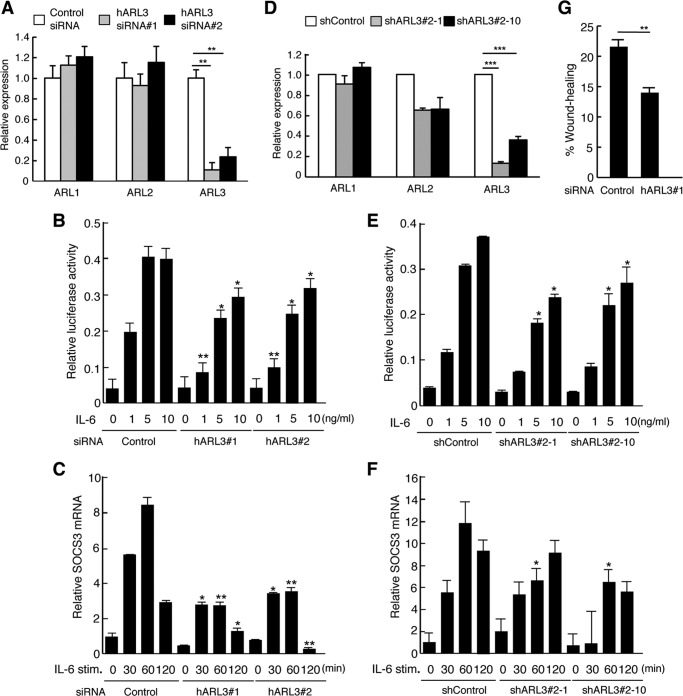
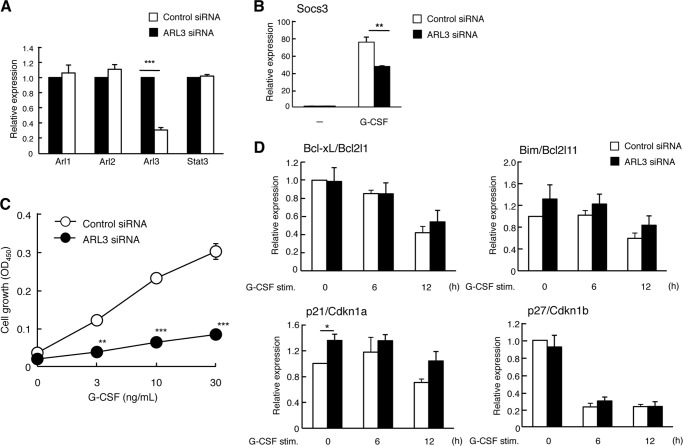
To further evaluate the physiological role of ARL3 on STAT3-mediated transactivation, we examined whether a reduction of endogenous ARL3 expression affects STAT3-mediated transactivation and gene expression using ARL3-specific siRNAs. Specific knockdown of ARL3 but not of ARL1 or ARL2 was observed, but this did not alter protein levels of STAT3 (Figs. 3, A and D, and 5, A and C). As shown in Fig. 3B, IL-6/STAT3-mediated transactivation was reduced in ARL3 siRNA-treated HeLa cells. Similarly, IL-6/STAT3-mediated SOCS3 mRNA levels were also reduced in ARL3 siRNA-treated HeLa cells (Fig. 3C). Furthermore, reduced STAT3-mediated transactivation and gene expression after IL-6 stimulation was observed in ARL3 knockdown cells (Fig. 3, E and F). These results indicate that ARL3 plays a role in the regulation of STAT3 transactivation in vivo. STAT3 has been shown to have a critical role in regulating cell migration (38, 39). Loss of STAT3 has exhibited impaired cell migration of keratinocytes or mouse embryonic fibroblasts in in vitro wound healing assay (38, 39). We tested the effect of ARL3 knockdown on cell migration in HeLa cells. As shown in Fig. 3G, ARL3 siRNA-treated HeLa cells demonstrated a significantly reduced capacity of cell migration. Therefore, ARL3 also plays a role in cell migration.
FIGURE 3.
