Abstract
DELLA proteins restrain the cell proliferation and enlargement that characterizes the growth of plant organs. Gibberellin stimulates growth via 26S proteasome–dependent destruction of DELLAs, thus relieving DELLA-mediated growth restraint. Here, we show that the Arabidopsis thaliana sleepy1gar2-1 (sly1gar2-1) mutant allele encodes a mutant subunit (sly1gar2-1) of an SCFSLY1 E3 ubiquitin ligase complex. SLY1 (the wild-type form) and sly1gar2-1 both confer substrate specificity on this complex via specific binding to the DELLA proteins. However, sly1gar2-1 interacts more strongly with the DELLA target than does SLY1. In addition, the strength of the SCFSLY1–DELLA interaction is increased by target phosphorylation. Growth-promoting DELLA destruction is dependent on SLY1 availability, on the strength of the interaction between SLY1 and the DELLA target, and on promotion of the SCFSLY1–DELLA interaction by DELLA phosphorylation.
INTRODUCTION
Regulated protein degradation plays an essential role in the normal development of all organisms. Although several different mechanisms for controlled proteolysis are known to exist, the central importance of ubiquitin-mediated proteolysis for many cellular processes has recently become apparent (Hershko and Ciechanover, 1998; Deshaies, 1999; Pickart, 2001). In ubiquitin-mediated proteolysis, activated ubiquitin is bound to the substrate protein by a ubiquitin-protein ligase (E3). Polyubiquitylated proteins are then recognized and degraded by the 26S proteasome (Patton et al., 1998). Because it controls the specificity of substrate ubiquitylation, the E3 ligase is a key regulatory component of the ubiquitin–proteasome pathway. Several different classes of E3 ligase activity have been identified. Some act as protein complexes (e.g., SCF [for skp1-cullin-F-box-Rbx1] and the anaphase-promoting complex), whereas others (such as the RING-finger domain E3 ligases) act as single polypeptides (Deshaies, 1999).
The importance of SCF-type E3 ligase activities in plant regulatory signaling pathways is now well established (Sullivan et al., 2003; Vierstra, 2003). Recent developments have suggested that the control of plant organ growth mediated by the gibberellin (GA)–DELLA signaling pathway is also proteasome dependent and involves an SCF E3 ligase activity (Fu et al., 2002; McGinnis et al., 2003; Sasaki et al., 2003). GA is a phytohormone that is essential for the normal growth of plants (Hooley, 1994). GA-deficient mutants of Arabidopsis thaliana exhibit a dwarfed, dark-green phenotype that can be corrected by the application of exogenous GA (Koornneef and van der Veen, 1980). GA promotes plant growth by overcoming the growth-restraining effects of a family of nuclear growth repressor proteins known as the DELLA proteins (Richards et al., 2001; Lee et al., 2002; Wen and Chang, 2002; Fu and Harberd, 2003). Although first identified in Arabidopsis, the DELLA proteins are now known to regulate the growth of a wide spectrum of higher plants, including maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), rice (Oryza sativa), and grape (Vitis vinifera) (Peng et al., 1999a; Boss and Thomas, 2002; Chandler et al., 2002; Gubler et al., 2002; Itoh et al., 2002).
In Arabidopsis, the DELLA proteins are encoded by a family of five genes (GIBBERELLIC ACID INSENSITIVE [GAI], REPRESSOR OF ga1-3 [RGA], and three different REPRESSOR OF ga1-3-LIKE genes [RGL1, RGL2, and RGL3]; Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; Lee et al., 2002). These five DELLA proteins have both overlapping and distinct functions in plant growth and development (Dill and Sun, 2001; King et al., 2001; Lee et al., 2002; Wen and Chang, 2002; Cheng et al., 2004). GAI was the first DELLA protein to be identified, following cloning of a dominant mutant allele (gai) via transposon-insertion inactivation (Peng et al., 1997). gai confers a dark-green, dwarfed phenotype that resembles the phenotype of GA-deficient mutants (Koornneef and van der Veen, 1980; Koornneef et al., 1985). However, unlike the phenotype of GA-deficient mutants, gai phenotype cannot be restored to normal by treatment with exogenous GA (Koornneef et al., 1985). The molecular cloning of the GAI and gai alleles revealed that gai encodes a mutant protein (gai) that lacks a particular segment of 17 amino acids (Peng et al., 1997). This segment is part of a conserved region of the DELLA proteins that is now known as domain I (Peng et al., 1999a; Lee et al., 2002). The molecular characterization of GAI and gai led to the hypothesis that GAI acts as a growth repressor whose repressing function can be overcome by GA, whereas the mutant gai protein continues to repress growth even in the presence of GA (because the mutation in domain I makes the mutant protein resistant to the effects of the hormone; Peng et al., 1997; King et al., 2001; Richards et al., 2001). Subsequently, mutations in domain I of DELLA proteins have been shown to confer reduced GA response in several species additional to Arabidopsis (Peng et al., 1999a; Boss and Thomas, 2002; Chandler et al., 2002; Itoh et al., 2002).
GA overcomes the growth-repressive effects of DELLA proteins, such as Arabidopsis RGA (Silverstone et al., 1998), by causing a reduction in their nuclear abundance (Dill et al., 2001; Silverstone et al., 2001; Fleck and Harberd, 2002; Gubler et al., 2002; Itoh et al., 2002; Fu and Harberd, 2003). However, the kinetics of the response of different DELLA proteins to GA varies. Whereas previous experiments did not detect destabilization of GAI or RGL1 in response to GA treatments (Fleck and Harberd, 2002; Wen and Chang, 2002), we describe in this article an effect of GA treatment on GAI level. In addition, in vitro mutagenesis of defined sections of the N-terminal regions of DELLA proteins (including the above-described domain I) results in mutant proteins that confer a dwarfed, reduced GA-response phenotype and that remain stable in the presence of GA (Dill et al., 2001; Itoh et al., 2002). Thus, GA overcomes the growth-repressing function of wild-type DELLA proteins by causing a reduction in their nuclear abundance, whereas the levels of some mutant DELLA proteins (including those lacking a functional domain I) remain resistant to the effects of GA.
Recent data indicates that the GA-induced reduction in DELLA levels is dependent on 26S proteasome function (Fu et al., 2002). Furthermore, a candidate F-box subunit of a proposed SCF complex that could target DELLA proteins for proteasome-mediated destruction has also been described (McGinnis et al., 2003; Sasaki et al., 2003). First identified via the characterization of novel reduced GA-response dwarf mutants of Arabidopsis and rice, the orthologous SLEEPY1 (SLY1) and GIBBERELLIN INSENSITIVE DWARF2 (GID2) genes encode proteins (SLY1 or GID2) containing the conserved F-box interaction domain (via which F-box proteins associate with the Skp1-like component of the SCF complex) (Steber et al., 1998; McGinnis et al., 2003; Sasaki et al., 2003). Lack of SLY1 or GID2 function (conferred by sly1-10 or gid2-1) causes accumulation of RGA or SLR1 (the rice DELLA protein), whereas lack of RGA or SLR1 suppresses the dwarf phenotype conferred by lack of SLY1 or GID2 (McGinnis et al., 2003; Sasaki et al., 2003). Additional results suggest that the rice SLR1 protein is phosphorylated in response to the GA signal (Sasaki et al., 2003). Taken together, these observations suggested an explanation for the above described GA-induced reduction in the level of nuclear DELLA proteins. According to this explanation, SLY1 and GID2 are F-box subunits of an SCF complex that targets phosphorylated DELLA proteins for destruction in the proteasome, thus releasing growth from DELLA-mediated restraint (Harberd, 2003; McGinnis et al., 2003; Sasaki et al., 2003). Recently, it has been shown that GID2 is indeed part of an SCFGID2 complex that interacts with phosphorylated SLR1 (Gomi et al., 2004). However, before the experiments reported here, robust demonstration that SLY1 is a subunit of a functional SCF E3 ubiquitin ligase complex was still lacking.
