Abstract
A common aspect of gene regulation in all developmental systems is the sustained repression of key regulatory genes in inappropriate spatial or temporal domains. To understand the mechanism of transcriptional repression of the floral homeotic gene AGAMOUS (AG), we identified two mutations in the BELLRINGER (BLR) gene based on a striking floral phenotype, in which homeotic transformations from sepals to carpels are found in flowers derived from old terminating shoots. Furthermore, this phenotype is drastically enhanced by growth at a high temperature and by combining blr with mutants of LEUNIG and SEUSS, two putative transcriptional corepressors of AG. We showed that the floral phenotype of blr mutants is caused by derepression of AG, suggesting that BLR functions as a transcription repressor. Because BLR encodes a BELL1-like (BELL) homeobox protein, direct binding of BLR to AG cis-regulatory elements was tested by gel-shift assays, and putative BLR binding motifs were identified. In addition, these putative BLR binding motifs were shown to be conserved in 17 of the 29 Brassicaceae species by phylogenetic footprinting. Because BELL homeobox proteins are a family of plant-specific transcription factors with 12 members in Arabidopsis thaliana, our findings will facilitate the identification of regulatory targets of other BELL proteins and help determine their biological functions. The age-dependent and high temperature–enhanced derepression of AG in blr mutants led us to propose that AG expression might be regulated by a thermal time-dependent molecular mechanism.
INTRODUCTION
An increasing number of signaling pathways in both plants and animals are found to function by overcoming transcription repression of target genes (Silverstone et al., 2001; Barolo and Posakony, 2002). Hence, transcription repression is emerging as a key regulatory strategy for both animal and plant development. However, our understanding of transcription repression in higher plants still lags behind animals and fungi. The floral homeotic gene AGAMOUS (AG) of Arabidopsis thaliana offers unique opportunities to study transcriptional repression in higher plants. AG encodes a MADS box transcription factor, and AG transcription is restricted to the inner two whorls of a flower and is turned on only in stage 3 floral meristems (Bowman et al., 1989; Yanofsky et al., 1990; Drews et al., 1991). Transcriptional regulation of AG has been under intensive study because of its pivotal importance in the specification of floral organ identity and floral meristem determinancy and its highly specific and precise expression pattern in floral meristems. Proper expression of AG requires sequences located within a 3-kb intron of AG (Sieburth and Meyerowitz, 1997; Busch et al., 1999; Deyholos and Sieburth, 2000), which is the target site of both positive and negative regulatory proteins. LEAFY (LFY), a novel transcription factor, and WUSCHEL (WUS), a novel homeodomain protein, were shown to bind to sequences within this intron and activate AG transcription (Busch et al., 1999; Lohmann et al., 2001). In addition, many negative regulators of AG have been identified, including LEUNIG (LUG), SEUSS (SEU), and APETALA2 (AP2) (Franks and Liu, 2001; Lohmann and Weigel, 2002). LUG and AP2 were shown to also act through this AG intronic sequence to exert their repressor activities (Sieburth and Meyerowitz, 1997; Bomblies et al., 1999; Busch et al., 1999; Deyholos and Sieburth, 2000). However, it is not known if any of the negative regulators directly or indirectly regulate AG transcription. Dissection of the 3-kb AG intron by reporter gene analyses or phylogenetic footprinting has been informative in pinpointing potential regions or conserved sites for repressor action (Bomblies et al., 1999; Busch et al., 1999; Deyholos and Sieburth, 2000; Hong et al., 2003).
In animals, homeodomain proteins are known to be crucial developmental regulators, especially important for pattern formation. The homeodomain contains three α helices with DNA binding activity (Gehring et al., 1994). BELL and KNOX proteins represent two major types of plant homeodomain proteins, with BELL1 and KNOTTED 1 (KN1) as founding members, respectively (Reiser et al., 1995, 2000; Chan et al., 1998). BELL proteins contain three highly conserved domains: SKY, BELL, and a single TALE-type homeodomain at the C terminus (Bellaoui et al., 2001). Specific BELL proteins were shown to interact or heterodimerize with specific KNOX proteins in both monocots and dicots (Bellaoui et al., 2001; Mueller et al., 2001; Smith et al., 2002). The cooperative interactions between KN1 and KIP (a maize [Zea mays] BELL protein) were reported to mediate high DNA binding affinity to the KN1 DNA binding motif TGACAG(G/C)T (Smith et al., 2002). Hence, BELL and KNOX homeodomain proteins may work together to regulate diverse developmental processes.
Recently, an Arabidopsis KNOX protein, BREVIPEDICELLUS (BP), was reported to directly bind to the KN1 DNA binding motif present in the promoters of several lignin biosynthesis genes, suggesting that BP regulates lignin pathway and cell wall deposition (Mele et al., 2003). A proposed partner of BP in Arabidopsis is a BELL gene with several names: BELLRINGER (BLR), PENNYWISE (PNY), REPLUMLESS (RPL), and LARSON (LSN) (Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003; X. Bao, R.G. Franks, and Z. Liu, unpublished results). For simplicity, we have chosen to use the name BLR in this article. blr mutants exhibited similar defects as bp mutants, including short internodes and slightly increased axillary branches. Furthermore, both genetic and physical interactions between BLR and BP were reported (Byrne et al., 2003; Smith and Hake, 2003), suggesting the significance of specific KNOX–BELL protein pairs in lignin deposition and phyllotaxy. In addition, genetic and physical interactions between BLR and another KNOX gene, SHOOTMERISTEMLESS (STM), were reported (Byrne et al., 2003), and BLR mRNA expression pattern in the embryonic and shoot apical meristem (SAM) closely resembles that of STM (Byrne et al., 2003; Smith and Hake, 2003), suggesting a role for BLR in shoot meristem development. The requirement of BLR in multiple developmental processes is revealed by yet another report, where blr was identified in a genetic screen for novel mutations affecting replum development in Arabidopsis fruits (Roeder et al., 2003). BLR was required to prevent ectopic expression of SHATTERPROOF (SHP1 and SHP2) genes in the replum region of the Arabidopsis fruit. Hence, BLR is emerging as a crucial developmental regulator with multiple roles in diverse tissues and organs.