ARL3 enhances the transcriptional activity of STAT3 after IL-6-stimulation. A, HeLa cells in a 24-well plate were transfected with control, huARL3#1, or huARL3#2 siRNAs (20 pmol). Total RNA was extracted at 48 h post-transfection, and target mRNA levels were quantified by qPCR. Data represent the levels of target mRNA normalized to that of a G3PDH internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. **, p < 0.01. B, HeLa cells in a 24-well plate were transfected with control, huARL3#1, or huARL3#2 siRNAs (20 pmol), and then transfected with STAT3-LUC (100 ng). At 24 h after transfection, cells were left untreated or treated with IL-6 for an additional 12 h. The cells were harvested and assayed for the luciferase activity. Relative luciferase activity represents the ratio of Firefly and Renilla luciferase activity for each sample. Data are representative of at least three independent experiments. Error bars represent the S.D. *, p < 0.05; **, p < 0.01. C, HeLa cells in a 24-well plate were transfected with control, huARL3#1, or huARL3#2 siRNAs (20 pmol). At 48 h after transfection, cells were treated with IL-6 (10 ng/ml) for the indicated periods. Total RNA samples isolated from these cells were also quantified by qPCR analysis. Data represent the levels of SOCS3 mRNA normalized to that of a G3PDH internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. *, p < 0.05; **, p < 0.01. D, total RNAs from HeLa shControl, shARL3#2-1, or shARL3#2-10 cells were extracted at 48 h post-transfection, and target mRNA levels were quantified by qPCR. Data represent the levels of target mRNA normalized to that of a G3PDH internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. ***, p < 0.001. E, HeLa shControl, shARL3#2-1, or shARL3#2-10 cells in a 24-well plate were transfected with STAT3-LUC (100 ng). At 24 h after transfection, cells were left untreated or treated with IL-6 for an additional 12 h. The cells were harvested and assayed for the luciferase activity. Relative luciferase activity represents the ratio of Firefly and Renilla luciferase activity for each sample. Data are representative of at least three independent experiments. Error bars represent the S.D. *, p < 0.05. F, HeLa shControl, shARL3#2-1 or shARL3#2-10 cells in a 24-well plate were treated with IL-6 (10 ng/ml) for the indicated periods. Total RNA samples isolated from these cells were also quantified by qPCR analysis. Data represent the levels of SOCS3 mRNA normalized to that of a glyceraldehyde-3-dehydrogenase (G3PDH) internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. *, p < 0.05. G, HeLa cells in a 24-well plate were transfected with control or huARL3#1 siRNAs (20 pmol). 48 h after siRNA-transfected cells were plated, and an artificial wound was created in the cell monolayer. Cells migrating into the open space from the wound edge were quantified. Data are representative of four independent experiments. Error bars represent the S.D. **, p < 0.01.
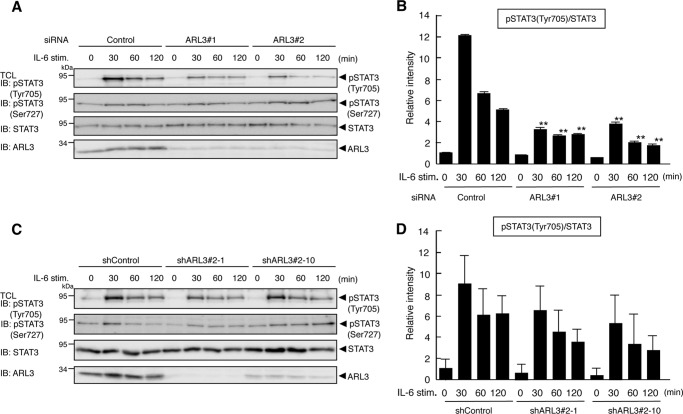
FIGURE 5.
