Abstract
The voltage-gated Na+ (Nav) channel provides the basis for electrical excitability in the brain. This channel is regulated by a number of accessory proteins including fibroblast growth factor 14 (FGF14), a member of the intracellular FGF family. In addition to forming homodimers, FGF14 binds directly to the Nav1.6 channel C-tail, regulating channel gating and expression, properties that are required for intrinsic excitability in neurons. Seeking amino acid residues with unique roles at the protein-protein interaction interface (PPI) of FGF14·Nav1.6, we engineered model-guided mutations of FGF14 and validated their impact on the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer formation using a luciferase assay. Divergence was found in the β-9 sheet of FGF14 where an alanine (Ala) mutation of Val-160 impaired binding to Nav1.6 but had no effect on FGF14:FGF14 dimer formation. Additional analysis revealed also a key role of residues Lys-74/Ile-76 at the N-terminal of FGF14 in the FGF14·Nav1.6 complex and FGF14:FGF14 dimer formation. Using whole-cell patch clamp electrophysiology, we demonstrated that either the FGF14V160A or the FGF14K74A/I76A mutation was sufficient to abolish the FGF14-dependent regulation of peak transient Na+ currents and the voltage-dependent activation and steady-state inactivation of Nav1.6; but only V160A with a concomitant alanine mutation at Tyr-158 could impede FGF14-dependent modulation of the channel fast inactivation. Intrinsic fluorescence spectroscopy of purified proteins confirmed a stronger binding reduction of FGF14V160A to the Nav1.6 C-tail compared with FGF14K74A/I76A. Altogether these studies indicate that the β-9 sheet and the N terminus of FGF14 are well positioned targets for drug development of PPI-based allosteric modulators of Nav channels.
Keywords: amino acid, electrophysiology, fibroblast growth factor (FGF), ion channel, protein-protein interaction, FGF14, Nav1.6, hot spots, split-luciferase assay, voltage-gated sodium channels
Introduction
Voltage-gated sodium (Nav)2 channels are responsible for the initiation and propagation of the action potential in excitable cells. Nine isoforms of Nav channels (Nav1.1–Nav1.9) have been characterized functionally, and evidence for a tenth one (Nax) has been provided (1–12). Nav channels are differentially expressed in organs, with Nav1.1, -1.2, -1.3, and -1.6 found primarily in the central and peripheral nervous systems, Nav1.4 in the adult skeletal muscle, Nav1.5 in cardiac muscle, and Nav1.7, -1.8, and -1.9 primarily in the peripheral nervous system (3, 4, 7, 12, 13). With such widespread expression, it is not surprising that numerous diseases have been ascribed to mutations of specific Nav channel isoforms (4, 14). These include the Dravet syndrome and other types of epilepsy (15–17); pain-related syndromes, such as congenital insensitivity to pain (18, 19), primary erythromelalgia (20), and paroxysmal extreme pain disorder (21, 22); and cardiac arrhythmias with congenital long QT syndrome (LQTS) type 3 (23, 24); and Brugada syndrome (25). Furthermore, SNPs and/or copy variants within Nav channel genes have been associated recently with autism (Nav1.2) (26). Nav channels blockers are currently used in combined therapy for bipolar disorder (27, 28), depression (29, 30), and schizophrenia (31), extending the role of Nav channels to virtually all brain disorders both neurological and psychiatric (14, 26, 32). Their centrality in the pathophysiology of so many disruptive diseases has made Nav channels key pharmacological target sites for antiepileptic, analgesic, antiarrhythmic, and psychiatric drugs (11, 14, 33, 34). Unfortunately, current Nav channel blockers lack specificity, as they are directed against molecular domains conserved across all Nav isoforms. As such, therapies based on these medications can result in severe side effects, such as Stevens-Johnson syndrome, blood dyscrasias, and ataxia (35). Although some success has been achieved in developing more targeted therapeutics against Nav channels (36), there is still an unmet need to develop safe and potent Nav isoform-specific compounds.
The pore-forming α-subunit of Nav channels is composed of four homologous domains (I–IV), each consisting of six transmembrane α-helices (S1–S6) and an additional pore loop located between the S5 and S6 segments (3). The S5 and S6 transmembrane segments from each domain make up a central pore when assembled within a tetrameric configuration. Upon depolarization, the pore of the channel allows Na+ to rapidly enter the cell; subsequently the channel inactivates and then closes (2). When expressed in heterologous systems, the α-subunit is sufficient to recapitulate the basic functional properties of the channel, but the kinetics, voltage dependence, gating, cellular targeting, and trafficking of the channel are modified by the many accessory proteins that compose the channel macromolecular complex in native conditions. Besides the β-subunits, other relevant regulatory proteins have been identified. As yet, caveolin-3, CaMKII, connexin-43, telethonin, plakophilin, ankyrins, NEDD4, SAPs, syntrophin-dystrophin complex, and intracellular fibroblast growth factors (iFGFs) have been identified as Nav channel accessory proteins (11, 24, 37–41). Some of these interactors have been confirmed as components of the proteome of native Nav1.2 in the brain (42). This rich macromolecular complex of native Nav channels offers a unique source of specific protein-protein interaction (PPI) sites that could serve as targets for drug development (43). This is a new direction in pharmacology that has paid off in cancer (44) and cardiovascular fields (45), but it is still at a nascent stage in neuroscience. In searching for PPI surfaces that could lead to the development of probes and drug-like molecules targeting Nav channels, we have identified FGF14, a member of the iFGF family, as a physiologically relevant accessory protein with implications for brain function and pathology in both animal models and humans (46–48). FGF14 is an emerging disease-relevant protein that was initially associated with neurological disorders such as ataxia (49) and, from more recent genome-wide association studies (GWAS), as a potential risk factor for schizophrenia (47) and depression (46). Binding of FGF14 to Nav1.1, Nav1.2, and Nav1.6 exerts powerful effects on Na+ currents, producing phenotypes that are Nav isoform-dependent and distinct from those associated with other iFGFs (39–41, 50–53).
In addition to binding to Nav channels, iFGF can form dimers. Previous structural studies have proposed the existence of a common interface of all iFGF responsible for both iFGF·Nav complexes and iFGF:iFGF dimer formation (51, 52). However, this hypothesis has never been tested systematically and might not hold for FGF14 given its unique primary sequence (at the N terminus) and modulation of Nav channels (54, 55).
To search for differences at the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer interface, we engineered model-guided mutations at the predicted FGF14 surface and applied the in-cell split-luciferase complementation assay (LCA) to evaluate the effects of these mutants on FGF14:FGF14 dimer formation and monomer binding to the Nav1.6 C-tail. Through patch clamp electrophysiology, we then showed that either a single alanine mutation at Val-160 or a double alanine mutation at Lys-74/Ile-76 is sufficient to abolish previously described functional modulations of Nav1.6 currents by FGF14 (54, 56) but that full functional activity of FGF14 requires an intact Val-160. Complementary studies using intrinsic fluorescence spectroscopy of purified proteins confirmed that Val-160 and Lys-74/Ile-76 are required for FGF14 binding to the Nav1.6 C-tail but that a single alanine mutation at Val-160 is structurally more disruptive. Overall, Lys-74/Ile-76 and Val-160 might be part of druggable pockets to be utilized for drug development against Nav channels.
Experimental Procedures
Materials
d-Luciferin was purchased from Gold Biotechnology (St. Louis, MO) and prepared as a 30 mg/ml stock solution in PBS and stored in a −20° freezer. Anti-luciferase antibodies against the C termini (aa 251–550) and N termini (aa 1–107) were purchased from Santa Cruz Biotechnology (Dallas, TX) and NovusBio (Littleton, CO), respectively.
DNA Construct Preparation
Plasmid DNA with cloned inserts encoding for FGF14K74F/I76R, FGF14L116K/R117F, FGF14N157D/Y1159H, FGF14L202R/K204M/P205S/V208S, FGF14Y158A,FGF14V160A, FGF14Y158A/V160A, and FGF14Y158N/V160N was synthesized by DNA2.0 (Menlo Park, CA) and transferred from the pJ204 shuttle vectors into mammalian expression vectors as described previously (55, 57). FGF14-GFP was generated and characterized as described previously (56). DNA with cloned inserts encoding for FGF14Y158A/V160A was synthesized by DNA2.0 (Menlo Park, CA) and transferred into the GFP plasmid (pQBI-fC2, Quantum Biotechnology Inc., Montreal, Canada). FGF14K74A-GFP, FGF14I76A-GFP, FGF14K74A/I76A-GFP, and FGF14V160A-GFP constructs were generated using FGF14WT-GFP as a template with Agilent Technologies QuikChange Lightning kits (Santa Clara, CA). CLuc-FGF14K74A, CLuc-FGF14I76A, and CLuc-FGF14K74A/I76A constructs were generated using CLuc-FGF14WT as a template, and FGF14K74A-NLuc, FGF14I76A-NLuc, and FGF14K74A/I76A-NLuc constructs were generated using FGF14WT-NLuc as a template with Agilent Technologies QuikChange Lightning kits (Santa Clara, CA).
Molecular Modeling
The FGF14·Nav1.6 homology model was generated using the FGF13·Nav1.5·CaM ternary complex crystal structure (4DCK) as a template. The FGF14 (aa 71–218) and Nav1.6 (aa 1790–1917) sequences were aligned with the crystal structure of the FGF13·Nav1.5 complex (PDB code, 4DCK), and a project Protein Data Bank file was created using DeepView/Swiss-PdbViewer (58). This file was submitted to the Swiss-Model Web server software (QMEAN is 0.808 of 1); subsequently the model was improved by energy minimization in the Chiron Web server (59) and validated by the MolProbity Web server (60) (MolProbity score is 1.56, 94th percentile). Similarly, the FGF14:FGF14 dimer model was generated using the FGF13:FGF13 dimer crystal structure (PBD code, 3HBW) as a template. The FGF14 target sequence (aa 71–218) and the FGF13 crystal structure were aligned using DeepView/Swiss-PDBViewer. The resulting PDB file (QMEAN is 0.652 of 1) was submitted to the Swiss-Model Web server to generate the FGF14 dimer homology model. The model obtained from the Swiss-Model Web server was further improved by energy minimization by the Chiron Web server (59) and subsequently validated by MolProbity (MolProbity score is 1.47, 96th percentile). FGF14K74A/I76A·Nav1.6, FGF14V160A·Nav1.6 C-tail, FGF14K74A/I76A·FGF14K74A/I76A, and FGF14Y158A/V160A·FGF14Y158A/V160A in silico mutations in FGF14 were generated by the USCF Chimera molecular modeling suite (61), and the best rotamers were selected according to their side-chain torsion as well as probability values in the rotamer library. Subsequently, energy minimization of the models was done using the Chiron Web server (59).
Cell Culture and Transient Transfections
HEK293 cells and HEK293 cells stably expressing Nav1.6 cells (HEK-Nav1.6) were maintained in medium composed of equal volumes of DMEM and F-12 (Invitrogen) supplemented with 0.05% glucose, 0.5 mm pyruvate, 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 80 μg/ml G418 (Invitrogen) for selection of Nav1.6 stably transfected cells and incubated at 37 °C with 5% CO2. Transfections were performed in 24-well CELLSTAR® tissue culture plates (Greiner Bio-One, Monroe, NC) at 4.5 × 105 cells/well and incubated overnight to produce monolayers at 90–100% confluence. The cells were then transiently transfected or co-transfected with the appropriate plasmids using Lipofectamine 2000 (Invitrogen). For co-transfections the DNA concentration of plasmid pairs was adjusted, based on previous studies, to achieve an equal ratio of protein production (51, 57, 62).
Split-luciferase Complementation Assay
Twenty-four hours after transfection, cells were replated from the 24-well plate using a 0.04% trypsin:EDTA mixture dissolved in PBS. Suspended cells were centrifuged and seeded in white, clear-bottom CELLSTAR® μClear® 96-well tissue culture plates (Greiner Bio-One) in 200 μl of medium. The cells were incubated for 24 h, and then the growth medium was replaced with 100 μl of serum-free, phenol red-free DMEM/F-12 medium (Invitrogen). The bioluminescence reaction was initiated by automatic injection of 100 μl of d-luciferin substrate (1.5 mg/ml dissolved in PBS) using a SynergyTM H4 multi-mode microplate reader (BioTek, Winooski, VT). Luminescence readings were initiated after 3 s of mild plate shaking and performed at 2-min intervals for 20 min with integration times of 0.5 s. Cells were maintained at 37 °C throughout the measurements. Detailed methods for LCA can be found in previous studies (57).
Western Blotting
Transfected HEK293 cells were washed with cold PBS. Subsequently, 50 μl of lysis buffer (20 mm Tris-HCl, 150 mm NaCl, and 1% Nonidet P-40) and 1 μl of protease inhibitor mixture (set 3, Calbiochem) were added. Cell extracts were collected, sonicated for 16 s, and centrifuged at 4 °C, 13,000 × g for 15 min, adding 4× sample buffer containing 50 mm tris(2-carboxyethyl)phosphine. Mixtures were heated for 10 min at 60 °C and resolved on 7.5% polyacrylamide gels (Bio-Rad). Resolved proteins were transferred to PVDF membranes (Millipore, Bedford, MA) for 2 h at 4 °C and blocked in TBS with 3% nonfat dry milk and 0.1% Tween 20. Membranes were then incubated overnight in blocking buffer containing anti-luciferase (epitope detects 251–550 aa) mouse polyclonal antibody (Santa Cruz Biotechnology) and anti-luciferase antibody (epitope detects 1–107 aa) mouse monoclonal antibody (NovusBio) or anti-calnexin rabbit polyclonal antibody (Cell Signaling Technology, Danvers, MA). The washed membranes were incubated with goat anti-mouse or goat anti-rabbit-HRP (1:5000) and visualized with an ECL Advance Western blotting detection kit (GE Healthcare). Protein bands were visualized using the FluorChem® HD2 system and analyzed with AlphaView 3.1 software (ProteinSimple, Santa Clara, CA).
