Abstract
Mortality due to snakebite is a serious public health problem, and available therapeutics are known to induce debilitating side effects. Traditional medicine suggests that seeds of Mucuna pruriens can provide protection against the effects of snakebite. Our aim is to identify the protein(s) that may be important for snake venom neutralization and elucidate its mechanism of action. To this end, we have identified and purified a protein from M. pruriens, which we have named MP-4. The full-length polypeptide sequence of MP-4 was obtained through N-terminal sequencing of peptide fragments. Sequence analysis suggested that the protein may belong to the Kunitz-type protease inhibitor family and therefore may potentially neutralize the proteases present in snake venom. Using various structural and biochemical tools coupled with in vivo assays, we are able to show that MP-4 does not afford direct protection against snake venom because it is actually a poor inhibitor of serine proteases. Further experiments showed that antibodies generated against MP-4 cross-react with the whole venom and provide protection to mice against Echis carinatus snake venom. This study shows that the MP-4 contributes significantly to the snake venom neutralization activity of M. pruriens seeds through an indirect antibody-mediated mechanism.
Keywords: antibody, crystal structure, humoral response, protease inhibitor, snake venom
Introduction
Death due to venomous snakes is an important public health problem in many tropical and subtropical countries. It is estimated that about 200,000 people die due to snakebite every year, and the majority of survivors suffer permanent disabilities (1–4). Snake venom is a complex mixture of enzymes, non-enzyme proteins, procoagulants, peptides, nucleotides, and inorganic ions. These components damage the central nervous system, vascular system, and muscular and cardiovascular system (5). The most common and effective method of treating snakebite victims is through the immediate administration of anti-venom. The anti-venom is composed of an immunized animal (horse, goat, or rabbit) blood product, such as blood sera (polyclonal antibodies), that does not contain any active components of snake venom (6). Monovalent anti-venom is generated in a hyperimmunized animal against a given species venom, whereas polyvalent anti-venom is generated against a mixture of various snake venoms, particularly cobra, krait, Russell's viper, and saw-scaled viper (7). Some patients are allergic to anti-venom and show sudden signs of neurotoxicity, edema at various body parts, shortness of breath, weak pulse, muscles tenderness, dizziness, fainting, and in some cases death due to hemorrhage (8–10).
Traditional medicine in many countries employs the extracts of certain plants to provide protection against snakebites. Since ancient times, several plant extracts have been known to protect human beings from the toxic effects of snakebite (8, 11–13). The characterization and isolation of effective ingredients in these plant extracts can pave the way for the development of safer prophylactic and therapeutic formulations against snake venom. The plant Mucuna pruriens is well known for its anti-snake venom properties, and it has been claimed that the oral intake of few seeds can protect an individual for a year against snakebites (14–18). M. pruriens seeds are also used for the treatment of Parkinson, neoplasty, diabetic, microbial, analgesic, and inflammatory diseases (19–24).
A number of studies have been done on extracts from M. pruriens to isolate the biochemical basis of snakebite protection. In one report, it was found that crude M. pruriens seed extract initiates a coagulation cascade and competes with the Echis carinatus venom components for common cellular targets (25). Other reports show that immunization with M. pruriens aqueous seed extract affords possible protection against venom of the snake families Elapidae and Viperidae (16, 26). One of the proteins present in the seed extract is a multiform glycoprotein (gpMuc) of apparent molecular mass 20–28 kDa. N-terminal sequences of seven glycosylated isoforms of this protein show the conserved signature sequence of Kunitz-type protease inhibitors (27, 28). This protein can inhibit proteolytic components of snake venom and thus may provide direct protection against the toxic effects of snakebite.
It was shown that antibodies raised in mice against M. pruriens seed proteins also react with venom components. This observation suggests that immunological neutralization of venom components provides protection against the toxic effects of snakebite (14, 29). However, the proteins in the M. pruriens extract that are responsible for antibody cross-reactivity remain to be identified and isolated. It is possible that immunization with the active protein(s) may be enough to afford long term protection against snakebite, and such a preparation can be used as a prophylactic agent. Moreover, these proteins can be used to generate polyclonal sera that may serve as an immediate and effective therapeutic for individuals suffering from the toxic effects of snakebite.
In the present study, we have identified one of the dominant proteins of the seed proteome of M. pruriens, named MP-4 (20.9 kDa). The full-length sequence of this protein was determined using the N-terminal Edman degradation method, and bioinformatic analysis of the derived sequence suggested that this protein may belong to the Kunitz-type protease inhibitor family. This observation raised the possibility that MP-4 may neutralize snake venom through direct inhibition of the proteases present in snake venom. However, in vivo and biochemical assays showed that the protein does not directly neutralize the toxic effects of snake venom. The structure of this protein (2.8 Å) showed that a residue critical for protease inhibition is missing in the reactive site loop. In line with the structural observation, the protein does not inhibit the proteolytic activity of trypsin and chymotrypsin. However, we observed that immunization of mice with this protein provided significant protection against the toxic effects of snake venom from E. carinatus. Overall, our studies show that the MP-4 protein contributes substantially to protection against snake venom by M. pruriens seeds through an antibody-mediated mechanism and not through direct inhibition of venom proteases. Our studies suggest that MP-4 can be utilized to develop prophylactic and therapeutic strategies against physiological effects of snake envenomation.
Experimental Procedures
Ethics Statement
Female BALB/c mice were obtained from the Small Animal Facility of the National Institute of Immunology (Delhi, India) and maintained in conventional environmental conditions throughout the experiment after due approval from the institutional animal ethical committee (approval 198). All experiments on animals were conducted according to relevant national and international guidelines.
Plant Materials
M. pruriens (family Fabaceae; subfamily: Faboideae; genus: Mucuna; species: pruriens) seeds were collected from a medicinal firm, M/S Shidh Seeds Sales Corp. (Dehradun District, India). Seeds were stored in an air-tight container in a dry and dark place at room temperature (25 °C).
Fractionation and Identification of Seed Proteome
M. pruriens seeds were washed thoroughly with milli-Q water and dried at room temperature (25 °C). The dried seeds were ground into fine powder using an electric grinder. Delipidification of 50 g of fine seed powder was done three times with 500 ml of petroleum ether for 3 h each, followed by air drying at room temperature (25 °C). 20 g of dried delipidified powder was homogenized in 400 ml of 50 mm sodium acetate buffer, pH 5.0, and stirred for 15 min at 4 °C in the dark. The homogenized mixture was centrifuged at 12,000 × g for 30 min at 4 °C. The resulting solubilized protein supernatant solution was then subjected to ammonium sulfate salt fractionation over the range of 0–80% (w/v) at 4 °C. The precipitated protein in each ammonium sulfate fraction was subjected to centrifugation at 12,000 × g for 1 h at 4 °C. The pellets corresponding to each fractionation step were resuspended in 25 ml of 50 mm phosphate buffer, pH 7.2, and analyzed by 12% SDS-PAGE. The major protein bands in the 40 and 60% ammonium sulfate fractions were transferred onto a polyvinylidene difluoride (PVDF) membrane using 10 mm CAPS buffer (pH 11.0). Each protein band from the PVDF membrane was subjected to N-terminal sequencing by the Edman degradation method on a Procise protein sequencer (Applied Biosystems). The N-terminal sequence obtained in this manner was utilized for preliminary identification of homologous protein sequences using the BLAST algorithm (30).
Protein Purification
The 60% ammonium sulfate fraction contained a dominant protein in the seed proteome of M. pruriens that was named MP-4. Resolubilized precipitate from the 60% fraction containing ∼60–100 mg of total protein was injected onto a Sephacryl-200 preparative size exclusion column (Amersham Biosciences). The column was pre-equilibrated with 50 mm phosphate buffer, pH 7.2, containing 140 mm NaCl, and the same buffer was also used as running buffer for elution of proteins at a 1 ml min−1 flow rate on ÄKTAprime (Amersham Biosciences). Fractions corresponding to different peaks were collected manually and analyzed by 12% SDS-PAGE for their homogeneity. The fractions of the third peak corresponding to MP-4 were pooled together and concentrated to 10 mg ml−1 using ultrafiltration (Amicon, 10 kDa cut-off; Millipore). All steps from fractionation to purification were done in the dark because the initial crude extract was found to be photosensitive. The concentration of purified protein was estimated by a BCA protein assay (Pierce) using BSA (Sigma-Aldrich) as a standard.