ARL3 is involved in phosphorylation of STAT3. A, HeLa cells in a 24-well plate were transfected with control, huARL3#1, or huARL3#2 siRNAs (20 pmol). 48 h after transfection, the cells were treated with IL-6 (10 ng/ml) for the indicated periods. Then the cells were lysed, and an aliquot of total cell lysates (1%) was blotted with anti-pSTAT3 (Tyr-705), anti-pSTAT3 (Ser-727), anti-STAT3, or anti-ARL3 antibody. IB, immunoblot. B, densitometric quantification of the above results was also shown. Relative intensity of pSTAT3 (Tyr-705) was normalized to the STAT3 protein of the same sample. The results are representative of three independent experiments, and the error bars represent the S.D. **, p < 0.01. C, HeLa shControl, shARL3#2-1, or shARL3#2-10 cells in a 24-well plate were treated with IL-6 (10 ng/ml) for the indicated periods. Then the cells were lysed, and an aliquot of total cell lysates (1%) was blotted with anti-pSTAT3 (Tyr-705), anti-pSTAT3 (Ser-727), anti-STAT3, or anti-ARL3 antibody. D, densitometric quantification of the above results was also shown. Relative intensity of pSTAT3 (Tyr-705) was normalized to the STAT3 protein of the same sample. The results are representative of three independent experiments, and the error bars represent the S.D.
We further investigated this effect in a murine hematopoietic cell line BaF-G133. G-CSF treatment of BaF-G133 cells induces dimerization of GCSFR/gp130 chimeric receptors causing activation of STAT3 (25). As shown in Fig. 4, A and B, G-CSF/STAT3-mediated SOCS3 expression was also decreased in ARL3 siRNA-treated BaF-G133 cells. BaF-G133 cells have also been used to measure gp130/STAT3-mediated cell growth by G-CSF treatment (25, 26, 40). We therefore tested the effect of reduced ARL3 expression on G-CSF/gp130/STAT3-mediated BaF-G133 cell growth. As shown in Fig. 4C, G-CSF/STAT3-mediated cell growth was decreased in ARL3 siRNA-treated BaF-G133 cells. In BaF-G133 cells, Bcl-xL expression is reported to be involved in gp130/STAT3-mediated cell survival signals (25). We examined the effect of reduced ARL3 expression on mRNA levels of Bcl-xL and proapoptotic Bim. Bcl-xL expression was not altered, but Bim expression was slightly enhanced in ARL3 siRNA-treated BaF-G133 cells. In these cells, STAT3 has been also been shown to regulate cell cycle progression by down-regulation of cyclin-dependent kinase inhibitors, such as p21 and p27 (40). Thus, we examined the effect of reduced ARL3 expression on mRNA levels of p21 and p27. p21 but not p27 expression was slightly enhanced in ARL3 siRNA-treated BaF-G133 cells. These results indicate that up-regulation of Bim and p21 may be involved in suppression of G-CSF/gp130/STAT3-mediated cell growth in ARL3 siRNA-treated BaF-G133 cells, although further detailed analyses are required to clarify this issue.
FIGURE 4.
ARL3 regulates the STAT3-mediated cell growth in BaF-G133 cells. A, BaF-G133 cells were transfected with control and muARL3 siRNAs (20 pmol). Total RNA was extracted at 48 h post-transfection, and target mRNA levels were quantified by qPCR. Data represent the levels of target mRNA normalized to that of a G3PDH internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. ***, p < 0.001. B, BaF-G133 cells were transfected with control and muARL3 siRNAs (20 pmol). 48 h after transfection, the cells were left untreated or treated with G-CSF (50 ng/ml) for 60 min. Total RNA was then extracted, and SOCS3 and mRNA levels were quantified by qPCR. Data represent the levels of SOCS3 mRNA normalized to that of a G3PDH internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. **, p < 0.01; ***, p < 0.001. C, BaF-G133 cells were transfected with control and muARL3 siRNAs (20 pmol). 24 h after transfection, siRNA-transfected BaF-G133 cells were harvested and seeded into 96-well plates (1 × 104/well). Cells were left untreated or were treated with G-CSF (3, 10, 30 ng/ml). At 48 h, viable cells were evaluated by a WST-8 assay. Shown is a representative experiment, which was repeated at least three times with similar results, and the error bars represent the S.D. **, p < 0.01; ***, p < 0.001. D, BaF-G133 cells were transfected with control and muARL3 siRNAs (20 pmol). 48 h after transfection, the cells were left untreated or treated with G-CSF (50 ng/ml) for the indicated periods. Total RNA was then extracted, and target mRNA levels were quantified by qPCR. Data represent the levels of target mRNA normalized to that of a G3PDH internal control and are expressed relative to the value of control siRNA-treated samples. The results are representative of three independent experiments, and the error bars represent the S.D. *, p < 0.05.