LCA Data Analysis
Relative luminescence units (in RLU) measured using a SynergyTM H4 multi-mode microplate reader were tabulated by well position and time point into Microsoft Excel. The signal intensity for each well was calculated as a mean value of peak luminescence measured at three adjacent time points; the calculated values were expressed as a percentage of mean signal intensity in the control samples from the same experimental plate. Statistical values were calculated as mean ± S.E. unless otherwise specified. The statistical significance (p < 0.05) of different groups was determined by Student's t test, one-way ANOVA with post hoc Bonferroni's method, or Kruskal-Wallis one-way ANOVA on ranks with post hoc Dunn's method using SigmaStat (San Jose, CA) and GraphPad Prism® (La Jolla, CA) software. Graphs were plotted in Origin 8.6 software (OriginLab Corp., Northampton, MA).
Protein Overexpression and Purification
cDNAs encoding FGF14WT (NCBI reference sequence accession number NP_787125, aa 64–252) or the C-terminal domain of Nav1.6 (NCBI reference sequence accession number NP_001171455, aa 1756–1939) were subcloned into suitable pET bacterial expression vectors (pET28a-FGF14 and pET30a-Nav1.6) with a His6 tag at the N-terminal site; these plasmids were a gift of Drs. Regina Goetz and Moosa Mohammadi (52). The mutation coding for FGF14K74A/I76A and FGF14V160A was generated by site-directed mutagenesis and PCR using FGF14 as a template. Upon transformation with the corresponding cDNA clones, recombinant proteins FGF14WT, FGF14K74A/I76A, FGF14V160A, and Nav1.6 C-tail were expressed in the bacterial strain Escherichia coli BL21(DE3) pLys (Invitrogen) after induction with 0.2 mm isopropyl thio-β-d-galactopyranoside for 8 h at 15 °C. After induction with isopropyl thio-β-d-galactopyranoside, bacterial cells were harvested and lysed by lysozyme and sonication at 4 °C in lysis/binding buffer containing 10 mm sodium phosphate (prepared from 1 m Na2HPO4 and NaH2PO4) + 0.1% CHAPS, pH 7.0 (for FGF14 proteins), 25 mm HEPES + 150 mm NaCl + 10% glycerol (Nav1.6), pH 7.5, containing 0.1 mm PMSF. The respective proteins were centrifuged at 18,000 × g for 30 min at 4 °C. For purification of FGF14WT, FGF14K74A/I76A,and FGF14V160A, the supernatant was applied to pre-equilibrated heparin, and the proteins were then eluted with0.2–2.0 mNaCl in the lysis/binding buffer. For purification of the Nav1.6 C-tail, the supernatant was applied first to a Ni2+-nitrilotriacetic acid column and eluted with 200 mm imidazole. The Nav1.6 C-tail was further purified using a HiTrap Q Sepharose Fast Flow column (GE Healthcare) with a buffer containing 50 mm Tris-HCl and eluted with 10–500 mm NaCl at pH 7.5. Finally, all concentrated proteins were purified on an AKTA purifier using Superdex 200 Hiload 16 × 60 columns (both products from GE Healthcare Bio-Sciences) and equilibrated in 50 mm Tris-HCl + 150 mm NaCl, pH 7.5.
Intrinsic Fluorescence Spectroscopy
The intrinsic fluorescence emission spectra of protein samples were recorded on a Spex FluoroMax (Spex Industries, Edison, NJ) in 25 mm Tris-HCl + 150 mm NaCl, pH 7.5. The spectra (300–450 nm) were recorded at an excitation wavelength of 295 nm in the proper spectral mode of the instrument using excitation and emission band passes of 5 nm each. The binding affinity of FGF14WT, FGF14K74A/I76A, and FGF14V160A with the Nav1.6-Ctail was determined by titrating the protein solutions with standard 5.5 μm concentration aliquots.
Electrophysiology Experiments and Data Analysis
HEK-Nav1.6 cells transfected with GFP, FGF14-GFP, FGF14V160A-GFP, FGF14Y158A/V160A, or FGF14Y158N/V160N-GFP were plated at low density on glass coverslips for 3–4 h and subsequently transferred to the recording chamber. Recordings were performed at room temperature (20–22 °C) 24 h post-transfection using a MultiClamp 700B amplifier (Molecular Devices, Sunnyvale, CA). The composition of the recording solutions consisted of extracellular salts (mm) (140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES, and 10 glucose, pH 7.3) and intracellular salts (mm) (130 CH3O3SCs, 1 EGTA, 10 NaCl, and 10 HEPES, pH 7.3). Membrane capacitance and series resistance were estimated by the dial settings on the amplifier and compensated for electronically by 70–80%. Data were acquired at 20 kHz and filtered at 5 kHz prior to digitization and storage. All experimental parameters were controlled by Clampex 9.2 software (Molecular Devices) and interfaced to the electrophysiological equipment using a Digidata 1200 analog-digital interface (Molecular Devices). Voltage-dependent inward currents for HEK-Nav1.6 cells were evoked by depolarizations to test potentials between −100 and +60 mV from a holding potential of −70 mV followed by a voltage pre-step pulse of −120 mV (Nav1.6). Steady-state (fast) inactivation of Nav channels was measured with a paired-pulse protocol. From the holding potential, cells were stepped to varying test potentials between −120 mV (Nav1.6) and +20 mV (pre-pulse) prior to a test pulse to −20 mV.
Current densities were obtained by dividing Na+ current (INa) amplitude by membrane capacitance. Current-voltage relationships were generated by plotting current density as a function of the holding potential. Conductance (GNa) was calculated by the following equation,
where INa is the current amplitude at voltage Vm and Erev is the Na+ reversal potential.
Steady-state activation curves were derived by plotting normalized GNa as a function of test potential and fitted using the Boltzmann equation,
where GNa,max is the maximum conductance, Va is the membrane potential of half-maximal activation, Em is the membrane voltage, and k is the slope factor. For steady-state inactivation, normalized current amplitude (INa/INa,max) at the test potential was plotted as a function of prepulse potential (Vm) and fitted using the Boltzmann equation,
where Vh is the potential of half-maximal inactivation, Em is the membrane voltage, and k is the slope factor.
Transient INa inactivation decay was estimated with a standard exponential equation. The inactivation time constant (τ) was fitted with the following equation,
where A1 and ƒ1 are the amplitude and time constant, respectively. The variable C is a constant offset term along the y axis. The goodness of fit was determined by the correlation coefficient (R), and the cutoff of R was set at 0.85.
Data analysis was performed using Clampfit 9 software (Molecular Devices) and Origin 8.6 software (OriginLab Corp.). Results were expressed as mean ± S.E. The statistical significance of observed differences among groups was determined by Student's t test or one-way ANOVA with post hoc Bonferroni or Dunnett test; p < 0.05 was regarded as statistically significant.
Results
Homology Model-based Characterization of Putative FGF14 PPI Surface Hot Spots
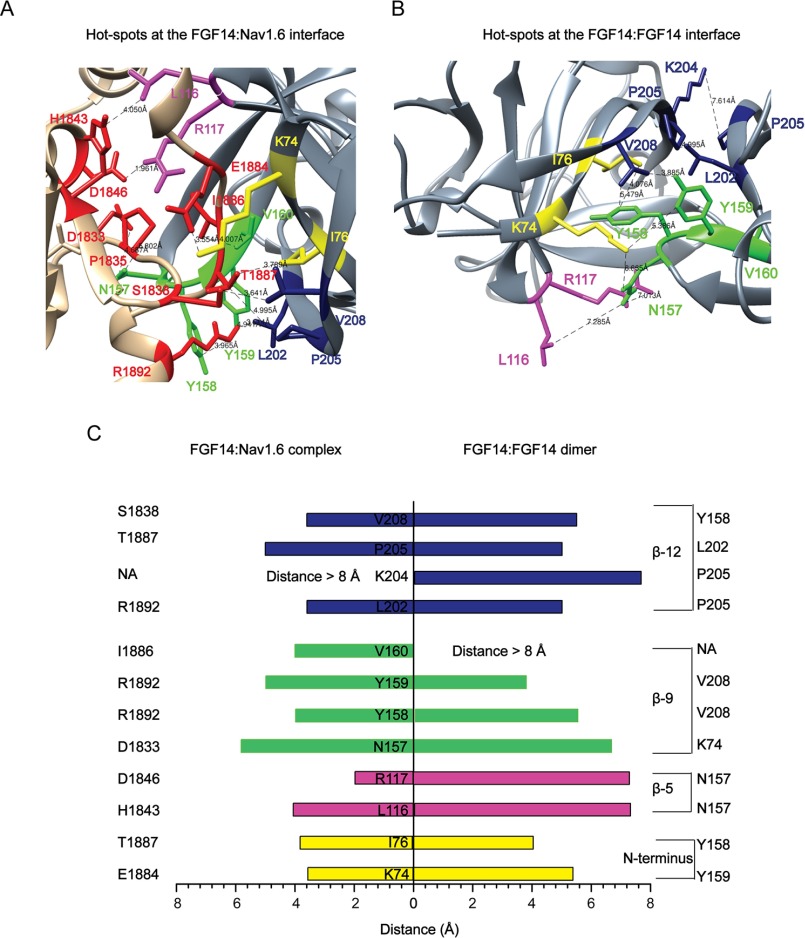
To compare putative amino acid residues at the FGF14 PPI interface of the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer formation, homology models based on other iFGFs (either in their dimeric form or in complex with the Nav1.5 channel C-tail) were created (Fig. 1, A and B, and Table 1). Inspection of the FGF14·Nav1.6 homology model revealed that in FGF14, residues Lys-74/Ile-76 (located at the N-terminal), Leu-116/Arg-117 (located at β-5), Asn-157/Tyr-158/Tyr-159/Val-160 (located at β-9), and Leu-202/Pro-205/Val-208 (located at β-12) were within a distance of <8 Å (63, 64) from the closest neighboring amino acid of the Nav channel, consistent with the putative hot spots (Fig. 1C). Although most of these residues seemed to exert a similar role in the FGF14:FGF14 dimer complex, Lys-204 and Val-160 appeared to be structurally divergent (Fig. 1C). Lys-204 interacted with the neighboring Pro-205 in FGF14 but had no close neighbors in Nav1.6, whereas Val-160 interacted with residue Ile-1886 in Nav1.6 but had no putative interactors in the FGF14:FGF14 dimer. Thus, homology modeling predicts some conserved residues at the FGF14 surface, but potential structural differences depend on the local microenvironment.
FIGURE 1.
Homology model-based predicted hot spots at the PPI interface of FGF14·Nav1.6 complex and FGF14:FGF14 dimer formation. A, the FGF14·Nav1.6 complex homology model (zoom view) was generated by FGF13·Nav1.5 (PDB code, 4DCK) crystal structure as a template. The C-tail of the Nav1.6 channel and FGF14 are shown as tan and gray colors, respectively. The critical amino acids Lys-74/Ile-76 (yellow), Leu-116/Arg-117 (magenta), Asn-157/Tyr-158/Tyr-159/Val-160 (green), and Leu-202/Lys-204/Pro-205/Val-208 (blue) are located at the N terminus, β5, β-9, and β-12 strands of FGF14. Critical amino acids of the C-tail of Nav1.6 channel are shown in red color. B, the FGF14:FGF14 dimer homology model (zoom view) was generated by FGF13:FGF13 (PDB code, 3HBW) dimer crystal structure as a template. C, the distance (less than 8 Å) between each critical amino acid of FGF14 to the neighboring critical amino acid of FGF14 or to the neighboring critical amino acid of the C-tail of the Nav channels was determined using UCSF Chimera software from homology models of the FGF14:FGF14 dimer and the FGF14·Nav1.6 complex.
TABLE 1.