Determination of Molecular Weight by Mass Spectrometry
For molecular weight determination, a Nano LC ultra 2D Plus system (AB Sciex) connected to a hybrid quadrupole-TOF LC/MS/MS mass spectrometer (AB Sciex) was used. The latter was equipped with a trap column and C8 RP analytical column followed by a nanoelectron spray ionization source (Nanosource II, AB/MDS Sciex). Protein (10–20 μg) was reconstituted in 10 μl of 0.1% (v/v) trifluoroacetic acid and was loaded onto a trap column. The column was equilibrated for 12 min in 0.1% formic acid/H2O. Further elution was carried out for 45 min, over a gradient of 13–32% solvent B (acetonitrile + 0.1% formic acid) at a flow rate of 550 nl min−1. The TOF-MS data were acquired using Analyst version 1.4.2 software (AB/MDS Sciex) and deconvoluted by BioAnalyst software for the intact MP-4 mass calculation.
Proteolytic Digestion and Sequencing of MP-4 by Edman Degradation Method
Internal sequences of MP-4 were derived using a variety of digestive enzymes (1% (w/w) MP-4). Tryptic digestion was carried out with 200 μg of MP-4 in 0.1 mm CaCl2 and 50 mm ammonium bicarbonate buffer, pH 8.0, with non-reducing and carboxymethylated MP-4. For V8 protease, Arg-C endoproteinase, and Lys-C endoproteinase enzymes, digestions were done with reduced and carboxymethylated MP-4 under similar digestion conditions. Incubation temperature (37 °C) was kept constant during all digestion steps. With each enzyme, samples were collected at several time points. Apart from enzymes, chemical digestion was also performed with cyanogen bromide (1:250 mol/mol) in 70% formic acid. All of the digested samples were run on 12% SDS-PAGE, transferred to PVDF membrane. The neatly excised peptide fragments were subsequently sequenced on a Procise protein sequencer (Applied Biosystems). The final 25 residues at the C terminus were built based on homologous sequences available in the sequence database. This endoproteinase Glu-C-digested peptide fragment was non-reproducible, and hence different combinations of amino acids of this peptide were used for homologous database search. The full-length polypeptide sequence obtained in this manner was utilized for identification of homologs using the BLAST algorithm (30).
Assay to Check Direct Neutralization of Snake Venom
The minimum lethal dose (MLD)3 of E. carinatus snake venom (EcV) was found to be 2 mg/kg. To check the direct neutralization efficiency of MP-4, experiments were carried out with three groups of eight mice each. 10 mg ml−1 of this protein was mixed thoroughly with EcV in 1:1 ratio (w/w). This mixture was incubated for 1 h at 4 °C, and 400 μl of this solution was administered intraperitoneally to each mouse in the sample group. The other two groups of animals were used for control experiments. In one control group, the MLD of EcV (2 mg/kg) was administrated, whereas in another group, saline was given. The number of mice surviving at the end of 24 h was recorded for each group. The entire experiment was repeated three times.
Crystallization and Data Collection
Cuboidal shape crystals were obtained by the hanging drop vapor diffusion method in the following conditions: 50 mm Tris-HCl, 1.5 m ammonium sulfate, pH 9.5, at 25 °C.
A single crystal was soaked in glycerol (33%) for 30 s and frozen directly in the cold nitrogen stream (−173 °C). X-ray diffraction data were collected and processed with MOSFLM (31), followed by scaling and merging with SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4). The data collection statistics are shown in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
| Parameters | Valuesa |
|---|---|
| Data collection statistics | |
| Space group | P21212 |
| Unit cell (a, b, c) (Å) | 51.00, 69.18, 45.32 |
| Resolution (Å) | 37.91–2.8 (2.95–2.8) |
| Rmergeb | 0.073 (0.162) |
| Completeness | 95.5 (94.0) |
| 〈I〉/σ(I) | 25.5 (14.5) |
| Multiplicity | 9.7 (10.1) |
| Refinement statistics | |
| Total no. of reflections used in refinement | 4035 |
| Rwork/Rfree (%)c | 26.7/28.4 |
| Average B-factors (Å2) | 21.83 |
| Macromolecule | 22.36 |
| Solvent | 15.65 |
| Root mean square deviation values | |
| Bond length (Å) | 0.0118 |
| Bond angle (degrees) | 1.6454 |
| Ramachandran plot (%) | |
| Favored | 87 |
| Allowed | 11 |
| Outliers | 2 |
a Values in parentheses are for the highest resolution shell.
b Rmerge = Σ|I − 〈I〉|/ΣI, where I is the integrated intensity of a given reflection.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|. Rfree was calculated using 5% of data excluded from refinement.
Structure Determination and Refinement
The closest homolog of MP-4 (44% sequence identity) for which structure was available was the trypsin inhibitor from Delonix regia (DrTI; Protein Data Bank entry 1R8N). 1R8N was therefore used as the search model to solve the MP-4 structure by molecular replacement (32). The coordinate file of 1R8N was rationally edited on the basis of sequence alignment. Non-conserved residues were either deleted or mutated into Ala. The regions corresponding to long loops in the structure, the first five N-terminal residues, and the last 25 C-terminal residues and water molecules were deleted. The top model of Phaser (CCP4) showed initial log likelihood gain, rotation factor Z score, and translation factor Z score for the MP-4 model of 234.13, 10.9, and 6.9, respectively. The molecular replacement solution was checked manually for symmetry-mate clashes within 8 Å range in PyMOL.
Rwork of 38.3% and Rfree of 42.8% were obtained with initial rigid body refinement that was carried out in the resolution range of 50–3.5 Å. B-group and positional refinement further lowered the Rwork to 36.5% and Rfree to 38.7%. The electron density maps at 3.5 Å resolution were examined, and the regions corresponding to residues 27–30, 37–44, 66–72, 100–107, 115–121, 157–161, 117–120, and 158–160 showed poor electron density. These poor density regions correspond to loops in the structure, whereas the core region showed good electron density. Further refinement was carried out in CNS initially for the core region. After each refinement cycle, the map quality was visualized in the COOT program (33), and model rebuilding was carried out based on electron density. Resolution of the data used for refinement was progressively increased by 0.05 Å, and missing regions were built gradually with the appearance of electron density. In the range of 37.9–2.8 Å resolution, the backbone was completely built, and side chains of most of the missing residues were constructed. Water molecules were initially modeled at 3 Å resolution and progressively added with the increment of resolution. The iterative cycles of gradual model building followed by refinement led to the appearance of clear electron density for the loop and C-terminal regions.
The entire MP-4 structure was built and refined at a resolution of 2.8 Å. Refinement statistics are given in Table 1. Quality of the model was evaluated by PROCHECK (CCP4 suite). Structure factors and the final coordinates of the crystal structure of the MP-4 were deposited in the Protein Data Bank under accession code 5DSS. All of the structure figures were prepared with PyMOL. Structural alignments were also carried out using PyMOL. The structural orthologs of MP-4 structure were identified using the Dali server (34).
Assays to Assess Protease Inhibition Activity
The protocol published by Erlanger et al. (35) was modified to estimate protease inhibitor activity of MP-4. Inhibition of trypsin by MP-4 was estimated by measuring the release of p-nitroaniline from substrate N-α-benzoyl-dl-arginine p-nitroanilide (Sigma-Aldrich). N-α-Benzoyl-dl-arginine p-nitroanilide was dissolved in a minimum volume of DMSO to obtain a 1 mm solution at room temperature and further diluted in buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mm CaCl2). In two different sets of experiments, a fixed concentration of trypsin (0.084 μm) was titrated against soybean trypsin inhibitor (STI; Sigma-Aldrich) (67.2–0.067 μm) and with MP-4 (67.2–0.067 μm). The mixtures were incubated for 10 min at room temperature, followed by the addition of 30 μl of 1 mm N-α-benzoyl-dl-arginine p-nitroanilide. Reaction mixture was incubated further for 40 min at room temperature, and reaction was quenched with 50 μl of 30% (v/v) acetic acid. Final volume of the total reaction mixture was 300 μl. Readings were taken at 405 nm in a microplate reader (Multiskan Ascent, Thermolab Systems). A similar experiment was also carried out with the standard trypsin inhibitor, STI, instead of MP-4. IC50 values were calculated by GraphPad Prism software.
Similarly, the inhibitory concentration of MP-4 was estimated for chymotrypsin. For comparison, experiments were also conducted with standard inhibitors, such as chymostatin (Sigma-Aldrich) and chymotrypsin·trypsin inhibitor (Sigma-Aldrich). For these experiments, N-benzoyl-l-tyrosine p-nitroanilide was used as a substrate (Sigma-Aldrich).