ARL3 Is Involved in Phosphorylation and Nuclear Accumulation of STAT3
We examined whether reduction of ARL3 expression affects phosphorylation of STAT3, which is related to its activation. As shown in Fig. 5, A and B, IL-6-induced tyrosine phosphorylation (Tyr-705), but not serine phosphorylation (Ser-727) of STAT3, was decreased by a reduction of ARL3 expression in HeLa cells, suggesting that ARL3 plays a role in the enhancement of the phosphorylation state of STAT3. Similarly, reduced STAT3 tyrosine phosphorylation after IL-6 stimulation was observed in ARL3 knockdown cells (Fig. 5, C and D).
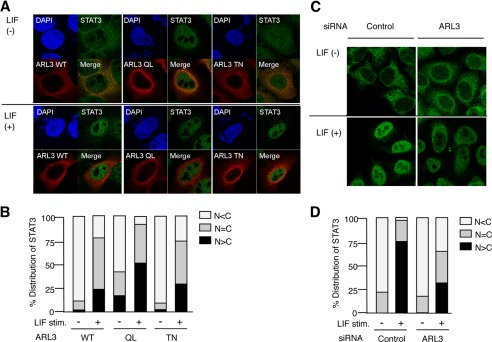
To further understand the molecular mechanisms underlying the regulation of STAT3 activities by ARL3, we analyzed the effect of overexpression of ARL3 on the intracellular localization of STAT3 using confocal microscopy. Interestingly, ARL3 QL exhibited strong perinuclear localization, but ARL TN was likely to diffuse in the cytoplasm even in the absence of LIF stimulation. Furthermore, when ARL3 QL, but not WT and TN, was expressed in HeLa cells, STAT3 accumulated in the nucleus, even in the absence of LIF stimulation (Fig. 6, A and B). Moreover, transfection of HeLa cells with ARL3 siRNA suppressed LIF-induced nuclear accumulation of STAT3 (Fig. 6, C and D). These results indicate that ARL3 is involved in the accumulation of STAT3 in the nucleus.
FIGURE 6.
ARL3 enhances nuclear accumulation of STAT3. A, HeLa cells in a six-well plate were transfected with Myc-ARL3 WT, QL, or TN (500 ng). 48 h after transfection, the cells were left untreated or treated with LIF (100 ng/ml) for 60 min. The cells were fixed and stained with anti-Myc or anti-STAT3 antibodies and then rhodamine-conjugated anti-mouse IgG (red) or FITC-conjugated anti-rabbit Ab (green). The same slide was also stained with DAPI for the nuclei staining. Original magnification ×600. B, quantitative analysis of the subcellular localization of STAT3 in A. Approximately 100 cells were classified according to FITC signals in the cytoplasm or nucleus. Results are representative of three independent experiments, in which 100 cells were counted. C, HeLa cells in a six-well plate were transfected with control or ARL3 siRNA (100 pmol). At 48 h after transfection, the cells were left untreated or treated with LIF (100 ng/ml) for 60 min. The cells were fixed and stained with anti-STAT3 antibody and then FITC-conjugated anti-rabbit antibody (green). D, quantitative analysis of the subcellular localization of STAT3 in C. Approximately 100 cells were classified according to FITC signals in the cytoplasm or nucleus. Results are representative of three independent experiments, in which 100 cells were counted.