Here, we report a novel function of BLR in flower and inflorescence meristem development. Specifically, BLR is required to prevent ectopic AG expression in the outer two whorls of a flower as well as in the reproductive SAM. Further, BLR was shown to act upon the 3-kb AG intronic sequence in vivo and to directly bind to the 3-kb intron of AG in vitro. The BLR DNA binding motif differs from the KN1 DNA binding motif, suggesting that BELL proteins may bind to a different DNA motif from KNOX proteins. The synergistic genetic interactions between blr and mutants of LUG and SEU suggest that BLR may be responsible for recruiting transcription corepressors, such as LUG and SEU, to the AG chromatin. The revelation of the underlying molecular mechanism for BLR function in floral and inflorescence meristems and the identification of the direct biological target of BLR reported here will help illuminate the mechanism of BLR function in other developmental processes.
RESULTS
blr-4 and blr-5 Mutants Form Terminal Carpelloid Flowers
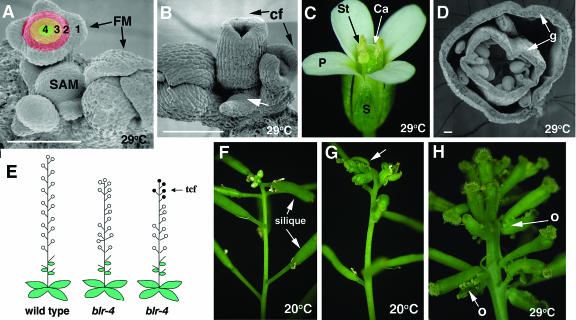
During the reproductive phase of Arabidopsis development, the SAM produces flowers in specific phyllotactic arrangement along the shoot axis (Figure 1A). Each floral meristem (FM) develops four types of floral organs (sepals, petals, stamens, and carpels) arranged in four concentric whorls (whorls 1 to 4) (Figures 1A and 1C). We have identified two ethyl methanesulfonate (EMS)-induced blr mutants, blr-4 and blr-5, which develop carpelloid flowers in the terminating shoots (Figures 1B and 1G). These carpelloid flowers develop carpels in place of sepals in whorl 1, and these whorl 1 carpels are often fused with each other to form a gynoecium that encloses the rest of the flower (Figures 1B, 1D, and 1H). Cross sections of a whorl 1 gynoecium revealed a complete absence of petals in whorl 2, a greatly reduced number of stamens in whorl 3, and a variable number of carpels in whorl 4 (Figure 1D). In addition, carpelloid bracts that subtend carpelloid flowers were often observed (Figure 1H). Strikingly, these carpelloid flowers are only formed in old terminating SAM (Figures 1E, 1G, and 1H). In other words, reproductive SAM first produces a large number of normal flowers and then switches to produce carpelloid flowers. Hence, we refer to this phenotype as terminal carpelloid flowers (tcf) (Figure 1E).
Figure 1.
Characterization of blr Mutant Phenotypes.
(A) A scanning electron microscopy photo of the tip of a wild-type shoot illustrating the SAM and FM. 1, 2, 3, and 4 indicate the four floral whorls.
(B) A scanning electron microscopy photo of a blr-4 SAM exhibiting the tcf phenotype. Carpelloid flowers (cf) are indicated. A smaller SAM (arrow) is evident.
(C) A wild-type flower showing the four floral organ types, sepal (S), petal (P), stamen (St), and carpel (Ca), in whorls 1, 2, 3, and 4, respectively.
(D) A cross section of a typical blr-4 carpelloid flower. Arrows indicate the formation of gyneocia (g) in whorls 1 and 4.
(E) A diagram summarizing the tcf and phyllotaxy defects of blr-4 mutants. Open circles indicate normal flowers, and closed circles indicate carpelloid flowers. Some blr-4 plants only exhibit phyllotaxy defects, whereas others exhibit both phyllotaxy and tcf.
(F) A wild-type terminating shoot. Siliques are remnant of normal flowers.
(G) A blr-4 plant exhibiting tcf (arrow). Note the abrupt transition from siliques to carpelloid flowers.
(H) A blr-4 terminating shoot exhibiting more carpelloid flowers at 29°C. Carpelloid bracts subtending each flower and bearing ovules (o) are indicated by arrows.
Bars in (A), (B), and (D) = 100 μm.
At 20°C, a standard Arabidopsis growth temperature, 36% of blr-4 and 14% blr-5 plants developed tcf (Table 1). In each of these tcf plants, only 5.6 to 7.9% of total flowers in a shoot were carpelloid (Table 1). When plants were grown at 29°C, a higher percentage of blr-4 (87%) and blr-5 (44%) plants developed tcf (Table 1). Furthermore, a higher percentage of flowers in a shoot are carpelloid (Figures 1G and 1H, Table 1). Hence, a higher temperature significantly enhances blr tcf phenotype. Like other previously reported blr alleles summarized in Table 2, both blr-4 and blr-5 exhibited a phyllotaxy defect, where flowers develop from seemingly random, instead of regularly spaced, positions on a stem (Figures 1E, 1H, and 3F). However, the phyllotaxy phenotype appeared the same for both blr-4 and blr-5 at 20°C and at 29°C.
Table 1.