Mutagenic studies of gai plants had previously identified intragenic loss-of-function alleles (gai-derivative alleles) that conferred a tall, rather than a dwarf, phenotype (Peng and Harberd, 1993; Wilson and Somerville, 1995; Peng et al., 1997). In addition, these screens led to the identification of extragenic suppressors, mutations in genes other than GAI itself, which cause suppression of the dwarf phenotype conferred by gai (Carol et al., 1995; Wilson and Somerville, 1995). One such extragenic suppressor mutation was gai revertant2-1 (gar2-1), a dominant extragenic suppressor of gai phenotype (Wilson and Somerville, 1995; Peng et al., 1997, 1999b). gar2-1 confers resistance to the GA biosynthesis inhibitor paclobutrazol (PAC) and acts additively with loss-of-function alleles of SPINDLY (a gene encoding an O-GlcNAc transferase with GA-signaling function; Jacobsen et al., 1996; Swain et al., 2002) in suppressing gai phenotype (Wilson and Somerville, 1995; Peng et al., 1997, 1999b). In addition, recent results show that gar2-1 increases the growth of GA-deficient pollen tubes (Swain et al., 2004). However, the nature of the gar2-1 gene product (or of the product of the presumed wild-type GAR2 allele) remained unknown, as did the relationship between gar2-1 and DELLA protein function in the regulation of plant growth. We therefore undertook further analysis of gar2-1 function. We found that gar2-1 affects DELLA protein accumulation and encodes a mutant SLY1 protein (sly1gar2-1). We also found that SLY1 is a subunit of an SCFSLY1 E3 ubiquitin ligase complex that binds directly to DELLA protein substrates. In further experiments, we found that plant growth is responsive to increases in the cellular level of SCFSLY1 E3 ubiquitin ligase activity because of increases in the availability of SLY1 subunits. Comparative studies of the sly1gar2-1 (mutant) and SLY1 (wild-type) proteins showed that sly1gar2-1 interacts more strongly with the DELLA protein target. In addition, we showed that phosphorylation of the DELLA target protein further strengthens the interaction between DELLA and either SLY1 or sly1gar2-1. Our results indicate that plant growth can vary in response to fluctuations in SCFSLY1 E3 ubiquitin ligase activity resulting from modulations of SLY1 availability or the strength of the SCFSLY1–DELLA interaction.
RESULTS
gar2-1 Promotes Plant Growth by Opposing gai-, GAI-, or RGA-Mediated Restraint
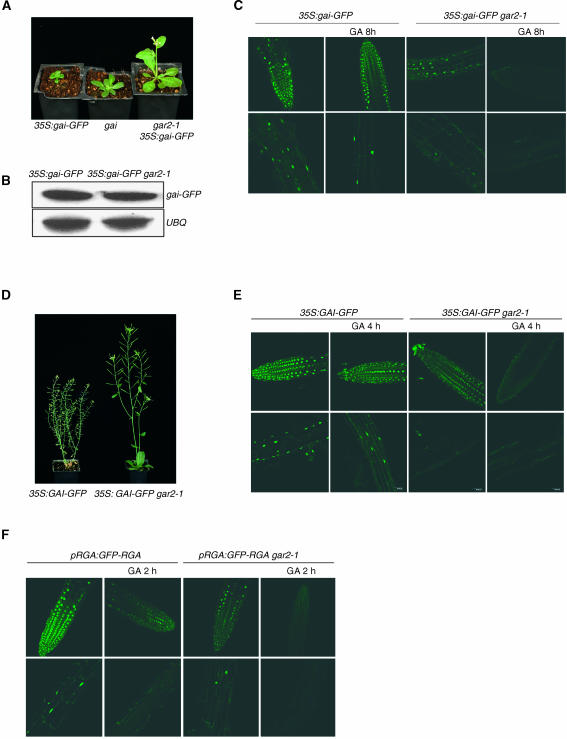
gai confers dwarfism because it encodes a mutant DELLA protein (gai) that is relatively resistant to the opposing effects of GA (Peng et al., 1997). Although gar2-1 was known to suppress gai phenotype (Wilson and Somerville, 1995; Peng et al., 1997, 1999b), it was not clear how gar2-1 affected the properties of the gai protein. We therefore further investigated the effect of gar2-1 on gai. Expression of a 35S:gai-GFP (green fluorescent protein) transgene confers dwarfism that is more severe than that conferred by the original gai mutant allele (Figure 1A; Fleck and Harberd, 2002). We found that 35S:gai-GFP gar2-1 plants were considerably less dwarfed than 35S:gai-GFP controls (Figure 1A). Thus, gar2-1 suppresses the phenotype conferred by 35S:gai-GFP. RNA gel blot analyses showed that the levels of gai-GFP transcripts in 35S:gai-GFP gar2-1 plants were not detectably different from those in 35S:gai-GFP controls (Figure 1B). This suggests that gar2-1 suppresses 35S:gai-GFP phenotype via a mechanism other than reduction of gai-GFP transcript levels.
Figure 1.
gar2-1 Reduces DELLA Protein Accumulation.
(A) Representative 21-d-old 35S:gai-GFP, gai, and 35S:gai-GFP gar2-1 plants.
(B) gai-GFP transcripts in 35S:gai-GFP and 35S:gai-GFP gar2-1 plants. UBQ, ubiquitin transcripts (loading control).
(C) gai-GFP fluorescence in primary roots of 35S:gai-GFP or 35S:gai-GFP gar2-1 seedlings (7 d old) treated or untreated with 100 μM GA3 for time shown. Top row, root tips; bottom row, elongation zone.
(D) Representative 42-d-old 35S:GAI-GFP or 35S:GAI-GFP gar2-1 plants.
(E) GAI-GFP fluorescence of 35S:GAI-GFP or 35S:GAI-GFP gar2-1 primary seedling roots treated or untreated with 100 μM GA3 for time shown. Top row, root tips; bottom row, elongation zone.
(F) GFP-RGA fluorescence in pRGA:GFP-RGA or pRGA:GFP-RGA gar2-1 primary seedling roots treated or untreated with 100 μM GA3 for time shown. Top row, root tips; bottom row, elongation zone.
We next investigated the effect of gar2-1 on gai-GFP fluorescence. gai-GFP was clearly detectable in 35S:gai-GFP root cell nuclei and was not detectably affected by GA treatment (Figure 1C; Fleck and Harberd, 2002). However, gai-GFP fluorescence was considerably less intense in 35S:gai-GFP gar2-1 than it was in 35S:gai-GFP controls (Figure 1C). In addition, the intensity of gai-GFP fluorescence in 35S:gai-GFP gar2-1 roots was further reduced by GA treatment (Figure 1C). Thus, gar2-1 causes both a reduction in initial gai-GFP levels and a further reduction in gai-GFP levels in response to exogenous GA.
We also investigated the possibility that gar2-1 suppression is not specific to phenotypes conferred by gai and that gar2-1 also affects the properties of GAI and RGA. 35S:GAI-GFP transgenes confer dwarfism because of high-level expression of GAI-GFP (Fleck and Harberd, 2002; Figure 1D). We found that 35S:GAI-GFP gar2-1 plants were less dwarfed than 35S:GAI-GFP controls (Figure 1D). Fluorescence because of GAI-GFP was clearly detectable in 35S:GAI-GFP root nuclei and not detectably changed by treatment with exogenous GA (Figure 1E; Fleck and Harberd, 2002). However, the intensity of GAI-GFP fluorescence was reduced in the nuclei of 35S:GAI-GFP gar2-1 roots compared with 35S:GAI-GFP roots. In addition, the intensity of GAI-GFP fluorescence in 35S:GAI-GFP gar2-1 roots was further reduced by GA treatment (Figure 1E). Thus, gar2-1 suppresses the dwarfism conferred by 35S:GAI-GFP, reduces GAI-GFP levels, and causes a further reduction in GAI-GFP levels in response to exogenous GA.