Discussion
We have demonstrated that ARL3 is a new STAT3-binding partner and that endogenous ARL3 enhances STAT3-mediated transactivation by influencing tyrosine phosphorylation and nuclear accumulation of STAT3. The interaction between STAT3 and a classical small GTPase, Rac1, has been reported previously (41); however, the biological significance of this interaction for STAT3 function was not fully understood. In our previous study, we reported BART as a novel STAT3-binding partner critical for the nuclear retention of STAT3 (24). Although BART was first reported to be an effector for an ARL (35), BART could also recognize ARL3 (36). Therefore, our findings further emphasize the involvement of the ARL family of GTPases on the regulation of STAT3 activation.
The strong interaction between STAT3 and ARL3 was detected only when 293T and HeLa cells were stimulated with LIF. Of importance, STAT3-F (a mutant lacking Tyr-705 phosphorylation) bound weakly to ARL3 as compared with wild-type STAT3. In addition, STAT3 bound strongly to dominant-active ARL3 QL and weakly to dominant-negative ARL3 TN. The treatment of cells with the non-hydrolyzable GTP analog GTPγS enhanced the interaction between STAT3 and ARL3. Taken together, STAT3 and ARL3 strongly interact when both are activated. ARL3 mainly recognized the DNA-binding domain of STAT3. BART, which is an effector of ARL and can bind to ARL3 (36), also recognizes the same domain of phosphorylated STAT3 (24). The two proteins seem to have similar mechanisms for the STAT3 recognition. Because both proteins are involved in the phosphorylation and the nuclear retention of STAT3, ARL3 and BART might work additively or synergistically on STAT3 function.
The transcriptional regulatory activity of STAT3 is in part dependent on its nuclear localization, although it also depends on STAT3 stability, STAT3 phosphorylation, and inhibitory molecules. In general, phosphorylated STAT3 dissociates from the receptor, forms homo- or heterodimers, and then translocates into the nucleus. However, recent work has indicated that STAT3 is continuously imported to the nucleus independently of tyrosine phosphorylation and that both phosphorylated and unphosphorylated STAT3 shuttle between nuclear and cytoplasmic compartments (12). Of note, the two states of STAT3 differently co-exist in the nucleus and regulate the expression of target genes. In addition, STAT3 phosphorylation transiently increases the nuclear accumulation, but this might be a result from retention by DNA binding or heterodimerization with other STATs. Our confocal microscopy clearly revealed that ARL3 enhanced the accumulation of phosphorylated STAT3 in the nucleus after LIF stimulation. Interestingly, ectopic expression of constantly active ARL QL induced the accumulation of unphosphorylated STAT3 into the nucleus, even in HeLa cells without LIF stimulation. Therefore, activated ARL3 is likely to be involved in STAT3 functions beyond cytokine signals. We also tested ARL3 siRNA knockdown on unphosphorylated STAT3-regulated genes, such as RANTES (regulated on activation normal T cell expressed and secreted) (42). However, we could not observe significant alteration on its expression by ARL3 siRNA knockdown because the expression was too low to evaluate (data not shown).
With regard to the mechanisms for the increase of STAT3 activity by activated ARL3, several interesting studies have been reported (36, 39, 43–45). Importin-α3 has been shown to be required for nuclear import of STAT3, independently of STAT3 tyrosine phosphorylation (43). Recent study has also demonstrated that nuclear import of STAT3 is dependent on the action of Ran GTPase and importin-β1, again independently of STAT3 tyrosine phosphorylation (44). Therefore, importin-α/importin-β1/Ran may be an important molecular mechanism by which STAT3 traffics to the nucleus, independently of STAT3 tyrosine phosphorylation. There might be a possibility that ARL3 influences the importin-α/importin-β1/Ran system, although ARL3 is involved in tyrosine phosphorylation of STAT3.