Summary of blr tcf Phenotype at 20 and 29°C
| Genotype | Wild Type | blr-4/+ | blr-4 | blr-5 | blr-2 | blr-4/blr-2 | blr-5/blr-2 | blr-4/blr-5 | |
|---|---|---|---|---|---|---|---|---|---|
| 20°C | |||||||||
| tcf %a | 0 | 0 | 36 | 14 | 8 | 4 | 4 | 33 | |
| cf %b | 0 | 0 | 7.9 ± 3.5 | 5.6 ± 0.8 | 5.8 ± 0.2 | 6.9c | 6.8c | 7.2 ± 4.1 | |
| No. of flowersd | 44.0 ± 5.9 | 51.0 ± 6.3 | 45.0 ± 6.6 | 48.0 ± 7.9 | 52.0 ± 5.7 | 51.0 ± 7.3 | 48.0 ± 5.9 | 48.0 ± 6.1 | |
| N | 24 | 17 | 25 | 28 | 25 | 25 | 26 | 27 | |
| 29°C | |||||||||
| tcf %a | 0 | 0 | 87 | 44 | 33.3 | 31 | 30 | 70 | |
| cf %b | 0 | 0 | 30.0 ± 12.4 | 22.0 ± 11.7 | 11.0 ± 7.5 | 19.0 ± 12.9 | 11.0 ± 2.8 | 22.0 ± 11.8 | |
| No. of flowersd | 28.0 ± 3.7 | 33.0 ± 6.3 | 35.0 ± 5.8 | 34.0 ± 4.9 | 38.0 ± 5.8 | 35.0 ± 4.7 | 33.0 ± 5.8 | 36.0 ± 5.8 | |
| N | 23 | 23 | 23 | 32 | 24 | 29 | 23 | 30 |
Percentage of plants with carpelloid flowers in the primary shoot.
Average percentage of carpelloid flowers per inflorescence shoot. Only primary shoot is used in the analysis.
No standard deviation is available because of the small number of plants exhibiting tcf phenotype.
Average of the number of total flowers made by each terminated shoot. Only primary shoot is used in the analysis.
N, number of plants scored; ±, standard deviation.
Table 2.
Summary of Different Alleles in At5g02030
| Allele | Ecotype | Other Name | Mutation Description | Reference |
|---|---|---|---|---|
| blr-1 | Ler | GT7797 | Ds insertion in intron 1 RNA null | Byrne et al. (2003) |
| blr-2 | Ler | ET6411 | Ds insertion in exon1 RNA null | Byrne et al. (2003); this study |
| blr-3a | Col | SALK_040126 | T-DNA insertion in intron 1 RNA null | Byrne et al. (2003) |
| blr-4 | Ler | larson-1 (lsn-1) | EMS-induced missense mutation Pro to Leu at residue 356 | This study |
| blr-5 | Col | larson-2 (lsn-2) | EMS-induced missense mutation Arg to Leu at residue 364 | This study |
| Pny-57747 | Col | SALK_057747 | T-DNA insertion in exon 1 RNA null | Smith and Hake (2003) |
| Pny-40126a | Col | SALK_040126 | T-DNA insertion in intron 1 RNA null | Smith and Hake (2003) |
| rpl-1 | Ler | EMS-induced nonsense mutation W > STOP in exon 2 | Roeder et al. (2003) | |
| rpl-2a | Col | SALK_040126 | T-DNA insertion in intron 1 RNA null | Roeder et al. (2003) |
| rpl-3 | Wsb | 197A05 | FST collection at INRA Veraillesc | Roeder et al. (2003) |
Same allele with different names.
Ws, Wassilewskija.
FST, flanking sequence tags.
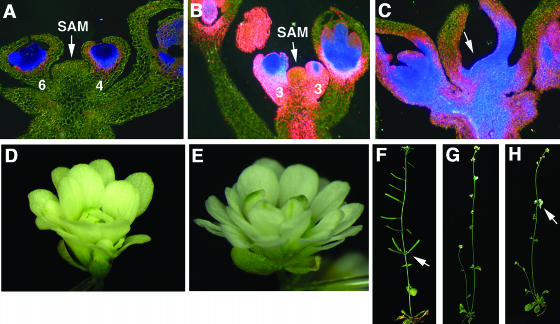
Figure 3.
Ectopic AG Expression Mediates tcf but Not Phyllotaxy Defects of blr-4.
All plants in this figure were grown at 29°C. Numbers indicate the stage of flowers. The AG:GUS reporter expression ([A] to [C]) is indicated by blue (strong) to pink (weak) staining.
(A) A longitudinal section of a blr-4 shoot before exhibiting tcf. The AG:GUS reporter expression reflects normal AG mRNA expression pattern in whorls 3 and 4 of flowers at stage 4 and stage 6.
(B) A blr-4 shoot at the beginning tcf phase. Ectopic AG expression is detected in all floral whorls, in SAM, and in the stem.
(C) A blr-4 shoot at the later phase of tcf. SAM (arrow) becomes flattened and difficult to identify. Ectopic AG is seen throughout the FM, SAM, and the stem.
(D) An ag-1 flower.
(E) An ag-1 blr-4 double mutant flower resembling ag-1 flowers.
(F) A blr-4 plant showing the abnormal phyllotaxy. A cluster of siliques (arrow) originates from the same position on a stem.
(G) An ag-1 plant with normal phyllotaxy.
(H) A blr-4 ag-1 plant showing clusters of flowers (arrow).
blr-4 and blr-5 Result from Missense Mutations in the Homeodomain of BLR
Using a map-based approach, we isolated BLR, which encodes a BELL protein on chromosome 5 (At5g02030). Like other BELL proteins, BLR also possesses three conserved domains, SKY, BELL, and a TALE-type homeodomain (Figure 2A). Both blr-4 and blr-5 are missense mutations affecting the homeodomain. blr-4 changes a conserved P in the N-terminal arm of the homeodomain, and blr-5 changes a conserved R in the first α helix of the homeodomain (Figure 2A). To identify the null phenotype of BLR in flower development, we obtained blr-2 from the Cold Spring Harbor Arabidopsis Genetrap database (Martienssen, 1998). blr-2 contains a Dissociation (Ds) transposon insertion 2 bp upstream from the ATG start codon (Figure 2A), resulting in an absence of BLR mRNA (Byrne et al., 2003). Thus, blr-2 is an RNA null. Surprisingly, blr-2 exhibited a much weaker tcf phenotype than blr-4 and blr-5 (Table 1). At 20°C, only 8% blr-2 plants exhibited tcf. At 29°C, blr-2 tcf phenotype was enhanced to 33%. The weaker tcf phenotype of blr-2 is unlikely because of its ecotype background because blr-2 is derived from the same (Landsberg erecta [Ler]) ecotype background as blr-4. Furthermore, blr-4 (Ler) and blr-5 (Columbia [Col]) are derived from different ecotypes but both caused a stronger tcf phenotype than blr-2 (Ler) (Table 1). Finally, blr-4(Ler)/blr-5(Col) and blr-2(Ler)/blr-5(Col) transheterozygous plants possess the same mix of ecotypes (Ler/Col) but differ significantly in the severity of the tcf phenotype (Table 1). Hence, it is the nature of the mutation (missense or null) rather than the ecotype background that determines the severity of the tcf phenotype in blr mutants.