Homology model-based hot spots at the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer PPI interface
| Location | FGF14·Nav1.6 | FGF14:FGF14 |
|---|---|---|
| N terminus | Lys-74, Ile-76 | Lys-74, Ile-76 |
| β-5 | Leu-116, Arg-117 | Leu-116, Arg-117 |
| β-9 | Asn-157,Tyr-158, Tyr-159, Val-160 | Asn-157,Tyr-158, Tyr-159 |
| β-12 | Leu-202, Pro-205, Val-208 | Leu-202, Lys-204, Pro-205, Val-208 |
In-cell Validation of Hot Spots at the FGF14·Nav1.6 and FGF14:FGF14 Dimer Interface
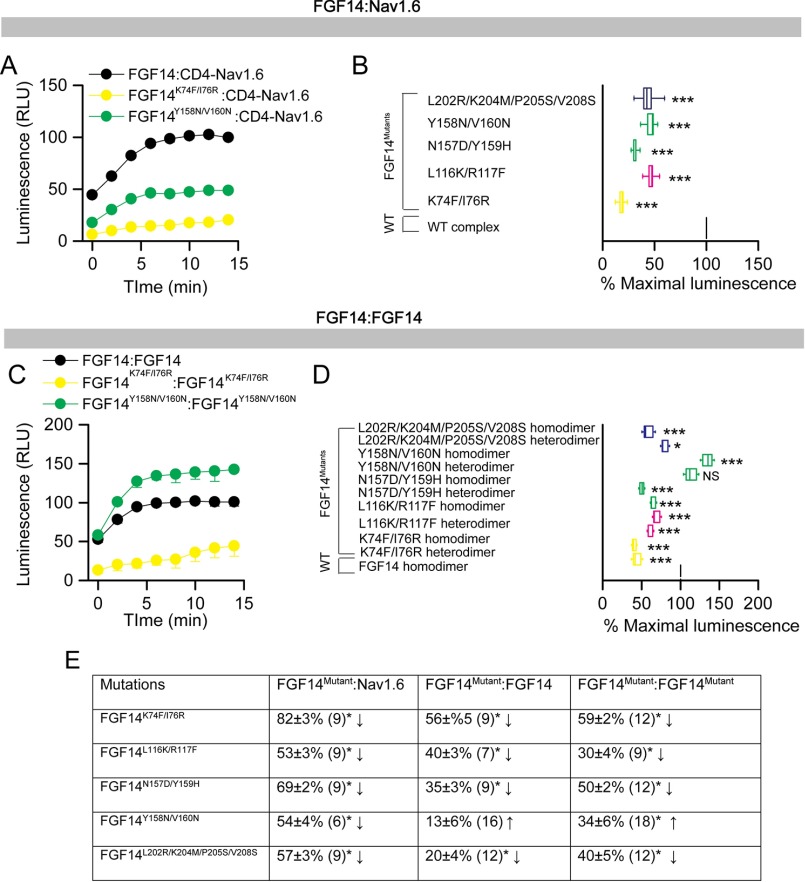
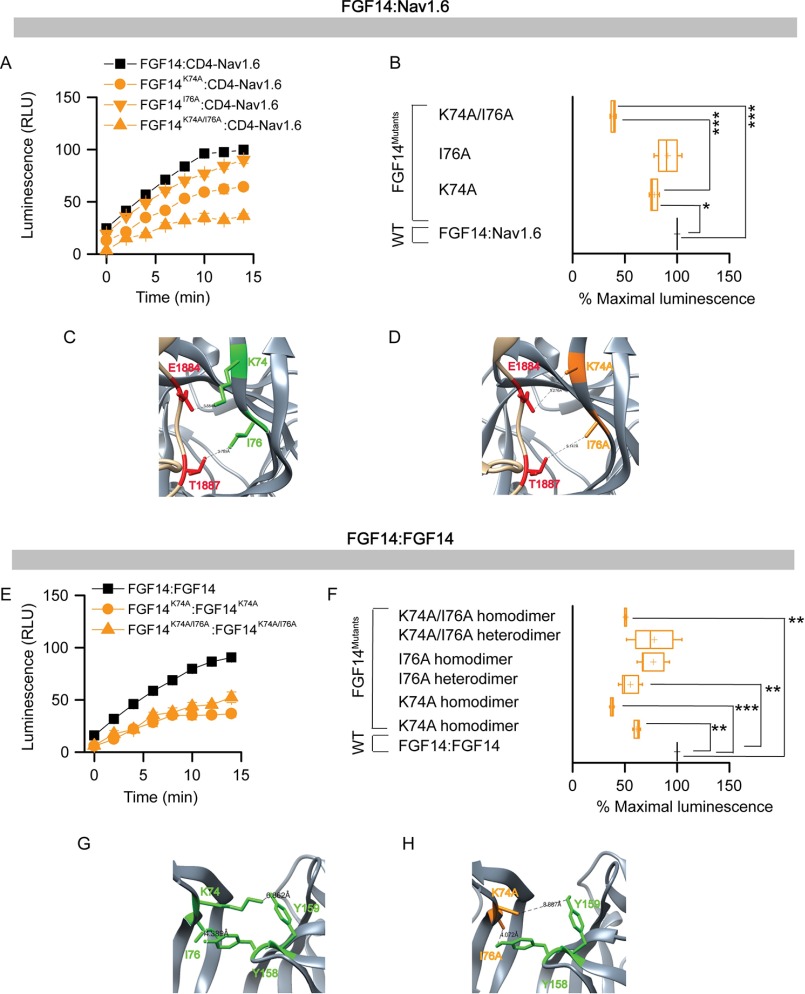
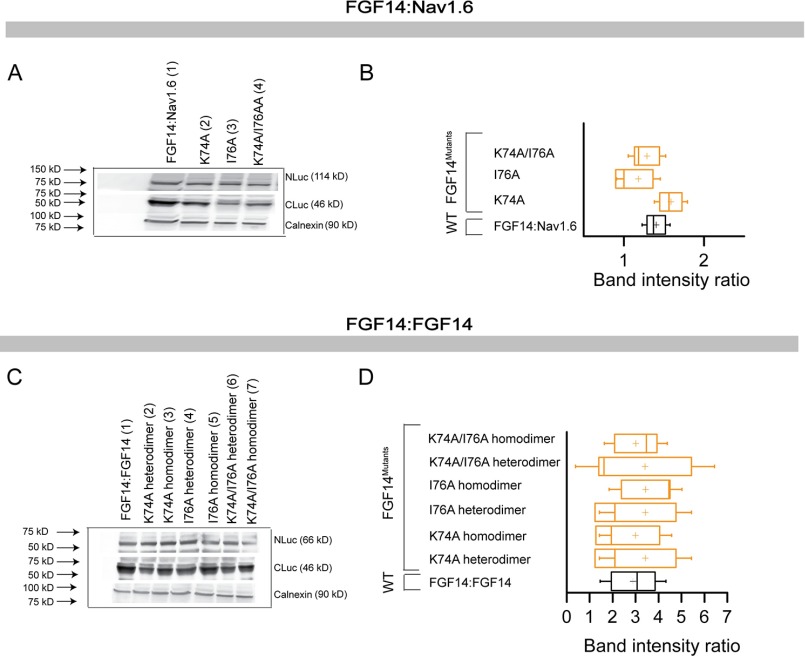
To evaluate the role of these model-based predicted hot spots experimentally, we engineered double/quadruple mutations in the FGF14 protein and examined their impact on the FGF14·Nav1.6 complex and FGF14:FGF14 dimer formation using our previously validated in-cell LCA (51, 55, 57, 62, 65). The FGF14 mutations were essentially grouped by β-sheet and/or N terminal location as in previous studies on FGF13 (52) and engineered to carry FGF14mutant proteins fused with either CLuc (fused to the 5′-terminal end of the cDNA of interest) or NLuc fragments (fused to the 3′-terminal end of the cDNA of interest) of the Photinus pyralis firefly enzyme, allowing for in-cell reconstitution of FGF14·Nav1.6 C-tail and/or FGF14·FGF14 protein pairs. Mutations of the FGF14 protein considered in this study were the following: FGF14K74F/I76R, FGF14L116K/R117F, FGF14N157D/Y159H, FGF14Y158N/V160N, and FGF14L202R/K204M/P205S/V208S. Combinations of FGF14 wild type (FGF14WT) or/and FGF14mutant constructs (tagged with either CLuc or NLuc fragments) were transiently co-expressed with either CD4-Nav1.6-C-tail-NLuc (a chimeric construct that allows the membrane presentation of the Nav1.6 C-tail) (51, 57, 62, 65) or with the same corresponding FGF14mutant proteins in HEK293 cells (Fig. 2); this latter set of experiments was designed to reconstitute hetero- and homodimer forms of each FGF14mutant. Upon binding of the respective protein pairs, the enzymatic activity of the luciferase enzyme was reconstituted by complementation of the full enzyme, giving rise to a robust luminescence response in the presence of the d-luciferin substrate. Representative luminescence responses of the assembly of CLuc-FGF14·CD4-Nav1.6-NLuc, CLuc-FGF14K74F/I76R·CD4-Nav1.6-NLuc, and CLuc-FGF14Y158N/V160N·CD4-Nav1.6-NLuc are shown in Fig. 2A. For each construct pair, the maximum luminescence response of the CLuc-FGF14mutant·CD4-Nav1.6-NLuc complex was normalized to the CLuc-FGF14WT·CD4-Nav1.6-NLuc complex (Fig. 2B). One-way ANOVA with post hoc Dunnett's analysis over a large data set (n = 6–9 independent experiments; n = 4 repetitions) revealed that the strength of interaction of all protein complexes carrying mutations within the FGF14 protein was significantly reduced (p < 0.001) compared with the CLuc-FGF14WT·Nav1.6-NLuc complex (Fig. 2B). LCA studies for both the FGF14mutant homo- and heterodimer complexes with representative traces and cumulative normalized luminescence responses are shown in Fig. 2, C and D. Importantly, as summarized in Fig. 2E, FGF14 mutations within the N terminus, β-5, and β-12 led to a relative decrease in PPI binding compared with control (Fig. 2, B–D, yellow, pink, and blue). Mutations of Val-160 and Tyr-158 resulted in reduced binding to the Nav1.6 C-tail (Fig. 2B), but they either had no significant effect on the relative binding strength (Y158N/V160N heterodimer) or augmented (Y158N/V160N homodimer) the relative binding strength when examined in the context of the FGF14 dimer (Fig. 2D). Furthermore, mutations at the Lys-74/Ile-76 residues had the greatest effect on both the FGF14·Nav1.6 complex and the FGF14 dimer, likely because of a strong interaction with neighboring amino acids as predicted by our molecular modeling. Western blotting analysis of total cell lysates derived from cells transfected with each pair of plasmids confirmed that the protein production across the experimental groups was comparable, confirming the validity of the LCA results (Fig. 3). Altogether, these data support our homology model predictions suggesting structural divergence at the FGF14 β-9 sheet with mutations of Tyr-158/Val-160 and structural conservation at the FGF14 N terminus with mutations of K74A/I76A having a significant role in both the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer formation.
FIGURE 2.
In-cell LCA characterization of hot spots at the FGF14·Nav1.6 channel and the FGF14:FGF14 dimer interface. A, representative luminescence response (RLU) corresponding to the assembly of CLuc-FGF14·CD4-Nav1.6-NLuc (black) and the respective mutants (Y158N/V160N, green circle; K74F/I76R, yellow circle). B, box plot representing percent maximal luminescence response of relative mutants normalized to the CLuc-FGF14·CD4-Nav-NLuc control (black). C, representative luminescence response (RLU) corresponding to the assembly of CLuc-FGF14·FGF14-NLuc (black) and the respective mutants (Y158N/V160N, green circle; K74F/I76R, yellow circle). D, box plot representing percent maximal luminescence response of relative mutants normalized to the CLuc-FGF14:FGF14-NLuc (black) homodimer response. Data are mean ± S.E. The statistical significance between the three groups was assessed using Kruskal-Wallis one-way ANOVA on ranks with post hoc Dunn's method. ***, p < 0.001; *, p < 0.05. NS, not significant. E, a data summary of the modulation of hot spots in FGF14 to form the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer is shown.
FIGURE 3.
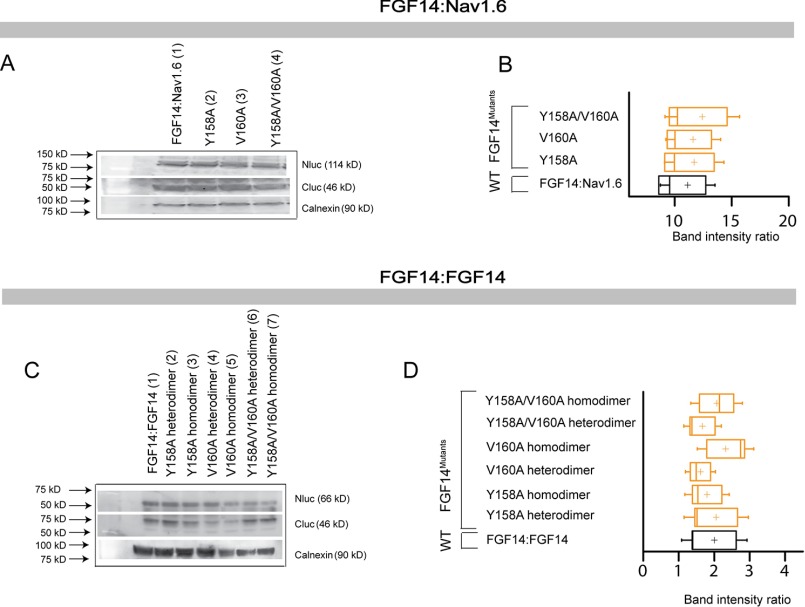
Protein production quantification from Western blotting analysis. A, Western blotting of whole-cell extracts from cells transfected with the indicated CLuc-FGF14 and CD4-Nav1.6-NLuc constructs. B, summary graph of densitometry analysis of CLuc and NLuc band intensity ratio of the respective protein products. C, Western blotting of whole-cell extracts from cells transfected with the indicated CLuc-FGF14 and FGF14-NLuc constructs. D, summary graph of densitometry analysis as described in C. Membrane were probed with anti-luciferase antibodies that recognize either the CLuc or the NLuc fragments (∼46 and ∼66/114 kDa, respectively). Immunodetection of calnexin was used as loading control.