Importantly, ARL3 has been shown to play a role in microtubule-dependent processes (36). siRNA knockdown of ARL3 in HeLa cells caused changes in cell morphology, increased acetylation of α-tubulin, failure of cytokinesis, and an increased number of binucleated cells. Notably, STAT3 also associates with microtubules (39), and the interaction between STAT3 and microtubules promotes migration by competing with the binding of the microtubule-associated protein Stathmin (39). Furthermore, STAT3 is required for the stabilization of microtubules. Most interestingly, treatment of breast cancer cells that constitutively express active STAT3 with microtubule-targeting drugs inhibited tyrosine phosphorylation of STAT3 and STAT3-dependent gene expression (45). Importantly, overexpression of a dominant-active ARL3 QL exhibits a very strong perinuclear staining in the absence of LIF stimulation, suggesting that this cellular location may be due to microtubule perturbations in cells. Moreover, perinuclear ARL3QL may stimulate tyrosine phosphorylation via a microtubule-dependent process. At the present time, we do not have data concerning this issue. Thus, ARL3 may regulate tyrosine phosphorylation and transactivation of STAT3 in a microtubule-dependent manner, although further analysis will clarify this issue.
Here, we propose a novel scheme in which a STAT3-ARL3 complex serves as machinery for STAT3 tyrosine phosphorylation and nuclear retention. Clarifying each step of STAT3 regulation is important because STAT3 is a key player in the pathogenesis of diverse human diseases and is a prime target for novel therapies. Specific inhibitors of the STAT3-ARL3 pathway are good candidates for the treatment of STAT3-related human diseases.
Author Contributions
S. T., R. M., K. H., Y. K., T. O., O. I., and N. M. performed the experiments. S. K. and Y. S. analyzed the data. K. O. wrote the paper, and T. M. designed the experiments, supervised the project, and wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- ARF
- ADP-ribosylation factor
- LIF
- leukemia inhibitory factor
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- BART
- binder of ADP-ribosylation factor-like 2
- TCL
- total cell lysate
- qPCR
- quantitative PCR
- QL
- Q71L
- TN
- T31N
- ARL
- ADP-ribosylation factor-like
- G3PDH
- glyceraldehyde-3-dehydrogenase.
References
- 1. Darnell J. E., Jr. (1997) STATs and gene regulation. Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 2. Jove R. (2000) Preface: STAT signaling. Oncogene 19, 2466–2467 [DOI] [PubMed] [Google Scholar]
- 3. Levy D. E., and Darnell J. E. Jr. (2002) Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 4. Bromberg J., and Darnell J. E. Jr. (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19, 2468–2473 [DOI] [PubMed] [Google Scholar]
- 5. Levy D. E., and Lee C. K. (2002) What does Stat3 do? J. Clin. Invest. 109, 1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., and Akira S. (1997) Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 94, 3801–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akira S. (2000) Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19, 2607–2611 [DOI] [PubMed] [Google Scholar]
- 8. Okita K., and Yamanaka S. (2006) Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr. Stem Cell Res. 1, 103–111 [DOI] [PubMed] [Google Scholar]
- 9. Yu H., Pardoll D., and Jove R. (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shuai K., and Liu B. (2003) Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3, 900–911 [DOI] [PubMed] [Google Scholar]
- 11. Yasukawa H., Sasaki A., and Yoshimura A. (2000) Negative regulation of cytokine signaling pathways. Annu. Rev. Immunol. 18, 143–164 [DOI] [PubMed] [Google Scholar]
- 12. Reich N. C., and Liu L. (2006) Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6, 602–612 [DOI] [PubMed] [Google Scholar]
- 13. Muromoto R., Nakao K., Watanabe T., Sato N., Sekine Y., Sugiyama K., Oritani K., Shimoda K., and Matsuda T. (2006) Physical and functional interactions between Daxx and STAT3. Oncogene 25, 2131–2136 [DOI] [PubMed] [Google Scholar]