We subsequently examined the effects of gar2-1 on GFP-RGA. Treatment with GA causes a rapid reduction in the intensity of nuclear GFP-RGA fluorescence (Figure 1F; Silverstone et al., 2001). We found that the GFP-RGA signal was reduced in the nuclei of pRGA:GFP-RGA gar2-1 roots compared with pRGA:GFP-RGA roots (Figure 1F). Furthermore, GFP-RGA became undetectable earlier in GA-treated pRGA:GFP-RGA gar2-1 roots than it did in GA-treated pRGA:GFP-RGA roots (Figure 1F). Thus, gar2-1 reduces initial GFP-RGA levels and decreases the treatment duration required for GFP-RGA to become undetectable in response to GA.
gar2-1 Is a Mutant SLY1 Allele and Encodes a Mutant SLY1 Protein (sly1gar2-1)
We next isolated gar2-1 via map-based cloning. GAR2 (the presumed wild-type allele of gar2-1) mapped to a region of Arabidopsis chromosome 4 that also contains SLY1 (see Supplemental Figure S1 online). We then detected a nucleotide substitution in the open reading frame of the SLY1 allele from gar2-1 plants (Figure 2A). Furthermore, expression of the mutant SLY1 open reading frame from gar2-1 (from a 35S:sly1gar2-1 transgene) caused complete suppression of gai phenotype (in gai 35S:sly1gar2-1 plants), resulting in plants that had bolt-stem internode lengths and plant color that resembled that of wild-type plants (Figures 2B and 2C). Expression of a control (35S:SLY1) transgene conferred only partial suppression of gai phenotype (Figure 2B; see also below). Thus, gar2-1 is a mutant SLY1 allele, henceforth designated as sly1gar2-1 (and its protein product as sly1gar2-1). The single nucleotide substitution in sly1gar2-1 results in an E138 to K138 amino acid residue substitution in a C-terminal portion (the LSL domain) of the sly1gar2-1 protein (Figure 2A).
Figure 2.
sly1gar2-1 Encodes a Mutant Form of SLY1.
(A) Illustration of SLY1 protein showing positions of previously defined F-box, GGF, and LSL domains (McGinnis et al., 2003) and the amino acid substitution conferred by sly1gar2-1.
(B) Representative 30-d-old gai, gai 35S:SLY1, and gai 35S:sly1gar2-1 plants (with wild-type control).
(C) SLY1/sly1gar2-1 transcripts in gai, gai 35S:SLY1, and gai 35S:sly1gar2-1 plants. Endogenous SLY1, endogenous SLY1 transcripts; total SLY1/sly1gar2-1, sum of endogenous SLY1 and 35S:SLY1 (or 35S:sly1gar2-1) transcripts; UBQ, ubiquitin transcripts (loading control).
SLY1 Affects Both RGA and GAI Function
Because sly1gar2-1 is a mutant SLY1 allele, we further investigated the role of SLY1 in plant growth regulation. Plants lacking SLY1 (e.g., sly1-10 homozygotes; Steber et al., 1998; Steber and McCourt, 2001) exhibit dwarfism, reduced GA response, and increased RGA accumulation (McGinnis et al., 2003). In addition, lack of RGA partially suppresses sly1-10 phenotype, indicating that the increased RGA accumulation in sly1-10 contributes to the dwarf phenotype that sly1-10 confers (McGinnis et al., 2003).
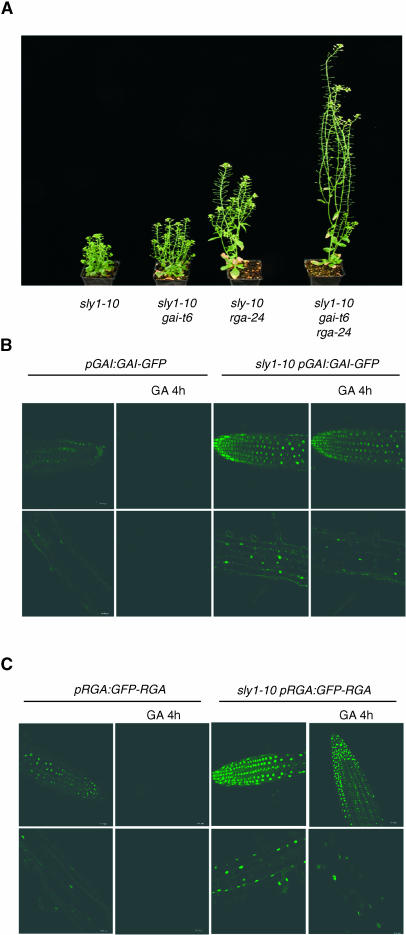
However, the relationship between SLY1 and GAI was unknown. We found that lack of GAI (conferred by gai-t6) slightly suppressed and lack of both GAI and RGA (conferred by gai-t6 rga-24) substantially suppressed the dwarfism conferred by sly1-10 (Figure 3A). Suppression from lack of both GAI and RGA was greater than that from lack of RGA alone (Figure 3A; McGinnis et al., 2003). Thus, the growth-repressing activity of both GAI and RGA is subject to modulation by SLY1.
Figure 3.
SLY1 Affects Both GAI and RGA Function.
(A) Representative 42-d-old sly1-10, sly1-10 gai-t6, sly1-10 rga-24, and sly1-10 gai-t6 rga-24 plants.
(B) GAI-GFP fluorescence in pGAI:GAI-GFP or sly1-10 pGAI:GAI-GFP primary roots treated (or control) with 100 μM GA3. Top row, root tips; bottom row, elongation zone.
(C) GFP-RGA fluorescence in pRGA:GFP-RGA or sly1-10 pRGA:GFP-RGA primary roots treated (or control) with 100 μM GA3. Top row, root tips; bottom row, elongation zone.
GAI-GFP is more resistant to GA-induced destabilization than GFP-RGA (Fleck and Harberd, 2002). However, in a line exhibiting a particularly low initial level of GAI-GFP fluorescence (pGAI:GAI-GFP), the intensity of GAI-GFP fluorescence was detectably reduced after GA treatment (Figure 3B). Interestingly, GAI-GFP fluorescence intensity was higher in sly1-10 pGAI:GAI-GFP plants than in pGAI:GAI-GFP controls (Figure 3B). In addition, GAI-GFP fluorescence in sly1-10 pGAI:GAI-GFP plants was not detectably reduced after GA treatment (Figure 3B). Similarly, GFP-RGA levels were higher in sly1-10 pRGA:GFP-RGA plants than in pRGA:GFP-RGA controls, and GFP-RGA fluorescence in sly1-10 pRGA:GFP-RGA plants was not detectably reduced after GA treatment (Figure 3C). Thus, SLY1 reduces the accumulation of both GAI-GFP and GFP-RGA, and SLY1 promotes the GA-mediated destabilization of both GAI-GFP and GFP-RGA.
