Abstract
The nuclear DELLA proteins are highly conserved repressors of hormone gibberellin (GA) signaling in plants. In Arabidopsis thaliana, GA derepresses its signaling pathway by inducing proteolysis of the DELLA protein REPRESSOR OF ga1-3 (RGA). SLEEPY1 (SLY1) encodes an F-box–containing protein, and the loss-of-function sly1 mutant has a GA-insensitive dwarf phenotype and accumulates a high level of RGA. These findings suggested that SLY1 recruits RGA to the SCFSLY1 E3 ligase complex for ubiquitination and subsequent degradation by the 26S proteasome. In this report, we provide new insight into the molecular mechanism of how SLY1 interacts with the DELLA proteins for controlling GA response. By yeast two-hybrid and in vitro pull-down assays, we demonstrated that SLY1 interacts directly with RGA and GA INSENSITIVE (GAI, a closely related DELLA protein) via their C-terminal GRAS domain. The rga and gai null mutations additively suppressed the recessive sly1 mutant phenotype, further supporting the model that SCFSLY1 targets both RGA and GAI for degradation. The N-terminal DELLA domain of RGA previously was shown to be essential for GA-induced degradation. However, we found that this DELLA domain is not required for protein–protein interaction with SLY1 in yeast (Saccharomyces cerevisiae), suggesting that its role is in a GA-triggered conformational change of the DELLA proteins. We also identified a novel gain-of-function sly1-d mutation that increased GA signaling by reducing the levels of the DELLA protein in plants. This effect of sly1-d appears to be caused by an enhanced interaction between sly1-d and the DELLA proteins.
INTRODUCTION
The hormone gibberellin (GA) tightly regulates many growth and developmental processes throughout the life cycle of a plant. The important roles of GA are illustrated by the dramatic defects of GA biosynthetic and signaling mutants in germination, leaf expansion, stem elongation, apical dominance, floral development, and fertility (Davies, 1995). The DELLA proteins are highly conserved negative regulators of GA signaling in Arabidopsis thaliana and several crop plants, including barley (Hordeum vulgare), grape (Vitis vinifera), maize (Zea mays), rice (Oryza sativa), and wheat (Triticum aestivum) (Boss and Thomas, 2002; Olszewski et al., 2002). These DELLA proteins were named after a conserved amino acid motif near their N termini (Olszewski et al., 2002; Peng and Harberd, 2002). The DELLA proteins form a subfamily within a family of putative transcriptional regulators known as GRAS (for GA INSENSITIVE [GAI], REPRESSOR OF ga1-3 [RGA], and SCR) (Pysh et al., 1999). In addition to GA signaling, these plant-specific GRAS family proteins also regulate other developmental processes, such as radial patterning (Di Laurenzio et al., 1996; Helariutta et al., 2000), control of axillary and shoot meristems (Stuurman et al., 2002; Greb et al., 2003; Li et al., 2003), and light signaling (Bolle et al., 2000). In Arabidopsis, there are >30 GRAS proteins, all of which demonstrate high sequence similarity in their C-terminal GRAS domain (Arabidopsis Genome Initiative, 2000). The N termini of GRAS proteins are in general divergent and probably specify their diverse roles in different cellular pathways. The DELLA proteins, however, contain two highly conserved motifs (named DELLA and VHYNP) within their N-terminal DELLA domain (Silverstone et al., 1998; Peng et al., 1999; Itoh et al., 2002). Sequence analysis of the DELLA proteins suggested that they are likely transcriptional regulators. They contain polymeric Ser/Thr motifs (possible target sites of phosphorylation or glycosylation), Leu heptad repeats that may mediate protein–protein interactions, nuclear localization signals, and a putative Src homology 2 phosphotyrosine binding domain. In support of their function in transcriptional regulation, several DELLA proteins direct the green fluorescent protein (GFP) fusion into plant cell nuclei (reviewed in Olszewski et al., 2002). Furthermore, transient expression of a fusion protein consisting of both the Gal4 DNA binding domain and the rice DELLA protein (Slender Rice1 [SLR1]) activates transcription of the reporter gene that contains a Gal4 binding site in spinach (Spinacia oleracea) leaf cells (Ogawa et al., 2000).
In Arabidopsis, five genes encoding the DELLA proteins (GAI, RGA, RGL1, RGL2, and RGL3) are present. With the exception of RGL3, all of them have been shown to function as negative regulators of GA signaling (Olszewski et al., 2002; Peng and Harberd, 2002). In the GA-deficient ga1-3 mutant background, a combination of rga and gai null alleles results in a complete suppression of a subset of defects of ga1-3 to wild-type or GA-overdose phenotype (Dill and Sun, 2001; King et al., 2001). These include leaf expansion, flowering time, apical dominance, and stem elongation. Therefore, RGA and GAI interact synergistically to repress these GA-induced growth processes, but they do not play a major role in regulating germination and floral development. By contrast, RGL1 and RGL2 have been implicated to control seed germination in studies using gene silencing or Ds insertion mutant lines (Lee et al., 2002; Wen and Chang, 2002).
The uniqueness of the N-terminal DELLA domain hints that this region may specify the role of the DELLA proteins in GA response. The initial evidence came from the finding that the gain-of-function gai-1 mutant allele encodes a gai protein lacking 17 amino acids of the DELLA motif (Peng et al., 1997). This mutant has a GA-insensitive dwarf phenotype (Koornneef et al., 1985). Peng et al. (1997) hypothesized that this mutation in the N-terminal regulatory domain produces a constitutively active repressor that is resistant to inactivation by the GA signal. Subsequently, it was shown that many GA-insensitive semidominant dwarf mutants in other plant species also contain mutations in DELLA protein genes (Peng et al., 1999; Boss and Thomas, 2002; Chandler et al., 2002). All of these mutations result in amino acid substitutions, deletions, or truncations in the DELLA domain of the encoded protein. In fact, this type of mutation in an Rht gene (encoding a DELLA protein) is the cause for the semidwarf phenotype of the wheat cultivars that were essential in improving grain yield during the Green Revolution in the 1960s and 1970s (Peng et al., 1999).
A previous genetic screen designed to identify suppressors of gai-1 resulted in the isolation of recessive spindly (spy) mutants and the dominant gar2 mutant (Wilson and Somerville, 1995). SPY encodes an O-linked N-acetylglucosamine transferase, which negatively regulates GA signaling probably by modifying its target proteins in the pathway (Thornton et al., 1999). The GAR2 gene had not been cloned. The dominant nature of gar2 could be because of a loss-of-function mutation that causes haplo-insufficiency or a gain-of-function mutation that either increases GAR2 function, generates a new function, or interferes with GAR2 function.
GA appears to derepress its signaling pathway by inducing rapid degradation of some of the DELLA proteins, including RGA in Arabidopsis (Silverstone et al., 2001), SLR1 in rice (Itoh et al., 2002), and SLN1 in barley (Gubler et al., 2002). The levels of SLN1 and RGA are significantly reduced after 5 to 10 min of GA treatment, indicating that this phenomenon is an early event in the GA-signaling cascade (Gubler et al., 2002; S.G. Thomas and T.-p. Sun, unpublished results). In the DELLA gain-of-function mutants (e.g., Arabidopsis rga-Δ17, barley sln-d, and rice slr1), the mutant protein is resistant to GA-dependent degradation (Dill et al., 2001; Gubler et al., 2002; Itoh et al., 2002). These results suggest that the N-terminal DELLA and VHYNP motifs are essential in the GA-induced degradation of these DELLA proteins. However, GFP protein fusions with RGL1 or GAI were reported to remain stable after GA treatment (Fleck and Harberd, 2002; Wen and Chang, 2002), suggesting that GA may regulate other DELLA proteins via different mechanisms.
Many eukaryotic proteins destined for degradation by the 26S proteasome are polyubiquinated by an E3 ubiquitin ligase enzyme complex (Hershko and Ciechanover, 1998; Pickart, 2001). These ubiquitin-proteasome pathways play crucial regulatory roles in a wide variety of cellular processes in yeast (Saccharomyces cerevisiae), animals, and plants (Conaway et al., 2002; Hellmann and Estelle, 2002; Hare et al., 2003; Vierstra, 2003). One group of E3 ubiquitin ligases that are important in plants is the SCF complex (named after the three core components Skp1, cdc53/cullin, and F-box proteins) (Gagne et al., 2002). The F-box protein binds to Skp1 through a degenerate F-box motif, which consists of ∼40 amino acids in the N-terminal region (Schulman et al., 2000). In addition, the F-box protein recruits the targets via its C-terminal protein–protein interaction domain and provides the substrate specificity to the SCF E3 (Kipreos and Pagano, 2000). The Arabidopsis SLY1 and rice GID2 genes encode highly homologous F-box proteins, which are positive regulators of GA signaling because loss-of-function sly1 and gid2 mutants are GA-insensitive dwarfs (Steber et al., 1998; Sasaki et al., 2003). The recessive sly1-10 and gid2 mutants accumulate markedly elevated levels of RGA and SLR1 proteins, respectively (McGinnis et al., 2003; Sasaki et al., 2003). Furthermore, the rga and slr1 loss-of-function mutations suppress the sly1-10 and gid2 mutant phenotypes, respectively. These results suggest that SLY1 and GID2 may be components of SCF E3 ligase complexes that ubiquitinate RGA and SLR1, respectively, and target their destruction via the 26S proteasome. In support of this model, GID2 interacts with a SCF Skp1 component OsSkp2 in a yeast two-hybrid assay (Sasaki et al., 2003). In addition, in the presence of 26S proteasome inhibitor, wild-type plants accumulated ubiquitinated forms of SLR1 protein in response to GA treatment, whereas gid2 plants did not. Furthermore, another DELLA protein, SLN1 in barley, can be stabilized by proteasome inhibitor treatment (Fu et al., 2002).