- 14. Sato N., Kawai T., Sugiyama K., Muromoto R., Imoto S., Sekine Y., Ishida M., Akira S., and Matsuda T. (2005) Physical and functional interactions between STAT3 and ZIP kinase. Int. Immunol. 17, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 15. Tsuruma R., Ohbayashi N., Kamitani S., Ikeda O., Sato N., Muromoto R., Sekine Y., Oritani K., and Matsuda T. (2008) Physical and functional interactions between STAT3 and KAP1. Oncogene 27, 3054–3059 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka T., Yamamoto Y., Muromoto R., Ikeda O., Sekine Y., Grusby M. J., Kaisho T., and Matsuda T. (2011) PDLIM2 inhibits T helper 17 cell development and granulomatous inflammation through degradation of STAT3. Sci. Signal. 4, ra85. [DOI] [PubMed] [Google Scholar]
- 17. Ohbayashi N., Taira N., Kawakami S., Togi S., Sato N., Ikeda O., Kamitani S., Muromoto R., Sekine Y., and Matsuda T. (2008) An RNA binding protein, Y14 interacts with and modulates STAT3 activation. Biochem. Biophys. Res. Commun. 372, 475–479 [DOI] [PubMed] [Google Scholar]
- 18. Muromoto R., Taira N., Ikeda O., Shiga K., Kamitani S., Togi S., Kawakami S., Sekine Y., Nanbo A., Oritani K., and Matsuda T. (2009) The exon-junction complex proteins, Y14 and MAGOH regulate STAT3 activation. Biochem. Biophys. Res. Commun. 382, 63–68 [DOI] [PubMed] [Google Scholar]
- 19. Minoguchi M., Minoguchi S., Aki D., Joo A., Yamamoto T., Yumioka T., Matsuda T., and Yoshimura A. (2003) STAP-2/BKS, an adaptor/docking protein, modulates STAT3 activation in acute-phase response through its YXXQ motif. J. Biol. Chem. 278, 11182–11189 [DOI] [PubMed] [Google Scholar]
- 20. Ikeda O., Sekine Y., Mizushima A., Nakasuji M., Miyasaka Y., Yamamoto C., Muromoto R., Nanbo A., Oritani K., Yoshimura A., and Matsuda T. (2010) Interactions of STAP-2 with Brk and STAT3 participate in cell growth of human breast cancer cells. J. Biol. Chem. 285, 38093–38103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kahn R. A., Volpicelli-Daley L., Bowzard B., Shrivastava-Ranjan P., Li Y., Zhou C., and Cunningham L. (2005) Arf family GTPases: roles in membrane traffic and microtubule dynamics. Biochem. Soc. Trans. 33, 1269–1272 [DOI] [PubMed] [Google Scholar]
- 22. Schrick J. J., Vogel P., Abuin A., Hampton B., and Rice D. S. (2006) ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am. J. Pathol. 168, 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakajima K., Yamanaka Y., Nakae K., Kojima H., Ichiba M., Kiuchi N., Kitaoka T., Fukada T., Hibi M., and Hirano T. (1996) A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 15, 3651–3658 [PMC free article] [PubMed] [Google Scholar]
- 24. Muromoto R., Sekine Y., Imoto S., Ikeda O., Okayama T., Sato N., and Matsuda T. (2008) BART is essential for nuclear retention of STAT3. Int. Immunol. 20, 395–403 [DOI] [PubMed] [Google Scholar]
- 25. Fukada T., Hibi M., Yamanaka Y., Takahashi-Tezuka M., Fujitani Y., Yamaguchi T., Nakajima K., and Hirano T. (1996) Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5, 449–460 [DOI] [PubMed] [Google Scholar]
- 26. Muromoto R., Kuroda M., Togi S., Sekine Y., Nanbo A., Shimoda K., Oritani K., and Matsuda T. (2010) Functional involvement of Daxx in gp130-mediated cell growth and survival in BaF3 cells. Eur. J. Immunol. 40, 3570–3580 [DOI] [PubMed] [Google Scholar]
- 27. Kamitani S., Togi S., Ikeda O., Nakasuji M., Sakauchi A., Sekine Y., Muromoto R., Oritani K., and Matsuda T. (2011) Krüppel-associated box-associated protein 1 negatively regulates TNF-α-induced NF-κB transcriptional activity by influencing the interactions among STAT3, p300, and NF-κB/p65. J. Immunol. 187, 2476–2483 [DOI] [PubMed] [Google Scholar]