Surface Plasmon Resonance (SPR)
All measurements were carried out on a BIAcore T200 system (GE Healthcare). 2 μm MP-4 (ligand) was prepared in 10 mm sodium acetate, pH 4.0, buffer and immobilized onto flow-cell format CM5 (carboxymethylated)-certified grade sensor chips by the amine coupling method using an equal mixture of N-ethyl-N-dimethylaminopropyl carbodiimide/N-hydroxysuccinimide (BIAcore amine coupling kit). Approximately 178 response units was achieved by immobilization on a CM5 sensor chip with a flow rate of 5 μl min−1 for 120 s, and the unreacted active sites were blocked with 1 m ethanolamine (BIAcore amine coupling kit). 16 μm to 125 nm porcine trypsin (Sigma-Aldrich) and 32 μm to 125 nm chymotrypsin (Sigma-Aldrich) were serially diluted in running buffer (10 mm HEPES, pH 7.4, containing 150 mm NaCl, 3 mm EDTA, and 0.005% surfactant P20) in two different sets of experiments wherein each cycle comprised a 3-min association phase and 15-min dissociation phase. Kinetic parameters were calculated by T200 Evaluation software. Interaction curves were simultaneously fitted by a 1:1 Langmuir model system (36). The T200 Evaluation program was used to determine the equilibrium dissociation constant (KD = kd/ka).
Immunization
The immune response against MP-4 protein was assessed in two sets of female BALB/c mice (8–10 weeks old), each group containing eight mice. The immunizing antigen was either MP-4 or whole venom of E. carinatus (Sigma-Aldrich). For primary immunization, each mouse was subcutaneously injected with 200 μl (60 μg) of emulsion of MP-4-CFA (complete Freund's adjuvant, Difco) in a ratio of 1:1 (v/v). Primary immunization was followed by two booster immunizations, after 14 days and 21 days of priming using incomplete Freund's adjuvant (Difco). After 10 days of each booster, the immunized animals were screened for the presence of antibodies raised against the antigen. Blood samples (∼0.2–0.4 ml) were collected from the retro-orbital sinus, allowed to coagulate for 1 h at 37 °C, and then centrifuged at 1500 × g for 30 min to separate the serum. 100 μl of the serum samples was diluted 1:500 to monitor the antibody titer by ELISA. Similarly, antibodies were generated against an optimized dose of whole E. carinatus venom (Sigma-Aldrich).
Characterization of Cross-reactivity
The antibody responses against respective antigens were monitored using antibody-specific indirect ELISA. In brief, 96-well plates (Greiner Bio-one) were coated in triplicates with 5 μg of protein in 100 μl of 50 mm carbonate buffer, pH 9.0, per well with overnight incubation at 4 °C. To avoid nonspecific binding, blocking was done with 5% lactogen (350 μl/well in PBS buffer) for 2 h at 37 °C. After complete blocking, the plates were washed three times with 1× phosphate-buffered saline with 0.05% Tween 20 (PBST). The serum samples were serially diluted (0.5×), 100 μl of each sample was added to the wells, and the plate was incubated for 2 h at 37 °C. Two sets of experiments were carried out, one with anti-MP-4 sera and the other with anti-EcV sera.
The plates were then washed three times with PBST, and 100 μl of 2 μg ml−1 solution of horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (Jackson ImmunoResearch) was added, followed by incubation at 37 °C for 1.5 h. The assay was developed with 100 μl of 0.5 mg ml−1 o-phenylenediamine dihydrochloride (Sigma-Aldrich) in 50 mm citrate phosphate buffer, pH 5.5, and 1 μl ml−1 hydrogen peroxide (Merck). o-Phenylenediamine dihydrochloride was used as a chromogenic substrate for HRP. Incubation was performed in the dark for 5–10 min. 50 μl of 1 n sulfuric acid (Thomas Baker) was used to inactivate the HRP enzyme, and immediately absorbance was measured at 490 nm using a microplate reader (Multiskan Ascent, Thermolab Systems).
In Vivo Protection Assay
In vivo protection assays were performed on four groups of BALB/c mice, each containing eight mice. Three groups of mice were immunized with MP-4 using the immunization protocol described above, whereas the last group was used as a control (injected with saline). The anti-MP4 antibody titers were checked after 28 days. Subsequently, the mice were challenged with a minimal lethal dose, LC50 2 (2 mg/kg/individual) and subminimal lethal dose <LC50 2 (1 mg/kg/individual) of crude EcV (Sigma-Aldrich) administrated intraperitoneally. One of the three groups immunized with MP-4 was used as the control experimental set, wherein these mice were administered saline instead of EcV. The unimmunized control group was also injected with EcV. The numbers of survivors and dead mice in all groups were recorded at the end of 24 h. The survival percentage for each group was calculated using the expression, Survival percentage = ((total mice − number of dead mice) × 100/total mice). The in vivo protection assay was repeated three times.
Results and Discussion
Fractionation of Proteins from Seeds of M. pruriens
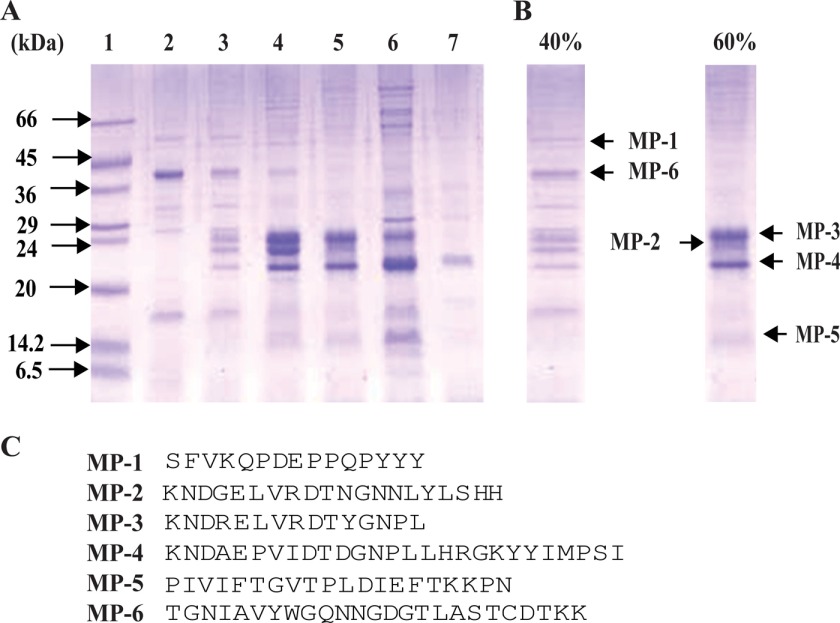
Ammonium sulfate fractionation over the range of 0–80% resulted in the identification of six major proteins: MP-1 and MP-6 in 40%, and MP-2, MP-3, MP-4, and MP-5 in 60% fractions (Fig. 1, A and B). All of them were subjected to N-terminal sequencing after transferring onto PVDF membrane, and the initial 15–27 amino acids were sequenced for each band (Fig. 1C). The plausible functions of these proteins were assigned based on homologous proteins present in the sequence database (supplemental Table S1). N-terminal sequence for MP-1 shows no significant match with known plant proteins. MP-2 shows sequence homology with water-soluble chlorophyll-binding protein (WSCP) of Lepidium virginicum and Kunitz-type trypsin inhibitors of winged bean with a high confidence value. MP-3 showed a significant match with WSCP of L. virginicum similar to MP-2. MP-4 showed significant homology with WSCP of L. virginicum and various Kunitz-type trypsin inhibitors of plant origin. In the case of MP-5, very high homology was found with the number of Kunitz-type trypsin inhibitors, water-soluble chlorophyll-binding protein and plant α-fucosidases. The N-terminal sequence of MP-6 showed 99% identity with plant chitinases.
FIGURE 1.
Analysis of seed proteins of M. pruriens by 12% SDS-PAGE in reducing conditions after ammonium sulfate fractionation. A, lanes 2–7, 0–30%, 30–40%, 40–50%, 50–60%, 60–70%, and 70–80% fractions, respectively. In lane 1, low molecular weight protein markers (Sigma-Aldrich) were loaded. B, 40% fraction highlights two major protein bands, MP-1 and MP-6; the 60% fraction shows four major protein bands, MP-2, MP-3, MP-4, and MP-5. C, N-terminal sequences by Edman degradation method of all of the major proteins present in 40 and 60% ammonium sulfate fractions.