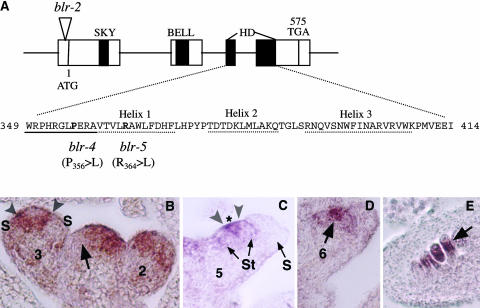
Figure 2.
Molecular Analyses of BLR, a BELL Class Homeodomain Protein.
(A) A diagram illustrating the BLR gene organization and the molecular lesions in blr-2, blr-4, and blr-5. The four exons are indicated by the four boxes connected by a line. The locations of the SKY, BELL, and homedomain (HD) are indicated by closed regions. The amino acid sequence in the homeodomain is shown. The three α helices and the N-terminal arm are underlined by dotted and solid lines, respectively. The amino acids affected by blr-4 and blr-5 are in bold. Numbers indicate the amino acid.
(B) to (E) In situ hybridizations of 8-μm longitudinal sections of inflorescences using a BLR 3′ probe. This probe detected no signal in blr-2 tissues. Numbers indicate the stage of each flower.
(B) In the shoot apex, an arrow indicates the exclusion of BLR mRNA from an emerging FM. In the stage 2 flower, BLR mRNA is present in the peripheral zone. In the stage 3 flower, BLR mRNA becomes excluded from the sepal primordia (S). A narrow band of BLR-expressing cells (marked by a pair of arrowheads) flanks the center of the stage 3 flower.
(C) A stage 5 flower showing a narrow BLR-expressing band (marked by a pair of arrowheads) situated between stamen (St) primordia and the remaining central zone marked by an asterisk.
(D) BLR is localized in the center (arrow) of a stage 6 flower.
(E) BLR mRNA is detected in the chalazal (arrow) domain of developing ovules.
Dynamic Expression of BLR in the Peripheral Zone of FM and SAM
Using in situ hybridization, BLR mRNA was detected both in reproductive SAM and in FM. Both SAM and FM are similarly organized into the central zone (C zone) and the peripheral zone (P zone) that surrounds the C zone (Baurle and Laux, 2003). Cells in the C zone divide infrequently and replenish their own stem cell population. By contrast, cells in the P zone divide rapidly and provide cells necessary for organ primordial initiation. In the SAM, BLR mRNA expression appears most abundant in the P zone but disappears temporarily from the emerging FMs (Figure 2B; Byrne et al., 2003; Smith and Hake, 2003). However, BLR mRNA reappears in late stage 1 and early stage 2 FMs (Figure 2B) and is quickly excluded from emerging sepal primordia in stage 3 flowers (Figure 2B). Soon after that, BLR-expressing cells encircle the undifferentiated center of the FM (Figure 2B). This BLR expression band moves progressively toward the center, as more inner whorl floral organ primordia emerge (Figure 2C). In stage 6 FM, BLR mRNA appears restricted to a small number of cells located in the center of the FM (Figure 2D). Later, BLR mRNA is detected in ovules (Figure 2E). Therefore, BLR expression in FM is dynamic and appears to differ from its expression in the SAM. However, this different expression pattern in FM versus SAM can be attributed to the fact that floral organs arise in whorls rather than in spirals and that FM is a determinant structure with a rapid disappearance of the C zone. The unifying theme of BLR expression in SAM and FM appears to be that BLR is expressed in the P zones of both SAM and FM and is quickly excluded from emerging FMs or floral organ primordia.
The tcf Phenotype of blr Is Mediated by Ectopic AG Expression
The development of stamen and carpels depends on the activity of AG, whose expression is normally restricted to whorls 3 and 4 of a flower. Mutants of AG repressors, such as ap2, lug, and seu, showed ectopic AG transcription in all floral whorls, resulting in the carpelloid sepals and reduced numbers of petals (Bowman et al., 1991; Drews et al., 1991; Liu and Meyerowitz, 1995; Franks et al., 2002). Therefore, we examined AG expression in blr-4 using a β-glucuronidase (GUS) reporter KB9 (Busch et al., 1999). In KB9 transgenic plants, the 3-kb AG intronic sequence directs GUS reporter expression in a pattern similar to endogenous AG mRNA. When blr-4 KB9 plants were grown at 29°C, AG:GUS was induced ectopically in the outer two whorls of flowers (Figures 3A and 3B). Most dramatically, AG:GUS was also detected in old SAM and in the stem (Figures 3B and 3C). Furthermore, the ag-1 mutation suppressed the tcf caused by blr-4. At 29°C, none of the blr-4 ag-1 plants showed tcf (Figure 4), and blr-4 ag-1 flowers developed normal sepals and petals (Figures 3D and 3E). These results indicate that the tcf phenotype of blr is mediated by ectopic AG activities. By contrast, the phyllotaxy defect of blr-4 was not suppressed in blr-4 ag-1 plants (Figures 3F to 3H). Thus, the phyllotaxy defect of blr-4 is not mediated by ectopic AG but by as yet unidentified genes.
