Abstract
Corticosteroid-binding globulin (CBG) was isolated from chicken serum and identified by mass spectrometry and genomic analysis. This revealed that the organization and synteny of avian and mammalian SerpinA6 genes are conserved. Recombinant zebra finch CBG steroid-binding properties reflect those of the natural protein in plasma and confirm its identity. Zebra finch and rat CBG crystal structures in complex with cortisol resemble each other, but their primary structures share only ∼40% identity, and their steroid-binding site topographies differ in several unexpected ways. Remarkably, a tryptophan that anchors ligands in mammalian CBG steroid-binding sites is replaced by an asparagine. Phylogenetic comparisons show that reptilian CBG orthologs share this unexpected property. Glycosylation of this asparagine in zebra finch CBG does not influence its steroid-binding affinity, but we present evidence that it may participate in protein folding and steroid-binding site formation. Substitutions of amino acids within zebra finch CBG that are conserved only in birds reveal how they contribute to their distinct steroid-binding properties, including their high (nanomolar) affinities for glucocorticoids, progesterone, and androgens. As in mammals, a protease secreted by Pseudomonas aeruginosa cleaves CBG in zebra finch plasma within its reactive center loop and disrupts steroid binding, suggesting an evolutionarily conserved property of CBGs. Measurements of CBG mRNA in zebra finch tissues indicate that liver is the main site of plasma CBG production, and anti-zebra finch CBG antibodies cross-react with CBGs in other birds, extending opportunities to study how CBG regulates the actions of glucocorticoids and sex steroids in these species.
Keywords: androgen, glucocorticoid, N-linked glycosylation, progesterone, protein crystallization, stress, birds, serine proteinase inhibitor
Introduction
Corticosteroid-binding globulin (CBG)2 is present in the blood of mammals, reptiles, amphibians, and birds (1). In these vertebrate subphyla, CBG transports glucocorticoids (cortisol and corticosterone) and regulates their access to tissues and cells (1, 2). To date, only mammalian CBGs have been characterized at the molecular level (3–7). The primary structure of human CBG defines it as a serine proteinase inhibitor (serpin) family member (3), and its gene (SERPINA6) is located in a cluster of related clade A SERPIN genes (8, 9) with synteny across other mammalian genomes (10). The crystal structures of rat (11) and human (12, 13) CBGs have been solved in complex with steroid ligands. These and other biochemical studies (14, 15) have revealed how specific residues in mammalian CBGs participate in steroid binding, including a conserved tryptophan that positions and holds steroids in their binding pocket (15). Importantly, this “signature tryptophan” residue distinguishes CBGs from other clade A serpins in mammals, and its absence in related SerpinA sequences of other terrestrial vertebrate species has hampered the identification of their SerpinA6 genes.
In mammals, plasma CBG has five or six sites for N-glycosylation, depending on the species (3–5, 7, 9). One of these sites is strictly conserved and appears essential for high affinity steroid-binding activity (15, 16). In addition, like other serpins, CBG has an unstructured reactive center loop (RCL) that is targeted by specific proteases (15). Proteolysis of the RCL in many serpins is an essential step in their inhibition of specific proteases. Proteolysis of the RCL of human CBG by neutrophil elastase (17) or chymotrypsin (18) or by exogenous bacterial proteases, such as LasB (19), causes a conformational rearrangement in its tertiary structure, as observed in other serpins (12). However, instead of inhibiting these proteases, proteolysis of the CBG RCL irreversibly disrupts its high affinity steroid-binding activity (17) and serves to promote the delivery of CBG-bound ligands to locations where these proteases are present (2, 17).
In birds, the affinity of CBG for progesterone is as high as for corticosterone if not higher (20). Avian CBGs also bind the androgens, testosterone and 5α-dihydrotestosterone (DHT), with nanomolar affinities (20, 21), and these steroid-binding properties distinguish them from mammalian CBGs. The high affinity of avian CBGs for androgens is considered important because birds lack a sex hormone-binding globulin, and CBG is thought to substitute for sex hormone-binding globulin in transporting androgens and regulating their actions in these species (21). Plasma corticosterone and CBG levels in birds are sexually dimorphic (22) and undergo marked seasonal changes in parallel with each other (23–25). Given the role of CBG in determining the biological activities of multiple classes of steroids in birds, which are widely used as models to study endocrine, neural, and behavioral responses to various stressors (26–28), we set out to identify and characterize the SerpinA6 genes and their products in avian species. In doing so, we also expected to gain insight into how SerpinA6 has evolved to accommodate the specialized functions of CBG in different vertebrate classes.
Experimental Procedures
Animals and Tissues
Blood and tissue samples from chickens (Gallus gallus), song sparrows (Melospiza melodia), and zebra finch (Taeniopygia guttata) were obtained under protocols approved by the University of British Columbia Animal Care Committee in compliance with regulations established by the Canadian Council on Animal Care. Chicken serum for CBG purification was obtained commercially (Sigma-Aldrich). Otherwise, trunk blood samples were taken after decapitation and prepared as heparinized plasma for storage at −80 °C until analysis. Tissues samples were snap-frozen on dry ice and also stored at −80 °C until analysis.
Isolation and Identification of Chicken CBG
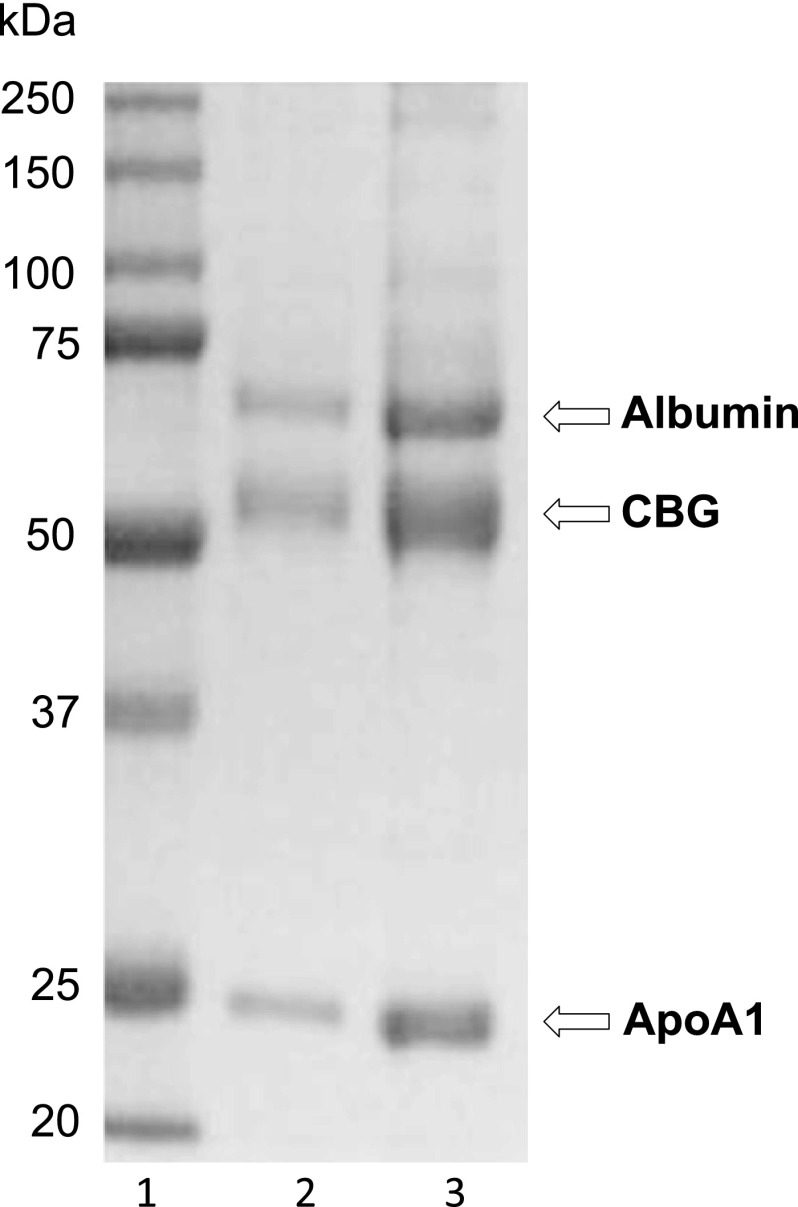
Chicken serum (10 ml) was diluted 1:1 in phosphate-buffered saline (PBS) for affinity chromatography using 3-oxo-17β-hydroxy-5α- androstane-17α-(6-hexyn-l-ol) linked to diaminoethyloxirane-Sepharose CL4B (29). Following extensive washing, 1-ml fractions were eluted from the steroid affinity column with 200 μm cortisol in PBS, and fractions containing cortisol-binding activity were pooled for SDS-PAGE. Protein bands were visualized in Coomassie Blue-stained SDS-polyacrylamide gels. Three major protein bands, with apparent molecular masses of ∼65, ∼55, and ∼24 kDa, were excised for mass spectrometry analysis (Fig. 1). Each band was in-gel-digested, as described (30), and analyzed by liquid chromatography-tandem mass spectrometry on an Agilent Q-TOF 6550, as described (31). Proteins were identified from tandem mass spectra using Mascot version 2.3 (Matrix Science) via Proteome Discoverer version 1.2 (Thermo Fisher) to search a database of all SWISS-PROT protein sequences under Metazoa taxonomy.
FIGURE 1.
Isolation and identification of chicken CBG. Coomassie Blue-stained SDS-polyacrylamide gel of proteins isolated by steroid affinity chromatography from chicken serum. Lane 1, molecular size markers (kDa values on the left); lane 2, 3 μl of affinity chromatography eluate; lane 3, 12 μl of affinity chromatography eluate. Protein bands were excised for mass spectrometric analysis, and their identifications are indicated on the right.
Recombinant Avian CBG Production and Purification
The ∼55-kDa chicken protein isolated using steroid affinity chromatography corresponded to that encoded by the chicken SerpinA4 gene (ENSGALG00000010969), and complete cDNA coding sequences for the chicken and zebra finch orthologs were obtained using gene- and species-specific oligonucleotides (supplemental Table S1), together with RNA templates extracted from chicken and zebra finch livers with an RNAeasy miniprep kit (Qiagen). These cDNAs were cloned into pcDNA3 (Life Technologies) for expression in Chinese hamster ovary (CHO) cells, as described for the production of recombinant human CBG (32).