Although previous studies suggested that SCFSLY1 and SCFGID2 target RGA and SLR1, respectively, for degradation in response to GA, there was no direct evidence for a physical interaction between SLY1-RGA and GID2-SLR1. In this article, we demonstrate that SLY1 interacts not only with RGA, but also with GAI through their GRAS domains. In contrast with a previous study (Fleck and Harberd, 2002), we show that GA induces degradation of the endogenous GAI protein and that the sly1-10 mutant accumulates a significantly higher amount of GAI than in the wild type. The rga and gai null alleles synergistically suppress several GA-mediated processes in sly1-10. These observations indicate a direct role for SLY1 in targeting GA-induced degradation of GAI as well as RGA. We further demonstrate that the dominant gar2 mutation is a sly1 gain-of-function allele (hence renamed sly1-d), which results in a single amino acid substitution in the protein. Similar to its effect on the gai-1 mutant, this novel sly1-d (gar2) allele partially suppresses the rga-Δ17 mutant phenotype. sly1-d interacts more strongly with RGA than SLY1 in yeast and causes reduced levels of RGA and rga-Δ17 proteins in plants. These observations are a case in which a gain-of-function mutation in an F-box protein gene perturbs plant development by causing a reduction in the levels of the F-box protein substrates.
RESULTS
Regulation of GA-Induced GAI Degradation by SLY1
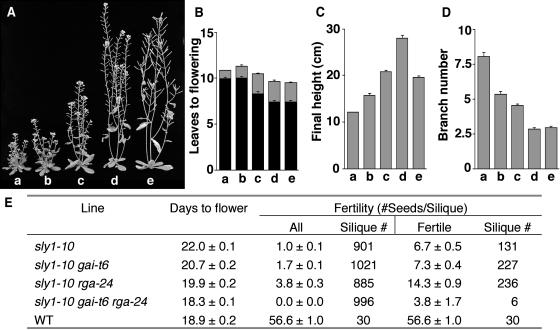
Our previous studies showed that SLY1 encodes an F-box–containing protein and that the loss-of-function sly1-10 mutant accumulates an elevated level of RGA protein (McGinnis et al., 2003). These results lead to the hypothesis that SLY1 is a component of the SCFSLY1 E3 ubiquitin ligase complex that targets RGA protein for degradation by the 26S proteasome. We further demonstrated previously that the rga-24 null mutation partially rescues the sly1-10 phenotype (McGinnis et al., 2003), suggesting that RGA is only one of the SLY1 targets (see Table 1 for molecular lesions of mutants). The other DELLA proteins, GAI and RGL, are candidates for SLY1 targets because these proteins are also repressors of GA signaling. To investigate whether GAI is a target of SLY1, we generated the double and triple homozygous mutants sly1-10 gai-t6 and sly1-10 rga-24 gai-t6. Similar to rga-24, the gai-t6 null allele alone also partially suppressed the sly1-10 phenotypic defects in stem height, flowering time, apical dominance, and fertility, although to a lesser extent than rga-24 (Figure 1, Table 1). Furthermore, the combination of rga-24 and gai-t6 alleles completely rescued most defects of sly1-10 to the wild type (Figure 1). One exception is that the triple mutant was less fertile than sly1-10 (Figure 1E), possibly because an elevated level of GA signaling occurs that mimics the effect of GA overdose in floral development. These results demonstrate that both rga-24 and gai-t6 are epistatic to sly1-10 and allow us to place RGA and GAI downstream of SLY1 in the GA signaling pathway. These observations support the model that both RGA and GAI are substrates of SLY1.
Table 1.
Gain-of-Function and Loss-of-Function Alleles of RGA, GAI, and SLY1
| Mutant Allele | Mutation in Protein | Loss-of-Function or Gain-of-Function |
|---|---|---|
| rga-1 | C-terminal 67-aa deletiona | Loss |
| rga-22 | Asn562 deletion | Loss |
| rga-24 | No protein (entire RGA open reading frame deleted) | Loss |
| rga-Δ17 | DELLA motif deletion | Gain |
| gai-t6 | C-terminal 350-aa deletion? (Ds insertion) | Loss |
| gai-1 | DELLA motif deletion | Gain |
| sly1-10 | C-terminal 8-aa truncation | Loss |
| sly1-d (gar2) | Glu138 to Lys | Gain |
aa, amino acid.
Figure 1.
Suppression of sly1-10 by rga-24 and gai-t6.
(A) Representative 42-d-old homozygous plants. a, sly1-10; b, gai-t6 sly1-10; c, rga-24 sly1-10; d, rga-24 gai-t6 sly1-10; e, the wild type. The rga-24 and gai-t6 single mutants were not shown because their phenotypes are similar to the wild type (Silverstone et al., 1997; Dill and Sun, 2001; King et al., 2001).
(B) to (E) Twenty-four plants per line were characterized except for days to flower in sly1-10 gai-t6 rga-24, for which 13 plants were studied. Shown are means ± se.
(B) Flowering time in leaf number. Black bars indicate rosette leaves; gray bars show cauline leaves.
(C) Final height.
(D) Branch number.
(E) Days to flower and fertility. Many siliques on sly1-10–containing plants were infertile. “All” indicates the average fertility of all siliques on the primary inflorescence; “fertile” indicates the average fertility of only those siliques that contained seeds.
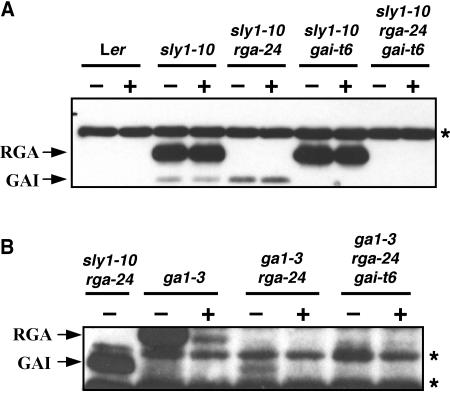
The genetic analyses presented above suggested that SLY1 plays a role in the GA-induced degradation of GAI. Although a previous study showed that levels of a GAI-GFP fusion were not reduced by GA treatment (Fleck and Harberd, 2002), we decided to use immunoblot analysis to examine whether endogenous GAI protein level is elevated in the sly1-10 mutant background and whether GA treatment induces a rapid reduction in GAI protein levels in the wild-type SLY1 background. The predicted molecular masses of RGA and GAI are 64 and 59 kD, respectively. By immunoblot analysis using polyclonal anti-RGA antibodies, we detected a protein in addition to RGA of ∼59 kD in both sly1-10 and sly1-10 rga-24, yet this second protein was not visible in the gai-t6 mutant backgrounds (Figure 2A). Similar to the RGA protein, this 59-kD protein (presumably GAI) is highly elevated in sly1-10, and the levels are unaffected after a 2-h GA treatment (Figure 2A). It should be noted that in rosette leaves of wild-type plants, the amounts of RGA and GAI proteins are below the detection level. These proteins are present at very low levels in rosette leaves and are not detectable even when using longer exposure times. Next, we explored whether GAI protein is responsive to GA-induced degradation in a SLY1 background. RGA protein levels are elevated in ga1-3 compared with wild-type plants (Silverstone et al., 2001). Although GAI protein was not clearly detectable in the ga1-3 mutant, a 59-kD protein (presumably GAI) was visible in the double mutant ga1-3 rga-24 (Figure 2B). This protein has the same electrophoretic mobility as GAI protein identified in sly1-10 rga-24 and was absent in the ga1-3 rga-24 gai-t6 mutant (Figure 2B). A 2-h GA treatment resulted in the disappearance of the GAI protein (Figure 2B). These results demonstrated that similar to RGA, GAI protein levels are elevated in the GA-deficient ga1-3 background and are rapidly reduced by GA treatment. Additionally, SLY1 is required for GA-induced degradation of GAI. Interestingly, when RGA protein is absent, GAI accumulates to a higher level as seen in ga1-3 rga-24 than in ga1-3 (Figure 2B), suggesting that the cell has a regulatory mechanism to sense and modulate different DELLA protein levels to achieve proper levels of GA signaling.