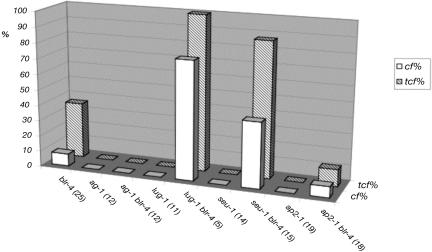
Figure 4.
blr-4 tcf Is Enhanced by lug and seu but Not by ap2.
tcf phenotype in various blr-4 double mutants. Hatched bars represent the percentage of plants with carpelloid flowers in the primary shoot. The open bars indicate the average percentage of carpelloid flowers per inflorescence shoot that exhibits a tcf phenotype. Only primary shoot is used in the analysis. Numbers in the parentheses indicate the number of plants scored. All plants were grown at 20°C except ag-1 and ag-1 blr-4 plants, which were grown at 29°C.
blr-4 tcf Is Enhanced by lug and seu but Not by ap2
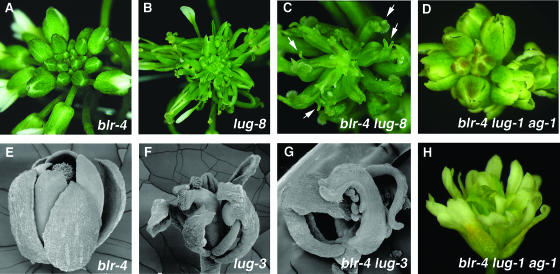
blr-4 was crossed into lug, seu, and ap2 mutants to test for genetic interactions. Whereas ap2-1 did not enhance blr-4 tcf, both lug-1 and seu-1 enhanced blr-4 tcf (Figure 4). At 20°C, whereas 36% blr-4 mutants showed tcf, 100% blr-4 lug-1 and 87% blr-4 seu-1 double mutants showed tcf (Figure 4). In addition, when only 7.9% blr-4 flowers per shoot were carpelloid, 76% blr-4 lug-1 and 42% blr-4 seu-1 flowers per shoot were carpelloid (Figure 4). At 20°C, young blr-4 inflorescences do not exhibit any tcf and are similar to the wild type (Figure 5A). Similarly, lug-8, a weak lug mutant, only produces narrow sepals and a slightly reduced number of petals in flowers (Figure 5B). However, blr-4 lug-8 double mutant inflorescences at the same developmental stage exhibit an enhanced phenotype, where almost all flowers are carpelloid flowers (Figure 5C). The carpelloid organs in blr-4 lug-8 double mutants are often topped with horns that are characteristic of lug carpels (Liu and Meyerowitz, 1995). Synergistic genetic interactions were also observed between blr-4 and strong lug alleles, including lug-3 (Figures 5E to 5G). However, ag-1 suppressed the carpelloid flower phenotype (Figures 5D and 5H) but not the phyllotaxy defect (data not shown) in ag-1 blr-4 lug-1 triple mutants. Therefore, the enhanced floral phenotype in blr-4 lug is mediated by an enhanced ectopic AG activity.
Figure 5.
blr-4 Exhibits Synergistic Genetic Interactions with lug.
All plants in this figure were grown at 20°C.
(A) A blr-4 young inflorescence. At this early stage and at 20°C, most plants do not exhibit any tcf and are similar to wild-type inflorescences.
(B) A lug-8 inflorescence with flowers exhibiting narrow sepals and reduced numbers of petals.
(C) A blr-4 lug-8 double mutant inflorescence showing a dramatically enhanced floral phenotype. Most carpelloid organs are topped with horns (arrows).
(D) A blr-4 lug-1 ag-1 triple mutant inflorescence showing no sign of tcf.
(E) A blr-4 flower developed from a young inflorescence at 20°C. It resembles wild-type flowers.
(F) A lug-3 flower showing partially carpelloid sepals and an absence of petals.
(G) A blr-4 lug-3 double mutant flower showing carpelloid whorl 1 organs.
(H) An ag-1 blr-4 lug-1 triple mutant flower resembling ag-1 flowers. However, floral organs of the triple mutant are narrower than those of ag-1.
Direct Binding of BLR to AG in Vitro
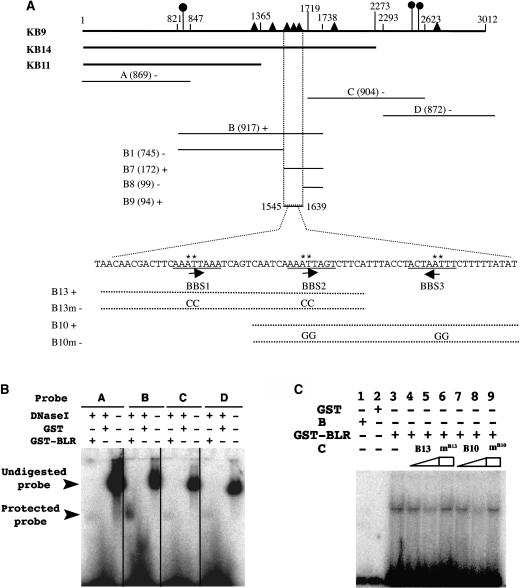
To test if BLR directly binds to the AG intronic sequences, electrophoretic mobility shift assay (EMSA) was used to test binding of BLR to four DNA fragments, A, B, C, and D, spanning the 3-kb AG intron (Figure 6A). GST-BLR was able to protect fragment B from DNase I digestion (Figure 6B), suggesting that fragment B may contain BLR binding sites. Several smaller DNA fragments within fragment B were tested, and a 94-bp fragment (B9) was the smallest fragment still capable of binding to GST-BRL (Figure 6A). This B9 fragment contains three 8-bp repeats spaced 19 to 20 bp apart with an ATTA core sequence in each repeat. The ATTA core was previously shown to be selectively bound by homeodomains encoded by distantly related genes such as ftz and Matα2 (Gehring et al., 1994). These three 8-bp repeats were named BLR binding sites (BBS1-3). Various cold oligonucleotides were used to compete with a radioactive fragment (B7) containing BBS1-3. Whereas a single BBS repeat did not compete with the B7 probe (data not shown), other oligos, such as B13 and B10, that contain BBS1-2 and BBS2-3, respectively, competed readily with the B7 probe (Figure 6C; data not shown). Furthermore, B13 and B10 oligos carrying mutations in either one or both of the two BBS repeats exhibited a significantly reduced ability to compete with the B7 probe (Figure 6C; data not shown). Hence, at least two adjacent BBS repeats are required for high affinity binding by BLR.