The cloned chicken and zebra finch cDNA sequences were compared with those reported at ensemble.org or GenBankTM. The chicken cDNA sequence (KU180444) is identical to BX935008.1 (GenBankTM), whereas the zebra finch cDNA sequence (KU180443) contained four non-synonymous single nucleotide variations when compared with ENSTGUT 00000013171.1. As indicated below, these cDNAs encode the chicken and zebra finch CBG precursors, respectively.
The cDNAs encoding chicken and zebra finch CBGs without their predicted signal polypeptide (see supplemental Fig. S1) were cloned into the pPIC9 expression vector (Invitrogen) after PCR amplification with specific oligonucleotides (supplemental Table S1) for expression in yeast (Pichia pastoris GS115 cells). Chicken and zebra finch CBG cDNA-pPIC9 constructs were all linearized with StuI before transformation of GS115 cells by electroporation. After selection on minimal medium, yeast colonies were grown to an A600 of 10 in BMGY medium, which was then replaced with methanol-containing BMMY medium to induce protein expression at 30 °C for 24 h. Yeast cells were removed by centrifugation, and supernatants were harvested. The steroid-binding properties of CBG secreted directly in the culture medium were initially determined without further purification. In studies of zebra finch CBG mutants expressed in yeast, a C-terminal His tag, which has no effect on steroid-binding activity, was added to aid in purification and concentration by nickel-nitrilotriacetic acid chromatography (Life Technologies) before analysis.
For expression in Escherichia coli BL21 cells, a cDNA encoding the predicted mature polypeptide sequence of zebra finch CBG was inserted in frame into the HTMpET vector (33). In brief, transformed E. coli were grown in LB with 50 μg/ml kanamycin until an A600 of 0.5, when 200 μm isopropyl 1-thio-β-d-galactopyranoside was added, and cultures were grown overnight at 30 °C. After centrifugation, cell pellets were resuspended in lysis buffer (5 mm imidazole, 50 mm NaH2PO4, 500 mm NaCl, pH 8) and left on ice for 30 min before sonication. Cell debris was then removed by centrifugation, and the lysate was clarified using a 0.45-μm filter and applied to a nickel-nitrilotriacetic acid affinity column (Life Technologies, Inc.). After elution with 500 mm imidazole, the His-maltose-binding protein-zebra finch CBG fusion protein was treated with tobacco etch virus protease to release the CBG, as confirmed by SDS-PAGE. The released CBG was purified by affinity column chromatography using 11β-hydroxy-androst-4-en-3-oxo-17β-carboxylic acid-diaminoethyloxirane-Sepharose CL4B (34)and used for rabbit antiserum production by ProSci Inc. (Poway, CA).
Mutagenesis
A QuikChange II kit (Agilent Technologies) was used for mutagenesis of cDNAs with oligonucleotide primer pairs (supplemental Table S1), and Sanger sequencing confirmed that only the targeted mutations occurred.
Phylogenetic Analyses
Putative chicken SerpinA6 orthologs were identified within the genomes of other submammalian species at ensemble.org using the ENSGALG00000010969 sequence or through BLAST searches against the predicted amino acid sequences of chicken and zebra finch CBG. Multiple-sequence alignments were made using Clustal ω (35) and default parameters with the BOX-SHADE program.
Steroid-binding Assays, Western Blotting, and Proteolysis
A radiolabeled steroid saturation assay (36) was used to detect and measure CBG in plasma, culture medium, or chromatographic fractions obtained during protein purification. In brief, samples were first diluted and incubated in a dextran-coated charcoal solution in PBS to remove steroids if necessary. Samples were then incubated (1 h at room temperature) with ∼10 nm [3H]cortisol or [3H]corticosterone (American Radiolabeled Chemicals) in the absence or presence of a 200-fold excess of the corresponding unlabeled steroids (to control for nonspecific binding) and then placed in ice water for 30 min. Ice-cold dextran-coated charcoal solution was then added for 10 min at 4 °C, followed by centrifugation to separate and measure CBG-bound [3H]steroid. This method was also used to determine affinity constants by Scatchard analysis as well as the steroid-binding specificities of CBGs in competitive ligand-binding assays (36). Affinity constants from Scatchard plots and IC50 determinations were obtained using Gnuplot software.
Western blotting was used to detect CBG in plasma and culture medium samples after SDS-PAGE. Rabbit anti-zebra finch CBG antiserum was used at appropriate dilutions or after affinity purification using yeast-expressed zebra finch CBG immobilized on a hiTrap NH-activated HP column, as recommended (GE Healthcare). Immunoreactive CBG on Western blots was detected using an enhanced chemiluminescence reagent (GE Healthcare) and an ImageQuant LAS4000 system (GE Healthcare).
For cleavage of plasma CBG, 199 μl of diluted (1:40) zebra finch plasma was incubated with 1 μl of Pseudomonas aeruginosa medium. After incubation at 37 °C for the indicated times, proteolysis was stopped by the addition of EDTA (final concentration, 10 mm). E. coli-expressed CBG (10 μg) was incubated with P. aeruginosa medium under the same conditions. Reaction products were examined by SDS-PAGE. Native and proteolytic fragments of E. coli-expressed zebra finch CBG were diluted in 5% acetonitrile and 0.1% formic acid and injected onto a Waters Xevo GS-2 QTOF mass spectrometer via a NanoAquity UPLC system. Deconvolution of MS spectra with the Waters MaxEnt algorithm revealed a single peak (42,890 Da) in the native sample, whereas cleaved CBG presented two peaks of 38,595 and 4,308 Da. MS/MS analysis of the 4,308-Da CBG fragment confirmed that it was the C-terminal peptide: ISFPPTIEFSHPFLMLIFDRDTNSTLFIGKIVNTITS.
Crystal Structure Determinations
Because the 23 N-terminal residues of mature zebra finch CBG were predicted as disordered, an N-terminally truncated CBG (i.e. from Glu-24) was also expressed in E. coli and purified, as described above, for crystallography. Attempts to crystalize this protein were unsuccessful, and the amino acid sequence was optimized for crystallization. For this, we introduced a C28A substitution and several other amino acid substitutions (i.e. E127A, K128A, and K130A), as suggested by the SERp server, into the zebra finch CBG sequence. This approach to aid crystallization relies on the concept of surface entropy reduction (i.e. the replacement of clusters of flexible, solvent-exposed residues with residues that have lower conformational entropy) (37). These amino acid substitutions did not affect the cortisol-binding activity of zebra finch CBG.
The zebra finch CBG produced in this way formed needle-shaped crystals overnight at room temperature. Crystals for x-ray diffraction were obtained from hanging drops by equilibrating 1 μl of CBG solution (10 mm Tris, 25 mm NaCl, 5 mm EDTA, 600 μm cortisol, pH 7.5, with CBG at 5 mg/ml) and 1 μl of reservoir solution (200 mm NaCl, 1.75 m (NH4)2SO4, pH 6.5) against 1 ml of reservoir solution. Before flash freezing, crystals were soaked in a cryoprotective solution (28.5% glucose, 200 mm NaCl, 1.75 m (NH4)2SO4, pH 6.5, 10 μm cortisol).
Diffraction experiments were performed at the Canadian Light Source (Saskatoon) beamline 08ID-1, and data sets were processed using XDS (38). Human SERPINA1 (Protein Data Bank code 1HP7) was used as a search model, where all side chains truncated to alanine residues and loops were deleted. Molecular replacement was performed using Phaser (39) with an LLG value of correct solution of 3,580 and LLG value of a second unrelated peak of 190. Successive rounds of manual model building in COOT (40) were used to complete the model with refinements using Phenix (41) and REFMAC5 (42). No residues were found in disallowed regions of the Ramachandran plot. Structure figures were prepared using PyMOL (DeLano Scientific) with secondary structures assigned through the DSSP plugin, and coordinates were deposited in the Protein Data Bank.
Measurement of CBG mRNA in Zebra Finch Tissues
Total RNA from adult zebra finch tissues was obtained with an RNeasy miniprep kit (Qiagen) and used for quantitative PCR measurements of CBG mRNA. In brief, 0.5 μg of RNA, 1 μl of dNTP mix, 2 μl of random primer mix, single strand buffer, and 1 μl of Reverse Transcriptase II was incubated at 42 °C for 1 h in 20 μl. The resulting cDNA templates (0.5 μl) were added to a Fast SYBR Green Master Mix (Applied Biosystems) and amplified using zebra finch-specific CBG (supplemental Table S1) and GAPDH primers (43) both at 500 μm using a real-time PCR system (Applied Biosystems) under the following conditions: 95 °C denaturation for 10 min, followed by 40 cycles of 95 °C denaturation (15 s), 60 °C annealing (40 s), and 72 °C extension (30 s). Melting curve analysis was performed to confirm the presence of a single amplification product, and PCR products were analyzed by 2% agarose gel electrophoresis to confirm the expected PCR product sizes. The relative abundance of CBG mRNA was calculated using the ΔΔ cycle threshold method.
Results
Isolation and Identification of Steroid-binding Proteins in Chicken Serum
In pilot experiments, a DHT-conjugated affinity matrix (29) proved more effective in isolating CBG from chicken serum, as compared with the corticosterone-based affinity matrix usually employed to isolate mammalian CBGs (34). Our DHT affinity column removed ∼70% of the high affinity cortisol-binding activity from chicken serum, and some of the chromatography fractions eluting from the column in the presence of either DHT or cortisol exhibited the steroid-binding properties of CBG. When these fractions were subjected to SDS-PAGE, the three major protein bands observed were identified as albumin, SerpinA4, and apoA1 by mass spectrometry (Fig. 1).
It was no surprise that albumin (∼65 kDa) was enriched using DHT affinity chromatography because it binds all steroid classes with low affinity. The enrichment of apoA1 (∼24 kDa) was unexpected, but human apoA1 interacts with steroids (44, 45), and chicken apoA1 probably bound the DHT affinity matrix with low affinity. The identification of the ∼55-kDa protein band as SerpinA4 was also surprising because SerpinA6 is the CBG-coding gene in mammals. However, the robust cortisol-binding activity in chromatography fractions containing the ∼55-kDa protein suggested that it is CBG (SerpinA6) rather than the serpin peptidase inhibitor, kallistatin (SerpinA4).