Purification, Sequencing, and Characterization of MP-4
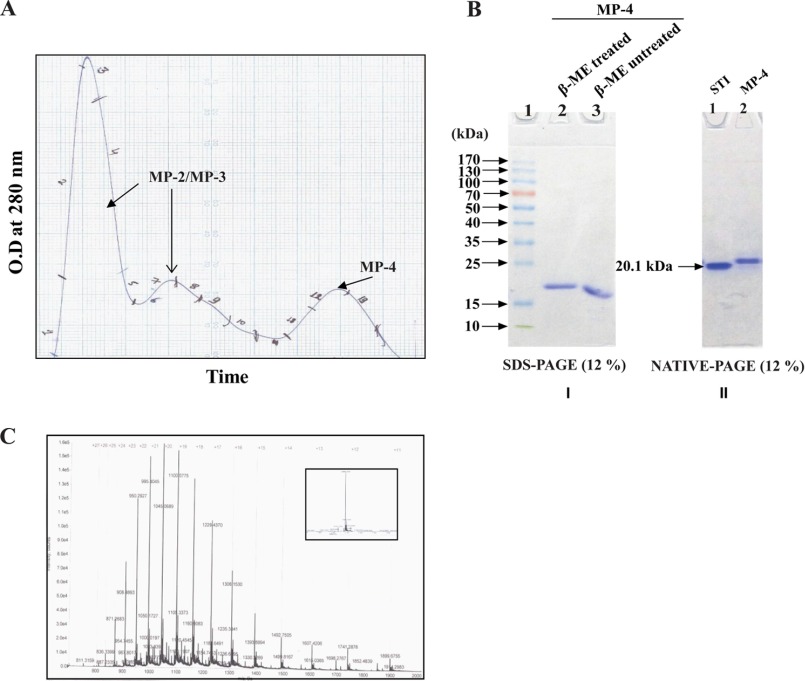
MP-4 appears to be a dominant protein based on the 12% SDS-PAGE profile in reducing conditions after ammonium sulfate fractionation of M. pruriens seeds. N-terminal sequencing of this protein provided identification of the first 27 residues. Sequence analysis of the N-terminal sequence by BLAST showed that this protein has 51% sequence identity with water-soluble chlorophyll-binding protein of L. virginicum and 41, 51, and 51% sequence identity with Kunitz-type protease inhibitors of D. regia, Medicago truncatula, and Cicer arietinum, respectively (supplemental Table S1). Gel filtration chromatography using Sephacryl-200 column facilitated purification of MP-4 from a 60% ammonium sulfate fraction. First and second peaks were identified as a mixture of MP-2 and MP-3, whereas the third peak was MP-4 (Fig. 2A). It was revealed that MP-4 is monomeric in nature, based on the comparison of reducing, non-reducing SDS-PAGE and native PAGE (Fig. 2B). It was therefore unlikely that MP-4 belongs to the WSCP class because these proteins are normally tetrameric (37). Using mass spectrometry, the molecular mass of MP-4 was found to be 20.9 kDa (Fig. 2C).
FIGURE 2.
Purification and characterization of MP-4. A, size exclusion chromatography profile of 60% ammonium sulfate fraction depicting the first two peaks, corresponding to MP-2 and MP-3. The third peak corresponds to MP-4. B (left), SDS-PAGE (12%) analysis shows the presence of a single band for MP-4 in reducing (lane 2) and non-reducing (lane 3) conditions. Lane 1, prestained molecular weight markers (Thermo Scientific). B (right), native PAGE (12%) shows a single band of STI (Sigma-Aldrich) in lane 1, which is used as a molecular weight marker (20.1 kDa), and lane 2 shows the band of MP-4. C, mass spectrometry profile of MP-4 with a deconvoluted single sharp peak of 20.884 kDa shown in the inset.
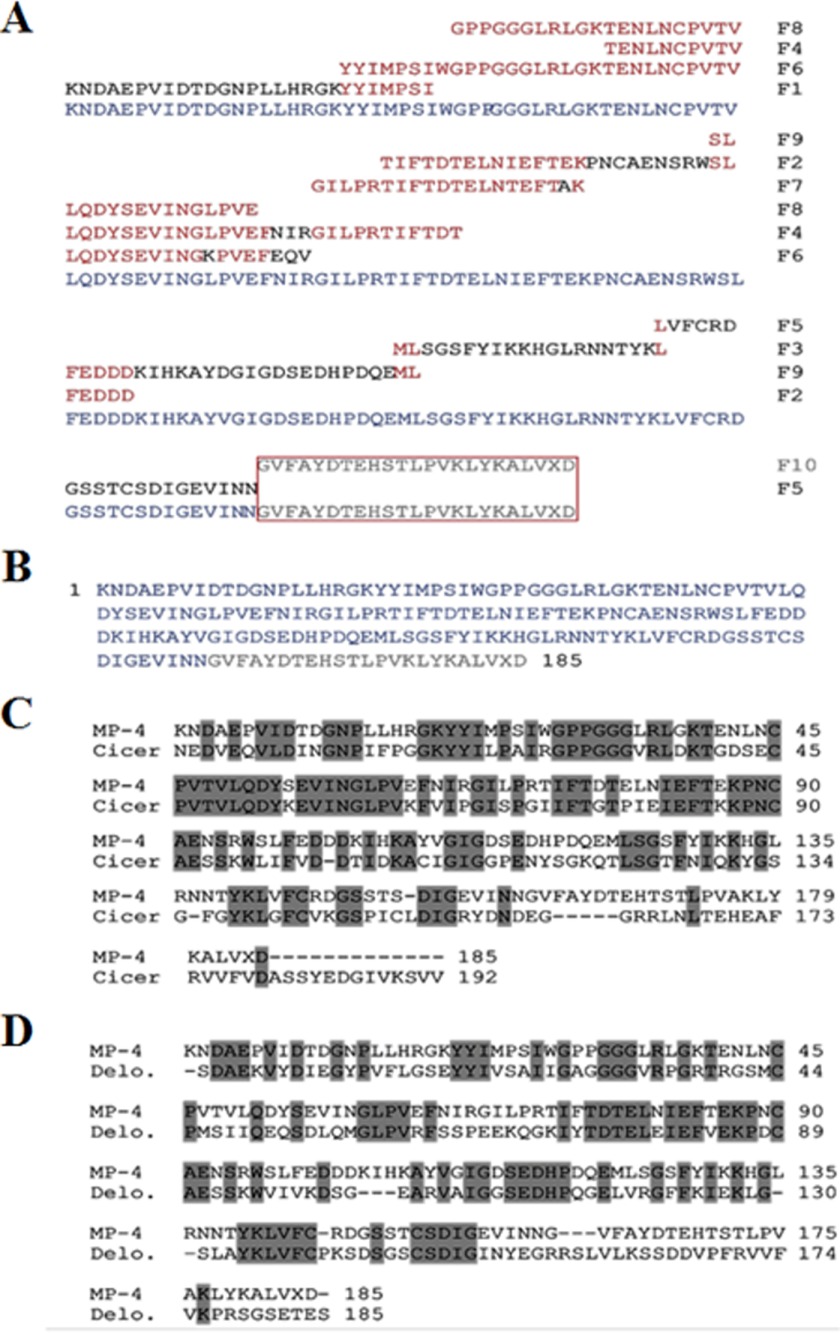
The internal peptide fragments of MP-4 were obtained by enzymatic and chemical digestion (Table 2) and were sequenced using N-terminal sequencing. All fragments were aligned manually to generate the 185-residue-long full-length protein sequence (Fig. 3, A and B). Sequence analysis showed that the closest homolog was a Kunitz-type trypsin inhibitor from C. arietinum that exhibited 57% sequence identity with MP-4 (Fig. 3C). The closest homolog for which structure was available is a Kunitz-type protease inhibitor from D. regia (Protein Data Bank code 1R8N) that showed 44% sequence identity (Fig. 3D). Overall, bioinformatics analysis of the MP-4 sequence suggested that it was a Kunitz-type protease inhibitor.
TABLE 2.
N-terminal sequencing of polypeptides obtained from enzymatic and chemical (CNBr) digestion of MP-4 by Edman degradation method
| Enzymes/chemical | Sequence | Fragment number |
|---|---|---|
| N-terminal | KNDAEPVIDTDGNPLLHRGKYYIMPSIWGPP | F1a |
| Trypsin | TIFTDTELNIEFTEKPNCAENSRWSLFEDDD | F2 |
| V8 protease | MLSGSFYIKKHGLRNNTYKL | F3 |
| Endoproteinase Lys-C | TENLNCPVTVLQDYSEVINGLPVEFNIRGILPRTIFTDT | F4 |
| LVFCRDGSSTCSDIGEVINN | F5 | |
| YYIMPSIWGPPGGGLRLGKTENLNCPVTVLQDYSEVINGKPVEFQV | F6 | |
| Endoproteinase Arg-C | GILPRTIFTDTELNTEFTAK | F7 |
| CNBr (chemical) | GPPGGGLRLGKTENLNCPVTVLQDYSEVINGLPVE | F8 |
| SLFEDDKIHKAYDGIGDSEDHPDQEML | F9 |
a Fragment obtained without digestion.
FIGURE 3.