Increased SLY1 Availability Overcomes the Phenotypic Effects of Stabilized DELLAs
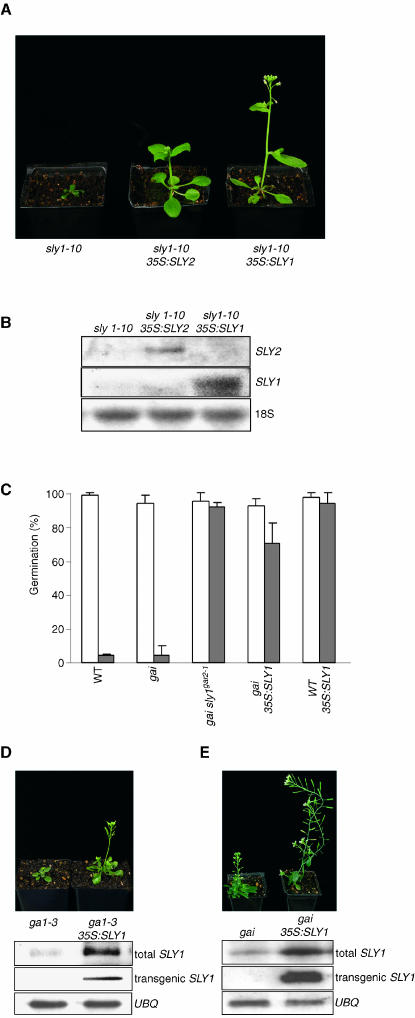
In addition to SLEEPY1 (SLY1; At4g24210), the Arabidopsis genome contains SLY2 (At5g48170), a gene that encodes a SLY1-like protein (McGinnis et al., 2003). Because lack of SLY1 confers dwarfism and reduced GA response (McGinnis et al., 2003), we next investigated the phenotypes conferred by transgenic overexpression of SLY1 or SLY2. Expression of 35S:SLY1 or 35S:SLY2 transgenes caused substantial or partial suppression, respectively, of sly1-10 phenotype (Figures 4A and 4B), suggesting that overexpression of SLY2 can at least partially compensate for lack of SLY1.
Figure 4.
Increased SLY1/SLY2 Availability Opposes DELLA Protein Function.
(A) Representative 21-d-old sly1-10, sly1-10 35S:SLY2, and sly1-10 35S:SLY1 plants.
(B) SLY1/SLY2 transcripts in plants in (A). 18S, 18S rRNA loading control.
(C) Germination of seeds (genotypes indicated) in the presence (closed bars) or absence (open bars) of 2 μM PAC. Results are mean ± se of three separate experiments (for each sample, n > 160).
(D) Representative 35-d-old ga1-3 and ga1-3 35S:SLY1 plants, together with SLY1 transcript levels in each. Transgenic SLY1, 35S:SLY1 transcripts; total SLY1, sum of endogenous SLY1 and 35S:SLY1 transcripts; UBQ, ubiquitin transcripts (loading control).
(E) Representative 35-d-old gai and gai 35S:SLY1 plants, together with SLY1 transcript levels in each (see [D]).
35S:SLY1 had no obvious effect on the growth and development of wild-type plants (data not shown). However, 35S:SLY1 conferred increased resistance to PAC, a GA biosynthesis inhibitor (Hedden and Graebe, 1985; Davis and Curry, 1991). 35S:SLY1 seeds germinated on PAC-containing medium (as did positive control gai sly1gar2-1 seeds; Peng et al., 1999b), whereas the germination of wild-type control seeds was substantially inhibited (Figure 4C). Furthermore, 35S:SLY1 suppressed the dwarf phenotype conferred by the GA-deficiency mutation ga1-3 (Koornneef and van der Veen, 1980; King et al., 2001; Figure 4D) and by gai (Figure 4E) and also conferred PAC-resistant germination on gai seeds (Figure 4C). All of the phenotypes shown here to be suppressed by 35S:SLY1 are attributable to stabilized DELLA proteins. Our results therefore indicate that the increased availability of SLY1 conferred by 35S:SLY1 overcomes the growth retardation or germination inhibition that these stabilized DELLA proteins would otherwise impose.
SLY1 Is the F-Box Subunit of an in Planta SCFSLY1 E3 Ubiquitin Ligase Complex
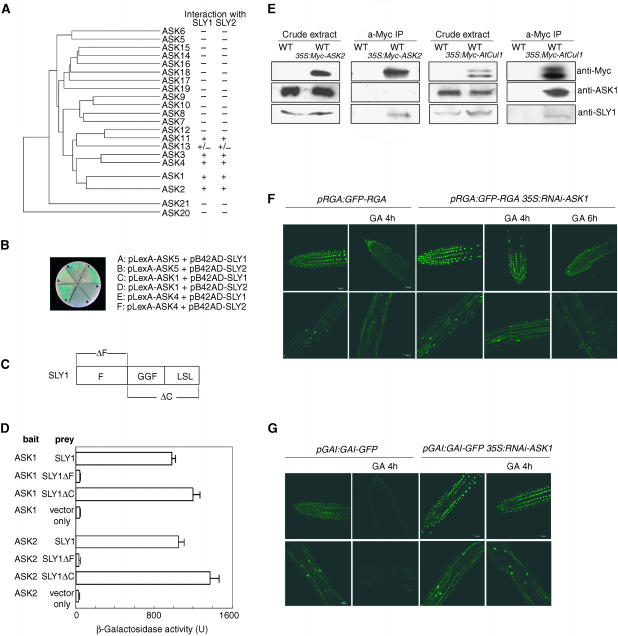
Because sly1gar2-1 encodes a mutant form of SLY1, it was important to determine if SLY1 is the F-box subunit of an SCFSLY1 complex. F-box subunits are anchored into SCF complexes via binding of the N-terminal F-box domain to SKP1 (Zheng et al., 2002). If Arabidopsis SLY1 or SLY2 is indeed part of a functional SCFSLY1/SCFSLY2 complex, SLY1 or SLY2 should interact with one or more members of the Arabidopsis family of SKP1 homologs (ASK1 to ASK21; Figure 5A; Gagne et al., 2002). We found that SLY1 and SLY2 both interacted strongly with ASK1, ASK2, ASK3, ASK4, and ASK11, more weakly with ASK13, and did not detectably interact with any of the remaining ASK family members in yeast two-hybrid assays (Figures 5A and 5B; X. Fu and N.P. Harberd, data not shown). This result is consistent with the previous demonstration of positive yeast two-hybrid interactions between a representative Arabidopsis C2 family (of which SLY1 and SLY2 are members) F-box protein and ASK1, ASK2, ASK4, ASK11, or ASK13 (Gagne et al., 2002).
Figure 5.
SLY1 Is the F-Box Subunit of an SCFSLY1 E3 Ubiquitin Ligase.
(A) Relatedness tree of ASK sequences. ASK subunits interacting with SLY1 or SLY2 are indicated (+, positive interaction; −, no detectable interaction; +/−, weak interaction).
(B) Representative yeast two-hybrid experiments: A and B, no detectable interaction; C to F, detectable interaction.
(C) Schematic indication of SLY1 derivative proteins (used in [D]; see also Figure 2A). SLY1ΔF lacks an N-terminal portion of SLY1 containing the conserved F-box sequence (F), whereas SLY1ΔC lacks a C-terminal portion of SLY1 (GGF and LSL domains; McGinnis et al., 2003).
(D) Quantitation of the interactions between ASK1 or ASK2 and SLY1, SLY1ΔF, or SLY1ΔC. Results are shown as mean and se of at least three independent experiments.
(E) Immunoblot assay of proteins coimmunoprecipitated by an anti-Myc antibody or in crude extracts from plants expressing 35S:Myc-ASK2 or 35S:Myc-AtCUL1 transgenes (or wild-type control).
(F) GFP-RGA fluorescence in pRGA:GFP-RGA or pRGA:GFP-RGA 35S:RNAi-ASK1 primary roots treated (or control) with 100 μM GA3. Top row, root tips; bottom row, elongation zone.
(G) GAI-GFP fluorescence in pGAI:GAI-GFP or pGAI:GAI-GFP 35S:RNAi-ASK1 primary roots treated (or control) with 100 μM GA3. Top row, root tips; bottom row, elongation zone.