Avian SerpinA6 Genes
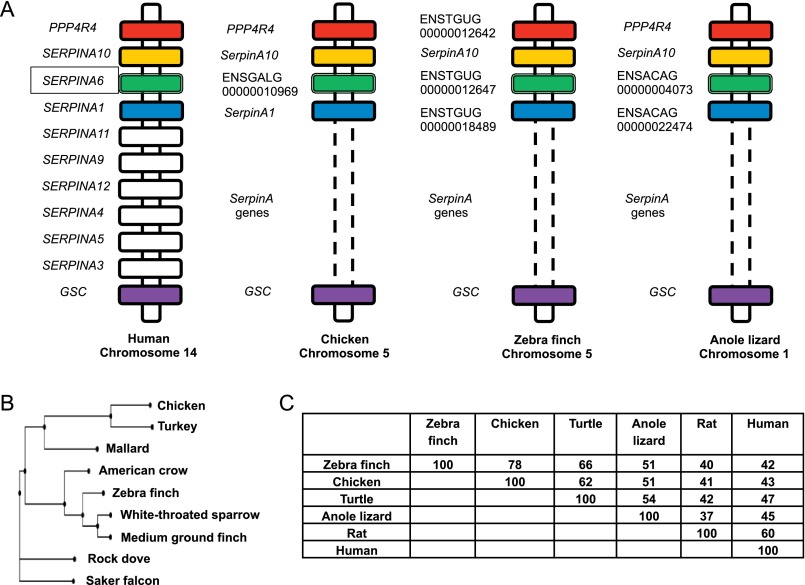
To date, only the mammalian SerpinA6 genes encoding CBG have been confirmed as being correctly annotated in genome databases. In the chicken genome, the protein that we suspected was chicken CBG is encoded by ENSGALG00000010969, currently annotated as SerpinA4 (ensemble.org). In mammals, SerpinA4 encodes kallistatin, which is not known to bind steroids. Like SERPINA6, SERPINA4 is located in close proximity to SERPINA1 on human chromosome 14 but is telomeric to SERPINA1, whereas SERPINA6 is centromeric to SERPINA1. This chromosomal arrangement of SERPINA genes in humans is conserved across mammalian species, and our findings indicate that synteny within this cluster of SerpinA genes extends to avian and reptilian species (Fig. 2A). These observations imply that SerpinA4 in the current chicken genome database is annotated incorrectly and that the zebra finch gene, ENSTGUG00000012647, as well as other annotated genes in several other avian species (supplemental Fig. S1) are the orthologs of the chicken gene encoding CBG (Fig. 2A).
FIGURE 2.
Phylogenetic comparisons of SerpinA6 genes and CBG sequences. A, chromosomal locations of SerpinA genes illustrating the syntenic conservation of SerpinA10, SerpinA6 (encoding CBG), and SerpinA1 in humans, chickens, zebra finch, and the anole lizard. In all species, SerpinA loci are flanked by PPP4R4 (protein phosphatase 4, regulatory subunit 4) and GSC (goosecoid homeobox) genes. Orthologs of PPP4R4, GSC, SerpinA10, SerpinA6, and SerpinA1 are colored identically. B, a neighbor-joining phylogram of CBG in avian species. Phylogenetic analyses were performed using ClustalW2 Phylogeny (65) with default settings, and the phylogram was created with the PHY.FI online tool (66). C, amino acid sequence identity of CBG precursor polypeptides from different species. Pairwise sequence alignments were produced using Clustal ω (35). Accession numbers for the sequences used in this figure are as follows: chicken (KU180444), turkey (XP_003206753.1), mallard (EOA99912.1), American crow (XP_008627951.1), zebra finch (KU180443), white-throated sparrow (XP_005483238.1), medium ground finch (XP_005417914.1), rock dove (XP_005506178.1), saker falcon (XP_005438961.1), Chinese softshell turtle (XP_006133744.1), anole lizard (XP_008105229.1), rat (NP_001009663.1), and human (P08185.1).
When compared with the corresponding sequence at ensemble.org, the zebra finch cDNA that we cloned contains four non-synonymous single nucleotide variations, resulting in the following amino acid differences in the mature zebra finch CBG sequence: A64T, N198S, S319N, and V363I. This was not unexpected because of genetic variations between zebra finch colonies (46). In addition, none of these amino acids are in regions that might influence steroid-binding activity, and CBG in plasma from the same colony of birds from which the mRNA was obtained for cDNA synthesis had normal steroid-binding properties.
Based on sequence comparisons and the positioning of SerpinA6 orthologs in the genomes of multiple avian species, we constructed a phylogram, which indicates that the chicken gene encoding CBG (SerpinA6) clusters most closely with its related genes in other species of fowl (turkey and duck), whereas the zebra finch gene is more closely related to the predicted orthologs in other perching birds (Fig. 2B). Extending this type of analysis also allows SerpinA6 orthologs to be identified in other non-mammalian vertebrates, including reptiles (Fig. 2, A and C).
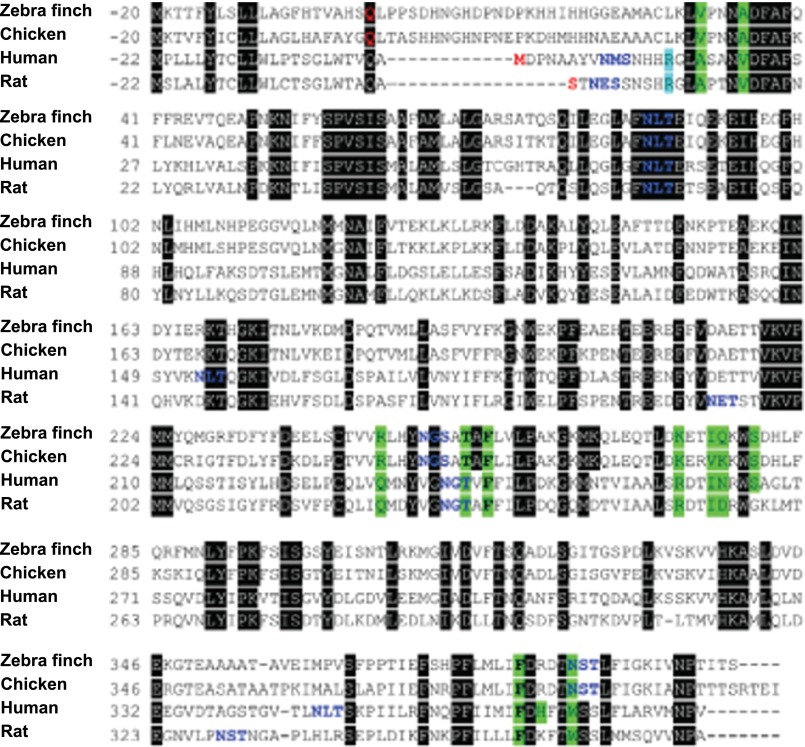
Importantly, the avian CBG sequences that we have identified display limited (<45%) sequence identity with mammalian CBGs, and alignment of mammalian and avian CBG primary structures indicated that only five of the 10–12 residues known to interact with steroids within mammalian CBG steroid-binding sites are conserved in avian CBGs (Fig. 3).
FIGURE 3.
Multiple sequence alignment of zebra finch, chicken, human, and rat CBGs. N-terminal residues (predicted or experimentally determined) are shown in red and are numbered as residue 1. Residues in the secretion signal polypeptide are numbered negatively with respect to residue 1. Amino acids whose side chains interact with cortisol are highlighted in green, and consensus sequences for N-linked glycosylation are shown in blue letters. Arginine, which forms cation-π interaction with Trp-362 in rat CBG and Trp-371 in human CBG, is highlighted in cyan. Black boxes highlight conserved amino acids. The three conserved amino acids that interact with cortisol are shown in boldface type.
Confirmation of Avian CBG Identity
To demonstrate conclusively that the chicken gene, ENSGALG00000010969, and its zebra finch ortholog, ENSTGUG00000012647, both encode CBG, we expressed their respective full-length cDNAs in CHO cells. Scatchard analysis of the recombinant proteins secreted by these cells was performed alongside plasma samples from both bird species, and this confirmed that their affinities for corticosterone are all in the low nanomolar range (Table 1). We also expressed the predicted mature polypeptide sequences encoded by these two bird genes in yeast, and the affinities of the yeast-expressed proteins are indistinguishable from CBG in the plasma for these birds (Table 1). As a further demonstration that the avian genes we had identified encode CBG, we compared the steroid-binding specificity of the zebra finch protein expressed in yeast with that of CBG in zebra finch plasma and found them to be virtually identical (Table 2).
TABLE 1.
Steroid-binding properties of avian CBGs; dissociation constants (Kd) of CBG for [3H]corticosterone in diluted (1:100) chicken and zebra finch plasma versus the corresponding CBGs expressed in yeast (P. pastoris) or CHO cells
| Species |
Kd |
||
|---|---|---|---|
| Plasma CBG | Yeast-expressed CBG | CHO-expressed CBG | |
| nm | |||
| Chicken | 1.9 ± 0.2 | 1.5 ± 0.2 | 0.9 ± 0.1 |
| Zebra finch | 2.6 ± 0.2 | 3.0 ± 0.3 | 3.2 ± 0.5 |
TABLE 2.
Steroid-binding properties of avian CBGs; competition of different steroids for zebra finch CBG in diluted (1:100) plasma versus recombinant zebra finch CBG expressed in yeast (P. pastoris)
The mean ± asymptotic S.E. IC50 values (nm) are presented for unlabeled steroids in competitive steroid binding assays using [3H]corticosterone as the labeled ligand.
| Unlabeled competitor | IC50 |
|
|---|---|---|
| Zebra finch CBG in plasma | Zebra finch CBG expressed in yeast | |
| nm | ||
| Corticosterone | 4.2 ± 0.3 | 4.0 ± 0.3 |
| Cortisol | 1.8 ± 0.2 | 1.7 ± 0.2 |
| Progesterone | 1.6 ± 0.1 | 2.0 ± 0.3 |
| Dexamethasone | 3.7 ± 0.8 | 2.4 ± 0.6 |
| DHT | 11.3 ± 2.7 | 16.4 ± 2.0 |
| Testosterone | 18.5 ± 3.0 | 20.8 ± 4.1 |
| Estradiol | >200 | >200 |
| Dehydroepiandrosterone | >200 | >200 |
| Aldosterone | >200 | >200 |
These steroid-binding specificity studies (Table 2) also indicated that zebra finch CBG has a greater affinity for cortisol and progesterone than for corticosterone, the major circulating glucocorticoid in this species. In addition, zebra finch CBG has a remarkably high affinity for the synthetic glucocorticoid, dexamethasone, and moderate affinity for both testosterone and DHT but does not bind dehydroepiandrosterone, estradiol, or aldosterone. This steroid-binding specificity profile reflects those reported for CBGs in avian species (1, 21, 27, 47) and proves that the chicken ENSGALG00000010969 and zebra finch ENSTGUG00000012647 genes encode CBG and should be annotated as SerpinA6.