Derivation of full-length sequence of MP-4 and sequence analysis. A, internal peptide fragments obtained from enzymatic and chemical digestion and their contig sequence alignment is displayed. The peptide fragment (F10) in the box was not clear in N-terminal sequencing, which was built on the basis of a homologous sequence present in the protein database (C-terminal 25 ambiguous residues highlighted in light gray). B, full-length amino acid sequence of MP-4 generated by fragment alignment. C, MP-4 sequence alignment with its closest ortholog from C. arietinum. This protein shows 57% sequence identity with MP-4. D, the closest MP-4 ortholog for which a structure is available is a Kunitz-type protease inhibitor from D. regia (Protein Data Bank code 1R8N). MP-4 shows 44% sequence identity to this protein. The identical residues are highlighted in dark gray.
Anti-snake Venom Activity of MP-4
Crude extract of seeds of M. pruriens showed an anti-venom property, and MP-4 is present in these seeds in large quantities. Hence, we hypothesized that MP-4 could be involved in the anti-snake venom property of M. pruriens seeds. The ability of MP-4 to directly neutralize the toxicity of snake venom from E. carinatus (saw-scaled viper) was evaluated. Because MP-4 is predicted to be a protease inhibitor, it is possible that MP-4 may inhibit the venom proteases and thus contribute to direct neutralization of snake venom.
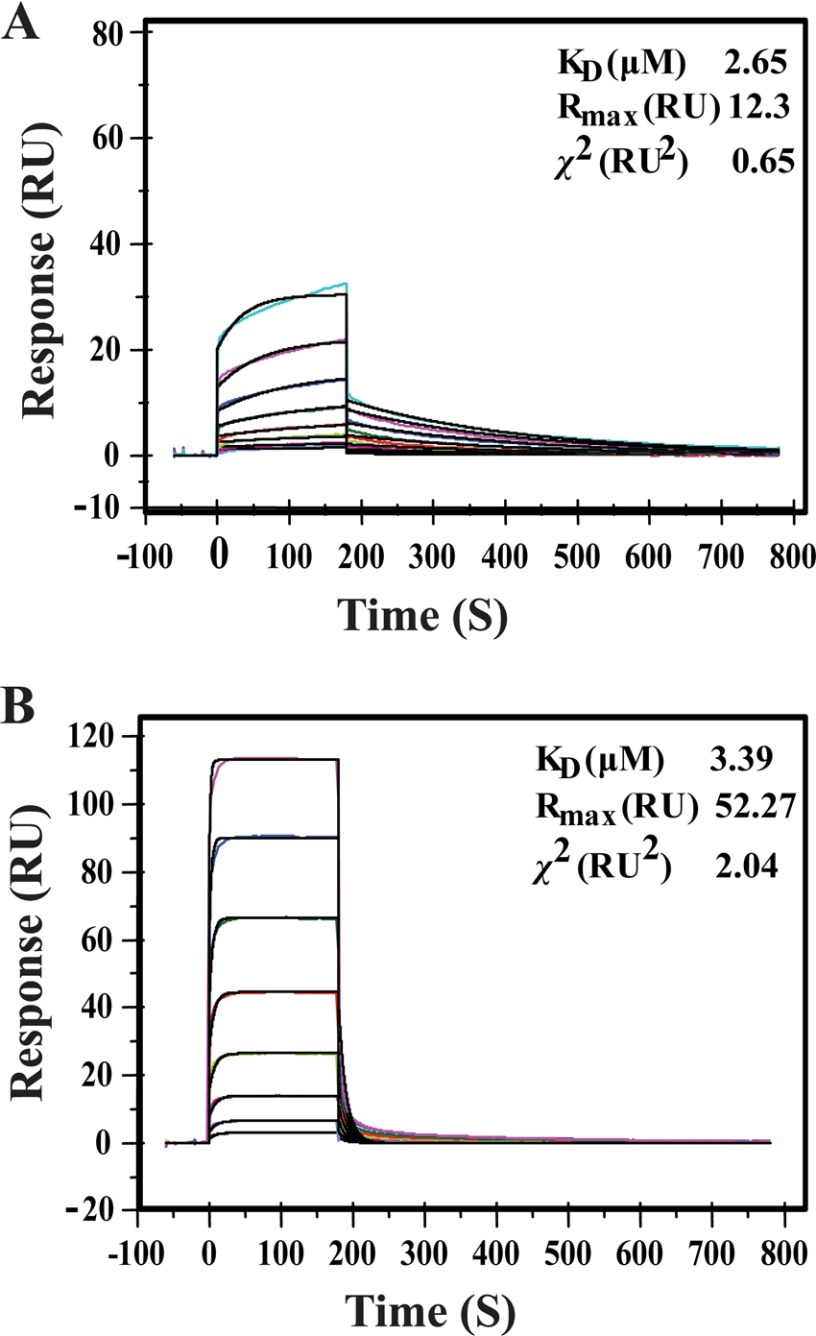
To test this hypothesis, initially, we optimized the LD50 of EcV on BALB/c mice. The MLD was found to be 2 mg/kg/individual, and this value is similar to one in an earlier report (Fig. 4A) (26). Subsequently, three groups of eight mice each were taken. The first group was injected with the MLD of venom intraperitoneally. MP-4 was mixed with EcV in a 1:1 ratio (w/w), and this mixture was incubated for 1 h at 4 °C. The second group of mice was treated with this mixture. Saline was administered intraperitoneally in the third group of mice. The first two groups, administered a minimal lethal dose of EcV and venom preincubated with MP-4, show an identical survival percentage of 16.7% (p < 0.001). This observation suggested that MP-4 does not interact with venom components and, therefore, may not contribute to neutralization of snake venom or utilize an alternate mechanism to do so (Fig. 4B).
FIGURE 4.
Optimization and Evaluation of anti-snake venom activity of MP-4. A, optimization of EcV LD50, which was found to be 2 mg/kg. B, three groups of mice were administrated with venom EcV, preincubated mixture EcV + MP-4, and saline, separately. The percentage survival in the first and second groups of mice were identical.
Crystal Structure of MP-4
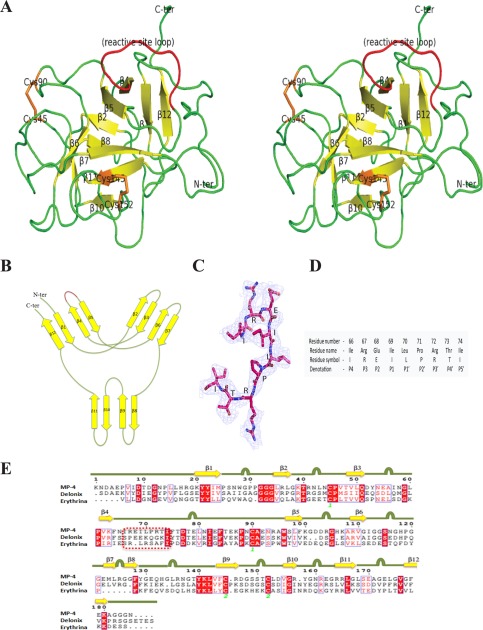
The overall structure of MP-4 adapts the β-trefoil fold. The structure consisted of 12 anti-parallel β-strands connected by long loops, resulting in a hairpin-like conformation (Fig. 5A). MP-4 structural features are coherent with homologous Kunitz-type protease inhibitors with 12 β-strands (β1–β12), six of which form a hairpin triplet (β2-β3, β4-β5, and β6-β7) capping a six-stranded antiparallel β-barrel (β8-β9, β10-β11, and β1-β12). The structure has pseudo-3-fold internal symmetry coinciding with the barrel axis (Fig. 5B). There are two internal disulfide bonds existing between Cys45–Cys90 and Cys145–Cys152 residues in the MP-4 structure.
FIGURE 5.
Overall structure of MP-4. A, stereo ribbon diagram of MP-4 structure. Yellow and green, β-strands and loops, respectively. The reactive site loop is highlighted in red. Orange, cysteine amino acid. B, a schematic of the secondary structure elements in MP-4. In this wiring diagram, the red region indicates the reactive site loop location. C, reactive site loop with 2Fo − Fc map at 1σ level. D, position, name, symbol, and denotation of the reactive site loop residues (P4–P5′) in a conventional representation. E, multiple sequence alignment of MP-4, Delonix, and Erythrina. Square dashed box indicates the reactive site loop in all of the three proteins. Yellow arrow for β-sheets and green line with bulge, loop regions in the MP-4 structure.