In further experiments, we tested if the interaction between ASK1 or ASK2 and SLY1 is dependent on the N-terminal F-box domain of SLY1 (Figures 5C and 5D). Low levels of reporter activation were conferred by DNA binding domain-ASK1/2 fusion proteins (encoded by pLexA-ASK1 or pLexA-ASK2 constructs) in the presence of the activation domain (AD) vector construct (Figure 5D). High levels of reporter activation (indicative of positive interaction) were obtained when the AD was fused with SLY1 derivatives containing the N-terminal F-box domain (SLY1; SLY1ΔC). A SLY1-derivative lacking this domain (SLY1ΔF) failed to interact with ASK1 or ASK2 (Figure 5D). These results suggest that the SLY1–ASK1/ASK2 interaction is dependent upon the F-box domain of SLY1.
We next determined if SLY1 is part of an in planta SCF complex. Polyclonal anti-ASK1 and anti-SLY1 antibodies detected ASK1 and SLY1 in crude extracts from wild-type plants and from plants expressing a Myc-tagged form of ASK2 (Myc-ASK2; Xu et al., 2002) (Figure 5E). SLY1 (but not ASK1) was detected in anti-Myc–immunoprecipitated proteins from the line expressing Myc-ASK2 but not in anti-Myc–immunoprecipitated proteins from wild-type control plants (Figure 5E). Similarly, both SLY1 and ASK1 were detected in anti-Myc–immunoprecipitated proteins from plants expressing Myc-AtCUL1 but not in anti-Myc–immunoprecipitated proteins from the wild-type control (Figure 5E). Thus, SLY1 is specifically associated with AtCUL1 and ASK1 or ASK2 in planta, indicating that it is indeed part of a functional SCFSLY1 complex.
If SLY1 functions as part of an SCFSLY1 E3 ubiquitin ligase that regulates the accumulation and GA-induced disappearance of DELLA proteins, then reduction in the availability of those ASK subunits that comprise SCFSLY1 should also affect DELLA stability. We tested this prediction in plants expressing GFP-RGA or GAI-GFP and an RNA interference (RNAi) construct that specifically reduced the levels of transcripts encoding ASK1 (data not shown). This construct (35S:RNAi-ASK1) conferred a dwarf phenotype on transgenic plants (see also Zhao et al., 2003; Liu et al., 2004), enhanced the (initial) nuclear fluorescence because of GFP-RGA or GAI-GFP, and inhibited reduction in levels of GFP-RGA or GAI-GFP in response to GA treatment (Figures 5F and 5G). Thus, reduction in the availability of ASK1 subunits affects the abundance and GA responses of nuclear GAI and RGA.
SLY1 Interacts with GAI, gai, and RGA
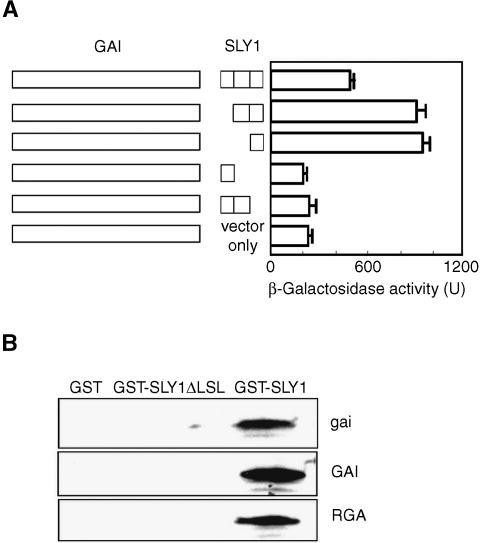
We next sought to determine if DELLA proteins are the substrate of the SCFSLY1 E3 ubiquitin ligase. Typically, SCF complexes target their substrates via binding of a C-terminal portion of the F-box subunit to a specific region of the substrate protein (Vierstra, 2003). We found that GAI interacts with SLY1 in yeast two-hybrid assays (Figure 6A). Low levels of reporter activation were conferred by a pLexA-GAI construct in the presence of the AD vector construct (Figure 6A). High levels of reporter activation (indicative of positive interaction) were obtained when the AD was fused with SLY1 (Figure 6A). We next identified the region of SLY1 that is the site of the GAI-SLY1 interaction and found that this interaction requires only a C-terminal portion of SLY1 (the LSL domain) (Figure 6A). In further experiments, we detected an interaction between the LSL domain of SLY1 and in vitro–translated GAI, gai, or RGA proteins. First, we immobilized Escherichia coli–expressed glutathione S-transferase (GST)-tagged SLY1 or SLY1ΔLSL (Figure 6B) on agarose beads. These beads were subsequently incubated with 35S-Met–labeled in vitro–translated GAI, gai, or RGA. None of GAI, gai, or RGA was specifically bound by immobilized GST (control) or by GST-SLY1ΔLSL (Figure 6B). However, GAI, gai, and RGA were all bound by GST-SLY1 (Figure 6B). Thus, GAI, gai, or RGA interacts with SLY1.
Figure 6.
The LSL Domain of SLY1 Interacts with GAI, gai, and RGA.
(A) Quantitation of the interactions between GAI and SLY1 or SLY1-derivative proteins lacking F-box, GGF, or LSL domains (McGinnis et al., 2003; Figure 2A). Results are shown as mean and se of three independent experiments.
(B) In vitro interaction of 35S-Met–labeled gai, GAI, or RGA with immobilized GST-SLY1. GST-SLY1ΔLSL lacks the LSL domain (Figure 2A).
Thus, the DELLA–SLY1 interaction proceeds via binding of the DELLA protein to the LSL domain of SLY1. A DELLA–SLY1 interaction is also detected in planta (see below), indicating that the DELLA proteins are direct targets of the SCFSLY1 E3 ubiquitin ligase.
sly1gar2-1 Interacts More Strongly Than SLY1 with Both Phosphorylated and Dephosphorylated DELLA Protein Targets
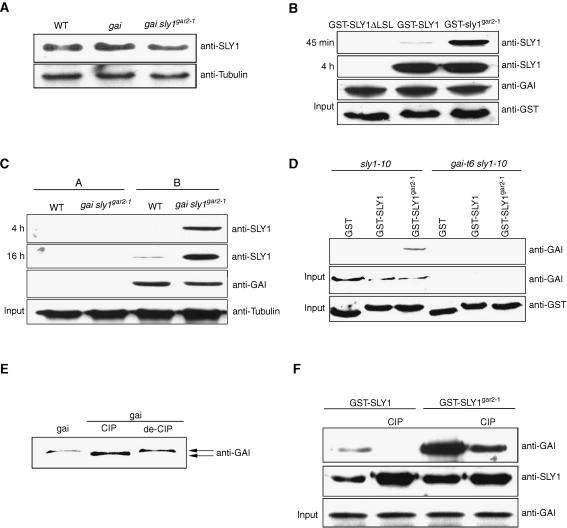
Because sly1gar2-1 suppresses gai phenotype (Wilson and Somerville, 1995; Peng et al., 1997, 1999b), we determined if the in planta abundance of sly1gar2-1 was different from that of SLY1. Protein gel blot experiments revealed no detectable difference in the levels of immunodetectable SLY1 protein in sly1gar2-1 mutant versus control plants (Figure 7A), suggesting that a change in SLY1 availability does not explain the phenotype conferred by sly1gar2-1.
Figure 7.
sly1gar2-1 Has Increased Affinity for DELLA Proteins.
(A) Immunoblot assay of SLY1/sly1gar2-1 levels in wild-type, gai, and gai sly1gar2-1 plants (with tubulin loading control).
(B) Immunoblot assay of E. coli–expressed proteins displaying specific interaction with gai. Anti-GST provides an input protein control. Anti-GAI provides control for amount of immobilized gai (subsequently released from beads by boiling).