Alanine-scanning Mutagenesis of Tyr-158 and Val-160 at the FGF14·Nav1.6 and the FGF14:FGF14 Dimer Complexes
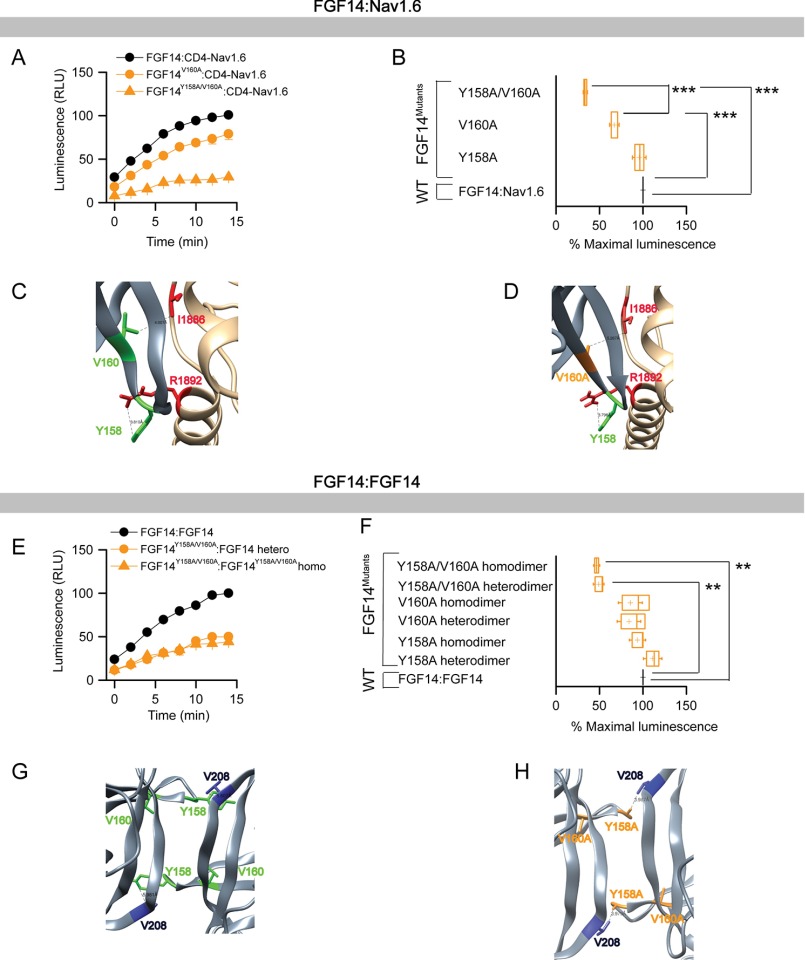
These results prompted us to investigate the impact of size and polarity at the Tyr-158 and Val-160 residues in the two PPI complexes. To this end, we engineered single and double Ala mutations of Tyr-158 and Val-160 (FGF14Y158A, FGF14V160A, and FGF14Y158A/V160A in both the CLuc and the NLuc format) and continued our screening evaluation with LCA for both FGF14·Nav1.6 and the FGF14:FGF14 dimer complexes. Representative traces and the maximal luminescence signal of the FGF14mutant·CD4-Nav1.6 complex (normalized to FGF14WT·CD4-Nav1.6 complex) are shown in Fig. 4, A and B, and Table 2. One-way ANOVA with Dunnett's post hoc analysis revealed that one single Ala mutation at Tyr-158 was not sufficient to disrupt binding (CLuc-FGF14Y158A·CD4-Nav1.6-NLuc; 95.80 ± 5.246%, n = 21, p > 0.05), but a single Val-160 to Ala disrupted the complex (CLuc-FGF14V160A·CD4-Nav1.6-NLuc; 67.11 ± 3.701%, n = 21, p < 0.001). However, the double mutant exhibited a much lower relative binding (CLuc-FGF14Y158A/V160A·CD4-Nav1.6-NLuc; 33.63 ± 2.0%, n = 6) when compared with the FGF14WT·Nav1.6 complex (p < 0.001) or with the single V160A mutation (p < 0.001). The expression of all single and double Ala mutant proteins was confirmed across all groups by Western blotting analysis (Fig. 5, A and B). Corresponding homology models of the FGF14WT·Nav1.6 and FGF14V160A·Nav1.6 complexes were built (Fig. 4, C and D) to inspect the role of Val-160 at the corresponding PPI interfaces. In the FGF14WT·Nav1.6 complex, Val-160 interacts with Ile-1886 (distance 4.1 Å) of Nav1.6 through a hydrophobic interaction (Fig. 4C). In the FGF14V160A·Nav1.6 model, the V160A mutation (orange) of FGF14 was further (red, 5.3 Å) from the Ile-1886 of Nav1.6 (Fig. 4D), suggesting fewer opportunities for interaction with Nav1.6 (66).
FIGURE 4.
Differential role of Tyr-158 and Val-160 at the FGF14·Nav1.6 channel and the FGF14:FGF14 dimer interface assessed by alanine-scanning mutagenesis and in-cell LCA. A, representative luminescence response (RLU) corresponding to the assembly of CLuc-FGF14·CD4-Nav1.6-NLuc (black circle) and respective mutants (V160A, orange circle; Y158A/V160A, orange triangle). B, box plot representing percent maximal luminescence response of relative mutants normalized to the CLuc-FGF14· CD4-Nav1.6-NLuc control (black). C, homology model of the FGF14·Nav1.6 complex (zoom view) in which FGF14 is shown as gray and C-tail of Nav1.6 is shown as tan. Tyr-158 (green) and Val-160 (green) interact respectively with Arg-1892 and Ile-1886 of C-tail of Nav1.6. D, interaction between V160A (orange) of FGF14 with Ile-1886 (red) of C-tail of Nav1.6 is shown in the FGF14V160A·Nav1.6 homology model. E, representative luminescence response (RLU) corresponding to the assembly of CLuc-FGF14·FGF14-NLuc (black) and the respective mutants (Y158A/V160A heterodimer, orange circle; Y158A/V160A homodimer, orange triangle). F, box plot representing percent maximal luminescence response of relative mutants normalized to the CLuc-FGF14:FGF14-NLuc (black) homodimer response. Data are mean ± S.E. The statistical significance between the three groups was assessed using Kruskal-Wallis one-way ANOVA on ranks with post hoc Dunn's method; ***, p < 0.001; **, p < 0.01. G, homology model of the FGF14:FGF14 homodimer (zoom view) in which Tyr-158 (green) from one FGF14 monomer interacts with Val-208 of the neighboring FGF14. H, interaction between Y158A (orange) of FGF14 with Val-208 (blue) of the neighboring FGF14 monomer is shown in the FGF14Y158A/V160A:FGF14Y158A/V160A homodimer model.
TABLE 2.
Impact of Tyr-158 and Val-160 at the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer interface
The number of independent experiments is shown in parentheses. **, p < 0.01; ***, p < 0.001; one-way ANOVA with post hoc Dunnett's multiple comparisons test compared with either FGF14WT·Nav1.6 or FGF14WT:FGF14WT.
| Mutation | FGF14mutant·Nav1.6 | FGF14mutant:FGF14 | FGF14mutant:FGF14mutant |
|---|---|---|---|
| FGF14Y158A | 95 ± 5% (21)↓ | 111 ± 7% (15)↑ | 93 ± 6% (18)↑ |
| FGF14V160A | 67 ± 4% (21)***↓ | 83 ± 9% (15)↓ | 85 ± 9% (18)↓ |
| FGF14Y158A/V160A | 33 ± 2% (6)***↓ | 49 ± 4% (6)**↓ | 49 ± 2% (12)**↓ |
FIGURE 5.
Protein production quantification from Western blotting analysis for FGF14Y158A, FGF14V160A, and FGF14Y158A/V160A. A, Western blotting of whole-cell extracts from cells transfected with the indicated CLuc-FGF14 and CD4-Nav1.6-NLuc constructs. B, summary graph of densitometry analysis of CLuc and NLuc band intensity ratio of the respective protein products. C, Western blotting of whole-cell extracts from cells transfected with the indicated CLuc-FGF14 and FGF14-NLuc constructs. D, summary graph of densitometry analysis as described in C. Membrane were probed with anti-luciferase antibodies that recognize either the CLuc or the NLuc fragments (∼46 and ∼66/114 kDa, respectively). Immunodetection of calnexin was used as loading control.
When examined in the context of the FGF14:FGF14 dimer (Fig. 4, E and F, and Table 2), neither the Y158A (CLuc-FGF14Y158A·FGF14WT-NLuc, 111.18 ± 4.89%, n = 25, p > 0.05; CLuc-FGF14Y158A·FGF14Y158A-NLuc, 93.69 ± 6.12%, n = 18, p > 0.05) nor the V160A mutation (CLuc-FGF14V160A·FGF14WT-NLuc, 83.19 ± 9.87%, n = 15, p > 0.05; CLuc-FGF14V160A·FGF14V160A-NLuc, 85.46 ± 9.23%, n = 18, p > 0.05) alone had a significant impact on complex stability. On the other hand, the FGF14Y158A/V160A double mutant (in both the hetero- and homodimers) exhibited reduced relative binding (CLuc-FGF14Y158A/V160A·FGF14WT, 49.04 ± 4.0%, n = 6, p < 0.01; CLuc-FGF14Y158A/V160A·FGF14Y158A/V160A-NLuc, 49.85 ± 2.05%, n = 12, p < 0.01). The expression of all single and double alanine mutant proteins was validated across all groups by Western blotting analysis (Fig. 5, C and D). These results indicate that Val-160 alone is not sufficient to mediate PPI in the FGF14·FGF14 complex but that Val-160 might work synergistically with Tyr-158 to stabilize it. Corresponding homology models of FGF14WT:FGF14WT and FGF14Y158A/V160A:FGF14Y158A/V160A dimers were built (Fig. 4, G and H). The model predicts that Tyr-158 and Val-160 are within protruding β-9 loops that connect the two monomers in the intertwined dimer (Fig. 4G). Notably, Tyr-158 in each monomer appears to interact with Val-208 of the neighboring monomer through hydrogen bonding, whereas Val-160 had no predicted interactions. Simultaneous replacement of Tyr-158 and Val-160 with a neutral alanine residue might disrupt hydrogen bonding, weakening the stability of the β-9 loop (Fig. 4H). Homology modeling predictions and LCA results together suggest that the PPI interface of the FGF14·Nav1.6 complex is controlled by Val-160 through hydrophobic interactions, whereas the FGF14:FGF14 dimer requires the synergistic action of Tyr-158 and Val-160 through hydrogen bonding.
Alanine-scanning Mutagenesis of Lys-74 and Ile-76 at FGF14·Nav1.6 and FGF14:FGF14 Dimer Complexes
The key role of Lys-74/Ile-76 in both the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer (Fig. 2) prompted us to investigate the role of these amino acid residues within single and double alanine mutations. To this end, FGF14K74A, FGF14I76A, and FGF14K74A/I76A in both the CLuc and the NLuc format were engineered and evaluated with LCA. Representative traces and the maximal luminescence signal of the FGF14mutant·Nav1.6 complex (normalized to FGF14WT·Nav1.6 complex) are shown in Fig. 6, A and B, and Table 3. One-way ANOVA (Kruskal-Wallis) with post hoc Dunn's multiple comparison test revealed that a single Ala mutation at Ile-76 was not sufficient to disrupt binding (CLuc-FGF14I76A·CD4-Nav1.6-NLuc; 91.80 ± 8.8%, n = 12, p > 0.05), but a single Ala mutation at Lys-74 moderately disrupted the complex (CLuc-FGF14K74A·CD4-Nav1.6-NLuc; 78.13 ± 3.22%, n = 12, p < 0.05). However, the double mutant showed a significantly lower relative binding affinity (CLuc-FGF14K74A/I76A·CD4-Nav1.6-NLuc; 38.08 ± 2.01%, n = 6) when compared with the FGF14WT·CD4-Nav1.6 complex (p < 0.001) or with the FGF14K74A·CD4-Nav1.6 complex (p < 0.001). The expression of all single and double alanine mutant proteins was confirmed across all groups by Western blotting analysis (Fig. 7, A and B). Corresponding homology models of the FGF14WT·Nav1.6 and FGF14K74A/I76A·Nav1.6 complexes were built (Fig. 6, C and D) to inspect the role of Lys-74 and Ile-76 at the corresponding PPI interfaces. In the FGF14WT·Nav1.6 complex, Lys-74 and Ile-76 interacted with Glu-1884 (distance 3.5 Å) and Thr-1887 (distance 3.71 Å) of Nav1.6, respectively, through a salt bridge and hydrophobic interactions (Fig. 6C). In the FGF14K74A/I76A·Nav1.6 model, the K74A and I76A mutations (orange) of FGF14 were further from the Ile-1884 and Thr-1887 residues of Nav1.6 (Fig. 6D), suggesting disruption of the salt bridge and hydrophobic interactions.
FIGURE 6.
Assessing the role of Lys-74 and Ile-76 at the FGF14·Nav1.6 channel and the FGF14:FGF14 dimer interface by alanine-scanning mutagenesis by in-cell LCA. A, representative luminescence response (RLU) corresponding to the assembly of CLuc-FGF14·CD4-Nav1.6-NLuc (black square) and respective mutants (K74A, orange circle; I76A, orange inverted triangle; K74A/I76A, orange triangle). B, box plot represents percent maximal luminescence response of relative mutants normalized to the CLuc-FGF14· CD4-Nav1.6-NLuc control (black). C, homology model of the FGF14·Nav1.6 complex (zoom view) in which FGF14 is shown as gray and the C-tail of Nav1.6 is shown as tan. Lys-74 (green) and Ile-76 (green) interact with Glu-1884 and Thr-1887 of the C-tail of Nav1.6, respectively. D, interaction of K74A (orange) and I76A (orange) of FGF14 with Glu-1884 (red) and Thr-1887 (red) of the C-tail of Nav1.6 is shown in the FGF14K74A/I76A·Nav1.6 homology model. E, representative luminescence response (RLU) corresponding to the assembly of CLuc-FGF14·FGF14-NLuc (black) and respective mutants (K74A homodimer, orange circle; K74A/I76A homodimer, upper orange triangle). F, box plot represents % maximal luminescence response of relative mutants normalized to the CLuc-FGF14:FGF14-NLuc (black) homodimer response. Data are mean ± S.E. The statistical significance between the three groups was assessed using Kruskal-Wallis one-way ANOVA on ranks with post-hoc Dunn's method; ***, p < 0.001 or **, p < 0.01. G, homology model of the FGF14:FGF14 homodimer (zoom view) in which Lys-74 (green) and Ile-76 (green) from one FGF14 monomer interacts with Tyr-159 and Tyr-158, respectively, with neighboring FGF14. H, interaction between K74A (orange) and I76A (orange) of FGF14 with Tyr-158 and Tyr-159, respectively, with neighboring FGF14 monomer is shown in the FGF14K74A/I76A:FGF14K74A/I76A homodimer model.