The MP-4 reactive site loop, which binds to the catalytic pocket of the target protease, was identified on the basis of sequence and structural alignment. The reactive site loop extends from residue 66 to 74 and is present between β4 and β5, protruding from one end of the structure (Fig. 5, A and C). The nine residues of the reactive site loop were denoted as P4–P5′ following the conventional nomenclature of Schechter and Berger (38) (Fig. 5D). To inhibit proteases, the reactive site loop is known to occupy the active site groove of the target enzyme. The central part of the loop is labeled as the P1–P1′ peptide and exposed to surrounding solvent molecules, known as the reactive site. The reactive site loop is flexible due to a lack of any secondary structure and disulfide bonds. The residue at the P1 position of the reactive site loop is the principal residue that decides the inhibitory efficiency of protease inhibitor (39, 40). This position is generally occupied by Arg or Lys in strong trypsin inhibitors and by Phe, Leu, Tyr, and Met in strong chymotrypsin inhibitors. However, in MP-4, this position is occupied by the branched chain aliphatic amino acid, Ile69.
The Dali server was used for the identification of MP-4 orthologs (34). The Kunitz-type protease inhibitor (DrTI) from D. regia (Protein Data Bank code 1R8N) and trypsin inhibitor (ETI) from Erythrina caffra (Protein Data Bank code 1TIE) were found to be close structural homologs of MP-4. The alignment of the MP-4 structure with that of DrTI and ETI gave a root mean square deviation of 1.16 Å (128 Cα pairs) and 1.09 Å (111 Cα pairs), respectively (supplemental Fig. S1A). In addition, the subunits of WSCP of L. verginicum (Protein Data Bank code 2DRE) were also found to be structural homologs of MP-4. The core region of MP-4 was found to be superimposed with these close homologous structures. The majority of the variations in structure and sequence are localized to the loop regions (Fig. 5E and supplemental Fig. S1B). The structure of the N-terminal 1–5 residues and last 25 residues of the C-terminal regions of MP-4 are different from that in the DrTI and ETI structures (supplemental Fig. S1A). The structure shows that, although the overall topology of MP-4 is similar to that of close structural homologs, there are substantial differences in the reactive site loop and the length and conformation of other loops.
Biochemical Evaluation of the Protease Inhibition Activity of MP-4
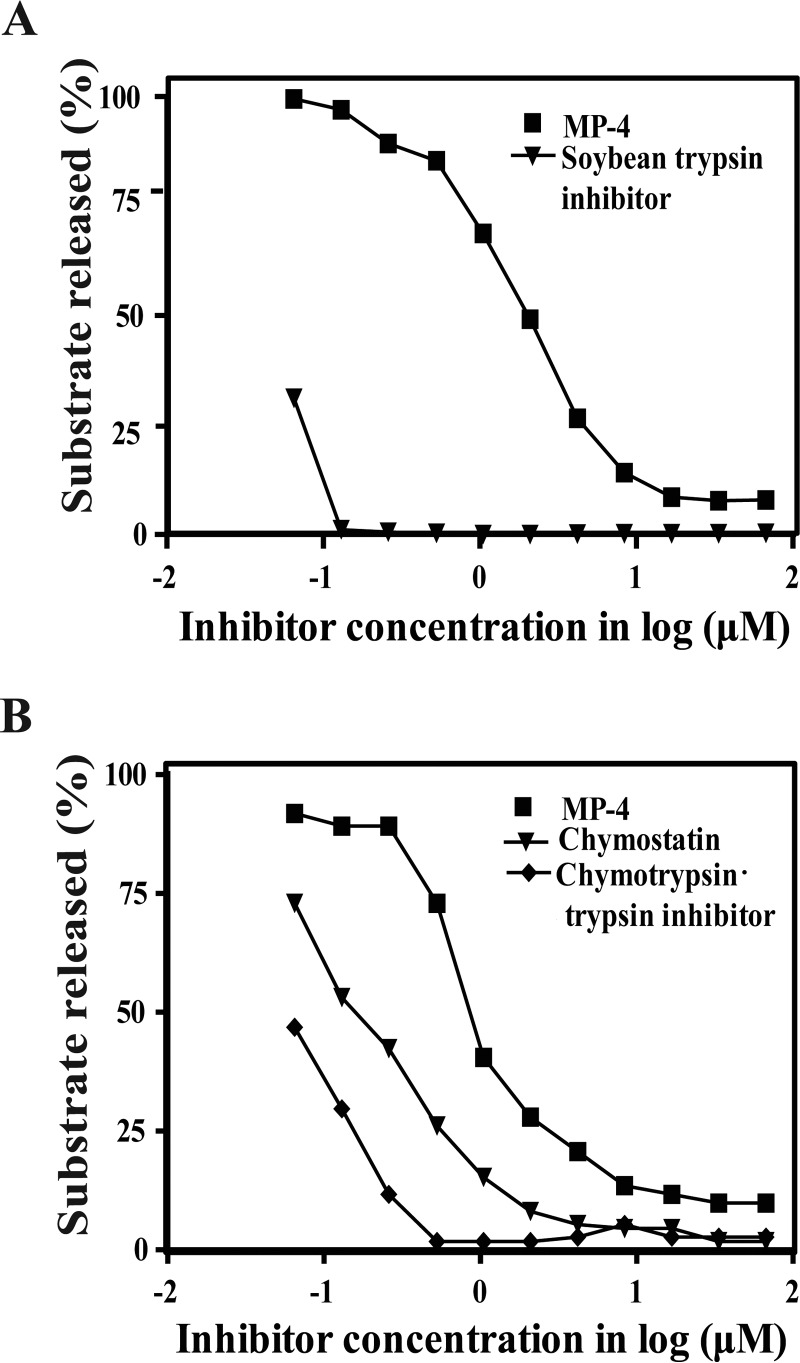
Although sequence analysis suggested that MP-4 may be a protease inhibitor, close inspection of the reactive site loop in the MP-4 structure suggested that the protease inhibition activity may not be optimal. The protease inhibition assays showed that the inhibitory efficiency (IC50) of MP-4 for trypsin is 1.96 μm, which is ∼100-fold less than the inhibitory efficiency of soybean trypsin inhibitor (IC50 0.016 μm) (Fig. 6A). When chymotrypsin was used as a substrate for MP-4, inhibitory efficiency (IC50) was found to be 1.03 μm, which shows 6-fold lower inhibitory activity than chymostatin (IC50 0.169 μm) and ∼20-fold lower inhibitory activity (IC50 0.048 μm) to chymotrypsin·trypsin inhibitor (Fig. 6B). For both of these proteases, MP-4 achieved inhibition comparable with known inhibitors only at very high concentrations (Table 3). These experiments clearly show that MP-4 is a weak inhibitor of serine proteases, such as trypsin and chymotrypsin.
FIGURE 6.
Protease inhibitory activity of MP-4. A, activity of MP-4 against trypsin. Lines marked with squares and triangles represent MP-4 and STI, respectively. B, activity of MP-4 against the chymotrypsin. Chymostatin and chymotrypsin·trypsin inhibitor are used as control. Lines marked with squares, triangles, and rhombi represent MP-4, chymostatin, and chymotrypsin·trypsin inhibitor, respectively.
TABLE 3.
Comparison of inhibitory efficiency of MP-4 with soybean trypsin inhibitor, chymostatin and chymotrypsin·trypsin
Top, comparison with STI. Bottom, comparison with chymostatin and chymotrypsin·trypsin for chymotrypsin. IC50 is given in μm.
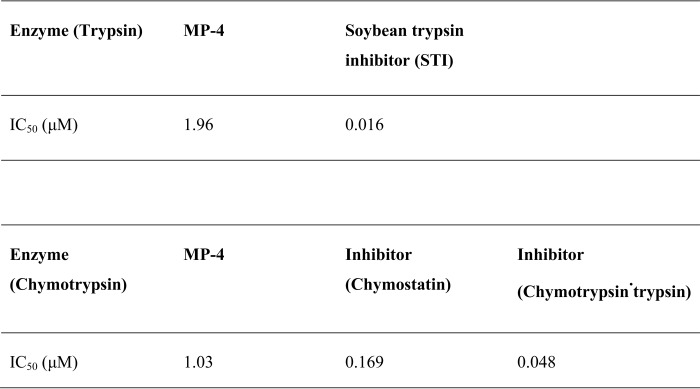
SPR was utilized to quantitate the binding affinity of MP-4 for trypsin and chymotrypsin. MP-4 shows a dissociation rate constant (KD) of 2.65 μm for trypsin, which is ∼50-fold less than that reported for soybean trypsin inhibitor (KD 48 nm) (Fig. 7A). Also, MP-4 has ∼5000-fold and ∼106-fold less affinity for bovine pancreatic trypsin inhibitor (KD 600 pm) and trypsin·squash trypsin inhibitor (KD 3.0 pm), respectively (41). Similarly, KD value for MP-4·chymotrypsin complex was calculated to be 3.4 μm, which is ∼32-fold less when compared with chymotrypsin·BPTI complex (KD 110 μm) (Fig. 7B) (41). The analyses of SPR data indicate that MP-4 binds to trypsin and chymotrypsin with weak affinity, and this may be the primary cause of poor inhibition of these serine proteases.