Figure 2.
SLY1 Regulates GA-Induced GAI Degradation.
The blots contain 20 μg of total proteins from rosette leaves of 24-d-old wild-type and homozygous mutant plants treated with 100 μM GA3 (+) or water (−) for 2 h. The protein blots were probed with anti-RGA antibodies from rabbit. Protein bands indicated with an asterisk represent nonspecific immunoreactive proteins. Blot (B) was exposed ∼10 times longer than blot (A).
Figure 2 also showed that the GAI signals were in general much lower than those of RGA in the tissues examined. Using recombinant His-tagged RGA and GAI proteins in immunoblot analysis, we found that the affinity of the polyclonal anti-RGA antibodies to RGA is approximately twofold of the affinity to GAI (data not shown). Therefore, GAI is present at a lower level than RGA, consistent with the previous finding that RGA plays a more predominant role than GAI in repressing GA signaling (Dill and Sun, 2001).
gar2, a Gain-of-Function sly1 Allele
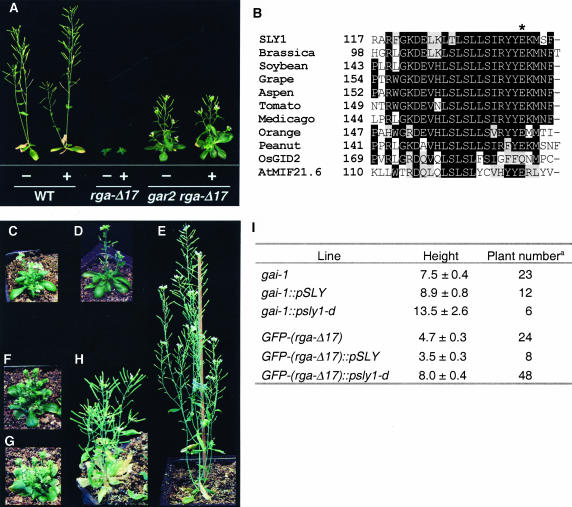
In addition to SLY1, another putative positive component of GA signaling in Arabidopsis is GAR2. Initial research led us to hypothesize that gar2 is a gain-of-function sly1 allele. First, the rga-Δ17 and gai-1 mutants are GA-insensitive dwarfs, presumably because the mutated rga and gai proteins (with DELLA-motif deletions) constitutively repress GA-mediated growth (Koornneef et al., 1985; Peng et al., 1997; Dill et al., 2001; Table 1). The dominant gar2 mutation partially suppresses the dwarf phenotype of gai-1 (Wilson and Somerville, 1995) and rga-Δ17 (Figure 3A; see also Supplemental Table 1S online). Second, the homozygous gar2 single mutant is resistant to the GA biosynthesis inhibitor paclobutrazol during seed germination (Peng et al., 1997), leaf expansion, and floral induction (data not shown). Therefore, the dominant gar2 mutant has elevated GA responses, which is opposite to the reduced GA-response phenotype of the loss-of-function sly1-10 mutant.
Figure 3.
gar2 Is a Gain-of-Function sly1 Allele That Partially Suppresses gai-1 and rga-Δ17 Dwarf Phenotypes.
(A) gar2 partially suppresses rga-Δ17 but does not restore GA responsiveness. Representative 36-d-old homozygous plants grown on soil are shown. Plants treated with GA (+) were sprayed with 100 μM GA3 weekly, beginning at 18 d after sowing.
(B) Sequence alignment of the LSL motif in SLY1 and other F-box proteins that contain both GGF and LSL motifs. Identical residues are shown in black boxes, and similar residues are in gray boxes. The asterisk indicates the residue mutated in sly1-d (gar2).
(C) to (H) Phenotype of 42-d-old control and T1 transgenic plants. Shown are gai-1 (C), gai-1∷pSLY1 number 4-1 (D), gai-1∷psly1-d number 6-1 (E), GFP-(rga-Δ17) (F), GFP-(rga-Δ17)∷pSLY1 number 4-1 (G), and GFP-(rga-Δ17)∷psly1-d number 5-1 (H).
(I) The average final height of parental control lines and transgenic lines containing pSLY1 or psly1-d transgene. a, The total number of T1 transgenic or control plants measured.
We performed DNA sequence analysis of the SLY1 locus in the gar2 mutant and found a G-to-A substitution in the gar2 mutant that results in the conversion of Glu-to-Lys in amino acid 138 near the C terminus of SLY1 protein (Figure 3B). This sly1 allele will be referred to as sly1-d. The SLY1 C-terminal domain consists of two motifs (GGF and LSL) that are highly conserved among closely related F-box proteins in plants, implying that these motifs play a functional role (McGinnis et al., 2003). The mutated residue in sly1-d is located in the LSL motif (Figure 3B).
To confirm that sly1-d confers the gar2 suppressor phenotype, a construct containing the SLY1 locus (pSLY1) or the sly1-d locus (psly1-d) was introduced into gai-1, GFP-(rga-Δ17), and wild-type plants. A GFP-(rga-Δ17) line that is partially rescued by gar2 (data not shown) was used for this experiment because rga-Δ17 is sterile, whereas this homozygous GFP-(rga-Δ17) line is fertile (Dill et al., 2001), which is necessary for generating transformants. Because gar2 is a dominant mutation, we examined the final heights of the T1 plants to determine whether the sly1-dtransgene rescues the dwarf phenotype of gai-1 and GFP-(rga-Δ17). The phenotype of the wild type transformed with either pSLY1 or psly1-d was indistinguishable from wild-type plants (data not shown). gai-1 and GFP-(rga-Δ17) transformed with pSLY1 were phenotypically similar to gai-1 and GFP-(rga-Δ17), respectively (Figures 3C, 3D, 3F, 3G, and 3I). However, psly1-d–containing gai-1 and GFP-(rga-Δ17) plants were, on average, much taller than their respective controls (Figures 3E, 3H, and 3I). These results confirmed that the gar2 suppressor phenotype is caused by the sly1-d mutation. Because SLY1 has been cloned previously (McGinnis et al., 2003), we propose changing the name of gar2 to sly1-d and will use this denotation for the remainder of the article.
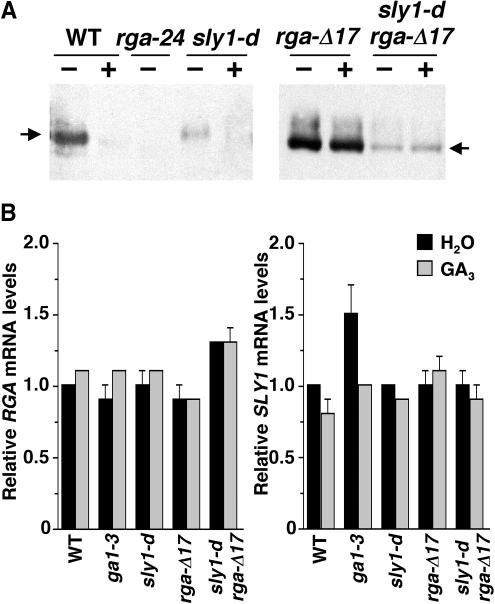
sly1-d Reduces the Levels of RGA and rga-Δ17 Proteins
We demonstrated previously that the RGA protein is degraded rapidly in response to the GA signal, whereas the rga-Δ17 protein is not responsive to GA and accumulates to a high level (Dill et al., 2001; Silverstone et al., 2001). Our previous studies also showed that the loss-of-function sly1-10 mutant accumulates an elevated level of RGA protein than the wild type (McGinnis et al., 2003), suggesting that SLY1 targets RGA protein for GA-induced degradation. Because sly1-d (gar2) suppresses rga-Δ17 morphologically (Figure 3A), we examined whether sly1-d affects RGA and rga-Δ17 protein accumulation by immunoblot analysis. Figure 4A shows that sly1-d dramatically reduced the amount of RGA protein (in the sly1-d mutant) and rga-Δ17 protein (in the sly1-d rga-Δ17 double mutant). However, the rga-Δ17 protein remained unresponsive to GA in the sly1-d mutant background. These results suggest that the lowered rga-Δ17 protein level causes the less severe phenotype of the sly1-d rga-Δ17 plants. The sly1-d mutation is unlikely to affect RGA expression at the transcript level because previous experiments showed that RGA mRNA levels are only slightly affected by GA treatment or in the loss-of-function sly1-10 mutant backgrounds (Silverstone et al., 1998; McGinnis et al., 2003). To verify this prediction, we examined RGA and rga-Δ17 transcript accumulation in the sly1-dand sly1-d rga-Δ17 mutants by quantitative RT-PCR using primers that specifically amplify both RGA and rga-Δ17 (Figure 4B). We found that RGA and rga-Δ17 mRNA levels in the sly1-d mutant background were similar to that in the wild type and did not change in response to GA. These results indicate that sly1-d affects RGA protein accumulation but is not involved in regulating RGA transcript levels.