Influence of Glycosylation on Avian CBG Steroid-binding Activity
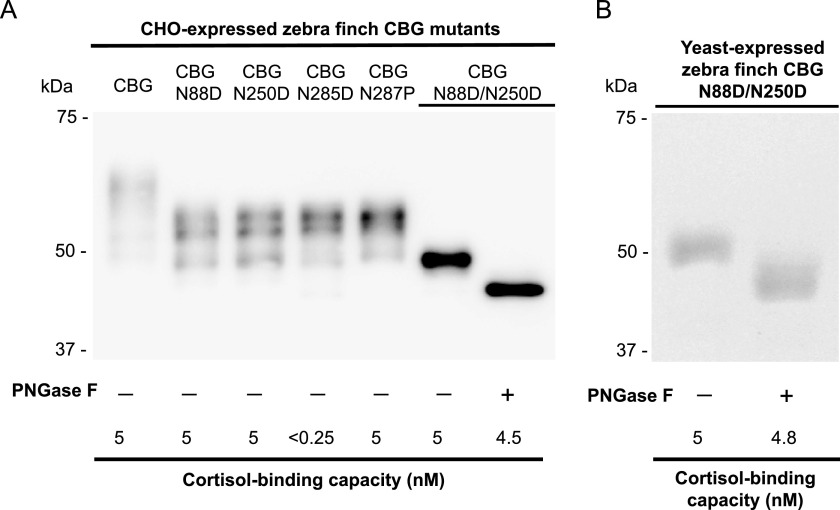
Avian CBGs have fewer consensus sites for N-glycosylation than do mammalian CBGs, and all three of them appear to be highly conserved (supplemental Fig. S1). Remarkably, the asparagine residue of one of these sites (Asn-385) is positioned in place of the tryptophan residue that plays a critical role within the CBG steroid-binding sites of all mammals (15). We therefore examined the impact of glycosylation on the steroid-binding properties of zebra finch CBGs expressed in yeast and CHO cells.
Each of the three N-glycosylation sites was first disrupted by substitution of the asparagine residues (Asn-88, Asn-250, and Asn-385) with aspartic acid (Table 3). When expressed in yeast, the N88D and N250D mutants bound steroids with the expected affinity, but the steroid-binding activity of the N385D mutant was undetectable. Although the N385D mutant also exhibited a decreased (<5% of unmutated CBG) cortisol-binding capacity (Fig. 4A) when expressed in CHO cells, its affinity (Kd) for cortisol appeared to be normal (Table 3). This suggests that only a small portion of the N385D mutant folds correctly and is capable of binding steroid. To examine this further, we disrupted the N-glycosylation consensus sequence at this site (i.e. Asn-Ser-Thr) by creating a T387P mutant. This also prevents N-linked glycosylation at Asn-385, and the T387P mutant bound steroid normally (Table 3). In addition, an N88D/N250D double mutant that only N-glycosylates at Asn-385 binds steroids normally. Deglycosylation of this N88D/N250D mutant with PNGase F results in a reduction in molecular size consistent with the loss of an N-linked oligosaccharide at Asn-385 (Fig. 4, A and B). However, by contrast to the almost complete loss of steroid-binding activity in the N385D mutant, its cortisol-binding activity was unaffected by deglycosylation in this way (Fig. 4, A and B), despite the fact that PNGase F treatment also converts Asn-385 into an aspartic acid (48). Thus, it appears that whereas glycosylation at Asn-385 does not affect steroid binding in zebra finch CBG, this asparagine residue may contribute in some way to the formation of a high affinity steroid-binding site.
TABLE 3.
Steroid-binding properties of avian CBGs; influence of N-glycosylation on zebra finch CBG steroid-binding activity
Yeast-expressed zebra finch CBG and CBG mutants with specific N-glycosylation sites disrupted were subjected to Scatchard analyses with [3H]cortisol to determine Kd values. Because the CBG N385D mutant expressed in yeast had no detectable cortisol-binding activity, the zebra finch CBG and CBG mutants were also produced in CHO cells. The amount of the CBG N385D mutant produced by CHO cells with cortisol-binding activity was low, but its affinity for cortisol was normal, as it was for the other mutants. Mean ± asymptotic S.E. Kd values are listed. ND, not detected.
| Expression system |
Kd |
|||||
|---|---|---|---|---|---|---|
| CBG | CBG N88D | CBG N250D | CBG N385D | CBG T387P | CBG N88D/N250D | |
| nm | ||||||
| Yeast | 2.2 ± 0.2 | 2.5 ± 0.3 | 2.5 ± 0.3 | ND | 2.5 ± 0.2 | 2.0 ± 0.3 |
| CHO cells | 2.8 ± 0.2 | 3.4 ± 0.1 | 3.3 ± 0.4 | 2.4 ± 0.2 | 3.6 ± 0.2 | 1.7 ± 0.1 |
FIGURE 4.
Glycosylation and cortisol-binding capacity of zebra finch CBG and its glycosylation-deficient mutants. A, Western blotting of media from CHO cells with a rabbit anti-zebra finch antiserum. Deglycosylation of the N88D/N250D mutant, which has only one N-glycosylation site at Asn-385, with PNGase F resulted in a ∼2–3-kDa reduction in apparent molecular size with only minor loss of cortisol-binding capacity. The molecular size of the deglycosylated N88D/N250D mutant is ∼45 kDa, consistent with a complete loss of N-linked oligosaccharides. B, the N88D/N250D mutant expressed in yeast examined by SDS-PAGE and Coomassie Blue staining before and after deglycosylation with PNGase F. Note that it also retained almost full cortisol-binding capacity after deglycosylation.
Proteolysis of Zebra Finch CBG and Effect on Its Steroid-binding Activity
P. aeruginosa infections are associated with high mortality in birds (49), and the metalloprotease protease, LasB, secreted by P. aeruginosa cleaves the RCL of human CBG and disrupts its cortisol-binding activity (19). To mimic a biologically relevant situation, we therefore incubated diluted zebra finch plasma in the absence or presence of P. aeruginosa culture medium (Fig. 5A). Analysis of the reaction products indicated that ∼50% of the zebra finch CBG underwent proteolysis within 2 h, with the appearance of a cleavage product that is ∼4 kDa smaller than the intact CBG together with a corresponding reduction in its corticosteroid-binding capacity. This reduction in the apparent molecular size of CBG is consistent with a single cleavage within the RCL sequence, as observed for human CBG treated in this way (19). This proteolysis appeared to continue over 6 h with further reductions in corticosteroid-binding capacity by as much as 90% (Fig. 4A). Although some additional proteolysis was evident when the reaction was extended for 17 h, no intact zebra finch CBG or detectable corticosteroid-binding activity was present at this time point (Fig. 5A). Importantly, when the reaction was performed for 17 h in the presence of EDTA, which is known to inhibit the bacterial metalloprotease LasB (19), the CBG was completely protected from proteolysis or loss of steroid-binding activity (Fig. 5A).
FIGURE 5.
Proteolysis within the RCL disrupts the corticosterone-binding activity of zebra finch CBG. A, diluted (1:40) zebra finch plasma (199 μl) was incubated with 1 μl of P. aeruginosa culture medium. Proteolysis was stopped by the addition of EDTA at predetermined times (0–17 h). In one 17-h reaction (*), EDTA was added before incubation with P. aeruginosa culture medium. Reaction products were analyzed by Western blotting and a corticosterone-binding capacity assay. The reduction in apparent molecular size of ∼4 kDa after incubations of 2–17 h is consistent with cleavage occurring in the RCL. At 17 h, additional proteolysis is evident, but proteolysis and loss of corticosterone-binding capacity was almost completely blocked by pretreatment with EDTA even after 17 h. B, incubation of unglycosylated N-terminally truncated zebra finch CBG (i.e. lacking the first 23 N-terminal residues) expressed in E. coli with P. aeruginosa medium for 2 h resulted in a molecular size reduction of ∼4 kDa, as shown by SDS-PAGE and Coomassie staining, together with a complete loss of corticosterone-binding activity. C, location of the proteolytic cleavage site within the RCL, as determined by mass spectrometric analysis of proteolytic fragments obtained after incubation of E. coli-expressed zebra finch CBG with P. aeruginosa medium, as in B.
When the N-terminally truncated zebra finch CBG produced in E. coli was incubated for 2 h with P. aeruginosa medium, a similar ∼4-kDa reduction in its molecular size was accompanied by a complete loss of its corticosteroid-binding activity (Fig. 5B). In addition, when the resulting proteolytic fragments were analyzed by mass spectrometry, the proteolytic cleavage site was identified between residues Pro-362 and Ile-363 within the C-terminal region of the RCL (Fig. 5C).
Crystal Structure of Zebra Finch CBG in Complex with Cortisol
Because avian CBG sequences display limited sequence identity with mammalian CBGs, and some of the residues that interact with steroids in mammalian CBGs are not conserved in avian CBG sequences (Fig. 3), it was important to obtain a bird CBG crystal structure in complex with a steroid ligand. To do this, zebra finch CBG was co-crystalized with its preferred glucocorticoid ligand, cortisol (Table 2). The crystal structure was solved to 2.4 Å resolution (Table 4). In this structure, the main chain is organized in a typical serpin fold with the RCL being fully exposed in the “stressed” or “active” serpin conformation (Fig. 6A). Only 7 of the 375 residues in the crystal structure failed to produce a clear electron density; five of these residues are within the unstructured RCL, and this was expected because the RCL is also disordered in the rat CBG crystal structure (11). The two other residues that could not be modeled are at the N terminus and C terminus of the protein. Moreover, an electron density for cortisol was clearly evident (Fig. 6B).
TABLE 4.