TABLE 3.
Impact of Lys-74 and Ile-76 at the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer interface
The number of independent experiments is shown in parentheses. ***, p < 0.001; **, p < 0.01; *, p < 0.05; one-way ANOVA with post hoc Dunn's/Dunnett's multiple comparisons test compared with either FGF14WT·Nav1.6 or FGF14WT:FGF14WT.
| Mutations | FGF14mutant·Nav1.6 | FGF14mutant:FGF14 | FGF14mutant:FGF14mutant |
|---|---|---|---|
| FGF14K74A | 78 ± 3% (12)*↓ | 62±%2 (6)**↓ | 36 ± 2% (6)***↓ |
| FGF14I76A | 91 ± 8% (12)↓ | 61 ± 8% (6)**↓ | 82 ± 9% (6)↓ |
| FGF14K74A/I76A | 38 ± 2% (6)***↓ | 78 ± 17% (3)↓ | 50 ± 1% (3)***↓ |
FIGURE 7.
Protein production quantification from Western blotting for FGF14K74A, FGF14I76A, and FGF14K74A/I76A. A, Western blotting analysis of whole-cell extracts from cells transfected with the indicated CLuc-FGF14 and CD4-Nav1.6-NLuc constructs. B, summary graph of densitometry analysis of CLuc and NLuc band intensity ration of the respective protein products. C, Western blotting analysis of whole-cell extracts from cells transfected with the indicated CLuc-FGF14 and FGF14-NLuc constructs. D, summary graph of densitometry analysis as described in C. Membrane were probed with anti-luciferase antibodies that recognize either the CLuc or the NLuc fragments (∼46 and ∼66/114 kDa, respectively). Immunodetection of calnexin was used as loading control.
When examined in the context of the FGF14:FGF14 dimer (Fig. 6, E and F, and Table 3), K74A alone or in combination with I76A was found to have the strongest impact on the complex stability in the hetero- and homodimer formation; I76A alone, on the other hand, was relatively inactive. Corresponding homology models of FGF14WT:FGF14WT and FGF14K74A/I76A:FGF14K74A/I76A dimers were built (Fig. 6, G and H). The model predicts that Lys-74 and Ile-76 interact with Tyr-159 and Tyr-158 of neighboring FGF14 through salt bridge and hydrogen bonding, respectively. These interactions were both disrupted upon alanine mutation, although K74A appears to have a stronger impact at the PPI interface (Fig. 6H). Overall, both LCA and homology modeling supported the idea that Lys-74 at the N terminus of FGF14 plays a more significant role in FGF14:FGF14 dimer formation compared with Ile-76. The expression of all single and double alanine mutant proteins was validated across all groups by Western blotting analysis (Fig. 7, C and D).
Val-160 is Required for Modulation of Nav1.6 Currents
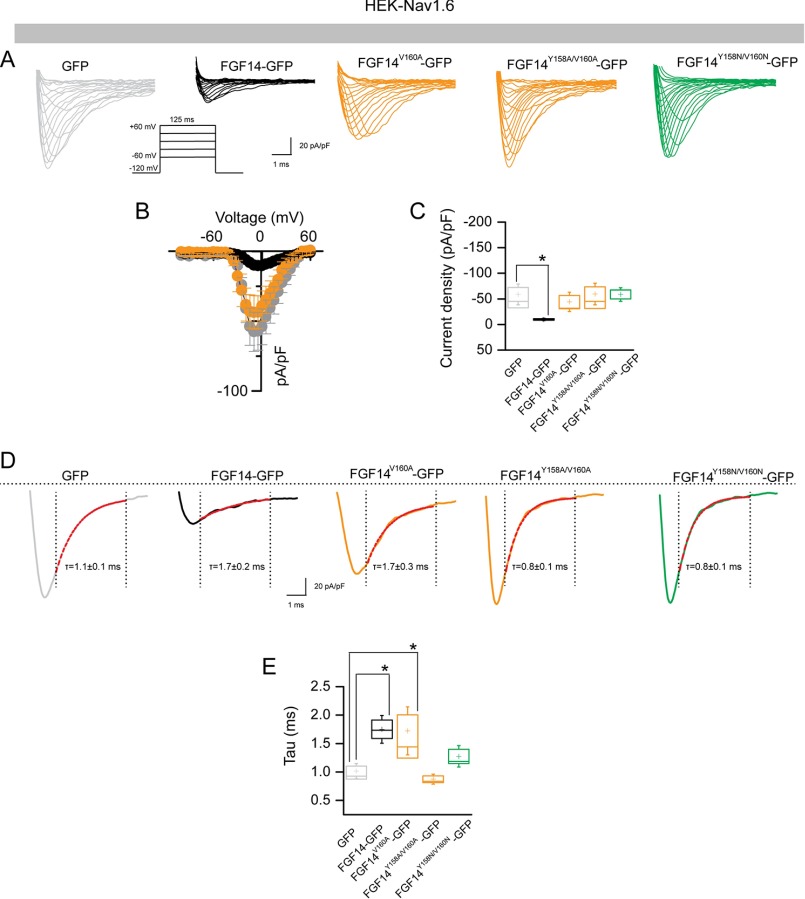
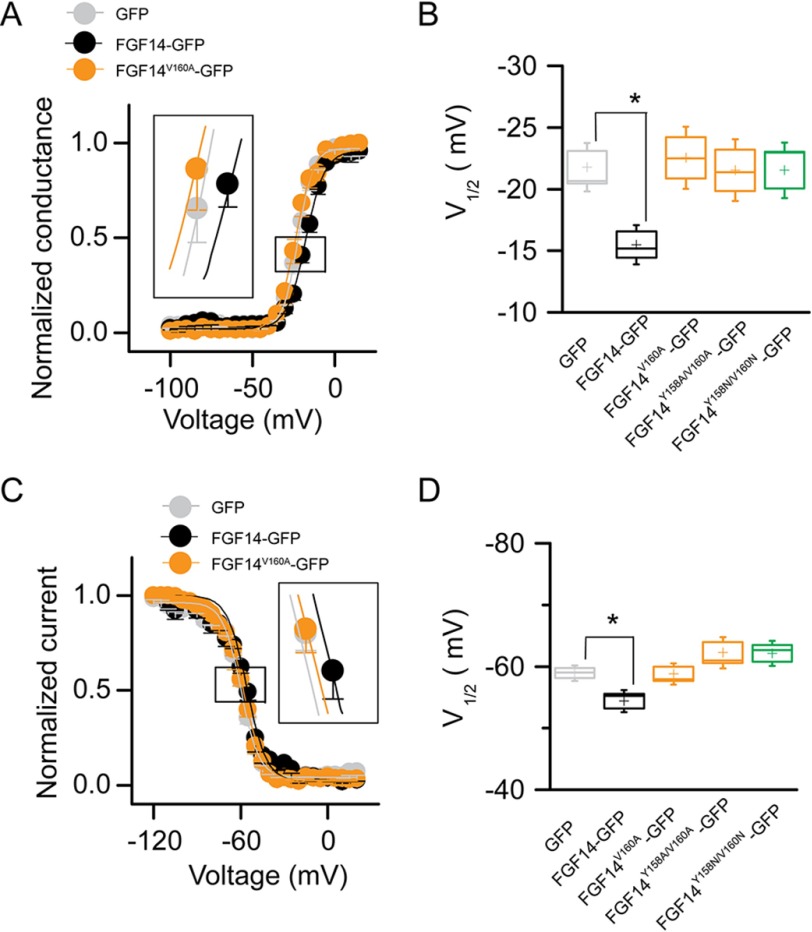
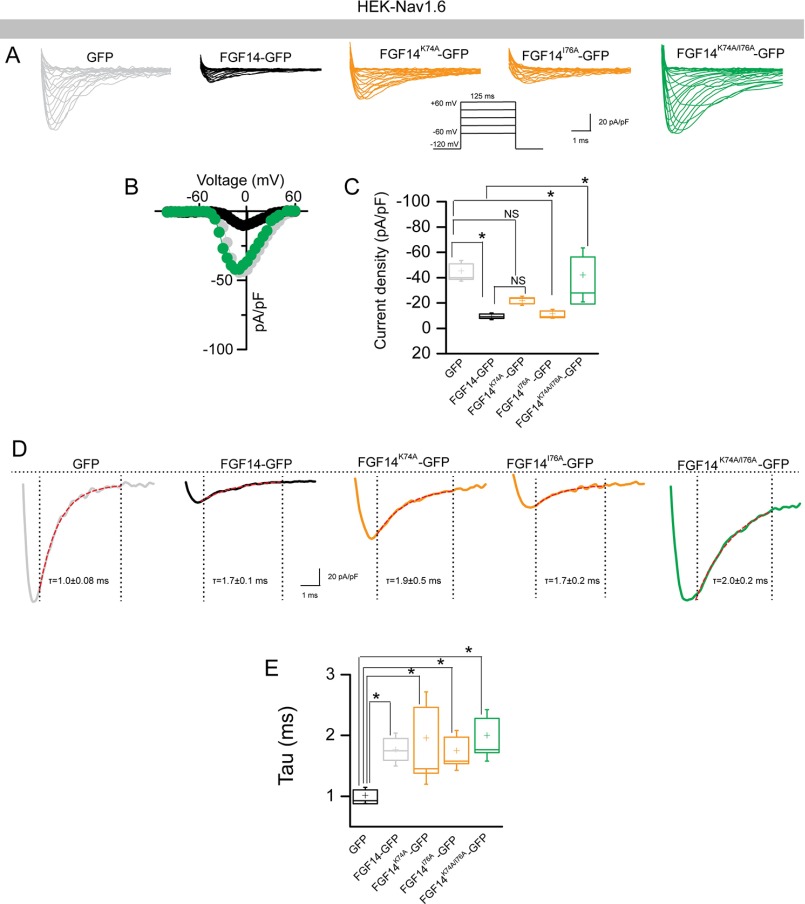
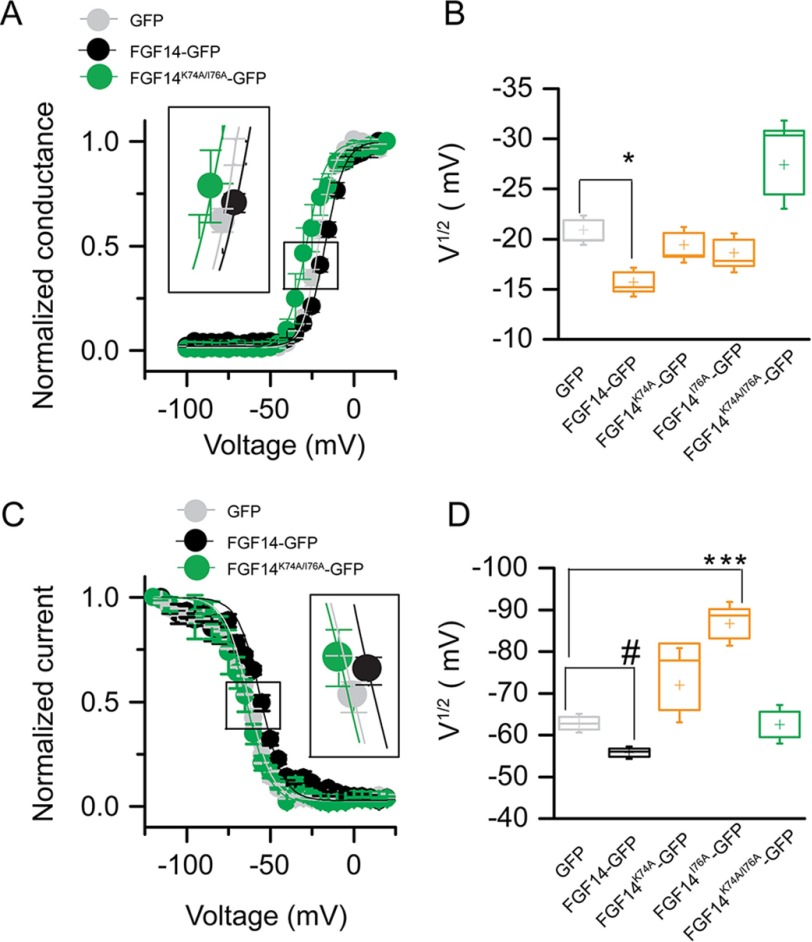
Abundant evidence exists for a role for FGF14 in regulating the current amplitude and biophysical properties of Nav1.6-mediated currents (54, 56). To investigate the functional impact of single Val-160 and combined Tyr-158/Val-160 mutations on the FGF14-dependent modulation of Nav1.6-encoded currents, we applied whole-cell patch clamp electrophysiology to HEK293 cells stably expressing Nav1.6 (HEK-Nav1.6 cells) that transiently expressed GFP, FGF14-GFP, FGF14V160A-GFP, FGF14Y158A/V160A-GFP, and/or FGF14Y158N/V160N-GFP constructs. The purpose of generating FGF14V160A, FGF14Y158A/V160A,and FGF14Y158N/V160N was to investigate the role of single amino acid polarity and size of Tyr-158 and Val-160 in modulating Na+ currents and to match our LCA studies. In agreement with previous studies (54, 55), we found that HEK-Nav1.6 cells expressing FGF14-GFP showed significantly lower peak INa+ densities (−9.8 ± 1.5 pA/pF, n = 14, p < 0.001, Kruskal-Wallis, post hoc Dunn test) than cells expressing GFP (−58.6 ± 13.4 pA/pF, n = 16 (Fig. 8, A–C, and Table 4)). Yet, we found that replacing the Val-160 residue with an alanine mutation led to no significant changes in peak INa+ densities and resulted in values comparable with GFP control (−44.14 ± 12.4 pA/pF, n = 10 for FGF14V160A-GFP (Table 4)). Similar phenotypes were observed also for FGF14 double mutants (−59.3 ± 14.1, n = 15 for FGF14Y158A/V160A and −58.5 ± 8.5 pA/pF, n = 11 for FGF14Y158N/V160N compared with GFP control). Further analysis revealed that the decay time constant (τ) of transient INa+ (which reflects the kinetics of open channel entry into the fast inactivation state) was significantly slower in FGF14-GFP (1.7 ± 0.2 ms, n = 10, p < 0.05) compared with GFP control (1.1 ± 0.1, n = 14). This phenotype persisted in the FGF14V160A-GFP group (1.7 ± 0.3, n = 10, p < 0.05) but was abolished in the FGF14Y158A/V160A-GFP group (0.8 ± 0.1, n = 15, p > 0.05) and FGF14Y158N/V160N-GFP (0.8 ± 0.1, n = 13, p > 0.05 (Fig. 8, D and E, and Table 4)). We also examined other basic biophysical properties of Nav1.6 channels in the presence of these FGF14 mutant proteins that were reported previously as targets of FGF14 modulation (54, 55). The voltage dependences of INa+ activation and steady-state inactivation were determined by plotting normalized conductance (activation) or normalized current amplitudes (inactivation) as a function of the test potential (activation) or the pre-pulse potential (inactivation) following previously described stimulation protocols (54, 55). In agreement with previous studies (54, 55), we found that expression of FGF14-GFP led to a significant depolarizing shift in both the voltage-dependent activation (p < 0.05) and steady-state inactivation of Nav1.6 (p < 0.05). Notably, we found that none of these parameters was significantly affected when cells expressed FGF14V160A-GFP, FGF14Y158A/V160A-GFP,and/or FGF14Y158N/V160N-GFP(Fig. 9, A–D, and Table 4). Thus, we concluded that Val-160, alone or in cooperation with Tyr-158, is required for FGF14-dependent functional modulation of Nav1.6 currents.