(C) Immunoblot assay of proteins coimmunoprecipitated by control immobilization substrate (A) (see Methods) or immobilized gai (B) from wild-type or gai sly1gar2-1 plant extracts. Anti-tubulin provides input protein control. Anti-GAI provides control for amount of immobilized gai (subsequently released from beads by boiling).
(D) Immunoblot assay of proteins from sly1-10 or gai-t6 sly1-10 plants specifically interacting with GST-SLY1 or GST-sly1gar2-1. Anti-GST and anti-GAI antibodies provide input controls.
(E) Immunoblot assay (anti-GAI antibody) of gai proteins from ethanol-induced alcA:gai plants (gai) together with gai incubated with CIP or denatured CIP (de-CIP).
(F) Immunoblot assay of specific interactions between native (treated with denatured CIP) or CIP-treated gai (CIP) and immobilized GST-SLY1 or GST-sly1gar2-1. The anti-GAI antibody provided input protein control. Anti-SLY1 provides control for amount of immobilized SLY1/sly1gar2-1 (subsequently released from beads by boiling).
We next tested the DELLA interaction of GST-tagged SLY1, SLY1ΔLSL, or sly1gar2-1 by incubating them with immobilized plant-derived gai. Previously, we have shown that gai accumulates to immunologically detectable levels in ethanol-induced alcA:gai plants (Ait-ali et al., 2003). Extracts from ethanol-induced alcA:gai plants were incubated with beads that had previously been cross-linked with polyclonal anti-GAI antibodies (see Methods), thus creating beads coated with immunocaptured gai (Figure 7B). There was no detectable interaction between this immobilized gai and SLY1ΔLSL (Figure 7B). However, strong interaction between sly1gar2-1 and gai and weaker interaction between SLY1 and gai was detected 45 min after the onset of incubation (Figure 7B). By 4 h these signals were indistinguishable from one another (Figure 7B).
In further tests of the interaction between gai and SLY1 or sly1gar2-1, immobilized gai was incubated with extracts from the wild type or gai sly1gar2-1 plants (Figure 7C). Control immobilization substrate (beads) had no detectable gai attached and failed to interact with SLY1 or sly1gar2-1 (Figure 7C). After 4 h of incubation, an interaction between sly1gar2-1 and gai was detected, but no interaction was detected between SLY1 and gai (Figure 7C). After 16 h, an interaction between SLY1 and gai was detected, but the signal was still weaker than that obtained with sly1gar2-1 (Figure 7C).
We next tested the interaction between sly1gar2-1 and GAI. Immobilized GST, GST-SLY1, or GST-sly1gar2-1 was incubated with extracts from sly1-10 or gai-t6 sly1-10 plants (Figure 7D). GAI accumulates in sly1-10 plants but not in plants that lack GAI (gai-t6 sly1-10; Figure 7D; X. Fu, unpublished data). An interaction was detected between plant-derived GAI and GST-sly1gar2-1 but not between GAI and GST or GST-SLY1 (Figure 7D). Thus, sly1gar2-1 interacts more strongly with GAI than does SLY1.
Previous work suggests that phosphorylation of rice DELLA proteins targets them for ubiquitylation via a putative rice SCFGID2 E3 ubiquitin ligase and, hence, for subsequent destruction in the proteasome (Sasaki et al., 2003; Gomi et al., 2004). We found that plant-derived gai is phosphorylated. Treatment with calf intestinal phosphatase (CIP) altered the electrophoretic mobility of gai (whereas treatment with denatured CIP did not; Figure 7E). In controlled immunoprecipitation experiments, we again found that GST-sly1gar2-1 interacted more strongly than did GST-SLY1 with the native (phosphorylated) form of gai. Interestingly, GST-sly1gar2-1 also bound dephosphorylated gai (obtained via CIP treatment), although less strongly than it did native gai (Figure 7F). An interaction between GST-SLY1 and dephosphorylated gai was not detected (Figure 7F), indicating either that this interaction does not occur or that it was below the limit of detection in our experiments. Taken together, these results suggest that phosphorylation of DELLA proteins potentiates the SLY1–DELLA interaction and that the sly1gar2-1 mutant protein binds both phosphorylated and nonphosphorylated forms of the DELLA proteins more strongly than does SLY1.
DISCUSSION
sly1gar2-1 was first identified as an extragenic mutant suppressor of gai phenotype (Wilson and Somerville, 1995). Subsequent analyses showed that sly1gar2-1 alters the GA dose–response relationship of a range of GA responses and thus affects a fundamental step in the regulation of plant growth and development by GA (Peng et al., 1997, 1999b). In this article, we have shown that sly1gar2-1 suppresses the dwarfism conferred by gai-GFP or GAI-GFP by reducing the levels of nuclear gai-GFP or GAI-GFP (also of GFP-RGA). In addition, we have shown that sly1gar2-1 causes further reductions in gai-GFP or GAI-GFP level in response to GA treatment and that GFP-RGA becomes undetectable earlier in GA-treated pRGA:GFP-RGA sly1gar2-1 plants than in controls. Previous experiments have shown that sly1gar2-1 increases the resistance of seeds to PAC-mediated inhibition of germination (Peng et al., 1999b), possibly by affecting the stability of RGL2 and/or RGL1 (responsible for GA regulation of Arabidopsis seed germination; Lee et al., 2002; Wen and Chang, 2002). Perhaps sly1gar2-1 reduces the nuclear levels of all five Arabidopsis DELLA proteins, thus causing a global reduction in DELLA levels, reducing DELLA-mediated restraint on multiple GA responses, and altering GA dose–response.
sly1gar2-1 is a dominant allele of SLY1 and encodes an altered product (sly1gar2-1) that has an altered function. Lack of SLY1 causes accumulation of RGA (McGinnis et al., 2003), GAI (this article), and perhaps other DELLA proteins. By contrast, either sly1gar2-1 or increased SLY1 availability (because of 35S:SLY1) reduce DELLA levels and suppress phenotypes conferred by stabilized DELLA proteins (such as are found in ga1-3 or gai mutant plants). Thus, SLY1 activity promotes growth because of release of DELLA restraint. Because increased SLY2 availability (because of 35S:SLY2) suppresses the phenotype conferred by lack of SLY1 (in sly1-10), it seems likely that SLY1 and SLY2 have similar activities.
Recent results have shown that GID2 is part of a rice SCFGID2 E3 ubiquitin ligase (Gomi et al., 2004). Here, we have shown that SLY1 is a subunit of an Arabidopsis SCFSLY1 E3 ubiquitin ligase complex that contains the highly conserved ASK1/2 and AtCUL1 subunits and promotes plant growth by targeting DELLA protein nuclear growth repressors for destruction in the proteasome. The likely redundancy of ASK1/2 function possibly explains why reduction in ASK1 levels resulted in delayed (rather than completely inhibited) GA-induced DELLA disappearance (Figures 5F and 5G). In addition, the fact that reduction in ASK3 or ASK4 levels had no discernible effect on plant phenotype or DELLA stability (data not shown) may also indicate redundancy of ASK function.
F-box proteins typically interact with SCF target substrates via a C-terminal domain (Vierstra, 2003). Thus, whereas the N-terminal F-box domain of SLY1 is required for incorporation of SLY1 into SCFSLY1, a C-terminal domain of SLY1 binds to the DELLA protein. These observations indicate that the DELLA proteins are substrates of the SCFSLY1 E3 ubiquitin ligase and that the C-terminal (LSL) domain of SLY1 provides specificity to the substrate interaction. An important but unanswered question concerns the nature of the specific domain of the DELLA proteins that interacts with the LSL domain of SLY1. One possible candidate for this domain is domain I of the DELLA proteins (Peng et al., 1999a; Lee et al., 2002). Mutations affecting domain I (such as gai; Peng et al., 1997) confer dwarfism, reduced GA response, and increased DELLA stability (Peng et al., 1999a; Dill et al., 2001; Chandler et al., 2002; Itoh et al., 2002). However, the fact that sly1gar2-1 was originally isolated as an extragenic suppressor of gai phenotype (Wilson and Somerville, 1995) and that SLY1 interacts with gai (this article) makes it unlikely that the LSL domain of SLY1 interacts with domain I of the DELLA proteins.