Data collection and refinement statistics
| Parameters | Values |
|---|---|
| Data collection | |
| Space group | P63 |
| Cell dimensions | |
| a, b, c (Å) | 116.2, 116.2, 72.7 |
| α, β, γ (degrees) | 90, 90, 120 |
| Resolution (Å) | 58.0–2.43 |
| Rsym or Rmerge | 19.9 (138.1)a |
| I/σI | 11.1 (1.9) |
| Completeness (%) | 100 (100) |
| Redundancy | 11.3 (11.1) |
| Wilson B factor | 32.6 |
| Refinement | |
| Resolution (Å) | 50.3–2.43 |
| No. of reflections | 21,312 |
| Rwork/Rfree | 16.2/21.7 |
| No. of atoms | |
| Protein | 2,996 |
| Water | 186 |
| Ligand | 26 |
| B-factors | |
| Protein | 39.29 |
| Water | 39.02 |
| Ligand | 34.11 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.017 |
| Bond angles (degrees) | 1.968 |
| Ramachandran plot (% residues) | |
| Most favored regions | 97.3 |
| Allowed regions | 2.7 |
| Outlier regions | 0 |
a Values in parentheses are for the highest resolution shell.
FIGURE 6.
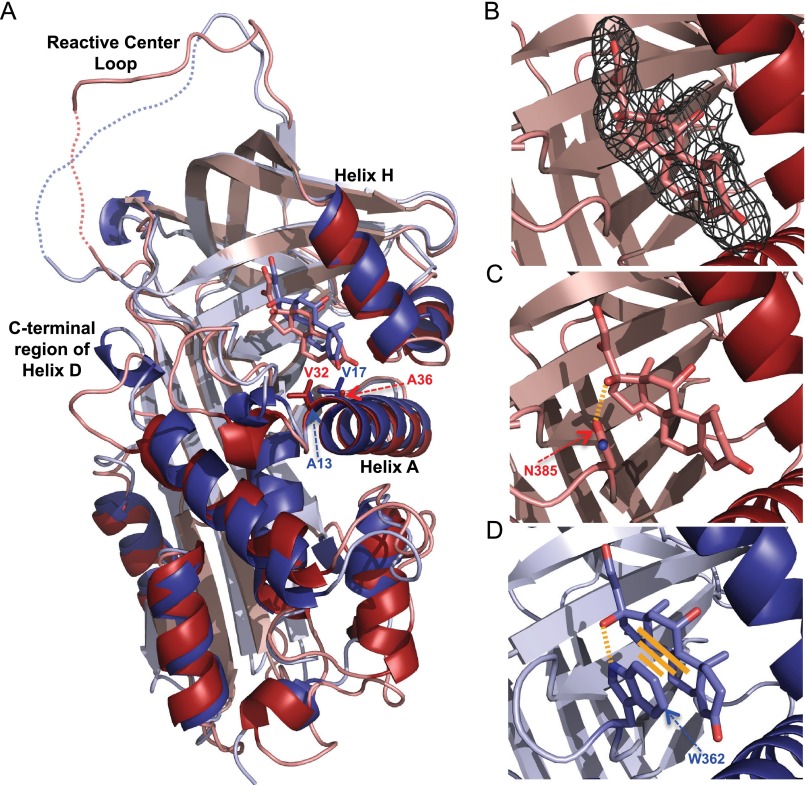
Comparisons of zebra finch (red; Protein Data Bank code 5HGC) and rat (blue; Protein Data Bank code 2V95) CBG crystal structures (A) and their steroid-binding sites (B–D). A, overall structural alignments of zebra finch and rat CBG with cortisol in the binding sites. Reactive center loop regions without clear electron density are shown with a dotted line. Structural alignment was performed in PyMOL using all residues. B, portion of the final 2Fo − Fc electron density map contoured at 1.0 σ shows electron density corresponding to cortisol. C, cortisol in the zebra finch CBG steroid-binding site showing a hydrogen bond between Asn-385 and the hydroxyl group at C17 of cortisol. D, cortisol in the rat CBG steroid-binding site showing how Trp-362 forms both a hydrogen bond and a stacking interaction with the ligand.
The cortisol in the zebra finch CBG crystal structure is positioned at the interface of helix A, helix H and β sheet B, as in the rat CBG structure (Fig. 6A). However, although the overall organization of the binding site is conserved in both species, there are some important differences. Cortisol in the zebra finch CBG steroid-binding site is slightly tilted when compared with the position of cortisol in the rat CBG structure (Fig. 6A). This can be explained by a different set of interactions with the steroid A ring. In rat CBG, the A ring of cortisol forms hydrophobic interactions with Ala-13 and Val-17, whereas in zebra finch CBG, it is held in place by the hydrophobic side chains of Val-32 and Ala-36. Furthermore, the most remarkable difference is the Asn-385 in zebra finch CBG (and other avian CBGs) that replaces the Trp-362 in rat CBG. In zebra finch CBG, Asn-385 is clearly a site for N-linked glycosylation, and our crystal structure data indicate that it is capable of forming a hydrogen bond with the hydroxyl group of cortisol at C17 (Fig. 6B). By contrast, in rat CBG, Trp-362 forms strong stacking interactions with the surface of the steroid, thus serving to anchor it within its relatively shallow binding site (Fig. 6C).
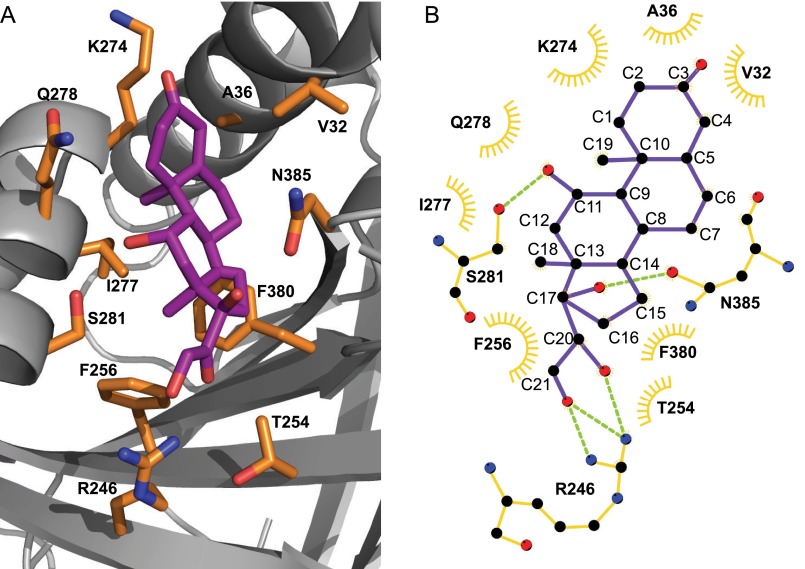
The zebra finch CBG crystal structure also shows how other key residues interact with oxygen atoms of cortisol via hydrogen bonds (i.e. Ser-281 (with the hydroxyl group at C11) and Arg-246 (with the hydroxyl group at C21 and the carbonyl group at C20)) as well as the positions of several residues that form hydrophobic contacts with the carbon atoms of cortisol (Fig. 7, A and B). It is also apparent that cortisol fits tightly into its binding pocket, as evidenced by the absence of water molecules bridging amino acid residues and functional groups of the steroid.
FIGURE 7.
Detailed structure of the zebra finch CBG steroid-binding site (A) and a schematic representation of how cortisol interacts with specific amino acid residues (B). A, cortisol is shown in purple with amino acids participating in cortisol binding in orange. B, a schematic diagram drawn with LigPlot (EMBL-EBI, Cambridge, UK) shows the carbon and oxygen atoms of cortisol (purple) as black and red circles, respectively. Amino acids participating in hydrophobic interactions with cortisol are shown as orange open half-circles. Amino acids forming hydrogen bonds (green dotted lines) with cortisol are depicted as schematic structures, where black, red, and blue circles represent carbon, oxygen, and nitrogen atoms of the amino acids, respectively.
Roles of Specific Amino Acid Residues in the Zebra Finch CBG Steroid-binding Site
Based on our new crystal structure data, we mutated several amino acids in the zebra finch CBG sequence that are probably involved in cortisol binding. Mutation of Ser-281 into alanine did not affect cortisol binding (Table 5). However, although this serine (Ser-281) appears to form a hydrogen bond with the cortisol hydroxyl group at C11, when it is substituted with alanine, the C11 hydroxyl group of cortisol is probably coordinated by water, as observed in rat CBG, in which the corresponding residue is Gly-259 (Fig. 3). In rat CBG, the side chain carboxyl group of Asp-256 hydrogen-bonds with the C11 hydroxyl of cortisol, and its substitution with alanine causes a loss of steroid-binding activity (11). The corresponding residue in zebra finch CBG is Gln-278 (Fig. 6), but it does not hydrogen-bond with the hydroxyl group at C11 of cortisol. Instead, it creates a surface for hydrophobic interactions with cortisol, and its substitution with alanine does not affect cortisol-binding activity (Table 5). Moreover, the fact that zebra finch CBG binds both cortisol and progesterone (which does not have a hydroxyl group at C11) with essentially equal affinity further suggests that interactions with a hydroxyl group at C11 have little influence on the affinity of avian CBGs for the glucocorticoids.
TABLE 5.
Steroid-binding properties of zebra finch CBG and its mutants expressed in P. pastoris; mean ± asymptotic S.E. Kd values for [3H]cortisol binding to zebra finch CBG and various zebra finch CBG mutants
| Protein | Kd |
|---|---|
| nm | |
| CBG | 2.2 ± 0.2 |
| CBG Q278A | 2.5 ± 0.2 |
| CBG S281A | 2.2 ± 0.2 |
| CBG R246Q | 2.8 ± 0.3 |
| CBG N385W | >30 |
The affinities of zebra finch CBG for progesterone and cortisol are almost identical, and this characteristic is shared by other avian CBGs (1, 21). In human CBG crystal structures, the hydroxyl group at C21 and the carbonyl group at C20 in cortisol both hydrogen-bond with Gln-232 (12), whereas progesterone hydrogen-bonds with Gln-232 through its carbonyl group at C20 only (13). In zebra finch CBG, the corresponding residue is Arg-246, and substitution of this residue with Gln caused a significant decrease in affinity for progesterone, whereas the affinities for cortisol and testosterone were unaffected (Table 6). Thus, although Arg-246 in zebra finch CBG may make similar contact with both cortisol and progesterone, substitution of this highly conserved residue in avian CBGs (supplemental Fig. S1) with glutamine explains why the affinities of avian CBGs for progesterone are much higher than for mammalian CBGs.