Figure 4.
sly1-d Reduces RGA and rga-Δ17 Protein Levels but Not the RGA mRNA.
(A) and (B) Proteins or mRNA were extracted from homozygous lines as labeled, except that the sample for the rga-Δ17 line was extracted from a mix of hemizygous and homozygous plants.
(A) The blots contain 25 μg of total proteins from 8-d-old seedlings after 2 h of treatment with water (−) or 100 μM GA3 (+) as labeled. Affinity-purified rabbit anti-RGA antibodies and a peroxidase-conjugated goat anti-rabbit IgG were used to detect the RGA (64-kD) and rga-Δ17 (62-kD) proteins (indicated by arrows). The blot at right was exposed for a shorter time than the blot at left because rga-Δ17 accumulates to a higher level than RGA in plants.
(B) Relative RGA and SLY1 mRNA levels determined by quantitative RT-PCR. Total RNA was isolated from the wild type and various mutants after 100 μM GA3 or water treatment. The relative RGA and SLY1 mRNA levels were determined by running three quantitative RT-PCR reactions for each sample and normalized using the housekeeping gene GAPC. The value of water-treated wild type was arbitrarily set to 1.0. Bars = means ± se.
The sly1-d mutation may enhance the SLY1 function by increasing the sly1 transcript stability or sly1 protein stability or activity. We investigated whether the sly1-d mutation caused a change in sly1 transcript levels by quantitative RT-PCR analysis using primers that specifically amplified SLY1. Figure 4B shows that sly1-d mRNA accumulated to a similar level as SLY1 mRNA, ruling out the possibility that the suppressor effect of sly1-d is caused by increased sly1-d transcripts. Therefore, sly1-d is likely to affect SLY1 gene function at the protein level (see next section).
DELLA Proteins Directly Interact with SLY1 in the Yeast Two-Hybrid Assay
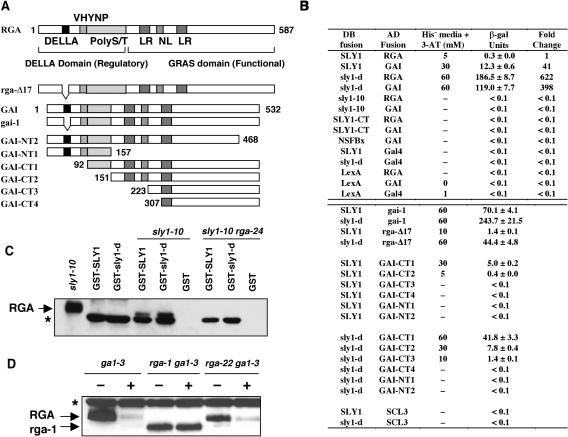
Our genetic and biochemical data strongly support a role for SLY1 in the GA-induced targeted degradation of the DELLA proteins RGA and GAI (McGinnis et al., 2003; Figures 1 and 2). From our work, and the study of the RGA and SLY1 orthologs in rice (SLR1 and GID2) (Sasaki et al., 2003), we predicted that GA promotes the direct interaction of RGA and GAI with SLY1, resulting in targeted degradation of these DELLA proteins. To test this model, we used the yeast two-hybrid system to determine whether SLY1 directly interacts with the DELLA proteins RGA and GAI. Previous studies using the yeast two-hybrid assay have demonstrated that DELLA proteins self-activate expression of reporter genes when they are expressed in yeast as DNA binding domain (DB) protein fusions (Itoh et al., 2002; S.G. Thomas and T.-p. Sun, unpublished results). Therefore, in this study, we only expressed the DELLA proteins in yeast as transactivation domain (AD) protein fusions.
Plasmids expressing LexA DB-SLY1 and Gal4 AD-RGA or GAI were cotransformed into the yeast strain L40 harboring the His3 and LacZ reporter genes. We found that yeast strains containing DB-SLY1 and AD-RGA constructs exhibited β-galactosidase (β-gal) activity 10-fold higher than the activity of the negative controls (DB-SLY1/Gal4 and LexA/AD-RGA, Figures 5A and 5B). This strain was also able to grow on His dropout (His−) plates containing up to 5 mM 3-aminotriazole (3-AT), as opposed to the negative controls, which were unable to grow in His− media even in the absence of 3-AT (Figure 5B). 3-AT is a competitive inhibitor of the His3 enzyme and served as an indicator of His3 expression level and, therefore, the strength of interactions between AD and DB fusions (Durfee et al., 1993). These results indicate that in yeast, SLY1 directly interacts with RGA. Similarly, we demonstrated that SLY1 also interacted with GAI (Figure 5B). Based on the levels of expression of the reporter genes in the L40 strains, the interaction between SLY1 and GAI was much stronger than that of SLY1 and RGA.
Figure 5.
Interaction of DELLA Proteins with SLY1 and sly1-d through the GRAS Domain.
(A) A schematic diagram showing the full-length RGA and GAI and the AD deletion constructs of GAI used in the yeast two-hybrid assay. The positions of conserved motifs (DELLA and VHYNP motif, poly Ser/Thr [polyS/T] sequence, nuclear localization signal [NL], and Leu heptad repeats [LR]) within the RGA and GAI full-length sequence are indicated. The numbers indicate the amino acid position at which the deletions start with regard to the full-length GAI sequence.
(B) DELLA proteins interact with SLY1 and sly1-d in yeast two-hybrid assays. Interactions of DB and AD fusion proteins in the L40 yeast cells were scored for the relative growth on His− plates containing 3-AT (0 to 60 mM) and β-gal activity (means ± se). A dash indicates no growth on His− plates at 0 mM 3-AT. The fold change indicates the relative β-gal activity with the activity of the DB-SLY1/AD-RGA L40 strain arbitrarily set as 1.0.
(C) Recombinant GST-SLY1, GST-(sly1-d), or GST was used in pull-down assays with lysates from sly1-10 and sly1-10 rga-24 leaves. The blot was probed with a rat anti-RGA antibody. The sly1-10 protein extract (2 μg) indicates the position of endogenous RGA on the blot. The asterisk indicates a nonspecific protein copurified from E. coli with GST-SLY and GST-(sly1-d), which is recognized by the RGA antibody.
(D) rga-1 protein is insensitive to GA-induced destabilization in the rga-1 ga1-3 mutant, whereas RGA and rga-22 proteins are degraded after GA treatment. Immunoblots contain 50 μg of total protein extracted from tissues of 8-d-old plants after a 30 min treatment with water (−) or 0.5 μM GA4 (+) as labeled. Blots were probed with a rabbit anti-RGA antibody. The asterisk represents a nonspecific immunoreactive protein.
To determine whether the interaction between the DELLA proteins and SLY1 is specific, we tested whether RGA and GAI could interact with a predicted nonspecific F-box protein, NSFBx (At5g04010). This control protein belongs to the same C2 group as SLY1 but has no obvious homology outside of the F-box motif (Gagne et al., 2002; Kuroda et al., 2002) and has not been shown to be involved in GA response. In yeast two-hybrid assays, we found that NSFBx (as a DB fusion) did not interact with RGA or GAI (Figure 5B).
Our current model predicts that the N-terminal F-box motif of SLY1 is required for binding the Skp1 component of the SCFSLY1, and the C terminus is responsible for binding the DELLA protein destined for degradation. The sly1-10 allele is predicted to encode a mutant sly1 protein lacking the last eight amino acids and containing an addition of 46 random amino acids at the C terminus (McGinnis et al., 2003). It is highly plausible that the elevated RGA accumulation and GA insensitivity in sly1-10 results from the inability of the mutant protein to interact with DELLA proteins. In support of this model, we found that thesly1-10 mutant protein did not interact with RGA or GAI in the yeast two-hybrid assays (Figure 5B). To determine whether the C terminus of SLY1 is sufficient for the interaction with GAI and RGA, the DB-(SLY1-CT) construct (consisting of SLY1 amino acid residues 73 to 151) was prepared. No interaction was observed between SLY1-CT and RGA or GAI in yeast (Figure 5B). Immunoblotting with LexA antibodies revealed similar levels of the various DB fusion proteins in the yeast strains shown in Figure 5B (data not shown). It is possible that additional N-terminal residues not present in SLY1-CT are necessary for the proper protein folding or the interaction with DELLA proteins. Our results support the hypothesis that SLY1 directly interacts with and targets RGA and GAI for degradation.