FIGURE 8.
The FGF14V160 residue is required for modulation of Nav1.6 currents. A, representative traces of voltage-gated Na+ currents (INa+) recorded from HEK-Nav1.6 cells transiently expressing GFP (gray), FGF14-GFP (black), FGF14V160A-GFP (orange), FGF14Y158A/V160A-GFP (orange), and FGF14Y158N/V160N-GFP (green) in response to voltage steps from −120 mV to +60 mV from a holding potential of −70 mV (inset). B, current-voltage relationships of INa+ from GFP (gray), FGF14-GFP (black), and FGF14V160A-GFP (orange). C, box plot represents peak current densities measured in individual HEK-Nav1.6 cells expressing GFP, FGF14, FGF14V160A-GFP, FGF14Y158A/V160A-GFP, and FGF14Y158N/V160N. D, representative traces of experimental groups described in A in which Tau (τ) of INa+ was estimated from a one-term exponential fitting function (red dotted line). Values are plotted as a function of amplitude and time constant. E, summary box plot of Tau calculated at the peak current density (−10 mV) in the indicated experimental groups. Data are mean ± S.E. *p < 0.05.
TABLE 4.
Nav1.6-mediated currents in the presence of FGF14 and Val-160 and Tyr-158 mutants
The number of independent experiments is shown in parentheses.
| Condition | Peak density | Activation | Kact | Inactivation | Kinact | τ |
|---|---|---|---|---|---|---|
| pA/pF | mV | mV | mV | mV | ms | |
| GFP | −58.6 ± 13.4 (16) | −20.4 ± 1.6 (12) | 5.3 ± 0.3 (12) | −58.9 ± 0.8(14) | 5.8 ± 0.3 (12) | 1.1 ± 0.1 (14) |
| FGF14-GFP | −9.8 ± 1.5 (14)a | −15.4 ± 1.1 (11)b | 6.3 ± 0.5 (11)c | −54.4 ± 1.4 (10)d | 6.3 ± 0.7(10) | 1.7 ± 0.2 (10)e |
| FGF14V160A-GFP | −44.14 ± 12.4 (10) | −22.4 ± 1.6 (10) | 4.6 ± 0.6 (10) | −58.8 ± 1.3 (10) | 6.4 ± 0.5 (10) | 1.7 ± 0.3 (10)f |
| FGF14Y158A/V160A-GFP | −59.3 ± 14.1 (15) | −21.5 ± 1.6 (15) | 4.6 ± 0.5 (15) | −62.2 ± 1.6 (15) | 7.8 ± 0.9 (15) | 0.8 ± 0.1 (15) |
| FGF14Y158N/V160N-GFP | −58.5 ± 8.5 (11) | −21.5 ± 1.5 (9) | 4.2 ± 0.4 (9) | −62.11 ± 1.3 (9) | 7.4 ± 0.6 (9) | 0.8 ± 0.1 (13) |
a p < 0.001, Kruskal-Wallis, post hoc Dunn's test compared with GFP; data are mean ± S.E.
b p < 0.05, one-way ANOVA, post hoc Dunnett's multiple comparisons test compared with GFP; data are mean ± S.E.
c p < 0.05, one-way ANOVA, post hoc Dunnett's multiple comparisons test compared with GFP; data are mean ± S.E.
d p < 0.001, Kruskal-Wallis, post hoc Dunnett's multiple comparisons test compared with GFP; data are mean ± S.E.
e p < 0.01, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
f p < 0.05, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
FIGURE 9.
The V160A mutation abolishes FGF14-dependent modulation of biophysical properties of Nav1.6 currents. A, voltage dependence of INa activation is plotted as a function of the membrane potential (mV); data (GFP, FGF14-GFP, and FGF14V160A-GFP) were fitted with the Boltzmann function as indicated under “Experimental Procedures.” B, box plot summary of V½ for voltage-dependent activation (voltage at which 50% of the channels are opened) in the indicated experimental groups. C, steady-state inactivation was measured using a two-step protocol, and values were plotted as a function of the membrane potential (mV). Data (GFP, FGF14-GFP, FGF14V160A-GFP) were fitted with the Boltzmann function as indicated under “Experimental Procedures.” The shift of voltage-dependent activation and steady-state inactivation is shown in the two insets in A and C, respectively. D, box plot summary of V½ for voltage-dependent steady-state inactivation (voltage at which 50% of the channels are closed) in the indicated experimental groups. Data are mean ± S.E.; *, p < 0.05.
Lys-74 and Ile-76 Are Required for Modulation of Nav1.6 Currents
We extended our whole-cell patch clamp studies to evaluate the impact of FGF14K74A-GFP, FGF14I76A-GFP, and/or FGF14K74A/I76A-GFP constructs on the FGF14-dependent modulation of Nav1.6 currents. We found that although the FGF14I76A mutant behaved similar to FGF14-GFP (−10.9 ± 2.4 pA/pF, n = 7 (Fig. 10, A–C, and Table 5)), the FGF14K74A single or FGF14K74A/I76A double mutant rescued peak INa+ densities to GFP control values either partially (−21.2 ± 2.5 pA/pF, n = 8 for FGF14K74A-GFP (Table 5)) or completely (−46.8 ± 13.6, n = 10 for FGF14K74A/I76A (Table 5)). However, none of the mutant combinations was effective in rescuing τ of transient INa+ (Fig. 10, D and E, and Table 5). Additional studies revealed that in the presence of these FGF14 mutants, the voltage-dependent activation and steady-state inactivation of Nav1.6 currents were either indistinguishable from GFP control or exhibited a gain-of-function phenotype (voltage-dependent steady-state inactivation in the presence of FGF14I76A-GFP), suggesting a complex and cooperative interaction of Lys-74 and Ile-76 in regulating the biophysical properties of Nav1.6 (Fig. 11 and Table 5).
FIGURE 10.
Functional validation of Lys-74 and Ile-76 in modulating Nav1.6 currents. A, representative traces of voltage-gated Na+ currents (INa+) recorded from HEK-Nav1.6 cells transiently expressing GFP (gray), FGF14-GFP (black), FGF14K74A-GFP (orange), FGF14I76A-GFP (orange), and FGF14K74A/I76A-GFP (green) in response to voltage steps from −120 mV to +60 mV from a holding potential of −70 mV (inset). B, current-voltage relationships of INa+ from GFP (gray), FGF14-GFP (black), and FGF14K74A/I76A-GFP (green). C, box plot represents peak current densities measured in individual HEK-Nav1.6 cells expressing GFP, FGF14, FGF14K74A-GFP, FGF14I76A-GFP, and FGF14K74A/I76A. D, representative traces of experimental groups described in A in which Tau (τ) of INa+ was estimated from a one-term exponential fitting function (red dotted line). Values are plotted as a function of amplitude and time constant. E, summary box plot of Tau calculated at the peak current density (−10 mV) in the indicated experimental groups. Data are mean ± S.E. *, p < 0.05.
TABLE 5.
Nav1.6-mediated currents in the presence of FGF14 and Lys-74 and Ile-76 mutants
The number of independent experiments is shown in parentheses.
| Condition | Peak density | Activation | Kact | Inactivation | Kinact | τ |
|---|---|---|---|---|---|---|
| pA/pF | mV | mV | mV | mV | ms | |
| GFP | −43.2 ± 5.1 (21) | −20.8 ± 0.9 (19) | 4.7 ± 0.2 (19) | −62 ± 1.4 (15) | 6.3 ± 0.3 (15) | 1.0 ± 0.08 (14) |
| FGF14-GFP | −9.3 ± 1.8 (12)a | −15.7 ± 0.9 (11)b | 6.2 ± 0.4 (11)c | −56.0 ± 1.0 (10)c | 7.1 ± 1.1 (10) | 1.7 ± 0.18 (10)d |
| FGF14K74A-GFP | −21.2 ± 2.5 (8) | −19.4 ± 1.1 (8) | 4.8 ± 0.2 (8) | −71.9 ± 5.9 (8) | 11.2 ± 1. 1 (8) | 1.9 ± 0.5 (8)d |
| FGF14I76A-GFP | −10.9 ± 2.4 (7)e | −18.6 ± 1.2 (7) | 6.2 ± 0.3 (7)c | −86.6 ± 3.5 (7)f | 15.7 ± 1.0 (7)g | 1.7 ± 0.2 (7)d |
| FGF14K74A/I76A-GFP | −46.8 ± 13.6 (10) | −27.4 ± 2.9 (8) | 3.9 ± 0.7 (8) | −62.5 ± 3.0 (9) | 8.8 ± 1.2 (9) | 2.0 ± 0.2 (9)d |
a p < 0.001, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
b p < 0.01, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
c p < 0.05, one-way ANOVA, post hoc Dunnett's multiple comparisons test compared with GFP; data are mean ± S.E.
d p < 0.05, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
e p < 0.01, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
f p < 0.05, student-t test compared with GFP; data are mean ± S.E.
g p < 0.01, one-way ANOVA, post hoc Dunn's multiple comparisons test compared with GFP; data are mean ± S.E.
FIGURE 11.
Role of Lys-74 and I76 in modulating biophysical properties of Nav1.6 currents. A, voltage dependence of INa activation is plotted as a function of the membrane potential (mV). Data (GFP, FGF14-GFP, and FGF14K74A/I76A-GFP) were fitted with the Boltzmann function as indicated in under “Experimental Procedures.” B, box plot summary of V½ for voltage-dependent activation (voltage at which 50% of the channels are opened) in the indicated experimental groups. C, steady-state inactivation was measured using a two-step protocol, and values were plotted as a function of the membrane potential (mV). Data (GFP, FGF14-GFP, and FGF14K74A/I76A-GFP) were fitted with the Boltzmann function as indicated under “Experimental Procedures.” The shift of voltage-dependent activation and steady-state inactivation is shown in the two insets in A and C, respectively. D, box plot summary of V½ for voltage-dependent steady-state inactivation (voltage at which 50% of the channels are closed) in the indicated experimental groups. Data are mean ± S.E.; *, p < 0.05.
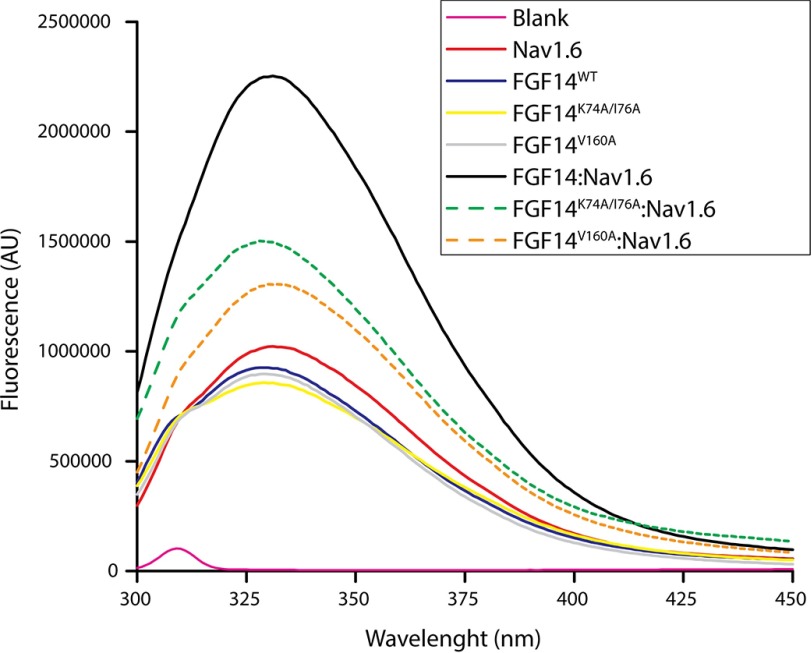
Intrinsic Fluorescence Spectroscopy Confirms a Key Role for Val-160 in the FGF14·Nav1.6 Complex
Our molecular modeling, LCA, and patch clamp data indicate that the Val-160 residue in FGF14 plays a unique and crucial role in modulating Nav1.6 currents. A single alanine switch at this site or a concomitant alanine mutation at Tyr-158 and Val-160 are the only changes that can fully restore Nav1.6 currents to the GFP control level. Lys-74 and Ile-76, on the other hand, might work more synergistically, and mutations at these sites cannot completely rescue changes in Nav1.6 currents mediated by FGF14. To provide correlative binding studies to our functional data, we used tryptophan-based fluorescence spectroscopy to probe energy transfer processes occurring in PPI. The tryptophan fluorescence spectra for individual FGF14WT, FGF14K74A/I76A, FGF14V160A, and Nav1.6-C tail proteins exhibited a λmax at 332 nm. Combining the FGF14WT and the Nav1.6 C-tail increased the fluorescence emission intensity by more than 2-fold without any shift in the λmax, indicating strong protein complex formation without a change in the local environment. Both FGF14K74A/I76A and FGF14V160A mutants disrupted the interaction with Nav1.6-C-tail, but FGF14V160A appeared to be the most damaging (Fig. 12). All mutant proteins had hydrodynamic radii identical to FGF14, as observed during gel filtration (data not shown). As evident from the emission spectra profiles (Fig. 12), none of the mutations lead to any major conformational changes in the protein complex, indicating that reduction in fluorescence intensity arises from decreased binding affinity (67).