TABLE 6.
Steroid-binding properties of zebra finch CBG and its mutants expressed in P. pastoris; IC50 values for different steroids measured for zebra finch CBG and the CBG R246Q mutant
Competition studies with unlabeled competitors were performed with [3H]cortisol as the labeled ligand.
| Unlabeled competitor | IC50 |
|
|---|---|---|
| CBG | CBG R246Q | |
| nm | ||
| Cortisol | 5.5 ± 0.7 | 6.3 ± 0.7 |
| Progesterone | 7.1 ± 0.4 | 27.0 ± 2.8 |
| Testosterone | 43.7 ± 3.0 | 44.5 ± 4.4 |
Given the importance of the tryptophan residue in the steroid-binding sites of rat and human CBGs (15), we also produced a zebra finch CBG N385W mutant. When expressed in yeast (Table 5) or CHO cells (not shown), it was secreted at similar levels to the unmutated CBG, as assessed by Western blotting (not shown), but its steroid-binding affinity was very low (Kd >30 nm) in a Scatchard analysis (Table 5).
Zebra Finch Liver Is the Major Site of SerpinA6 Expression and Anti-zebra Finch CBG Antibodies Detect Plasma CBG in Other Avian Species
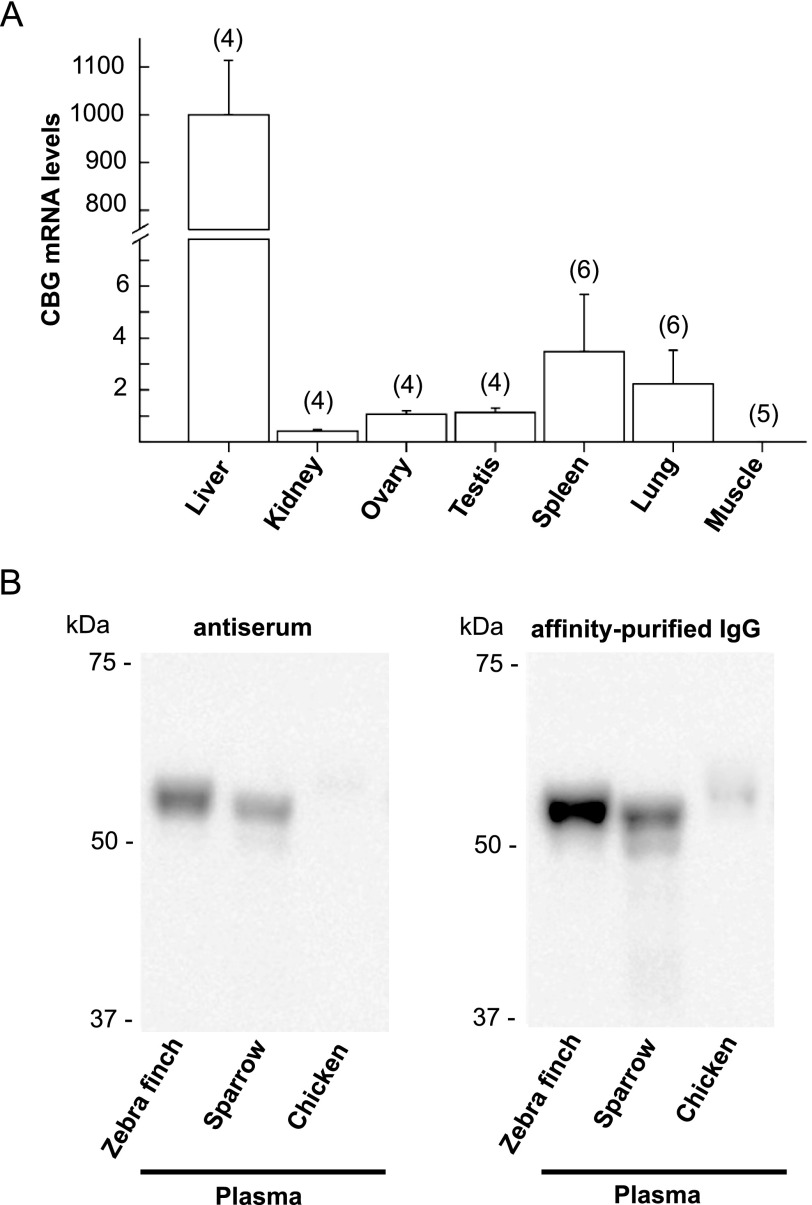
Quantitative RT-PCR was used to determine the relative abundance of CBG mRNA in adult male and female zebra finch tissues (Fig. 8A). Single RT-PCR products of the expected sizes were detected in all tissues except for skeletal muscle (not shown), but measurements of CBG mRNA in relation to GAPDH mRNA levels in these tissues indicated that the relative abundance of CBG mRNA in liver is >300-fold higher than in other tissues (Fig. 8A). The amounts of CBG mRNA in kidney, ovary, and testis are uniformly low and are undetectable in skeletal muscle. Moreover, although spleen and lung showed greater variability in CBG mRNA levels, they were also very low (Fig. 8A).
FIGURE 8.
A, levels of CBG mRNA in adult zebra finch tissues (A) and immunodetection of CBG in plasma from different avian species (B). A, CBG mRNA levels are expressed relative to GAPDH mRNA levels. Error bars, S.E.; numbers of tissues from different animals are indicated in parenthesis. B, plasma samples were diluted for SDS-PAGE (0.3–0.4 μl) to ensure that similar amounts of CBG were analyzed based on their corticosterone-binding capacity measurements. Western blotting was performed using anti-zebra finch CBG antiserum (1:5,000 dilution) or immunoaffinity-purified anti-zebra finch CBG antibodies (1:250 dilution).
A rabbit anti-zebra finch CBG antiserum recognizes CBG in zebra finch plasma with high specificity on Western blots (Fig. 5A) and also recognizes plasma CBG in other avian species (Fig. 8B). For this experiment, plasma from the different species was diluted so that equal amounts of CBG, as determined in corticosterone-binding capacity assays, were used for Western blotting. This showed that both anti-zebra finch antiserum and affinity-purified antibodies recognized CBG in song sparrow plasma slightly less well than in zebra finch plasma, whereas chicken CBG was only recognized clearly by concentrated affinity-purified antibodies. This is consistent with a higher amino acid sequence identity between zebra finch and sparrow CBGs (95%) than between zebra finch and chicken CBGs (78%).
Discussion
We have found that a chicken gene (ENSTGUG00000012647), currently annotated as SerpinA4, encodes the mammalian CBG (SerpinA6) ortholog, and sequence alignments allowed us to identify ENSTGUG00000012647 in a syntenic region of the zebra finch genome as the gene that encodes CBG. Moreover, we can also now predict with confidence the identities of SerpinA6 genes within the genomes of other avian species as well as several reptiles. The proximity and synteny of SerpinA6 and SerpinA1 in the genomes of these species is intriguing, because the proteins that they encode are considered the most closely related among SerpinA family members (50). It is also of interest that fish lack CBG, whereas it is present in the blood of amphibians (1). This is consistent with the concept that the SerpinA6 genes originated as a result of SerpinA gene duplications within a syntenic region of early terrestrial vertebrate genomes.
As anticipated, the structural organization of avian and mammalian SerpinA6 genes is identical, with four similarly sized exons encoding the CBG precursor polypeptide. However, chicken SerpinA6 (ENSTGUG00000012647) appears to comprise a larger 5′ non-coding exon than in mammalian SerpinA6 genes (ensembl.org). As in mammals (8, 51, 52), this 5′ exon in birds is probably flanked by a conserved promoter sequence that controls the tissue-specific and physiologic expression of their SerpinA6 genes, and the identification of avian SerpinA6 genes provides opportunities to study their regulation at the transcriptional levels.
The protein encoded by zebra finch SerpinA6 possesses all of the steroid-binding properties previously reported for avian CBGs (1, 20), but its identity was surprising for several reasons. In particular, it did not share several key features of its mammalian orthologs, and one of the three conserved N-glycosylation sites in avian CBGs is located within the steroid-binding site. The latter was most remarkable because the asparagine that is N-glycosylated in this position replaces a tryptophan in mammalian CBGs that interacts directly with steroid ligands (53). Positioning of this tryptophan in mammalian CBGs (11, 13) relies on a cation-π interaction with an N-terminal arginine that is also absent in the avian CBG sequences (Fig. 3). By contrast, the asparagine within the avian CBG steroid-binding sites is incapable of forming similar interactions with other residues or the steroid ligand. However, although an N-linked oligosaccharide at Asn-385 does not influence the steroid-binding activity of avian CBG, our results suggest that this asparagine may participate in the formation of the steroid-binding site during synthesis in eukaryotic cells.
Our crystal structure and mutagenesis studies explain why avian CBGs have greater affinities for progesterone than mammalian CBGs, and an asparagine instead of a tryptophan residue in the steroid-binding site may allow avian CBGs to bind dexamethasone with high affinity. In addition to these functionally relevant differences in the structures of mammalian and avian CBGs, only ∼50% of residues that interact with steroids in mammalian CBGs are conserved in avian CBG sequences. The mechanisms that control the binding and release of steroids may therefore differ between avian and mammalian CBGs.
When the zebra finch and rat CBG crystal structures in complex with cortisol were aligned, the structural similarity was evident. We used the crystal structure of rat CBG for this comparison because it also has cortisol in the binding site, and it is the only available native CBG structure in which the RCL is not cleaved and inserted in the β sheet A (11). This revealed differences in their main chain conformations, especially within the C-terminal part of helix D, which is unstructured in zebra finch CBG, most likely because of a proline at position 111. In a human CBG crystal structure, in which the RCL was cleaved, the protein had undergone the typical “stressed to relaxed” serpin structural conformational rearrangement, and the C-terminal part of helix D was unwound (14). This suggested that the unwinding of helix D causes allosteric rearrangements in the steroid-binding site conformation with a resulting loss of steroid- binding affinity (14), but this has not been formally demonstrated. However, the fact that this region of helix D in the zebra finch CBG structure is unwound, whereas the RCL appears to be intact, suggests that the conformation of helix D may not influence steroid binding in bird CBGs.