Enhanced Interaction Between sly1-d and DELLA Proteins
As indicated above, the sly1-d mutation results in both reduced levels of RGA and rga-Δ17 protein (Figure 4A). A possible explanation for this effect could be that the mutant sly1-d protein has a higher affinity for DELLA proteins. We would predict that the higher affinity would lead to a reduction in DELLA protein levels through more efficient ubiquitination by the SCFSLY ubiquitin E3 ligase (and subsequent degradation by the 26S proteasome). To explore this possibility, we compared the interaction of sly1-d and SLY1 with both RGA and GAI using the yeast two-hybrid assay. We found that the yeast strain expressing DB-(sly1-d) and AD-RGA demonstrated β-gal activity levels that were >600-fold higher than the corresponding strain expressing DB-SLY1 and AD-RGA, indicating a much stronger interaction between sly1-d and RGA (Figure 5B). The interaction between DB-(sly1-d) and AD-GAI was also stronger than that of DB-SLY1 and AD-GAI, although less dramatic with a 10-fold increase in β-gal reporter gene activity. Immunoblot analysis using LexA antibodies indicated that DB-SLY1 and DB-(sly1-d) accumulated to similar levels in yeast (data not shown). Therefore, the decreased accumulation of DELLA proteins in the sly1-d mutant is likely because sly1-d protein has a higher affinity for DELLA proteins than SLY1.
RGA and SLY1 Interaction in Pull-Down Assays
To provide additional evidence indicating a direct interaction between SLY1 and RGA, we performed in vitro pull-down assays. Both SLY1 and sly1-d were expressed in Escherichia coli as glutathione S-transferase (GST) fusion proteins and purified using glutathione-sepharose. The purified fusion proteins were incubated with a crude lysate prepared from sly1-10 and sly1-10 rga-24 rosette leaves, washed, and then immunoblotted with anti-RGA antibodies. RGA was pulled down from a sly1-10 lysate by both the GST-SLY1 and GST-(sly1-d) fusion proteins but not by GST alone (Figure 5C). The interaction between a GST-SLY1 fusion protein and endogenous RGA from a sly1-10 lysate provides strong support that RGA and SLY1 proteins physically interact.
GRAS Domain Is Necessary for the Interaction with SLY1 and sly1-d
The gai-1 and rga-Δ17 mutants have a GA-unresponsive dwarf phenotype (Koornneef et al., 1985; Dill et al., 2001). In the case of rga-Δ17, the phenotype is attributable to the rga-Δ17 mutant protein being resistant to GA-induced degradation (Dill et al., 2001). One possible mechanism for this GA insensitivity is that the rga-Δ17 mutant protein is unable to interact with SLY1. This does not appear to be the case because in yeast, both AD-(gai-1) and AD-(rga-Δ17) interact more strongly with DB-SLY1 than wild-type controls (Figures 5A and 5B). It was also demonstrated that the sly1-d mutation dramatically enhanced the interaction with both AD-(gai-1) and AD-(rga-Δ17) (Figure 5B). To determine which regions of the DELLA proteins are necessary for interacting with SLY1 and sly1-d, we made a series of AD-GAI N-terminal and C-terminal truncations (Figure 5A) and performed interaction tests with DB-SLY1 and sly1-d in the yeast two-hybrid assay. GAI was used for these studies because it demonstrates a much stronger interaction with SLY1 in yeast (Figure 5B). Immunoblot analyses indicated that expression of AD fusions was roughly equal (data not shown). A C-terminal truncation of only 64 amino acids (GAI-NT2) completely abolished the interaction with both SLY1 and sly1-d (Figures 5A and 5B). N-terminal truncations removing the regulatory domain did not completely abolish the interaction, although there was a significant reduction in the strength of the interaction as more of the N terminus was removed. Using both DB-SLY1 and DB-(sly1-d), the GAI GRAS domain (GAI-CT2) was sufficient for an interaction to occur. Next, we tested whether a GRAS family member that is not a DELLA protein (SCL3; Pysh et al., 1999) can interact with SLY1 or sly1-d. Figure 5B showed that no reporter activity was detected in cells expressing DB-SLY1 (or sly1-d) and AD-SCL3, suggesting that SLY1 specifically interacts with the GRAS domain of DELLA proteins. As was previously observed, the DB-(sly1-d) fusion displayed a stronger interaction with all of the AD-GAI N-terminal truncations compared with DB-SLY1. These results indicated that the GRAS domain, but not the DELLA domain, is necessary for the interaction with SLY1 in yeast.
The rga-1 Mutant Protein Is Resistant to GA-Mediated Degradation
The yeast two-hybrid assays indicated that RGA and GAI proteins interact with SLY1 via their GRAS domain (Figures 5A and 5B). In addition, deletion of the last 64 amino acids of GRAS domain in GAI (GAI-NT2) completely abolished its interaction with SLY1 or sly1-d. Therefore, we would predict that in the plant, a similar C-terminal truncation in the DELLA proteins will make them unable to interact with SLY1 and resistant to GA-induced degradation. The loss-of-function rga-1 allele contains a premature stop codon, which is predicted to encode a mutant rga protein that lacks 67 amino acids from the C terminus and has a molecular mass of 57 kD (Silverstone et al., 1998; Table 1). Immunoblot analysis shows that a protein of ∼57 kD was recognized by the anti-RGA antibodies in protein extracts prepared from the homozygous rga-1 ga1-3 mutant (Figure 5D). Unlike RGA, this 57-kD protein (presumably rga-1) was insensitive to GA treatment (Figure 5D). This GA-resistant property of rga-1 is unlikely to be an indirect effect of inactivation of the protein because another loss-of-function rga mutant protein (rga-22) with a single amino acid (Asn562) deletion in the GRAS domain still underwent GA-induced degradation (Figure 5D, Table 1). These in vivo observations further support the hypothesis that the C-terminal GRAS domain of RGA plays an important role in the GA-mediated degradation of RGA.
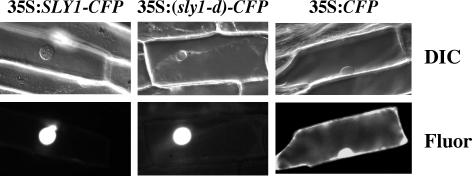
Nuclear-Localization of SLY1-Cyan Fluorescent Protein in Onion Cells
The GFP protein fusions with RGA and GAI are localized to the plant cell nuclei in transgenic Arabidopsis (Silverstone et al., 2001; Fleck and Harberd, 2002). Although SLY1 does not contain a predicted nuclear localization signal by the PSORT II program (Horton and Nakai, 1997), we demonstrated that transiently expressed SLY1-cyan fluorescent protein (CFP) and (sly1-d)-CFP fusion proteins were localized exclusively to the nuclei of onion (Allium cepa) epidermal cells (Figure 6). By contrast, the CFP protein was detected in both the nucleus and cytoplasm because of its small size (Haseloff et al., 1997). Nuclear localization of SLY1 supports its role in targeting the DELLA proteins for degradation in the plant cell nucleus.
Figure 6.
Nuclear Localization of the SLY1- and (sly1-d)-CFP Fusion Proteins in Onion Cells.
Individual cells were imaged by differential interference contrast (DIC) and epifluorescence (fluor) microscopy. The CFP protein (control) is present in both the cytoplasm and the nucleus because of its small size.
DISCUSSION
Our data provide multiple lines of evidence to support the model that SLY1 is a nuclear-localized F-box component of the SCFSLY1 E3 ubiquitin ligase, which regulates GA responses by binding directly and triggering GA-induced degradation of the DELLA proteins RGA and GAI. Moreover, the interaction between GAI and SLY1 requires the GRAS domain of GAI, as opposed to the regulatory N-terminal DELLA domain. In addition, we demonstrated that the dominant gar2 mutation is a gain-of-function allele of SLY1 (named sly1-d), which caused a much stronger interaction with RGA and GAI in the yeast two-hybrid system and suppressed the dwarf phenotype of rga-Δ17 by reducing the levels of RGA and rga-Δ17 proteins in plants.
SLY1 Targets GAI for Degradation in Response to GA
We previously speculated that in addition to its effect on RGA, SLY1 might play a role in targeting the entire DELLA family for degradation in response to GA (McGinnis et al., 2003). In this article, we provide support for this model by showing that SLY1 plays a role in the GA-mediated degradation of GAI. A previous study by Fleck and Harberd (2002) suggested that GAI was not subject to GA-mediated degradation. Their conclusions were based on GFP fluorescence of GAI-GFP reporter proteins rather than immunological detection of the native GAI protein, which may account for the differences in results. It remains to be determined whether SLY1 also plays a role in GA-mediated degradation of RGL1, RGL2, and RGL3.