Figure 6.
BLR Directly Binds to the 3-kb AG Intron.
(A) A diagram summarizing the mobility-shift assay results. The ∼3-kb AG HindIII fragment used in the KB9 reporter roughly coincides with the AG second intron. Numbers above the KB9 line indicate the nucleotide sequence with the 5′ HindIII site designated 1. AG intron fragments used in KB14 and KB11 reporters (Busch et al., 1999) are indicated. Closed circles indicate the location of previously identified LFY/WUS binding sites (Hong et al., 2003). Closed triangles indicate the location of BBS. The A, B, C, D, B1, B7, B8, and B9 fragments served as probes. The size of each fragment is indicated by a number within parentheses. Plus and minus signs indicate positive and negative mobility shifts, respectively. BBS1-3 is underlined beneath the sequence within the B9 fragment. The orientation of each BBS is indicated by an arrow. The sequence of B13 and B10 oligos are indicated by dotted lines. B13m and B10m are mutant oligos with mutations introduced in the ATTA core sequence. The asterisk marks the specific nucleotide changed in B13m and B10m.
(B) A representative gel-shift using the A, B, C, and D DNA fragments as probes. After incubating the corresponding hot probe with GST or GST-BLR proteins, DNase I was added to digest naked DNA. Only fragment B showed a band protected from DNase I digestion by GST-BLR.
(C) A gel-shift assay using 32P-labeled B7 DNA as a probe and cold B13 or B10 oligos as competitors. B, protein extracts that do not contain GST-BLR; C, cold competitors. The presence or absence of protein extracts or oligo competitors is indicated by plus or minus signs above each lane. Lanes 4, 5, 7, and 8 show the effect of increasing amounts of cold B13 and B10 oligos, respectively, at 0.02 pmol (lanes 4 and 7) and 2 pmol (lanes 5 and 8). mB13 (lane 6) and mB10 (lane 9) represent mutant B13 and B10 oligos as cold competitors at 2 pmol. GST alone (lane 2) and protein extracts from E. coli that did not express GST-BLR (lane 1) served as negative controls.
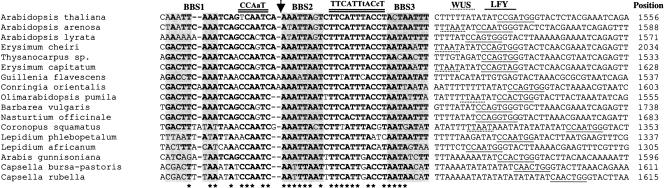
If BBS1 to BBS3 are functionally important, they are likely to be evolutionarily conserved. The already extensive phylogenetic analyses of the AG second intron and the availability of the AG second intron sequences from 29 Brassicaceae species (including Arabidopsis) and from 12 non-Brassicaseae species (Hong et al., 2003) served as the framework for our analyses. Using the sliding window analyses, we found extensive sequence conservation in the AG intronic region containing BBS1-3 in 17 of the 29 Brassicaceae species (Figure 7). However, similar conservation was not found in the other 12 Brassicaceae species nor in any of the non-Brassicaceae species. Several conserved features of the BBS1-3 region are significant. First, the 20-bp spacing between BBS2 and BBS3 is conserved in all 17 species. The spacing between BBS1 and BBS2 varies from 18 to 20 bp. Second, whereas BBS2 and BBS3 are highly conserved, BBS1 is not. BBS1 in Arabidopsis results from deleting a C from a more conserved AAATTCAAA motif. Third, the sequences located between BBS1 and BBS2 and between BBS2 and BBS3 are also highly conserved (Figure 7). Fourth, 3′ to the BBS3 are AT-rich sequences of variable length (8 to 25 bp). In 16 of the 17 species, the AT-rich sequences are immediately followed by CCANTGG, the binding site for LFY (Figure 7). In five of the 17 species, the LFY binding sites are preceeded by a TTAAT, the consensus binding site for WUS, a homeodomain protein known to partner with LFY (Lohmann et al., 2001). Finally, the relative position of BBS1-3 within the intron in each of the 17 species remains relatively constant, ranging from 1305 to 2034 bp (Figure 7). These conserved features strongly support the functional significance of the BBS1-3 region identified by the EMSA.
Figure 7.
DNA Sequence Alignment in the BBS1-3 Region of the AG Intron in 17 Brassicaceae Species.
Conserved nucleotides are in bold, and the asterisk marks the most conserved positions. BBS1, BBS2, and BBS3 are shaded. The CCAAT box and the conserved TTCATTtACc motif are underlined twice. Putative WUS and LFY binding sites are underlined with dotted and solid lines, respectively. The number at the end of each sequence indicates the position of the first nucleotide in BBS2 (indicated by an arrow) relative to the 5′ end of the AG intron (44 bp downstream of the 5′ HindIII site shown in Figure 6).