FIGURE 7.
Binding analysis of MP-4 with trypsin and chymotrypsin by SPR. A, SPR sensorgram for the binding of MP-4 (ligand) to trypsin (analyte) immobilized on the surface of a CM5 chip. The ligand was tested in the concentration range from 16 to 0.125 μm. B, binding of MP-4 (ligand) to chymotrypsin (analyte), where chymotrypsin was used in the range from 32 to 0.125 μm. Shown are the equilibrium rate constant, Rmax, and χ2 value for MP-4·trypsin and MP-4·chymotrypsin complex over the corresponding sensorgram. RU, response units.
We tried to identify the possible cause of weak inhibitory activity of MP-4 on the basis of a sequence/structural comparison with closely related proteins. Comparative analysis was done with DrTI (Protein Data Bank code 1R8N) and STI (Protein Data Bank code 1AVW). STI was taken for studies because the mechanistic information of its inhibitory efficiency is well known. Sequence alignment analysis of MP-4 with D. regia and STI indicates that there is an insertion of one residue, Arg70, in MP-4 and Gly66 in Delonix between the P2′ and P4′ positions (supplemental Fig. S2A). Comparison of the MP-4 structure with that of DrTI and STI shows that the Arg70 insertion results in substantial divergence of the reactive loop conformation (supplemental Fig. S2, B and C). The presence of aliphatic residue Ile at the P1 position and insertion of the residue in the reactive site loop explains the reason for weak inhibitory activity of MP-4 for trypsin and chymotrypsin.
On the basis of our data, it is clear that MP-4 is a poor protease inhibitor and does not directly neutralize the toxicity of snake venom. Therefore, it is possible that the contribution of MP-4 in snake venom neutralization by M. pruriens seeds, if any, occurs through an indirect mechanism.
Antibody-mediated Anti-snake Venom Activity of MP-4
We hypothesized that MP-4 might contribute to anti-snake venom activity of M. pruriens seeds through generation of cross-reactive antibodies. To confirm this, polyclonal antibodies were raised against purified MP-4 in mice, and ELISA was performed to assess cross-reactivity of these polyclonal antibodies with EcV (Fig. 8A). Similarly, antibodies were raised against EcV, and ELISA was performed to probe its cross-reactivity with MP-4 (Fig. 8B). Anti-MP-4 antibodies were found to cross-react with venom proteins, and anti-venom antibodies (anti-EcV) were found to cross-react with MP-4. These observations suggested that EcV and MP-4 may exhibit similar epitopes and that immunization with MP-4 may protect against toxicity of EcV.
FIGURE 8.
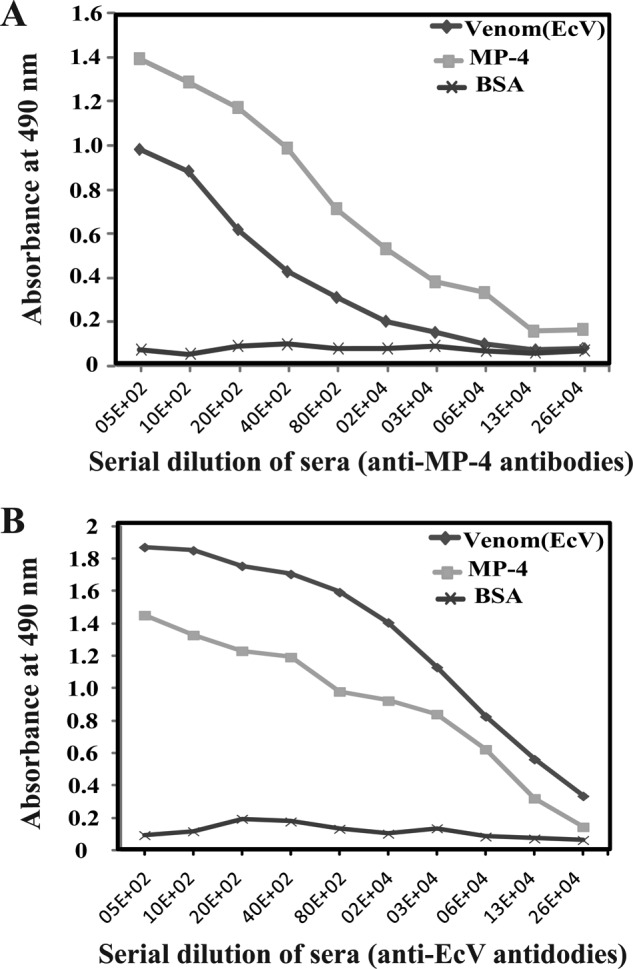
Evaluation of cross-reactivity of antibodies generated against MP-4 and EcV by ELISA. A, antibodies generated against MP-4 cross-react with whole EcV. B, antibodies generated against EcV proteins cross-react with MP-4 protein. BSA was used as a control.
To elucidate whether MP-4 acts against snake venom through a mechanism involving the immune system, we performed in vivo protection assays in BALB/c mice (Fig. 9). Four groups of mice were taken having eight mice each. Three groups of female BALB/c mice were immunized with MP-4, whereas a fourth group was maintained as a negative control that was not preimmunized with MP-4. When the first group of mice that were preimmunized with MP-4 were challenged with a subminimal lethal dose (1 mg/kg) of EcV, the survival rate was 41.7%. The second group of preimmunized mice was administered MLD of EcV (2 mg/kg), and the survival rate was 25%. The third group of preimmunized mice were administered saline and, as expected, did not show any fatalities. The fourth group of mice were not preimmunized with MP-4 protein, and when challenged with the minimum lethal dose of EcV (2 mg/kg), a 100% death rate was observed. In these experiments, p values are <0.001. The results of these experiments clearly showed that antibodies generated against MP-4 provide significant protection against EcV by cross-reacting with venom proteins.
FIGURE 9.
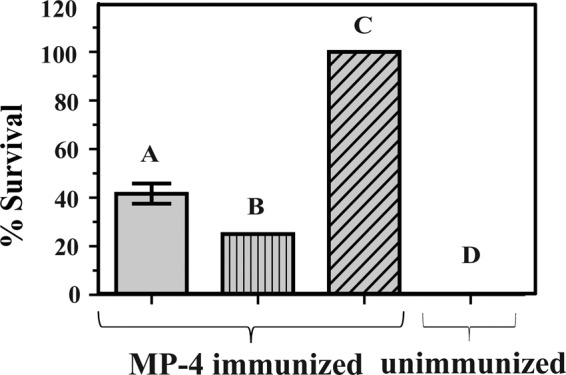
In vivo protection of MP-4 against EcV. Bar A, mice immunized with MP-4 and administered 1 mg/kg EcV; bar B, MP-4-immunized mice administered the MLD of EcV (2 mg/kg); bar C, MP-4-immunized mice administered saline; bar D, unimmunized mice that were subjected to the MLD of EcV (2 mg/kg). Each bar is a representation of the percentage survival of mice for experiments conducted three times with eight mice in each group. Error bar, S.D.
Conclusion
Although MP-4 showed significant sequence and structural homology to protease inhibitors, sequence divergence at key positions in the reactive site loop drastically attenuated its ability to effectively inhibit proteases. Consequently, MP-4 did not directly neutralize the toxicity of snake venom. However, this protein does contribute to protection against snake venom by M. pruriens seeds through an antibody-mediated indirect mechanism. From these observations, we can conclude that MP-4 can be added as an effective adjuvant in prophylactic preparations for protection against snake envenomation.
Author Contributions
D. M. S. and A. K. conceived and designed the experiments. A. K. and C. G. performed the experiments. A. K., D. T. N., and D. M. S. analyzed the data. A. K., D. T. N., and D. M. S. wrote the manuscript.
Acknowledgments
We thank H. S. Sarna and Sushma Nagpal for technical assistance.
This work was supported by funds provided by the Department of Biotechnology (Government of India) to the Regional Centre for Biotechnology (Faridabad, India) and the National Institute of Immunology (New Delhi, India). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 5DSS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MLD
- minimal lethal dose
- EcV
- E. carinatus snake venom
- STI
- soybean trypsin inhibitor
- WSCP
- water-soluble chlorophyll binding protein
- DrTI
- D. regia trypsin inhibitor
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- SPR
- surface plasmon resonance
- ETI
- E. caffra trypsin inhibitor
- contig
- group of overlapping protein fragments.