- 28. Ikeda O., Sekine Y., Yasui T., Oritani K., Sugiyma K., Muromoto R., Ohbayashi N., Yoshimura A., and Matsuda T. (2008) STAP-2 negatively regulates both canonical and non-canonical NF-κB activation induced by Epstein-Barr virus-derived LMP1. Mol. Cell. Biol. 28, 5027–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sekine Y., Ikeda O., Tsuji S., Yamamoto C., Muromoto R., Nanbo A., Oritani K., Yoshimura A., and Matsuda T. (2009) Signal-transducing adaptor protein-2 regulates stromal cell-derived factor-1a-induced chemotaxis in T cells. J. Immunol. 183, 7966–7974 [DOI] [PubMed] [Google Scholar]
- 30. Katanaev V. L., and Wymann M. P. (1998) GTPγS-induced actin polymerisation in vitro: ATP- and phosphoinositide-independent signalling via Rho-family proteins and a plasma membrane-associated guanine nucleotide exchange factor. J. Cell Sci. 111, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 31. Sekine Y., Togi S., Muromoto R., Kon S., Kitai Y., Yoshimura A., Oritani K., and Matsuda T. (2015) STAP-2 protein expression in B16F10 melanoma cells positively regulates protein levels of tyrosinase, which determines organs to infiltrate in the body. J. Biol. Chem. 290, 17462–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muromoto R., Ishida M., Sugiyama K., Sekine Y., Oritani K., Shimoda K., and Matsuda T. (2006) Sumoylation of Daxx regulates IFN-induced growth suppression of B-lymphocytes and the hormone receptor-mediated transactivation. J. Immunol. 177, 1160–1170 [DOI] [PubMed] [Google Scholar]
- 33. Cavenagh M. M., Breiner M., Schurmann A., Rosenwald A. G., Terui T., Zhang C., Randazzo P. A., Adams M., Joost H. G., and Kahn R. A. (1994) ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J. Biol. Chem. 269, 18937–18942 [PubMed] [Google Scholar]
- 34. Zhong Z., Wen Z., and Darnell J. E. Jr. (1994) Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc. Natl. Acad. Sci. U.S.A. 91, 4806–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharer J. D., and Kahn R. A. (1999) The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J. Biol. Chem. 274, 27553–27561 [DOI] [PubMed] [Google Scholar]
- 36. Zhou C., Cunningham L., Marcus A. I., Li Y., and Kahn R. A. (2006) Arl2 and Arl3 Regulate different microtubule-dependent processes. Mol. Biol. Cell 17, 2476–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feig L. A., and Cooper G. M. (1988) Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol. Cell. Biol. 8, 2472–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sano S., Itami S., Takeda K., Tarutani M., Yamaguchi Y., Miura H., Yoshikawa K., Akira S., and Takeda J. (1999) Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ng D. C., Lin B. H., Lim C. P., Huang G., Zhang T., Poli V., and Cao X. (2006) Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 172, 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fukada T., Ohtani T., Yoshida Y., Shirogane T., Nishida K., Nakajima K., Hibi M., and Hirano T. (1998) STAT3 orchestrates contradictory signals in cytokine-induced G1 to S cell-cycle transition. EMBO J. 17, 6670–6677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simon A. R., Vikis H. G., Stewart S., Fanburg B. L., Cochran B. H., and Guan K. L. (2000) Regulation of STAT3 by direct binding to the Rac1 GTPase. Science 290, 144–147 [DOI] [PubMed] [Google Scholar]
- 42. Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., and Stark G. R. (2007) Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 21, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu L., McBride K. M., and Reich N. C. (2005) STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. U.S.A. 102, 8150–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cimica V., Chen H. C., Iyer J. K., and Reich N. C. (2011) Dynamics of the STAT3 transcription factor: nuclear import dependent on Ran and importin-β1. PLoS ONE 6, e20188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walker S. R., Chaudhury M., Nelson E. A., and Frank D. A. (2010) Microtubule-targeted chemotherapeutic agents inhibit signal transducer and activator of transcription 3 (STAT3) signaling. Mol. Pharmacol. 78, 903–908 [DOI] [PubMed] [Google Scholar]