FIGURE 12.
Intrinsic fluorescence emission spectra reveal reduced assembly of FGF14K74A/I76A and FGF14V160A to Nav1.6 C-tail. Shown are the fluorescence spectra of the indicated purified proteins alone or combined; blank, Nav1.6, FGF14WT, FGF14K74A/I76A, FGF14V160A, FGF14WT·Nav1.6, FGF14K74A/I76A·Nav1.6, and FGF14V160A·Nav1.6 are shown as pink, red, blue, yellow, gray, black, green broken line, and orange broken line, respectively.
Discussion
Previous studies have proposed that all iFGF might utilize a common interface for PPI with specific Nav isoform C-tail or other iFGFs (iFGF:iFGF dimer complexes); a hypothesis not yet directly tested. Through molecular, cellular, functional, and structural studies focused on FGF14, a disease-associated protein (46, 48, 49, 68) and potent regulator of Nav1.6 channels (54, 56), we identified a significant structure-function similarity and divergence between the PPI interface within the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer.
Using homology models we compared in silico the FGF14·Nav1.6 and FGF14·FGF14 complexes (Fig. 1) and proposed Lys-74, Ile-76, Leu-109, Leu-116, Arg-117, Asn-157, Tyr-158, Tyr-159, Leu-202, Pro-205, and Val-208 at the FGF14 surface as potential hot spots shared by the two complexes. These residues are part of the N terminus, β-5, β-9, and β-12 strand of FGF14, and corresponding residues in FGF13 are already recognized as key structural amino acids (24, 52, 69). Yet, our in silico models predict potential structural divergence at the FGF14 Val-160 residue of the β-9.
To provide experimental evidence for our model studies, we designed single, double, and quadruple mutations at the in silico predicted hot spots and tested FGF14 mutant activities using LCA to reconstitute PPI complexes in live cells (51, 57, 62, 65). LCA studies confirmed our in silico predictions showing that most mutations destabilized both the FGF14·Nav1.6 and the FGF14·FGF14 complex but that the FGF14Y158N/V160N double mutant led to opposite phenotypes depending on the structural context. The FGF14Y158N/V160N mutant increased the stability of the FGF14:FGF14 dimer, but it impaired FGF14·Nav1.6 complex formation. Other amino acid residues that deserved attention were Lys-74 and Ile-76 in the N terminus of the FGF14. Mutations at these residues strongly impaired the FGF14·Nav1.6 complex formation, but the effect was preserved in the FGF14 dimer complex, indicating a potential conserved role for the N terminus at the two PPI interfaces.
This result prompted us to examine the role of Tyr-158, Val-160, Lys-74, and Ile-76 in the FGF14·Nav1.6 and FGF14:FGF14 dimer complex using targeted alanine-scanning mutations in combination with LCA. When examined in the context of the FGF14·Nav1.6 complex, I76A and Y158A did not lead to any phenotypes (Figs. 4B and 6B), whereas K74A and/or V160A were sufficient to disrupt FGF14·Nav1.6 complex formation (Fig. 4B and 6B). In the FGF14WT·Nav1.6 homology model, we observed that Lys-74 and Val-160 interacted with Glu-1884 and Ile-1886 of Nav1.6, respectively, through salt bridge and hydrophobic interactions. These findings corroborate the critical role of Lys-74 and Val-160 residues in holding PPI interfaces through salt bridge (69) and hydrophobic interactions, respectively (70). Replacement of Lys-74 and Val-160 with a smaller alanine residue might increase the distance between the two neighboring residues, resulting in a less favorable structural environment for PPI (66). Importantly, we showed that FGF14Y158A or FGF14V160A alone are not sufficient to disrupt the FGF14:FGF14 dimer formation. In the FGF14:FGF14 dimer homology model, Tyr-158 directly interacts with Val-208 of the neighboring monomer via hydrogen bonding (71). Replacing Tyr-158 with Ala is not sufficient to interfere structurally with the dimer, but if combined with V160A the stability of the β-9 strand might be weakened and monomer affinity reduced. Simultaneous mutations of Tyr-158 and Val-160 to alanine can work synergistically to disrupt both the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer, but the single V160A mutant has a different impact on the FGF14:FGF14 dimer versus the FGF14·Nav1.6 complex. Replacing both Tyr-158 and Val-160 with an Asn in each FGF14 monomer increases FGF14:FGF14 dimer formation (Fig. 2D). Both Tyr and Asn are polar residues; however, Asn is smaller than Tyr. Replacing both the bulky Tyr-158 and the Val-160 residues in the two FGF14 monomers with a smaller Asn residue might facilitate interactions and increase the stability of the mutant homodimer (FGF14Y158N/V160N:FGF14Y158N/V160N). These predictions and results are in agreement with previous in silico and LCA studies from our group (51).
At the N terminus of FGF14, we found that Lys-74 directly interacts with Tyr-159 through a strong salt bridge and that replacing Lys-74 with an alanine disrupts this interaction, impairing the FGF14:FGF14 dimer formation. Lys-74 and Ile-76 acted synergistically in the FGF14·Nav1.6 complex formation but not in the FGF14 dimer (Fig. 6, A, B, E, and F), further supporting structural divergence at the two PPI interfaces.
Our molecular modeling and LCA studies were corroborated by whole-cell patch clamp electrophysiology. We investigated whether mutations at Lys-74 and Val-160 had any functional impact on the well described effect of FGF14 on Nav1.6-mediated currents. In agreement with previous studies, we found that FGF14 suppresses transient peak INa+ density and affects voltage-dependent activation and steady-state inactivation compared with control. The single FGF14V160A completely rescued peak current density to the control (GFP) (Fig. 8 and Table 4), whereas FGF14K74A was only partially effective (Fig. 10 and Table 5). Ile-76 worked synergistically with Lys-74 in that the FGF14K74A/I76A mutant fully rescued Nav1.6-mediated currents to the control (GFP), supporting the LCA results (Fig. 6B). A more thorough analysis of Nav1.6 currents revealed a previously unreported effect of FGF14 on fast inactivation. This phenotype persisted with expression of FGF14V160A and required the double mutations at Val-160 and Tyr-158 (FGF14Y158A/V160Aand FGF14Y158N/V160N) to be abolished. Notably, single lys-74 or Ile-76 or double Lys-74/Ile-76 mutations were unable to abolish the changes in Tau (Fig 10, D and E). Intrinsic fluorescence spectroscopy (72) based on purified proteins confirmed this model, indicating that the single V160A mutation was more disruptive than K74A/I76A in impairing the FGF14·Nav1.6 complex formation.
Collectively, our studies demonstrate that amino acid residues located at the N terminus and at the β-9 of FGF14 are crucial for the FGF14·Nav1.6 complex and the FGF14:FGF14 dimer formation. Yet, the Val-160 residue is a point of divergence between the two complexes and is required for full FGF14 functional activity toward Nav1.6 channels. Although Val-160 is conserved in other iFGFs, its role varies depending on the structural environment provided by the specific iFGF and Nav channel isoforms. In the FGF13·Nav1.5 complex, for instance, Tyr-98 (the FGF13 residue corresponding to FGF14 Tyr-158) appears to have a more prominent role in the PPI complex formation, suggesting high precision and fidelity at each iFGF·Nav channel complex interface (69). Chemical probes that could leverage these unique structure-function features might provide an unprecedented opportunity for targeted interventions against excitability-driven brain and heart pathologies.
Author Contributions
S. R. A., A. K. S., and F. L. all wrote the manuscript. S. R. A. designed and performed the split-luciferase complementation assay, Western blotting analysis, and patch clamp electrophysiological experiments, analyzed the data, and built the homology models. A. K. S. purified the proteins and performed intrinsic fluorescence experiments. F. L. designed and supervised the work and the analysis and interpretation of the data. All authors read and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Neli Panova for technical support, Dr. Miroslav Nenov for critical feedback on analyzing patch clamp data and for reviewing the manuscript. We acknowledge the Sealy Center for Structural Biology and Molecular Biophysics at the University of Texas Medical Branch at Galveston for providing research resources and Dr. Heather Lander for proofreading the manuscript.
This work was supported by National Institutes of Health Grants R01MH095995 (to F. L.) and 1 T32 GM089657-04 (to S. R. A.) from NIGMS (a training fellowship from the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia). This work was also supported by the John Sealy Memorial Endowment Fund (to F. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Nav
- voltage-gated sodium
- iFGF
- intracellular fibroblast growth factor
- PPI
- protein-protein interaction
- LCA
- split-luciferase complementation assay
- aa
- amino acid(s)
- PDB
- Protein Data Bank
- RLU
- relative luminescence unit
- ANOVA
- analysis of variance.