Divergence of CBG structures, while maintaining key functional characteristics, probably reflects how the protein evolved to control steroid transport and bioavailability in vertebrate subphyla in response to environmental pressures or differences in their physiology. During vertebrate evolution, aldosterone first appeared in amphibians (54), and CBG may have appeared at about the same time to protect tissues rich in mineralocorticoid receptors from unnecessary exposures to glucocorticoids. Identification of an amphibian SerpinA6 gene would therefore help in understanding the phylogenetic divergence of CBG structures.
The degree of evolutionary change with respect to the primary structures and steroid- binding properties of CBGs within vertebrate subphyla is remarkable compared, for instance, with the steroid hormone receptors, including the ligand-binding domain of glucocorticoid receptor, which tends to be more highly conserved across the vertebrates (55, 56). It might be argued that evolutionary changes in CBG structure and function compensated for the lack of sex hormone-binding globulin in avian species, but reptiles have genes encoding both plasma CBG and sex hormone-binding globulin, yet their CBG appears to be more closely related to avian CBGs than mammalian CBGs (Fig. 2C). In addition to sequence identity, the similarity between avian and reptile CBGs extends to the conservation of residues within their steroid-binding sites, the locations of N-glycosylation consensus sequences, and their RCL sequences (supplemental Fig. S1).
In addition to CBG, other members of the SERPIN clade A family bind hormones (e.g. thyroxine-binding globulin (SERPINA7) and the protein C inhibitor (SERPINA5) that has been reported to have a binding site for retinoic acid) (57). If avian orthologs of these other hormone-binding serpins exist, it would be of interest to determine how their structures and functions may have evolved across vertebrate species, as we have shown for CBG.
In addition to binding anti-inflammatory steroids or steroids involved in stress responses, our data indicate that the irreversible loss of high affinity steroid-binding activity, which occurs when the RCL of CBG is targeted and cleaved by specific classes of proteases, is conserved across vertebrate species. In particular, it is remarkable that the zebra finch CBG RCL appears to be targeted by the virulence factor, LasB, secreted by P. aeruginosa. It has been proposed that environmental pressures, such as exposures to different subsets of pathogens, may explain the apparent accelerated evolutionary change in SerpinA RCL sequences (58). Thus, our data suggest that opportunistic pathogens like P. aeruginosa may have been a driving force behind how Serpina6 genes evolved in terrestrial vertebrates and that RCL cleavage of CBG by exogenous proteases released by pathogens has been central to the role of CBG in controlling the actions of inflammatory steroids during infectious diseases throughout evolution.
As expected, a survey of adult zebra finch tissues indicated that the liver is the main site of CBG production. However, as in mammals, several other zebra finch tissues also contained low levels of CBG mRNA, the significance of which remains to be determined. In this regard, some extrahepatic tissues (exocrine pancreas and the convoluted tubules of the kidney) contain remarkably high levels of CBG mRNA at early developmental stages in rodents, and it has been proposed that this may influence the local actions of glucocorticoids on tissue morphogenesis and development (59, 60). It will therefore be of interest to examine the tissue-specific expression of SerpinA6 throughout development in avian species. The identification of avian SerpinA6 genes and the production of antibodies against zebra finch CBG, which cross-react with CBGs in other avian species, therefore also provide opportunities to examine the physiological impact of CBG in modulating the actions of multiple classes of steroids in birds.
Birds, particularly songbirds like the zebra finch, are commonly used as models across diverse fields in the biological sciences (61, 62). Many studies of birds have focused on neural and behavioral responses to environmental and social stressors and reproductive pressures (23–27). Steroid hormones mediate many of the biological responses studied using these models (63), and CBG serves as a primary gatekeeper of the actions of both the major stress (corticosterone) and reproductive (progesterone and testosterone) steroid hormones in birds by transporting them and regulating their access to target tissues (20–22). The identification and characterization of avian CBGs therefore helps to explain their distinct steroid-binding properties when compared with the CBG in mammalian species and provides the tools to study how CBG controls plasma concentrations and activities of steroid hormones in a range of biological studies in birds. Furthermore, recent advances in avian genomic biodiversity promise to provide unprecedented insight into vertebrate evolution (64). In this regard, differences in the topography of the mammalian and avian CBG steroid-binding sites are shared at least by reptilian CBGs, suggesting that evolutionary adaptations in CBG functions occurred to accommodate physiological and endocrine changes during the evolution of mammals, and our work provides a foundation for studies to further explore this.
Author Contributions
G. V. designed the study, performed the majority of the experiments, and prepared the paper; S. D. performed the crystallographic data collection and solved the structure together with G. V.; F. V. P. supervised the x-ray crystallographic part; K.-M. M. and J. C. R. performed the mass spectrometry and interpreted those data; M. D. T. collected tissue samples, provided reagents, and assisted with preparation of the paper; K. K. S. provided reagents and assisted with preparation of the paper; L. J. F. designed and supervised the mass spectrometry; G. L. H. supervised the project and guided the experimental design and the preparation of the paper.
Acknowledgments
We thank Caroline Underhill and Cathy Ma for technical assistance. We also thank the support staff at the Canadian Light Source (Saskatoon, Canada), which is supported by the Natural Sciences and Engineering Research Council of Canada, Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
This work was supported by Canadian Institutes of Health Research Grants MOP111102 (to G. L. H.), MOP77688 (to L. J. F.), and MOP119404 (to F. V. P.) and Natural Sciences and Engineering Research Council Grant RGPIN-2014-04884 (to K. K. S). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (code 5HGC) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CBG
- corticosteroid-binding globulin
- RCL
- reactive center loop
- PNGase F
- peptide:N-glycosidase F.
References
- 1. Westphal U. (1986) Steroid-protein interactions II. Monogr. Endocrinol. 27, 1–603 [PubMed] [Google Scholar]
- 2. Hammond G. L. (1990) Molecular properties of corticosteroid binding globulin and the sex-steroid binding proteins. Endocr. Rev. 11, 65–79 [DOI] [PubMed] [Google Scholar]
- 3. Hammond G. L., Smith C. L., Goping I. S., Underhill D. A., Harley M. J., Reventos J., Musto N. A., Gunsalus G. L., and Bardin C. W. (1987) Primary structure of human corticosteroid binding globulin, deduced from hepatic and pulmonary cDNAs, exhibits homology with serine protease inhibitors. Proc. Natl. Acad. Sci. U.S.A. 84, 5153–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith C. L., and Hammond G. L. (1988) The amino acid sequence of rat CBG deduced from a cDNA, and identification of CBG mRNA in the liver under different physiological states. Steroids 52, 331–332 [DOI] [PubMed] [Google Scholar]
- 5. Berdusco E. T., Hammond G. L., Jacobs R. A., Grolla A., Akagi K., Langlois D., and Challis J. R. (1993) Glucocorticoid-induced increase in plasma corticosteroid-binding globulin levels in fetal sheep is associated with increased biosynthesis and alterations in glycosylation. Endocrinology 132, 2001–2008 [DOI] [PubMed] [Google Scholar]
- 6. Seralini G. E., Smith C. L., and Hammond G. L. (1990) Rabbit corticosteroid-binding globulin: primary structure and biosynthesis during pregnancy. Mol. Endocrinol. 4, 1166–1172 [DOI] [PubMed] [Google Scholar]
- 7. Ousova O., Guyonnet-Duperat V., Iannuccelli N., Bidanel J. P., Milan D., Genêt C., Llamas B., Yerle M., Gellin J., Chardon P., Emptoz-Bonneton A., Pugeat M., Mormède P., and Moisan M. P. (2004) Corticosteroid binding globulin: a new target for cortisol-driven obesity. Mol. Endocrinol. 18, 1687–1696 [DOI] [PubMed] [Google Scholar]
- 8. Underhill D. A., and Hammond G. L. (1989) Organization of the human corticosteroid binding globulin gene and analysis of its 5′-flanking region. Mol. Endocrinol. 3, 1448–1454 [DOI] [PubMed] [Google Scholar]
- 9. Seralini G. E., Bérubé D., Gagné R., and Hammond G. L. (1990) The human corticosteroid binding globulin gene is located on chromosome 14q31-q32.1 near two other serine protease inhibitor genes. Hum. Genet. 86, 73–75 [DOI] [PubMed] [Google Scholar]
- 10. Forsyth S., Horvath A., and Coughlin P. (2003) A review and comparison of the murine α1-antitrypsin and α1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics 81, 336–345 [DOI] [PubMed] [Google Scholar]
- 11. Klieber M. A., Underhill C., Hammond G. L., and Muller Y. A. (2007) Corticosteroid-binding globulin, a structural basis for steroid transport and proteinase-triggered release. J. Biol. Chem. 282, 29594–29603 [DOI] [PubMed] [Google Scholar]
- 12. Zhou A., Wei Z., Stanley P. L., Read R. J., Stein P. E., and Carrell R. W. (2008) The S-to-R transition of corticosteroid-binding globulin and the mechanism of hormone release. J. Mol. Biol. 380, 244–251 [DOI] [PubMed] [Google Scholar]
- 13. Gardill B. R., Vogl M. R., Lin H. Y., Hammond G. L., and Muller Y. A. (2012) Corticosteroid-binding globulin: structure-function implications from species differences. PLoS One 7, e52759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin H. Y., Underhill C., Gardill B. R., Muller Y. A., and Hammond G. L. (2009) Residues in the human corticosteroid-binding globulin reactive center loop that influence steroid binding before and after elastase cleavage. J. Biol. Chem. 284, 884–896 [DOI] [PubMed] [Google Scholar]