DISCUSSION
BLR Directly and Negatively Regulates AG Expression
In animals, homeodomain proteins play fundamental roles in development and evolution, in particular in the specification of body plan, pattern formation, and cell fate determination. Our results showed that the plant homeodomain protein BLR also plays fundamental roles in cell fate specification by regulating proper temporal and spatial expression pattern of the floral homeotic gene AG. BELL1, the founding member of the BELL family, has previously been shown to regulate ovule development and was proposed to regulate AG expression in ovules (Modrusan et al., 1994; Ray et al., 1994; Reiser et al., 1995). However, the AG mRNA expression pattern was not affected in ovules of bell1 mutants, excluding the possibility of BELL1 as a direct regulator of AG (Reiser et al., 1995). Our demonstration of BLR as a transcriptional repressor of AG parallels with the study of BLR as a transcription repressor of SHP (Roeder et al., 2003). Because AG and SHP both encode MADS box transcription factors (Yanofsky et al., 1990; Liljegren et al., 2000), it will be interesting to determine if BLR also directly binds to the SHP cis-elements and if repression of MADS box transcription factors is a common role for BLR as well as other BELL proteins. The identification of the BLR binding sequence motif reported here will facilitate the identification of other direct targets of BLR as well as targets of other BELL proteins and increase our understanding of their biological functions.
In addition to BBS1-3 identified by the EMSA analyses, four other sequences identical to BBS are found within the 3-kb AG intron (Figure 6A). However, unlike BBS1-3, these four sequences are not clustered together and may thus have a lower binding affinity for BLR because of a lack of cooperativeness. BBS does not share any sequence similarity with the KN1 DNA binding motif. Examination of the 3-kb AG intron identifies a putative KN1 binding site (with one mismatch from KN1 binding consensus) located ∼121 bp 5′ to the BBS1. It remains to be seen if this putative KN1 binding site could be bound by a KNOX-type partner of BLR.
The functional significance of BBS1-3 is supported by previous reporter analyses. Specifically, the smallest AG intronic sequences (KB14, KB17, and PMD983) that confer wild-type AG expression patterns all contain BBS1-3 (Busch et al., 1999; Deyholos and Sieburth, 2000). Furthermore, KB11, whose GUS expression is driven by the 5′ 1355-bp AG intronic sequence that lacks BBS1-3 (Figure 6A), showed GUS expression ectopically throughout early flowers, shoot apexes, and in the stem (Busch et al., 1999).
Using the method of phylogenetic footprinting, we found strong sequence conservations in the AG intronic region spanning BBS1-3 among 17 of the 29 Brassicaceae species. Based on the phylogenetic relatedness of the 29 Brassicaceae species determined by Hong et al. (2003), these 17 species do not show specific phylogenetic grouping, but all four Arabidopsis species are among those that possess the conserved BBS1-3 region. Second, our analyses differed from Hong et al. (2003) in that we did not ask for motif conservation across all 29 Brassicaceae species and that we used different parameters in the sliding window analyses. Thus, the BBS1 to BBS3 are not among the six conserved motifs previously identified by Hong et al. (2003). Third, the high level of conservation both in the sequence and in the spacing between BBS1, BBS2, and BBS3 are striking. The 19- to 20-bp spacing between each BBS correlates with two DNA helical turns and may reflect a stringent spatial arrangement for protein–DNA and protein–protein interactions. In addition, there appears to be a conserved pattern of multiple binding site clustering. Specifically, the CCAAT box located between BBS1 and BBS2 could serve as the recognition site of the NF-Y factors belonging to multimember families in Arabidopsis (Gusmaroli et al., 2002). Two highly conserved CCAAT boxes were previously identified near the 3′ end of the AG second intron, and both were shown to be required for maintaining AG expression (Hong et al., 2003). Our identification of a third conserved CCAAT box in the BBS1-3 region further supports a role of NF-Y factors in AG regulation and possible functional interactions between NF-Y and BLR. TTCATTtACcT is a highly conserved motif situated between BBS2 and BBS3 (Figure 7). By searching Match (http://www.gene-regulation.com), we found that aTTTAC is the binding site core consensus for Oct-1, a ubiquitously expressed mammalian POU homeodomain protein (reviewed in Phillips and Luisi, 2000). Because the POU domain proteins are absent from Arabidopsis, the identities of the factors that bind to this motif remain to be elucidated. Finally, a LFY binding site or a WUS/LFY double binding site can be almost certainly located 3′ to the BBS3. Future reporter analyses using various mutated motifs in the BBS1-3 region will help determine the significance of such a conservation in the number, type, order, and relative position of these regulatory binding sites.
BLR May Act as a General Repressor of AG in SAM and FM
If BLR function is required in the outer two whorls of a flower to repress AG, why is BLR mRNA transiently expressed in all four floral whorls? Although we do not know where BLR protein is located, it is likely that BLR protein is also transiently present in all floral whorls and hence may repress AG expression in all four whorls. Perhaps, rather than specifically repressing AG in the outer two whorls, BLR may repress AG in all four whorls. Because lug and seu both genetically enhanced blr-4 tcf (Figures 4 and 5), BLR may act as the DNA binding partner of LUG and SEU, both of which encode putative transcriptional corepressors with no DNA binding motifs, and both are widely expressed (Conner and Liu, 2000; Franks et al., 2002). Therefore, inner whorl–specific AG expression might come from both inner whorl–specific activators, such as WUS, that antagonize BLR activity (Lohmann et al., 2001) and the outer whorl–specific AG repressors, such as AP2, whose translation was recently shown to be inhibited by an inner whorl–specific microRNA (Aukerman and Sakai, 2003; Chen, 2004).
A striking phenotype of blr is the ectopic expression of AG in the SAM. This phenotype was not observed for lug, seu, or ap2-1 single mutants nor for lug seu double mutants, where ectopic AG and carpelloid floral organs were only confined to the FM (Figure 4; Liu and Meyerowitz, 1995; Franks et al., 2002). However, mutants of the strong ap2 allele ap2-2 also exhibited a terminal carpelloid phenotype indicative of ectopic AG in the SAM (X. Bao and Z. Liu, unpublished data). At least two alternative hypotheses could explain the different ability between blr and lug/seu in causing ectopic AG in the SAM. First, LUG/SEU are floral-specific repressors, but BLR is a general repressor of AG. For example, BLR may bind to AG intronic sequences and recruit SAM-specific or FM-specific repressors. Alternatively, this unique phenotype of blr may indicate BLR as an initiating factor for AG repression, whereas LUG and SEU may be involved in maintaining AG in a repressed state. This latter hypothesis draws on studies of transcriptional repression of engrailed (en) in Drosophila melanogaster (Wheeler et al., 2002). Whereas Runt and Tramtrack, two DNA binding transcription factors, cooperate to initiate transcription repression of en at the blastoderm stage, maintenance of en repression after the blastoderm stage requires the recruitment of Groucho, a LUG homolog, by Runt. The maintenance of en repression by Groucho continues into germband extension stage even when Runt and Tramtrack are no longer present in the cell. This latter hypothesis is consistent with our observation that BLR mRNA is only transiently present in each floral organ primordia.