The Role of SCFSLY1/GID2 in the GA Signaling Cascade
The Arabidopsis genome contains >700 genes encoding proteins with F-box domains (Gagne et al., 2002; Kuroda et al., 2002; Risseeuw et al., 2003), suggesting that targeted degradation mediated by SCF E3s regulates many aspects of plant growth and development. In support of this conclusion, mutations in F-box protein encoding genes TIR1, COI1, EBF1, EBF2, UFO, and EID1 have been demonstrated to affect auxin signaling, jasmonic acid signaling, ethylene signaling, floral development, and light signaling, respectively (Xie et al., 1998; Gray et al., 1999; Samach et al., 1999; Dieterle et al., 2001; Guo and Ecker, 2003; Potuschak et al., 2003). TIR1, COI1, EBF1, EBF2, and UFO have been shown to exist as components of SCF E3 complexes (Gray et al., 2001; Xu et al., 2002; Guo and Ecker, 2003; Potuschak et al., 2003; Wang et al., 2003). In the yeast two-hybrid system, many of the uncharacterized F-box proteins interact with Arabidopsis Skp1 homologs, supporting the idea that they are also components of SCF E3 complexes (Gagne et al., 2002; Kuroda et al., 2002; Risseeuw et al., 2003). The Arabidopsis F-box protein SLY1 and its rice ortholog GID2 are positive regulators of the GA response (Steber et al., 1998; McGinnis et al., 2003; Sasaki et al., 2003). Several lines of evidence support that the SCFSLY1/GID2 E3 complex modulates the levels of DELLA proteins, repressors of the GA signaling pathway. In the loss-of-function sly1 and gid2 mutants, DELLA proteins are elevated and unresponsive to GA treatment. In the yeast two-hybrid system, GID2 interacts with the rice Skp1-like protein, OsSkp2 (Sasaki et al., 2003), and there is a direct interaction between SLY1 and the DELLA proteins (RGA and GAI) (Figure 5B). Further in vivo studies will be necessary to demonstrate the presence and predicted roles of putative SCFSLY1/GID2 complexes in GA signaling. Recent studies showed that auxin regulates Arabidopsis root growth at least in part by enhancing the GA-mediated degradation of RGA (Fu and Harberd, 2003). This raises the intriguing possibility that the interaction between auxin and GA signaling pathways could be through the regulation of SCF E3 ubiquitin ligase activity.
The Domains of the DELLA Proteins Involved in Their Interaction with SLY1
The C-terminal GRAS domain present in all GRAS family members is believed to be a functional domain, probably involved in transcriptional regulation (Pysh et al., 1999; Olszewski et al., 2002). In support of this hypothesis, most of the loss-of-function rga mutations are located within the GRAS domain (Silverstone et al., 1998; A.L. Silverstone and T.-p. Sun, unpublished results). The interaction between SLY1 and GAI in yeast allowed us to map the interaction domain to the GRAS domain of GAI. In Arabidopsis, we further demonstrated that a predicted 67–amino acid C-terminal truncation in rga-1 prevents GA-mediated degradation (Figure 5D). Studies of RGA orthologs in barley (SLN1) and rice (SLR1) also showed similar results. The sln1c mutant protein, lacking 18 amino acids from the C terminus, and a slr1(ΔC-Ter)-GFP fusion protein that is missing most of the GRAS domain are resistant to GA-dependent degradation (Gubler et al., 2002; Itoh et al., 2002). Therefore, the GRAS domain is essential for F-box protein–targeted degradation of the DELLA protein.
In yeast, the N-terminal DELLA domain of GAI and RGA appears to be dispensable in their interactions with SLY1, although we cannot rule out the possibility that in planta, the N-terminal DELLA domain plays a role in the interaction with SLY1. Our finding is surprising based on previous data demonstrating that N-terminal deletions in DELLA proteins produce constitutively active repressors that are not degraded in response to GA (Dill et al., 2001; Itoh et al., 2002). If the N-terminal regulatory domain is not necessary for recognition by the degradation machinery, what is its role? The simplest model predicts that the N terminus of the DELLA proteins is necessary to perceive the GA signal, which in turn triggers a conformational change in the protein and allows recognition by the SCFSLY1 E3 complex.
Studies of SCF-mediated signaling pathways in yeast and mammalian cells have identified phosphorylation as the predominant posttranslational modification of the substrate that promotes its interaction with the SCF E3 complex (Deschaies, 1999). There is some evidence to suggest that phosphorylation of DELLA proteins might target their degradation in response to GA. In rice, GA treatment promotes the accumulation of a phosphorylated form of SLR1 in the gid2 mutant (Sasaki et al., 2003). The phosphorylated SLR1 has a slower mobility than the unphosphorylated form on the SDS-PAGE gel. In Arabidopsis, we have been unable to detect the presence of a phosphorylated form of RGA, even in the sly1 mutants using the standard SDS-PAGE gel conditions (McGinnis et al., 2003; Figure 2A). Two-dimensional PAGE may be needed to separate the phosphorylated and unphosphorylated forms of RGA.
Enhanced Interaction between sly1-d and the DELLA Proteins
We demonstrated that GA response can be perturbed by the gain-of-function sly1-d allele that reduces DELLA protein levels. The sly1-d mutation (Glu to Lys) is located within the conserved LSL motif of the protein. This Glu residue is absolutely conserved in several putative SLY1 homologs, although it is not conserved in GID2 (Figure 3B). The C terminus of F-box proteins commonly contains protein–protein interaction domains that mediate interaction and confer specificity to the substrate (Deschaies, 1999). Although SLY1 does not contain a well-characterized C-terminal protein–protein interaction domain, we predict that the GGF and LSL motifs perform this role. This is supported by our finding that the interaction with DELLA proteins in yeast is enhanced by the sly1-d mutation and abolished by the sly1-10 mutation.
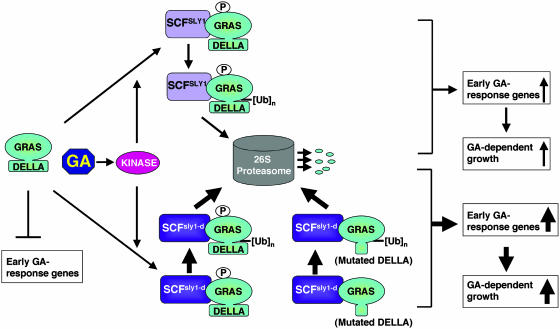
Updated Model of GA Signaling in Plants
Based on previous studies and this report, we propose a model of GA signaling in plants (Figure 7). DELLA proteins repress GA response genes in a quantitative fashion. GA modulation of plant growth and development is achieved through regulation of DELLA protein levels by rapidly inducing their degradation. We propose that GA activates a signaling cascade, including a protein kinase. The kinase phosphorylates DELLA proteins, promoting their direct interaction with the SLY1 subunit of a SCFSLY1 E3 ubiquitin ligase through the GRAS domain. The DELLA proteins are then polyubiquitinated by the SCFSLY1 E3 and subsequently recognized and degraded by the 26S proteasome. We also propose that the DELLA domain is essential for GA-induced phosphorylation and subsequent conformational change of DELLA proteins. The gain-of-function sly1-d mutation causes a reduction in the levels of wild-type and GA-resistant mutant DELLA proteins by an increased affinity between the sly1-d F-box protein and its substrates (DELLA proteins), leading to increased ubiquitination (by the SCFsly1-d compared with SCFSLY1) and subsequent degradation by the 26S proteasome.
Figure 7.
An Updated Model of GA Signaling in Plants.
GA induced phosphorylation (P) of DELLA proteins via an unidentified kinase and the SCFSLY1 complex interacts with the GRAS domain of DELLA proteins and targets their polyubiquitination ([Ub]n) and degradation via the ubiquitin-26S proteasome pathway. The novel sly1-d mutation results in an elevated affinity of SCFsly1-d with both wild-type and GA-resistant mutant DELLA proteins.
Our work provides further insight into the mechanism of targeted degradation of DELLA proteins in Arabidopsis. However, there are still many questions regarding the GA-signaling cascade. Further in vivo studies of the SCFSLY1/GID2 complex and its interaction with DELLA proteins should help to answer some of these questions. In particular, these studies should help in the identification of GA-induced posttranslational modifications of DELLA proteins that promote their interaction with SCF and also the identity of the enzyme(s) that catalyze the modifications. Elucidating the mechanism of GA-induced proteolysis of the DELLA proteins will shed light on our understanding of the SCF E3 ligase–targeted protein degradation in modulating growth and development in plants and animals.
METHODS
Isolation of Mutant Plant Lines
In this report, all of the Arabidopsis thaliana plants are in the Landsberg erecta (Ler) ecotype background, and Ler is the wild-type control. The homozygous gar2 mutant was isolated from crosses between gar2 gai-1 angustifolia (an) (Ohio State Stock Center; Wilson and Somerville, 1995) and the wild type. The transgenic line containing homozygous rga-Δ17 transgene did not bolt and was sterile, whereas the hemizygous rga-Δ17 plants are semidwarf and partially fertile (Dill et al., 2001). The double homozygous gar2 rga-Δ17 mutant was generated by crosses between hemizygous transgenic rga-Δ17 plants (Dill et al., 2001) and gar2 gai-1 an. gai-1 contains a 51-bp deletion relative to GAI. Primers that span this deletion were used to identify GAI versus gai-1 alleles by PCR (see Supplemental Table 2S online). The an mutation results in small, narrow, thick leaves (Rédei, 1962). AN homozygotes were identified by finding plants whose progeny were all wild type in leaf shape. The rga-Δ17 transgene is linked to the kanamycin resistance gene. The rga-Δ17 homozygotes were identified by finding plants with progeny that were all resistant to kanamycin. gar2 was originally identified phenotypically and subsequently verified by PCR using derived cleaved-amplified polymorphic sequence markers (Neff et al., 1998; see Supplemental Table 2S online) after we discovered that gar2 is sly1-d.