- 15. Lin H. Y., Muller Y. A., and Hammond G. L. (2010) Molecular and structural basis of steroid hormone binding and release from corticosteroid-binding globulin. Mol. Cell. Endocrinol. 316, 3–12 [DOI] [PubMed] [Google Scholar]
- 16. Avvakumov G. V., Warmels-Rodenhiser S., and Hammond G. L. (1993) Glycosylation of human corticosteroid-binding globulin at aspargine 238 is necessary for steroid binding. J. Biol. Chem. 268, 862–866 [PubMed] [Google Scholar]
- 17. Hammond G. L., Smith C. L., Paterson N. A., and Sibbald W. J. (1990) A role for corticosteroid-binding globulin in delivery of cortisol to activated neutrophils. J. Clin. Endocrinol. Metab. 71, 34–39 [DOI] [PubMed] [Google Scholar]
- 18. Lewis J. G., and Elder P. A. (2014) The reactive centre loop of corticosteroid-binding globulin (CBG) is a protease target for cortisol release. Mol. Cell. Endocrinol. 384, 96–101 [DOI] [PubMed] [Google Scholar]
- 19. Simard M., Hill L. A., Underhill C. M., Keller B. O., Villanueva I., Hancock R. E., and Hammond G. L. (2014) Pseudomonas aeruginosa elastase disrupts the cortisol-binding activity of corticosteroid-binding globulin. Endocrinology 155, 2900–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wingfield J. C., Matt K. S., and Farner D. S. (1984) Physiologic properties of steroid hormone-binding proteins in avian blood. Gen. Comp. Endocrinol. 53, 281–292 [DOI] [PubMed] [Google Scholar]
- 21. Deviche P., Breuner C., and Orchinik M. (2001) Testosterone, corticosterone, and photoperiod interact to regulate plasma levels of binding globulin and free steroid hormone in dark-eyed juncos, Junco hyemalis. Gen. Comp. Endocrinol. 122, 67–77 [DOI] [PubMed] [Google Scholar]
- 22. Malisch J. L., and Breuner C. W. (2010) Steroid-binding proteins and free steroids in birds. Mol. Cell. Endocrinol. 316, 42–52 [DOI] [PubMed] [Google Scholar]
- 23. Romero L. M., Cyr N. E., and Romero R. C. (2006) Corticosterone responses change seasonally in free-living house sparrows (Passer domesticus). Gen. Comp. Endocrinol. 149, 58–65 [DOI] [PubMed] [Google Scholar]
- 24. Romero L. M. (2006) Seasonal changes in hypothalamic-pituitary-adrenal axis sensitivity in free-living house sparrows (Passer domesticus). Gen. Comp. Endocrinol. 149, 66–71 [DOI] [PubMed] [Google Scholar]
- 25. Li D., Zhang X., Li Y., Hao C., Zhang J., and Wu Y. (2012) Stress responses of testosterone and corticosterone-binding globulin in a multi-brooded species, Eurasian Tree Sparrows (Passer montanus): does CBG function as a mediator? Horm. Behav. 61, 582–589 [DOI] [PubMed] [Google Scholar]
- 26. Lynn S. E., Stamplis T. B., Barrington W. T., Weida N., and Hudak C. A. (2010) Food, stress, and reproduction: short-term fasting alters endocrine physiology and reproductive behavior in the zebra finch. Horm. Behav. 58, 214–222 [DOI] [PubMed] [Google Scholar]
- 27. Malisch J. L., Satterlee D. G., Cockrem J. F., Wada H., and Breuner C. W. (2010) How acute is the acute stress response? Baseline corticosterone and corticosteroid-binding globulin levels change 24 h after an acute stressor in Japanese quail. Gen. Comp. Endocrinol. 165, 345–350 [DOI] [PubMed] [Google Scholar]
- 28. Charlier T. D., Underhill C., Hammond G. L., and Soma K. K. (2009) Effects of aggressive encounters on plasma corticosteroid-binding globulin and its ligands in white-crowned sparrows. Horm. Behav. 56, 339–347 [DOI] [PubMed] [Google Scholar]
- 29. Hammond G. L., Robinson P. A., Sugino H., Ward D. N., and Finne J. (1986) Physicochemical characteristics of human sex hormone binding globulin: evidence for two identical subunits. J. Steroid Biochem. 24, 815–824 [DOI] [PubMed] [Google Scholar]
- 30. Chan Q. W., Howes C. G., and Foster L. J. (2006) Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics 5, 2252–2262 [DOI] [PubMed] [Google Scholar]
- 31. Parker R., Melathopoulos A. P., White R., Pernal S. F., Guarna M. M., and Foster L. J. (2010) Ecological adaptation of diverse honey bee (Apis mellifera) populations. PLoS One 5, e11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simard M., Hill L. A., Lewis J. G., and Hammond G. L. (2015) Naturally occurring mutations of human corticosteroid-binding globulin. J. Clin. Endocrinol. Metab. 100, E129–E139 [DOI] [PubMed] [Google Scholar]
- 33. Lobo P. A., Kimlicka L., Tung C. C., and Van Petegem F. (2011) The deletion of exon 3 in the cardiac ryanodine receptor is rescued by β strand switching. Structure 19, 790–798 [DOI] [PubMed] [Google Scholar]
- 34. Robinson P. A., Langley M. S., and Hammond G. L. (1985) A solid-phase radioimmunoassay for human corticosteroid binding globulin. J. Endocrinol. 104, 259–267 [DOI] [PubMed] [Google Scholar]
- 35. Sievers F., and Higgins D. G. (2014) Clustal ω. Curr. Protoc. Bioinformatics 10.1002/0471250953.bi0313s48 [DOI] [PubMed] [Google Scholar]
- 36. Hammond G. L., and Lähteenmäki P. L. (1983) A versatile method for the determination of serum cortisol binding globulin and sex hormone binding globulin binding capacities. Clin. Chim. Acta 132, 101–110 [DOI] [PubMed] [Google Scholar]
- 37. Goldschmidt L., Cooper D. R., Derewenda Z. S., and Eisenberg D. (2007) Toward rational protein crystallization: a Web server for the design of crystallizable protein variants. Protein Sci. 16, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zinzow-Kramer W. M., Horton B. M., and Maney D. L. (2014) Evaluation of reference genes for quantitative real-time PCR in the brain, pituitary, and gonads of songbirds. Horm. Behav. 66, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Panin L. E., Tuzikov F. V., and Gimautdinova O. I. (2003) Tetrahydrocortisol-apolipoprotein A-I complex specifically interacts with eukaryotic DNA and GCC elements of genes. J. Steroid Biochem. Mol. Biol. 87, 309–318 [DOI] [PubMed] [Google Scholar]
- 45. Panin L. E., Knyazev R. A., Sumenkova D. V., and Polyakov L. M. (2007) Effect of complexes of apolipoprotein A-I with tetrahydrocortisol and pregnenolone on protein biosynthesis in rat hepatocytes culture. Bull. Exp. Biol. Med. 144, 291–293 [DOI] [PubMed] [Google Scholar]
- 46. Forstmeier W., Segelbacher G., Mueller J. C., and Kempenaers B. (2007) Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol. Ecol. 16, 4039–4050 [DOI] [PubMed] [Google Scholar]
- 47. Schmidt K. L., Malisch J. L., Breuner C. W., and Soma K. K. (2010) Corticosterone and cortisol binding sites in plasma, immune organs and brain of developing zebra finches: intracellular and membrane-associated receptors. Brain Behav. Immun. 24, 908–918 [DOI] [PubMed] [Google Scholar]
- 48. Maley F., Trimble R. B., Tarentino A. L., and Plummer T. H. Jr. (1989) Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 49. Walker S. E., Sander J. E., Cline J. L., and Helton J. S. (2002) Characterization of Pseudomonas aeruginosa isolates associated with mortality in broiler chicks. Avian Dis. 46, 1045–1050 [DOI] [PubMed] [Google Scholar]
- 50. Heit C., Jackson B. C., McAndrews M., Wright M. W., Thompson D. C., Silverman G. A., Nebert D. W., and Vasiliou V. (2013) Update of the human and mouse SERPIN gene superfamily. Hum. Genomics 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Underhill D. A., and Hammond G. L. (1995) Cis-regulatory elements within the proximal promoter of the rat gene encoding corticosteroid-binding globulin. Gene 162, 205–211 [DOI] [PubMed] [Google Scholar]
- 52. Verhoog N., Allie-Reid F., Vanden Berghe W., Smith C., Haegeman G., Hapgood J., and Louw A. (2014) Inhibition of corticosteroid-binding globulin gene expression by glucocorticoids involves C/EBPβ. PLoS One 9, e110702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hill L. A., Vassiliadi D. A., Simard M., Pavlaki A., Perogamvros I., Hadjidakis D., and Hammond G. L. (2012) Two different corticosteroid-binding globulin variants that lack cortisol-binding activity in a Greek woman. J. Clin. Endocrinol. Metab. 97, 4260–4267 [DOI] [PubMed] [Google Scholar]
- 54. Colombo L., Dalla Valle L., Fiore C., Armanini D., and Belvedere P. (2006) Aldosterone and the conquest of land. J. Endocrinol. Invest. 29, 373–379 [DOI] [PubMed] [Google Scholar]
- 55. Baker M. E., Funder J. W., and Kattoula S. R. (2013) Evolution of hormone selectivity in glucocorticoid and mineralocorticoid receptors. J. Steroid Biochem. Mol. Biol. 137, 57–70 [DOI] [PubMed] [Google Scholar]
- 56. Harms M. J., and Thornton J. W. (2014) Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature 512, 203–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huntington J. A., Kjellberg M., and Stenflo J. (2003) Crystal structure of protein C inhibitor provides insights into hormone binding and heparin activation. Structure 11, 205–215 [DOI] [PubMed] [Google Scholar]
- 58. Hill R. E., and Hastie N. D. (1987) Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326, 96–99 [DOI] [PubMed] [Google Scholar]
- 59. Scrocchi L. A., Hearn S. A., Han V. K., and Hammond G. L. (1993) Corticosteroid-binding globulin biosynthesis in the mouse liver and kidney during postnatal development. Endocrinology 132, 910–916 [DOI] [PubMed] [Google Scholar]
- 60. Scrocchi L. A., Orava M., Smith C. L., Han V. K., and Hammond G. L. (1993) Spatial and temporal distribution of corticosteroid-binding globulin and its messenger ribonucleic acid in embryonic and fetal mice. Endocrinology 132, 903–909 [DOI] [PubMed] [Google Scholar]
- 61. Konishi M., Emlen S. T., Ricklefs R. E., and Wingfield J. C. (1989) Contributions of bird studies to biology. Science 246, 465–472 [DOI] [PubMed] [Google Scholar]
- 62. Wingfield J. C. (2005) Historical contributions of research on birds to behavioral neuroendocrinology. Horm. Behav. 48, 395–402 [DOI] [PubMed] [Google Scholar]
- 63. Goodson J. L., Saldanha C. J., Hahn T. P., and Soma K. K. (2005) Recent advances in behavioral neuroendocrinology: insights from studies on birds. Horm. Behav. 48, 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang G., Jarvis E. D., and Gilbert M. T. (2014) A flock of genomes. Science 346, 1308–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fredslund J. (2006) PHY.FI: fast and easy online creation and manipulation of phylogeny color figures. BMC Bioinformatics 7, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]