Typically, specific phosphorylation of target sequences is required for efficient F-box–mediated recruitment of target proteins to SCF E3 ubiquitin ligase complexes (Patton et al., 1998; Jackson and Eldridge, 2002; Busino et al., 2003). The rice DELLA protein SLR1 is phosphorylated in planta, and it has recently been shown that the GID2 F-box component of rice SCFGID2 interacts specifically with this phosphorylated form (Sasaki et al., 2003; Gomi et al., 2004). During recruitment to SCF E3 ubiquitin ligases, it is often amino acid residues from within the specific domain of the target protein with which the F-box protein interacts that are targeted for phosphorylation (e.g., Busino et al., 2003). Here, we have shown that the Arabidopsis DELLA–SLY1 interaction occurs with in vitro–translated DELLA proteins (Figure 6B; where specific phosphorylations are unlikely) and in yeast (Saccharomyces cerevisiae) (Figure 6A; where specific phosphorylations may or may not occur) and is reduced after dephosphorylation of plant-derived gai (Figure 7F). These observations suggest that there is an inherent (phosphorylation-independent) level of interaction between the DELLA and SLY1 (LSL domain) proteins and that the strength of this interaction is further increased by phosphorylation (perhaps phosphorylation of specific amino acid residues within the target domain of DELLA with which the LSL domain of SLY1 interacts).
SCF E3 ubiquitin ligase activity is commonly controlled via regulation at the level of the F-box subunit. For example, levels of the Skp1, Roc1/Rbx1, and Cul1 subunits of the cell-cycle regulatory SCFSkp2 complex do not change significantly during the cell cycle, whereas expression of Skp2 is cell-cycle regulated (Lisztwan et al., 1998). Inappropriate Skp2 expression changes cell-cycle regulation and is oncogenic (Gstaiger et al., 2001). Furthermore, the existence of mechanisms for regulating rapid turnover of F-box subunits (e.g., Zhou and Howley, 1998) highlights the importance of precise regulation of their availability. As shown here, SLY1 availability limits the capacity for release of the growth restraint imposed by stabilized DELLA proteins, presumably by limiting the levels of SCFSLY1 E3 ubiquitin ligase activity. Thus, modulation of SCFSLY1 activity via differential regulation of SLY1 expression/function is a possible mechanism for the regulation of plant growth via environmental or other factors.
We have also shown that sly1gar2-1 encodes a dominant mutant sly1gar2-1 protein that interacts more strongly than does the wild-type SLY1 protein with the DELLA protein target. Presumably, this increased strength of interaction results from an alteration in the three-dimensional structure of the SLY1 LSL domain (the domain that interacts with the DELLA protein and in which the amino acid substitution in sly1gar2-1 occurs) and explains the phenotypic properties of sly1gar2-1. We conclude that the increased capacity to bind DELLA target proteins that is characteristic of sly1gar2-1 increases the specific activity of the SCFSLY1 E3 ubiquitin ligase, thus enhancing the ubiquitylation of DELLA proteins, increasing rates of DELLA destruction and promoting the release of the DELLA protein restraint on plant growth. To our knowledge, mutations specifically enhancing the substrate binding of F-box proteins have not been described previously.
Although first identified as GA-signaling components (Peng et al., 1997; Silverstone et al., 1998), the DELLA proteins are now known to be fundamental to the regulation of growth via several plant developmental regulatory pathways and are thought to serve as integrators of multiple growth regulatory signaling inputs (Achard et al., 2003; Fu and Harberd, 2003). Thus, DELLA proteins restrain the cell proliferation and enlargement that underlies plant growth, GA promotes growth by opposing DELLA function, and other growth regulators (e.g., auxin and ethylene) modulate the GA–DELLA relationship. Our results point to the existence of at least two distinct mechanisms for influencing the efficiency of DELLA ubiquitylation via SCFSLY1. First, phosphorylation of the DELLA substrate increases the efficiency with which it is bound by SCFSLY1 complexes. Second, the level of SCFSLY1 E3 ubiquitin ligase activity may itself be regulated via change in SLY1 subunit availability or control of the strength with which SLY1 binds the DELLA protein substrate. These distinct mechanisms identify separate routes by which different signaling pathways may control plant growth via release of DELLA restraint.
METHODS
Plant Materials, Growth, and Genetic Analysis
Genotypes are written in italics (e.g., GAI), with mutant alleles represented in lower case (e.g., gai). Protein products are represented in nonitalic script (e.g., GAI and gai). Plants were grown as previously described (Peng et al., 1997; King et al., 2001). All plants shown were of homozygous genotype, determined by PCR analysis (e.g., King et al., 2001) or via tests of genetic segregation in subsequent generations (data not shown). Except for plants used in experiments shown in Figure 5E, all plants were of the Landsberg erecta laboratory strain or were mutant or transgenic derivatives thereof. Plants containing gar2-1 (Figures 1C, 1E, and 1F) or sly1-10 (Figures 3B and 3C) were F2 progeny of a cross between plants containing the respective mutant allele and the relevant (control) transgenic line. Plants containing 35S:RNAi-ASK1 (Figures 5F and 5G) were obtained via transformation of the (control) pRGA:GFP-RGA or pGAI:GAI-GFP lines. Assays of the effect of PAC on seed germination were performed as previously described (Peng et al., 1999b).
Generation of Transgenic Plants and Subsequent Analysis
Plant transformations and confocal microscopic analysis of GFP fluorescence in nuclei of primary root cells of GA-treated (and control) transgenic plants were as described previously (Fleck and Harberd, 2002; Fu and Harberd, 2003). GFP fluorescence levels were compared visually. Previous experiments have correlated differences in level of immunologically detectable GFP-RGA with visually detected differences in GFP-RGA fluorescence (Achard et al., 2003). Transgene constructs, further construct making, RT-PCR, and RNA gel blot analyses of transcripts were also as described previously (Fu et al., 2001; Fleck and Harberd, 2002). The DNA sequence of novel constructs (e.g., 35S:SLY1 and 35S:sly1gar2-1) was confirmed before plant transformation. For details of 35S:RNAi-ASK1, see the supplemental data online.
Yeast Two-Hybrid and β-Galactosidase Assays
The ASK relatedness tree was constructed using the GCG sequence analysis package (Phylip program). Sequences encoding GAI, SLY1, SLY2, and ASK1-ASK21 (or deletion derivatives thereof, see also supplementary data online) were cloned into the BamHI/NotI sites of the pLexA vector or the EcoRI/XhoI sites of pB42AD vector (Clontech, Palo Alto, CA). Vectors carrying candidate insertions (or empty vector) were transformed into the yeast EGY48 strain, and transformants plated on SD/-His/-Trp/-Ura medium. Interactions were further confirmed via replating of transformants on SD/-His/-Trp/-Leu/-Ura and SD/Raf/-Gal/-His/-Trp/-Leu/-Ura 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside media. In tests of β-galactosidase activities, overnight cultures of EGY48 carrying prey and bait (or control plasmids) were diluted 1:10 into fresh SD (-His-Trp-Ura) and placed in a shaking incubator at 250 rpm and 30°C for 3 h. Samples were assayed as described for the MATCHMAKER LexA two-hybrid system with o-nitrophenyl-β-d-galactopyranoside as a substrate (Clontech). Each assay was performed a minimum of three times.