The homozygous sly1-10 rga-24 double mutant was isolated previously (McGinnis et al., 2003). The sly1-10 rga-24 gai-t6 and sly1-10 gai-t6 homozygous mutant lines were isolated from a cross between sly1-10 and rga-24 gai-t6 tt1-1 (Dill and Sun, 2001). Genotyping of the rga-24,gai-t6, and sly1-10 alleles was performed as described previously (Dill and Sun, 2001; McGinnis et al., 2003) except that new improved primers were designed for rga-24 and RGA (see Supplemental Table 2S online).
Identification of the gar2 Mutation
A 1.3-kb DNA fragment that spans the SLY1 locus was amplified by PCR using primers SLY1-5 and SLY1-6, and genomic DNA isolated from the wild type (Ler) and the gar2 mutant. DNA sequence analysis using primer SLY1-7 revealed a G-to-A substitution at nucleotide 412 from the ATG start codon of SLY1 in the gar2 mutant DNA.
Accession Numbers of SLY1 and Other Highly Similar F-Box Protein Genes
Homology searches for protein sequences that are similar to SLY1 were performed in the GenBank database using the tBLASTn program. Accession numbers are as follows: AtSLY1, NM_118554; Brassica napus, CD829466; soybean (Glycine max), BI785351; grape (V. vinifera), CB971820; aspen (Populus tremuloides), BU888340; tomato (Lycopersicon esculentum), BG643332; Medicago truncatula, BG452802; orange (Citrus sinensis), CF836247; peanut (Arachis hypogaea), CD038695; OsGID2, AB100246; AtMIF21.6, AB023039.
Plasmid Constructs
Sequences of all primers used in cloning and sequencing are listed in Supplemental Table 2S online. The PCR-amplified regions in all constructs were analyzed by DNA sequence analysis to ensure that no mutations had been introduced.
Two plasmids (pSLY1-300 and pSLY1-307) containing the SLY1 or sly1-d genomic DNA were generated for plant transformation. The SLY1 locus was amplified from wild-type genomic DNA using primers SLY1-8 and SLY1-9 and cut with BamHI. The resulting 3.5-kb DNA fragment was ligated into the binary vector pDHB321.1 (a gift from David Bouchez, Institut National de la Recherche Agronomique, Versailles, France) to create pSLY1-300. The BamHI fragment was also ligated into pUC18 (Gibco BRL, Carlsbad, CA) and the resulting plasmid named pSLY1-301. The 753-bp ClaI DNA fragment in pSLY1-301 was replaced with the corresponding DNA fragment amplified from gar2 genomic DNA, which contains the sly1-d mutation, creating pSLY1-306. The 3.5-kb BamHI fragment (from pSLY1-306) that contains the sly1-d locus was ligated into the binary vector pDHB321.1, generating plasmid pSLY1-307.
For the yeast two-hybrid assay, bait and prey protein fusions were expressed as LexA DB and Gal4 AD fusions using the yeast plasmid expression vectors pLexA-NLS (Vojtek et al., 1993) and pACTII (Li et al., 1994), respectively. The DB plasmid expression constructs were prepared by designing PCR primers incorporating EcoRI and BamHI restriction sites in the correct reading frame and then PCR amplifying the coding regions of SLY1, gar2, SLY-CT1 (residues 73 to 151), sly1-10 (encodes the first 143 residues of SLY1 followed by 46 nonsense residues), and a predicted nonspecific F-box protein (At5g04010) using genomic DNA from the wild type or the mutants. The EcoRI- and BamHI-digested DNA fragments were subcloned into pLexA-NLS. The AD fusion constructs (except for RGA and rga-Δ17) were prepared by designing PCR primers incorporating BamHI and EcoRI restriction sites in the correct reading frame and then PCR amplifying the coding regions of GAI (from pACT-GAI, a gift from Caren Chang), gai-1 (from gai-1 genomic DNA), and SCL3 (from wild-type genomic DNA; Pysh et al., 1999). The BamHI- and EcoRI-digested DNA fragments were subcloned into pACTII. The AD-RGA and AD-rga-Δ17 constructs were made by amplifying the coding regions of RGA (from pRG20; Silverstone et al., 1998) and rga-Δ17 (from pRG41; Dill et al., 2001) with PCR primers, which incorporate BamHI and BgIII sites in the correct reading frame. The BamHI- and BglII-digested DNA fragments were subcloned into the BamHI site of pACTII. The series of constructs in pACTII that encode AD fusions with N-terminal or C-terminal truncations of GAI were similarly prepared by designing PCR primers incorporating BamHI and EcoRI restriction sites in the correct reading frame and then PCR amplifying the coding regions from pACT-GAI. The BamHI- and EcoRI-digested DNA fragments were subcloned into pACTII. The AD fusions with GAI truncations include the following: GAI-NT1 (amino acids 1 to 157); GAI-NT2 (amino acids 1 to 468); GAI-CT1 (amino acids 92 to 532); GAI-CT2 (amino acids 151 to 532); GAI-CT3 (amino acids 223 to 532), and GAI-CT4 (amino acids 307 to 532).
For the pull-down assays, the SLY1 and gar2 coding sequences were cloned in frame into the GST fusion vector pGEX2-TK (Amersham Pharmacia Biotech, Piscataway, NJ). The bacterial expression constructs were prepared by designing PCR primers incorporating BamHI and EcoRI restriction sites in the correct reading frame and then PCR amplifying the coding regions of SLY1 and gar2. The BamHI- and EcoRI-digested DNA fragments were subcloned into pGEX2-TK.
For nuclear localization studies of SLY1, the following constructs were generated. The 0.5-kb SLY1 and sly1-d coding regions were PCR amplified from pSLY1-300 and pGST-(sly1-d), respectively, cut with NcoI and KpnI, and then ligated into pRTL2 behind the 35S promoter (named pSLY1-401 and pgar2-31). The 0.7-kb CFP coding sequence was PCR amplified from pECFP (Clontech, Palo Alto, CA), cut with KpnI, and ligated into pSLY1-401 and pgar2-31 to generate constructs that contain 35S:SLY1-CFP (pSLY1-402) and 35S:gar2-CFP (pgar2-33). The pCFP1 plasmid (containing 35S:CFP) was generated by amplifying 0.7-kb CFP DNA from pECFP, cutting with NcoI and KpnI, and ligating into pRTL2.
Nuclear Localization Studies
The onion (Allium cepa) epidermal layers were prepared and bombarded as previously described (Varagona et al., 1992) using tungsten particles (Bio-Rad, Hercules, CA) coated with plasmid DNA expressing CFP, SLY1-CFP, or (sly1-d)-CFP. The cells were viewed using a Leica DMRB microscope (Heerbrugg, Switzerland) equipped with a fluorescence module. For each construct, ∼20 cells that showed CFP fluorescence were scored.
Transformation and Isolation of Transgenic Lines
Using Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998), pSLY1-300 and pSLY1-307 (abbreviated as pSLY1 and psly1-d in Results) were each transformed into the wild type, gai-1 (Koornneef et al., 1985), and GFP-(rga-Δ17) line B (Dill et al., 2001). T1 transformants were selected on MS media (Invitrogen, Carlsbad, CA) containing 10 μg/mL of gluphosinate ammonium (Cresent Chemical Company, Happauge, NY), and BASTA resistant plants were transferred to soil after 10 to 14 d.
Plant Growth Conditions for Phenotypic Analyses
Plants were grown on soil at 22°C with a 16-h-light and 8-h-dark cycle supplied under a light intensity of 140 μE. To determine whether gar2 rga-Δ17 plants were responsive to GA treatment, the soil-grown plants were sprayed weekly with 100 μM GA3 starting at 18 d.
Because of a germination defect when sown on soil, seeds of mutant lines homozygous for the sly1-10 allele were germinated on MS agar plates. Seedlings were then transplanted to soil 7 d after germination. Seeds sterilizations were performed by washing with 95% ethanol for 1 min and then bleach for 2 min. Seeds were then rinsed five times with sterile water and imbibed for 4 d at 4°C before sown on MS plates.
The flowering time in days was scored when the flower bud was first visible without manipulation or magnification. Rosette diameter was obtained by measuring the diameter of the plant in two directions and averaging these measurements.