References
- 1. Catterall W. A. (2012) Voltage-gated sodium channels at 60: structure, function, and pathophysiology. J. Physiol. 590, 2577–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Catterall W. A. (2014) Structure and function of voltage-gated sodium channels at atomic resolution. Exp. Physiol. 99, 35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Catterall W. A., Goldin A. L., and Waxman S. G. (2005) International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57, 397–409 [DOI] [PubMed] [Google Scholar]
- 4. Chahine M., Chatelier A., Babich O., and Krupp J. J. (2008) Voltage-gated sodium channels in neurological disorders. CNS Neurol. Disord. Drug Targets 7, 144–158 [DOI] [PubMed] [Google Scholar]
- 5. Cusdin F. S., Clare J. J., and Jackson A. P. (2008) Trafficking and cellular distribution of voltage-gated sodium channels. Traffic 9, 17–26 [DOI] [PubMed] [Google Scholar]
- 6. Denac H., Mevissen M., and Scholtysik G. (2000) Structure, function and pharmacology of voltage-gated sodium channels. Naunyn Schmiedebergs Arch. Pharmacol. 362, 453–479 [DOI] [PubMed] [Google Scholar]
- 7. Goldin A. L., Barchi R. L., Caldwell J. H., Hofmann F., Howe J. R., Hunter J. C., Kallen R. G., Mandel G., Meisler M. H., Netter Y. B., Noda M., Tamkun M. M., Waxman S. G., Wood J. N., and Catterall W. A. (2000) Nomenclature of voltage-gated sodium channels. Neuron 28, 365–368 [DOI] [PubMed] [Google Scholar]
- 8. Leterrier C., Brachet A., Fache M. P., and Dargent B. (2010) Voltage-gated sodium channel organization in neurons: protein interactions and trafficking pathways. Neurosci. Lett. 486, 92–100 [DOI] [PubMed] [Google Scholar]
- 9. Marban E., Yamagishi T., and Tomaselli G. F. (1998) Structure and function of voltage-gated sodium channels. J. Physiol. 508, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drendall C. I., Pham Q. H., and Dietze E. C. (2010) Purification and characterization of recombinant CH3 domain fragment of the CREB-binding protein. Protein Expr. Purif. 70, 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Savio-Galimberti E., Gollob M. H., and Darbar D. (2012) Voltage-gated sodium channels: biophysics, pharmacology, and related channelopathies. Front. Pharmacol. 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu F. H., and Catterall W. A. (2003) Overview of the voltage-gated sodium channel family. Genome Biol. 4, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laedermann C. J., Pertin M., Suter M. R., and Decosterd I. (2014) Voltage-gated sodium channel expression in mouse DRG after SNI leads to re-evaluation of projections of injured fibers. Mol. Pain 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mantegazza M., Curia G., Biagini G., Ragsdale D. S., and Avoli M. (2010) Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 9, 413–424 [DOI] [PubMed] [Google Scholar]
- 15. Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., and De Jonghe P. (2001) De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 68, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mantegazza M., Gambardella A., Rusconi R., Schiavon E., Annesi F., Cassulini R. R., Labate A., Carrideo S., Chifari R., Canevini M. P., Canger R., Franceschetti S., Annesi G., Wanke E., and Quattrone A. (2005) Identification of an Nav1.1 sodium channel (SCN1A) loss-of-function mutation associated with familial simple febrile seizures. Proc. Natl. Acad. Sci. U.S.A. 102, 18177–18182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mullen S. A., and Scheffer I. E. (2009) Translational research in epilepsy genetics: sodium channels in man to interneuronopathy in mouse. Arch. Neurol. 66, 21–26 [DOI] [PubMed] [Google Scholar]
- 18. Wright G. E. (2015) Genomic study of congenital insensitivity to pain provides new avenues for the development of analgesics. Clin. Genet. 88, 342–343 [DOI] [PubMed] [Google Scholar]
- 19. Woods C. G., Babiker M. O., Horrocks I., Tolmie J., and Kurth I. (2015) The phenotype of congenital insensitivity to pain due to the NaV1.9 variant p.L811P. Eur. J. Hum. Genet. 23, 1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang Z., Chen Z., Tang B., and Jiang H. (2015) Primary erythromelalgia: a review. Orphanet J. Rare Dis. 10, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dib-Hajj S. D., Binshtok A. M., Cummins T. R., Jarvis M. F., Samad T., and Zimmermann K. (2009) Voltage-gated sodium channels in pain states: role in pathophysiology and targets for treatment. Brain Res. Rev. 60, 65–83 [DOI] [PubMed] [Google Scholar]
- 22. Lampert A., O'Reilly A. O., Reeh P., and Leffler A. (2010) Sodium channelopathies and pain. Pflugers Arch. 460, 249–263 [DOI] [PubMed] [Google Scholar]
- 23. Wang Q., Shen J., Splawski I., Atkinson D., Li Z., Robinson J. L., Moss A. J., Towbin J. A., and Keating M. T. (1995) SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell 80, 805–811 [DOI] [PubMed] [Google Scholar]
- 24. Musa H., Kline C. F., Sturm A. C., Murphy N., Adelman S., Wang C., Yan H., Johnson B. L., Csepe T. A., Kilic A., Higgins R. S., Janssen P. M., Fedorov V. V., Weiss R., Salazar C., et al. (2015) SCN5A variant that blocks fibroblast growth factor homologous factor regulation causes human arrhythmia. Proc. Natl. Acad. Sci. U.S.A. 112, 12528–12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Probst V., Kyndt F., Potet F., Trochu J. N., Mialet G., Demolombe S., Schott J. J., Baró I., Escande D., and Le Marec H. (2003) Haploinsufficiency in combination with aging causes SCN5A-linked hereditary Lenegre disease. J. Am. Coll. Cardiol. 41, 643–652 [DOI] [PubMed] [Google Scholar]
- 26. Emmett M. R., Kroes R. A., Moskal J. R., Conrad C. A., Priebe W., Laezza F., Meyer-Baese A., and Nilsson C. L. (2014) Integrative biological analysis for neuropsychopharmacology. Neuropsychopharmacology 39, 5–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Post R. M., Frye M. A., Denicoff K. D., Leverich G. S., Kimbrell T. A., and Dunn R. T. (1998) Beyond lithium in the treatment of bipolar illness. Neuropsychopharmacology 19, 206–219 [DOI] [PubMed] [Google Scholar]
- 28. Farber N. B., Jiang X. P., Heinkel C., and Nemmers B. (2002) Antiepileptic drugs and agents that inhibit voltage-gated sodium channels prevent NMDA antagonist neurotoxicity. Mol. Psychiatry 7, 726–733 [DOI] [PubMed] [Google Scholar]
- 29. Prakriya M., and Mennerick S. (2000) Selective depression of low-release probability excitatory synapses by sodium channel blockers. Neuron 26, 671–682 [DOI] [PubMed] [Google Scholar]
- 30. Papale L. A., Paul K. N., Sawyer N. T., Manns J. R., Tufik S., and Escayg A. (2010) Dysfunction of the Scn8a voltage-gated sodium channel alters sleep architecture, reduces diurnal corticosterone levels, and enhances spatial memory. J. Biol. Chem. 285, 16553–16561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Large C. H., Bison S., Sartori I., Read K. D., Gozzi A., Quarta D., Antolini M., Hollands E., Gill C. H., Gunthorpe M. J., Idris N., Neill J. C., and Alvaro G. S. (2011) The efficacy of sodium channel blockers to prevent phencyclidine-induced cognitive dysfunction in the rat: potential for novel treatments for schizophrenia. J. Pharmacol. Exp. Ther. 338, 100–113 [DOI] [PubMed] [Google Scholar]
- 32. Eijkelkamp N., Linley J. E., Baker M. D., Minett M. S., Cregg R., Werdehausen R., Rugiero F., and Wood J. N. (2012) Neurological perspectives on voltage-gated sodium channels. Brain 135, 2585–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clare J. J., Tate S. N., Nobbs M., and Romanos M. A. (2000) Voltage-gated sodium channels as therapeutic targets. Drug Discov. Today 5, 506–520 [DOI] [PubMed] [Google Scholar]
- 34. Dib-Hajj S. D., Black J. A., and Waxman S. G. (2009) Voltage-gated sodium channels: therapeutic targets for pain. Pain Med. 10, 1260–1269 [DOI] [PubMed] [Google Scholar]
- 35. Bath K. G., and Scharfman H. E. (2013) Impact of early life exposure to antiepileptic drugs on neurobehavioral outcomes based on laboratory animal and clinical research. Epilepsy Behav. 26, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Theile J. W., and Cummins T. R. (2011) Recent developments regarding voltage-gated sodium channel blockers for the treatment of inherited and acquired neuropathic pain syndromes. Front. Pharmacol. 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olsen S. K., Garbi M., Zampieri N., Eliseenkova A. V., Ornitz D. M., Goldfarb M., and Mohammadi M. (2003) Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J. Biol. Chem. 278, 34226–34236 [DOI] [PubMed] [Google Scholar]
- 38. Schoorlemmer J., and Goldfarb M. (2001) Fibroblast growth factor homologous factors are intracellular signaling proteins. Curr. Biol. 11, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Cj., Dib-Hajj S. D., and Waxman S. G. (2001) Fibroblast growth factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNav1.9a (NaN). J. Biol. Chem. 276, 18925–18933 [DOI] [PubMed] [Google Scholar]
- 40. Liu C. J., Dib-Hajj S. D., Renganathan M., Cummins T. R., and Waxman S. G. (2003) Modulation of the cardiac sodium channel Nav1.5 by fibroblast growth factor homologous factor 1B. J. Biol. Chem. 278, 1029–1036 [DOI] [PubMed] [Google Scholar]
- 41. Wittmack E. K., Rush A. M., Craner M. J., Goldfarb M., Waxman S. G., and Dib-Hajj S. D. (2004) Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J. Neurosci. 24, 6765–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wildburger N. C., Ali S. R., Hsu W. C., Shavkunov A. S., Nenov M. N., Lichti C. F., LeDuc R. D., Mostovenko E., Panova-Elektronova N. I., Emmett M. R., Nilsson C. L., and Laezza F. (2015) Quantitative proteomics reveals protein-protein interactions with fibroblast growth factor 12 as a component of the voltage-gated sodium channel 1.2 (nav1.2) macromolecular complex in mammalian brain. Mol. Cell. Proteomics 14, 1288–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stoilova-McPhie S., Ali S., and Laezza F. (2013) Protein-protein interactions as new targets for ion channel drug discovery. Austin J. Pharmacol. Ther. 1, pii5. [PMC free article] [PubMed] [Google Scholar]
- 44. Pitteri S., and Hanash S. (2010) A systems approach to the proteomic identification of novel cancer biomarkers. Dis. Markers 28, 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teiwes J., and Toto R. D. (2007) Epithelial sodium channel inhibition in cardiovascular disease: a potential role for amiloride. Am. J. Hypertens. 20, 109–117 [DOI] [PubMed] [Google Scholar]
- 46. Verbeek E. C., Bakker I. M., Bevova M. R., Bochdanovits Z., Rizzu P., Sondervan D., Willemsen G., de Geus E. J., Smit J. H., Penninx B. W., Boomsma D. I., Hoogendijk W. J., and Heutink P. (2012) A fine-mapping study of 7 top scoring genes from a GWAS for major depressive disorder. PLoS One 7, e37384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez-Murillo L., Xu B., Roos J. L., Abecasis G. R., Gogos J. A., and Karayiorgou M. (2014) Fine mapping on chromosome 13q32–34 and brain expression analysis implicates MYO16 in schizophrenia. Neuropsychopharmacology 39, 934–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiao M., Xu L., Laezza F., Yamada K., Feng S., and Ornitz D. M. (2007) Impaired hippocampal synaptic transmission and plasticity in mice lacking fibroblast growth factor 14. Mol. Cell. Neurosci. 34, 366–377 [DOI] [PubMed] [Google Scholar]
- 49. van Swieten J. C., Brusse E., de Graaf B. M., Krieger E., van de Graaf R., de Koning I., Maat-Kievit A., Leegwater P., Dooijes D., Oostra B. A., and Heutink P. (2003) A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia [corrected]. Am. J. Hum. Genet. 72, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rush A. M., Wittmack E. K., Tyrrell L., Black J. A., Dib-Hajj S. D., and Waxman S. G. (2006) Differential modulation of sodium channel Na(v)1.6 by two members of the fibroblast growth factor homologous factor 2 subfamily. Eur. J. Neurosci. 23, 2551–2562 [DOI] [PubMed] [Google Scholar]
- 51. Ali S., Shavkunov A., Panova N., Stoilova-McPhie S., and Laezza F. (2014) Modulation of the FGF14:FGF14 homodimer interaction through short peptide fragments. CNS Neurol. Disord. Drug Targets 13, 1559–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goetz R., Dover K., Laezza F., Shtraizent N., Huang X., Tchetchik D., Eliseenkova A. V., Xu C. F., Neubert T. A., Ornitz D. M., Goldfarb M., and Mohammadi M. (2009) Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 284, 17883–17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tempia F., Hoxha E., Negro G., Alshammari M. A., Alshammari T. K., Panova-Elektronova N., and Laezza F. (2015) Parallel fiber to Purkinje cell synaptic impairment in a mouse model of spinocerebellar ataxia type 27. Front. Cell. Neurosci. 9, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laezza F., Lampert A., Kozel M. A., Gerber B. R., Rush A. M., Nerbonne J. M., Waxman S. G., Dib-Hajj S. D., and Ornitz D. M. (2009) FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 42, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shavkunov A. S., Wildburger N. C., Nenov M. N., James T. F., Buzhdygan T. P., Panova-Elektronova N. I., Green T. A., Veselenak R. L., Bourne N., and Laezza F. (2013) The fibroblast growth factor 14.voltage-gated sodium channel complex is a new target of glycogen synthase kinase 3 (GSK3). J. Biol. Chem. 288, 19370–19385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lou J. Y., Laezza F., Gerber B. R., Xiao M., Yamada K. A., Hartmann H., Craig A. M., Nerbonne J. M., and Ornitz D. M. (2005) Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J. Physiol. 569, 179–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shavkunov A. S., Ali S. R., Panova-Elektronova N. I., and Laezza F. (2015) Split-luciferase complementation assay to detect channel-protein interactions in live cells. Methods Mol. Biol. 1278, 497–514 [DOI] [PubMed] [Google Scholar]
- 58. Guex N., Peitsch M. C., and Schwede T. (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, Suppl. 1, S162–S173 [DOI] [PubMed] [Google Scholar]
- 59. Ramachandran S., Kota P., Ding F., and Dokholyan N. V. (2011) Automated minimization of steric clashes in protein structures. Proteins 79, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 62. Shavkunov A., Panova N., Prasai A., Veselenak R., Bourne N., Stoilova-McPhie S., and Laezza F. (2012) Bioluminescence methodology for the detection of protein-protein interactions within the voltage-gated sodium channel macromolecular complex. Assay Drug Dev. Technol. 10, 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cohen M., Potapov V., and Schreiber G. (2009) Four distances between pairs of amino acids provide a precise description of their interaction. PLoS Comput. Biol. 5, e1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tobi D., and Elber R. (2000) Distance-dependent, pair potential for protein folding: results from linear optimization. Proteins 41, 40–46 [PubMed] [Google Scholar]
- 65. Hsu W. C., Nenov M. N., Shavkunov A., Panova N., Zhan M., and Laezza F. (2015) Identifying a kinase network regulating FGF14:Nav1.6 complex assembly using split-luciferase complementation. PLoS One 10, e0117246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gregoret L. M., and Sauer R. T. (1998) Tolerance of a protein helix to multiple alanine and valine substitutions. Fold. Des. 3, 119–126 [DOI] [PubMed] [Google Scholar]
- 67. Möller M., and Denicola A. (2002) Protein tryptophan accessibility studied by fluorescence quenching. Biochem. Mol. Biol. Educ. 30, 175–178 [Google Scholar]
- 68. Wang Q., Bardgett M. E., Wong M., Wozniak D. F., Lou J., McNeil B. D., Chen C., Nardi A., Reid D. C., Yamada K., and Ornitz D. M. (2002) Ataxia and paroxysmal dyskinesia in mice lacking axonally transported FGF14. Neuron 35, 25–38 [DOI] [PubMed] [Google Scholar]
- 69. Wang C., Chung B. C., Yan H., Lee S. Y., and Pitt G. S. (2012) Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure 20, 1167–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ochiai K., Yoshikawa Y., Yoshimatsu K., Oonuma T., Tomioka Y., Takeda E., Arikawa J., Mominoki K., Omi T., Hashizume K., and Morimatsu M. (2011) Valine 1532 of human BRC repeat 4 plays an important role in the interaction between BRCA2 and RAD51. FEBS Lett. 585, 1771–1777 [DOI] [PubMed] [Google Scholar]
- 71. Jiang L., and Lai L. (2002) CH.O hydrogen bonds at protein-protein interfaces. J. Biol. Chem. 277, 37732–37740 [DOI] [PubMed] [Google Scholar]
- 72. Chen Y., and Barkley M. D. (1998) Toward understanding tryptophan fluorescence in proteins. Biochemistry 37, 9976–9982 [DOI] [PubMed] [Google Scholar]