In Vitro Protein Interaction Assays, Coimmunoprecipitation Assays, and Antibodies
SLY1, SLY1ΔLSL, and sly1gar2-1 coding sequences were cloned in frame into the BamHI/NotI sites of the pGEX-5X-1 GST fusion vector (Amersham, Buckinghamshire, UK), verified by DNA sequencing, and introduced into Escherichia coli strain BL21 (DE3) pLysS. Overnight cultures were diluted 100-fold and grown for 2 h at 37°C. Subsequently, isopropylthio-β-galactoside was added and growth continued for 2 h at 21°C. Soluble GST-fusion proteins were extracted and purified using a GST purification kit (BD Biosciences, Franklin Lakes, NJ). For in vitro translation, GAI, gai, and RGA coding sequences were cloned into the BamHI/NotI sites of the luciferase T7 vector (Promega, Madison, WI). Proteins were then in vitro translated using a TNT7-coupled reticulocyte lysate system (Promega) with 35S-Met labeling.
Coimmunoprecipitations were performed using the Seize primary immunoprecipitation kit (Pierce, Rockford, IL) as described previously (Xu et al., 2002). GST pull-down assays were performed with 6 μg of purified GST or GST-fusion proteins fixed to GST-Mag agarose beads (Novagen, Madison, WI) and incubated with in vitro–translated proteins, total proteins extracted from 7-d-old wild-type or mutant Arabidopsis seedlings, or from ethanol-treated alcA:gai plants (Ait-ali et al., 2003). Extracts from ethanol-treated alcA:gai plants were also incubated with protein A beads (Pierce) to which polyclonal anti-GAI antibodies previously had been cross-linked, resulting in beads coated with immunocaptured gai protein. Control beads were incubated with extract from untreated alcA:gai plants. Treatments with CIP in essence were performed as described previously (Sasaki et al., 2003).
Anti-SLY1 polyclonal antibodies were raised against a KRSTTDSDLAGDAHN peptide fragment, and anti-GAI polyclonal antibodies were raised against soluble E. coli–expressed GAI (both from Eurogenetics, Herstal, Belgium; in both cases further antibody purification was performed using the AminoLink Plus immobilization trial kit; Pierce). The anti-Myc monoclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), anti-GST polyclonal antibodies were from Amersham, anti-tubulin antibodies (YOL1/34) were from ABCOM (Cambridge, UK) and anti-GFP monoclonal antibodies were from Convance (Berkeley, CA).
Supplementary Material
Acknowledgments
The mapping of sly1gar2-1 was undertaken in collaboration with Jon Clarke and Jane Kirby from the Genotyping and Mapping Service of the John Innes Genome Lab (jicgenotyping@jicgenomelab.co.uk; http://www.jicgenomelab.co.uk). We thank Camille Steber for providing sly1-10, Tai-ping Sun for pRGA:GFP-RGA plants, and Yoshie Hanzawa for advice on yeast two-hybrid experiments. This work was supported by a Gatsby Foundation studentship to B.F., by Biotechnology and Biological Sciences Research Council Grant 208/P18610 to N.P.H., and by the Core Strategic Grant to the John Innes Centre from the Biotechnology and Biological Science Research Council.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nicholas P. Harberd (nicholas.harberd@bbsrc.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021386.
References
- Achard, P., Vriezen, W.H., Van Der Straeten, D., and Harberd, N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-ali, T., Rands, C., and Harberd, N.P. (2003). Flexible control of plant architecture and yield via switchable expression of Arabidopsis gai. Plant Biotechnol. J. 1, 337–343. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., and Thomas, M.R. (2002). Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416, 847–850. [DOI] [PubMed] [Google Scholar]
- Busino, L., Donzelli, M., Chiesa, M., Guardavaccaro, D., Ganoth, D., Dorello, N.V., Hershko, A., Pagano, M., and Draetta, G.F. (2003). Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 426, 87–91. [DOI] [PubMed] [Google Scholar]
- Carol, P., Peng, J., and Harberd, N.P. (1995). Isolation and preliminary characterization of gas1-1, a mutation causing partial suppression of the phenotype conferred by the gibberellin-insensitive (gai) mutation in Arabidopsis thaliana (L.) Heyhn. Planta 197, 414–417. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya: Molecular and physiological characterization. Plant Physiol. 129, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Davis, T.D., and Curry, E.A. (1991). Chemical regulation of vegetative growth. Crit. Rev. Plant Sci. 10, 151–188. [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–476. [DOI] [PubMed] [Google Scholar]
- Dill, A., Jung, H.S., and Sun, T.-p. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T.-p. (2001). Synergistic derepression of gibberellin signalling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck, B., and Harberd, N.P. (2002). Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J. 32, 935–947. [DOI] [PubMed] [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743. [DOI] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Ait-ali, T., Hynes, L.W., Ougham, H., Peng, J., and Harberd, N.P. (2002). Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Sudhakar, D., Peng, J., Richards, D.E., Christou, P., and Harberd, N.P. (2001). Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 13, 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, S.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi, K., Sasaki, A., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Kitano, H., and Matsuoka, M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37, 626–634. [DOI] [PubMed] [Google Scholar]
- Gstaiger, M., Jordan, R., Lim, M., Catzavelos, C., Mestan, J., Slingerland, J., and Krek, W. (2001). Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. USA 98, 5043–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P.M., White, R.G., Llewellyn, D.J., and Jacobsen, J.V. (2002). Gibberellin signalling in barley aleurone cells: Control of SLN1 and GAMYB expression. Plant Physiol. 129, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd, N.P. (2003). Relieving DELLA restraint. Science 299, 1853–1854. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Graebe, J.E. (1985). Inhibition of gibberellin biosynthesis by paclobutrazol in cell-free homogenates of Cucurbita maxima endosperm and Malus pumilla embryos. J. Plant Growth Regul. 4, 111–122. [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hooley, R. (1994). Gibberellins: Perception, transduction and responses. Plant Mol. Biol. 26, 1529–1555. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, P.K., and Eldridge, A.G. (2002). The SCF ubiquitin ligase: An extended look. Mol. Cell 9, 923–925. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.E., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen-Martinet, E.P., van Rijn, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65, 33–39. [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heyhn. Theor. Appl. Genet. 58, 257–263. [DOI] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan, J., Marti, A., Sutterlüty, H., Gstaiger, M., Wirbelauer, C., and Krek, W. (1998). Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): Evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 17, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Ni, W., Griffith, M.E., Huang, Z., Chang, C., Peng, W., Ma, H., and Xie, D. (2004). The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16, 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.-p., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (1993). Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999. a). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Peng, J., Richards, D.E., Moritz, T., Caño-Delgado, A., and Harberd, N.P. (1999. b). Extragenic suppressors of the Arabidopsis gai mutation alter the dose–response relationship of diverse gibberellin responses. Plant Physiol. 119, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.-p. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S.E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., Shirasu, K., and Deng, X.-W. (2003). The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nature Rev. Gen. 4, 948–958. [DOI] [PubMed] [Google Scholar]
- Swain, S.M., Muller, A.J., and Singh, D.P. (2004). The gar2 and rga alleles increase the growth of gibberellin-deficient pollen tubes in Arabidopsis. Plant Physiol. 134, 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Tseng, T.S., Thornton, T.M., Gopalraj, M., and Olszewski, N.E. (2002). SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 129, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. [DOI] [PubMed] [Google Scholar]
- Wen, C.K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.N., and Somerville, C.R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 108, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate responses in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D., Ni, W., Feng, B., Han, T., Petrasek, M.G., and Ma, H. (2003). Members of the Arabidopsis-SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol. 133, 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
- Zhou, P., and Howley, P.M. (1998). Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2, 571–580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.