Immunoblot Analyses and Pull-Down Assays
Seeds of the wild type, rga-24, rga-Δ17, gar2, and gar2 rga-Δ17 were sterilized and imbibed for 3 d at 4°C. All seeds were plated on MS plates (100 × 15 mm) and grown under continuous light of 100 μE at 22°C. The seeds of the rga-Δ17 line were produced from hemizygous parents. The seedlings that did not contain the transgene had a wild-type phenotype (longer hypocotyls and larger leaves) and were discarded from the plate after 7 d. Seedlings (8 d old) were treated with 3 mL of 100 μM GA3 or water for 2 h before harvesting. For GA response experiments with sly1-10, 24-d-old rosette plants were sprayed with 100 μM GA3 or water 3 h before harvesting. Total plant proteins were extracted and analyzed by immunoblot analysis using affinity-purified anti-RGA antibodies from a rabbit (DU176) as described (Silverstone et al., 2001). Ponceau staining was used to confirm equal loading.
For the pull-down assays, GST and GST-SLY1 and GST-(sly1-d) fusion proteins were expressed in the Escherichia coli strain XL1-Blue. Cells were grown to mid-log phase at 30°C and then GST fusion protein expression was induced by adding 0.4 mM isopropylthio-β-galactoside for 3 h. Cells were collected, resuspended in buffer A (PBS buffer containing 0.5% Igepal CA-630) and lysed using a French press. The GST fusion proteins were affinity purified using glutathione-sepharose (Amersham-Pharmacia Biotech) and washed three times with buffer A. Pull-down protocol is similar to that previously described (Gray et al., 2001) with some modifications. Arabidopsis tissue used in the pull-down assays was finely ground in liquid nitrogen, resuspended in buffer A containing 1 mM DTT, 20 μM MG132, 1 mM NaF, 10 mM β-glycerophosphate, 1 mM orthovanadate, and a protease inhibitor cocktail (Roche, Indianapolis, IN), and cleared by centrifugation. For each pull-down assay, 4 μg of purified GST or fusion protein (bound to glutathione-sepharose) was added to the Arabidopsis extract prepared from 50 mg of tissue and incubated at 4°C for 1.5 h. Glutathione-sepharose was washed three times in the pull-down buffer, resuspended in SDS-PAGE sample buffer, and analyzed by SDS-PAGE electrophoresis and immunoblotting using anti-RGA antibodies from rat (DUR18) as described previously (McGinnis et al., 2003).
Measurements of Transcript Levels by Quantitative RT-PCR
Thirteen-day-old seedlings that were grown on MS plates were treated with water or 100 μM GA3 as described in the section for preparing tissues for protein extractions. Total RNA was isolated from 0.1 g of tissue using the RNeasy plant mini prep kit (Qiagen, Valencia, CA) and then treated with the RNase free DNase set (Qiagen) to remove genomic DNA contamination. The RGA and SLY1 message levels were analyzed by quantitative RT-PCR using a Roche LightCycler and the LightCycler RNA amplification kit SYBR Green I (Roche) according to the manufacturer's instructions. Gene-specific primers for RGA, SLY1, and the GAPC gene for glyceraldehyde-3-phosphate dehydrogenase C subunit (see Supplemental Table 2S online) were used in the quantitative RT-PCR with the annealing temperature at 55°C in 6 mM MgCl2. A no-template control was included in each set of reactions to confirm the absence of DNA or RNA contamination. Relative transcript levels of SLY1 and RGA in all samples were normalized using GAPC, whose transcript levels are not affected by the GA treatment.
Yeast Two-Hybrid Assay
Saccharomyces cerevisiae strain L40 [MATa his3-200 trp1-901 leu2-3, -112 ade2 LYS:(lexAop)4-HIS3 URA3:(lexAop)8-lacZ GAL4; Vojtek et al., 1993] was used for the studies. Yeast transformations were performed as previously described (Gietz et al., 1992). The yeast strain L40 was cotransformed with DB and AD plasmid expression constructs and transformants selected on synthetic complete medium–Leu, Trp (Qbiogene, Carlsbad, CA). The ability to drive expression of the HIS3 reporter gene was tested by plating strains on synthetic complete medium–His, Leu, Trp containing increasing concentrations (0, 2, 5, 10, 30, and 60 mM) of 3-AT. Growth of yeast strains was scored after 5 d at 30°C. LacZ reporter gene activity was determined quantitatively by measuring β-gal activity in log-phase liquid cultures as described (Ausubel et al., 1990). β-gal activity (units) was calculated as follows: OD420 of the supernatents × 1000/reaction time (min) × culture volume used for assay (mL) × OD600 of the culture. For each pairwise combination, three independent enzyme assays were performed. Relative levels of DB and AD fusion protein expression were determined by growing yeast strains to mid log-phase in selective liquid media. Pelleted cells were lysed using Yeastbuster reagent (Novagen, Madison, WI) following the manufacturer's protocol. Yeast protein extracts from equivalent cell numbers were loaded and separated on 8% or 15% SDS-PAGE gels. Immunoblot analysis was performed using anti-LexA and an-HA (Roche) antibodies to detect the DB and AD fusion proteins, respectively.
Supplementary Material
Acknowledgments
We thank Shelley Cockrell for technical assistance. We also thank Philip Benfey, Xinnian Dong, and Aron Silverstone for helpful comments on the manuscript. This work was supported by the National Science Foundation (IBN-0078003) to T.-p.S. and by the USDA (2002-01351) to C.M.S.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tai-ping Sun (tps@duke.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020958.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., and Smith, J.A. and Struhl, K., eds (1990). Current Protocols in Molecular Biology. (New York: Green Publishing Associates/Wiley-Interscience).
- Bolle, C., Koncz, C., and Chua, N.-H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Boss, P.K., and Thomas, M.R. (2002). Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416, 847–850. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya: Molecular and physiological characterization. Plant Physiol. 129, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conaway, R.C., Brower, C.S., and Conaway, J.W. (2002). Emerging roles of ubiquitin in transcription regulation. Science 296, 1254–1258. [DOI] [PubMed] [Google Scholar]
- Davies, P.J., ed (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Deschaies, R.J. (1999). SCF and cullin/RING-H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Zhou, Y.-C., Schafer, E., Funk, M., and Kretsch, T. (2001). EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 15, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Jung, H.-S., and Sun, T.-p. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T.-p. (2001). Synergistic de-repression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S.J. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Fleck, B., and Harberd, N.P. (2002). Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilized by gibberellin. Plant J. 32, 935–947. [DOI] [PubMed] [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743. [DOI] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Ait-ali, T., Hynes, L.W., Ougham, H., Peng, J., and Harberd, N.P. (2002). Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shin-Han, S., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Greb, T., Clarenz, O., Schäfer, E., Müller, D., Herrero, R., Schmitz, G., and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17, 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P., White, R., Llewellyn, D., and Jacobsen, J. (2002). GA signaling in barley aleurone cells: Control of SLN1 and GAMYB expression. Plant Physiol. 129, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, R.D. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. [DOI] [PubMed] [Google Scholar]
- Hare, P.D., Seo, H.S., Yang, J.-Y., and Chua, N.-H. (2003). Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 6, 453–462. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M.-T., and Benfey, P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Horton, P., and Nakai, K. (1997). Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 147–152. [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K., Moritz, T., and Harberd, N. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. 1, 3002.1–3002.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen, M.E.P., van Rijn, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant 65, 33–39. [Google Scholar]
- Kuroda, H., Takahashi, N., Shimada, H., Seki, M., Shinozaki, K., and Matsui, M. (2002). Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Elledge, S.J., Peterson, C.A., Bales, E.S., and Legerski, R.J. (1994). Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 91, 5012–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., et al. (2003). Control of tillering in rice. Nature 422, 618–621. [DOI] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.-p., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Kusano, T., Katsumi, M., and Sano, H. (2000). Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245, 21–29. [DOI] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (2002). The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 5, 376–381. [DOI] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. [DOI] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. [DOI] [PubMed] [Google Scholar]
- Rédei, G.P. (1962). Single locus heterosis. Z. Vererbungs 93, 164–170. [Google Scholar]
- Risseeuw, E.P., Daskalchuk, T.E., Banks, T.W., Liu, E., Cotelesage, J., Hellmann, H., Estelle, M., Somers, D.E., and Crosby, W.L. (2003). Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34, 753–767. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.-H., An, G., Kitano, J., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R.E., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.-S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.-p. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y.A., Casamitjana Martínez, E., and Sun, T.-p. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman, J., Jäggi, F., and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16, 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, T.M., Swain, S.M., and Olszewski, N.E. (1999). Gibberellin signal transduction presents the SPY who O-GlcNAc'd me. Trends Plant Sci. 4, 424–428. [DOI] [PubMed] [Google Scholar]
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4, 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. [DOI] [PubMed] [Google Scholar]
- Vojtek, A.B., Hollenberg, S.M., and Cooper, J.A. (1993). Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Wang, X., Feng, S., Nakayama, N., Crosby, W.L., Irish, V., Deng, X.W., and Wei, N. (2003). The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell 15, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C.-K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.N., and Somerville, C.R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 108, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.