References
- 1. Warrell D. A. (2010) Snake bite. Lancet 375, 77–88 [DOI] [PubMed] [Google Scholar]
- 2. Kasturiratne A., Wickremasinghe A. R., de Silva N., Gunawardena N. K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D. G., and de Silva H. J. (2008) The global burden of snakebite: a literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 5, e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chippaux J. P. (1998) Snake-bites: appraisal of the global situation. Bull. World Health Organ. 76, 515–524 [PMC free article] [PubMed] [Google Scholar]
- 4. Hsu J. (2015) Defanging snakebites. Sci. Am. 313, 14–16 [Google Scholar]
- 5. Koh D. C., Armugam A., and Jeyaseelan K. (2006) Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 63, 3030–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson E. K., Kardong K. V., and Mackessy S. P. (1987) Electric shocks are ineffective in treatment of lethal effects of rattlesnake envenomation in mice. Toxicon 25, 1347–1349 [DOI] [PubMed] [Google Scholar]
- 7. Lake S. (2004) Pit Vipers: Friends or Foe? NC Herps, the newsletter of the North Carolina Herpetological Society, Vol. 27, No. 3 [Google Scholar]
- 8. Rita P., Animesh D. K., Aninda M., Benoy G. K., and Halder S. (2011) Snake bite, snake venom, anti-venom and herbal antidote: a review. Int. J. Res. Ayurveda Pharm. 2, 1060–1067 [Google Scholar]
- 9. Amin M. R., Mamun S. M., Rashid R., Rahman M., Ghose A., Sharmin S., Rahman M. R., and Faiz M. A. (2008) Anti-snake venom: use and adverse reaction in a snake bite study clinic in Bangladesh. J. Venom. Anim. Toxins Incl. Trop. Dis. 10.1590/S1678-91992008000400009 [DOI] [Google Scholar]
- 10. Reimers A. R., Weber M., and Müller U. R. (2000) Are anaphylactic reactions to snake bites immunoglobulin E-mediated? Clin. Exp. Allergy 30, 276–282 [DOI] [PubMed] [Google Scholar]
- 11. Dey A., and De J. N. (2012) Traditional use of plants against snakebite in Indian subcontinent: a review of the recent literature. Afr. J. Tradit. Complement Altern. Med. 9, 153–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makhija I. K., and Khamar D. (2010) Anti-snake venom properties of medicinal plants. Pharm. Lett. 2, 399–411 [Google Scholar]
- 13. Dhananjaya B. L., Zameer F., Girish K. S., and D'Souza C. J. (2011) Anti-venom potential of aqueous extract of stem bark of Mangifera indica L. against Daboia russellii (Russell's viper) venom. Indian J. Biochem. Biophys. 48, 175–183 [PubMed] [Google Scholar]
- 14. Tan N. H., Fung S. Y., Sim S. M., Marinello E., Guerranti R., and Aguiyi J. C. (2009) The protective effect of Mucuna pruriens seeds against snake venom poisoning. J. Ethnopharmacol. 123, 356–358 [DOI] [PubMed] [Google Scholar]
- 15. Fung S. Y., Tan N. H., Liew S. H., Sim S. M., and Aguiyi J. C. (2009) The protective effects of Mucuna pruriens seed extract against histopathological changes induced by Malayan cobra (Naja sputatrix) venom in rats. Trop. Biomed. 26, 80–84 [PubMed] [Google Scholar]
- 16. Aguiyi J. C., Igweh A. C., Egesie U. G., and Leoncini R. (1999) Studies on possible protection against snake venom using Mucuna pruriens protein immunization. Fitoterapia 10.1016/S0367-326X(98)00004-5 [DOI] [Google Scholar]
- 17. Kavitha C., and Thangamani C. (2014) Amazing bean “Mucuna pruriens”: a comprehensive review. J. Med. Plants Res. 10.5897/JMPR2013.5036 [DOI] [Google Scholar]
- 18. Scirè A., Tanfani F., Bertoli E., Furlani E., Nadozie H. O., Cerutti H., Cortelazzo A., Bini L., and Guerranti R. (2011) The belonging of gpMuc, a glycoprotein from Mucuna pruriens seeds, to the Kunitz-type trypsin inhibitor family explains its direct anti-snake venom activity. Phytomedicine 18, 887–895 [DOI] [PubMed] [Google Scholar]
- 19. Iauk L., Galati E. M., Kirjavainen S., Forestieri A. M., and Trovato A. (1993) Analgesic and antipyretic effects of Mucuna pruriens. Int. J. Pharmacogn. 31, 213–216 [Google Scholar]
- 20. Manyam B. V., Dhanasekaran M., and Hare T. A. (2004) Neuroprotective effects of the antiparkinson drug Mucuna pruriens. Phytother. Res. 18, 706–712 [DOI] [PubMed] [Google Scholar]
- 21. Adepoju G. K. A., and Odubena O. O. (2009) Effect of Mucuna pruriens on some haematological and biochemical parameters. J. Med. Plants Res. 3, 73–76 [Google Scholar]
- 22. Siddhuraju P., and Becker K. (2003) Studies on antioxidant activities of mucuna seed (Mucuna pruriens var. utilis) extracts and certain non-protein amino/imino acids through in vitro models. J. Sci. Food Agric. 83, 1517–1524 [Google Scholar]
- 23. Grover J. K., Yadav S., and Vats V. (2002) Medicinal plants of India with anti-diabetic potential. J. Ethnopharmacol. 81, 81–100 [DOI] [PubMed] [Google Scholar]
- 24. Salau A. O., and Odeleye O. M. (2007) Antibacterial activities of Mucuna pruriens on selected bacteria. Afr. J. Biotechn. 6, 2091–2092 [Google Scholar]
- 25. Guerranti R., Aguiyi J. C., Errico E., Pagani R., and Marinello E. (2001) Effects of Mucuna pruriens extract on activation of prothrombin by Echis carinatus venom. J. Ethnopharmacol. 75, 175–180 [DOI] [PubMed] [Google Scholar]
- 26. Guerranti R., Aguiyi J. C., Leoncini R., Pagani R., Cinci G., and Marinello E. (1999) Characterization of the factor responsible for the antisnake activity of Mucuna pruriens seeds. J. Prev. Med. Hyg. 40, 25–28 [Google Scholar]
- 27. Guerranti R., Aguiyi J. C., Ogueli I. G., Onorati G., Neri S., Rosati F., Del Buono F., Lampariello R., Pagani R., and Marinello E. (2004) Protection of Mucuna pruriens seeds against Echis carinatus venom is exerted through a multiform glycoprotein whose oligosaccharide chains are functional in this role. Biochem. Biophys. Res. Commun. 323, 484–490 [DOI] [PubMed] [Google Scholar]
- 28. Hope-Onyekwere N. S., Ogueli G. I., Cortelazzo A., Cerutti H., Cito A., Aguiyi J. C., and Guerranti R. (2012) Effects of Mucuna pruriens protease inhibitors on Echis carinatus venom. Phytother. Res. 26, 1913–1919 [DOI] [PubMed] [Google Scholar]
- 29. Guerranti R., Aguiyi J. C., Neri S., Leoncini R., Pagani R., and Marinello E. (2002) Proteins from Mucuna pruriens and enzymes from Echis carinatus venom: characterization and cross-reactions. J. Biol. Chem. 277, 17072–17078 [DOI] [PubMed] [Google Scholar]
- 30. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 31. Leslie A. G. W. (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 32. Crowther A. R., and Blow M. D. (1967) A method of positioning a known molecule in an unknown crystal structure. Acta Crystallogr. D 10.1107/S0365110X67003172 [DOI] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holm L., Kääriäinen S., Rosenström P., and Schenkel A. (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Erlanger B. F., Kokowsky N., and Cohen W. (1961) The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 95, 271–278 [DOI] [PubMed] [Google Scholar]
- 36. O'Shannessy D. J., Brigham-Burke M., Soneson K. K., Hensley P., and Brooks I. (1993) Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal. Biochem. 212, 457–468 [DOI] [PubMed] [Google Scholar]
- 37. Horigome D., Satoh H., Itoh N., Mitsunaga K., Oonishi I., Nakagawa A., and Uchida A. (2007) Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein. J. Biol. Chem. 282, 6525–6531 [DOI] [PubMed] [Google Scholar]
- 38. Schechter I., and Berger A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 39. Bode W., and Huber R. (2000) Structural basis of the endoproteinase-protein inhibitor interaction. Biochim. Biophys. Acta 1477, 241–252 [DOI] [PubMed] [Google Scholar]
- 40. Otlewski J., Jaskólski M., Buczek O., Cierpicki T., Czapińska H., Krowarsch D., Smalas A. O., Stachowiak D., Szpineta A., and Dadlez M. (2001) Structure-function relationship of serine protease-protein inhibitor interaction. Acta Biochim. Pol. 48, 419–428 [PubMed] [Google Scholar]
- 41. Kastritis P. L., Moal I. H., Hwang H., Weng Z., Bates P. A., Bonvin A. M., and Janin J. (2011) A structure-based benchmark for protein-protein binding affinity. Protein Sci. 20, 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]