blr-4 and blr-5 May Represent Recessive Antimorphic Alleles
One obvious difference between blr-4/blr-5 and previously reported alleles is the nature of blr-4 and blr-5 mutations. All other previously reported alleles for this gene were caused either by Ds transposon/T-DNA insertions or by a nonsense mutation (rpl-1) (Table 2; Byrne et al., 2003; Roeder et al., 2003; Smith and Hake, 2003). The lack of detectable RNA transcripts in the Ds/T-DNA insertion alleles (Table 2) and the truncation of BLR protein in rpl-1 suggest that all previously reported alleles are null or near-null alleles. Although these null alleles were reported to cause defects in phyllotaxy (Byrne et al., 2003; Smith and Hake, 2003) and replum development (Roeder et al., 2003), they were not reported to cause tcf. It is highly likely that these blr/pny/rpl null or near-null alleles may only exhibit a weak tcf phenotype that is difficult to notice under normal growth conditions.
Why do blr-4 and blr-5 cause a more severe tcf phenotype than the null alleles? We propose that the blr-4 and blr-5 missense alleles are recessive antimorphic alleles. BLR-4 and BLR-5 mutant proteins not only lose their normal functions but also interfere with the function of redundant family members by competing for the same partner in a protein complex or competing for the same DNA binding sites. Because blr/pny/rpl null alleles could be partially compensated by redundant family members but blr-4 and blr-5 could not, blr-4 and blr-5 may cause a more severe tcf phenotype than the null alleles. Because blr-4 and blr-5 are recessive (Table 1), the dosage of BLR-4 and BLR-5 mutant proteins may determine the severity of tcf. This is consistent with the observation that plants transheterozygous for blr-4/blr-2 and blr-5/blr-2 exhibited a tcf phenotype weaker than blr-4 and blr-5 homozygous plants (Table 1). Our study indicates that antimorphic alleles are likely to be particularly informative on gene function in cases where the phenotype is not revealed or only partially revealed by null alleles because of genetic redundancy.
AG Expression May Be Regulated by the Thermal Clock
Two intriguing aspects of the blr tcf phenotype are the temperature sensitivity (ts) and the old age–dependent expression of the mutant phenotype. Temperature sensitivity is often attributed to the formation of temperature-sensitive mutant proteins or temperature-sensitive processes. Because blr-4 and blr-5 missense mutations and blr-2 RNA null all exhibited temperature sensitivity, it is unlikely that the ts phenotype is caused by ts mutant proteins. Alternatively, an inhibitory activity for AG expression weakens at higher temperatures. This can be accomplished either by a decrease in the expression or activity of AG repressors, such as a redundant BELL protein, or by a change in AG chromatin.
Second, why are carpelloid flowers only derived from old SAM? One attractive explanation that integrates both the ts and the old age–dependent property of blr tcf is that AG transcription is modulated by thermal time, which can be simply put as the cumulative amount of heat to which a plant is exposed (Poethig, 2003). As thermal time increases (either because of old age or growth at a high temperature), AG is more easily derepressed. This hypothesis is supported by the observation that ap2 mutants exhibited a more severe carpelloid transformation at a higher temperature (Bowman et al., 1991), and mutants of several AG repressors, including lug, seu, and ap2, also exhibited a more complete homeotic transformation from sepals to carpels in flowers developed from old SAM than from young SAM (Bowman et al., 1991; Liu and Meyerowitz, 1995; R.G. Franks and Z. Liu, unpublished results). Whereas the acropetal increase in the severity of carpelloidy in lug, seu, and ap2 are gradual, the change from morphologically normal to severely carpelloid flowers in blr mutants is rather abrupt, perhaps reflecting a threshold response to the accumulating changes in AG regulation. Nevertheless, the underlying molecular mechanism for the age- and high temperature–enhanced AG derepression is likely the same for all AG repressors. Future experiments will be necessary to test these hypotheses. Our analyses suggest that one way the plant SAM keeps track of its age or temporal identity is through thermal time-dependent modulation of gene expression.
METHODS
Genetic Analyses
blr-4 was isolated from an EMS-mutagenized population in Ler ecotype (Levin et al., 1998). blr-5 was isolated by the method of targeting induced local lesions in genomes (TILLING) in ecotype Col (McCallum et al., 2000). This Col carries an erecta mutation. blr-2, in Ler ecotype, was identified by screening the Cold Spring Harbor Arabidopsis Genetrap database (http://genetrap.cshl.org) (Martienssen, 1998). Plants were grown at either 20 or 29°C under 16 h light. Because of poor germination rate at 29°C, plants were first germinated at 20°C and then shifted to 29°C after germination.
The blr-4 mutation resulted in a new TaqI site. Primer pair 5′-GCCACAACAACAAGATCCAACA-3′ and 5′- AGAAATGATCGAAGAGCCAAGC-3′ was used to PCR-genotype blr-4. blr-4/blr-2 and blr-4/blr-5 transheterozygotes were verified using this blr-4 cleaved-amplified polymorphic sequence marker in the F1 progeny from crosses using blr-4 pollen. blr-5/blr-2 transheterozygotes were similarly verified using primer pair 5′-ACCCGGGACTTTTCACTTTT-3′ and 5′-TACGATAACGGTCGGTACGC-3′ that amplifies the 5′ junction of the blr-2 Ds insertion. In constructing blr lug, blr seu, and blr ap2 double mutants, lug-1, lug-8, seu-1, and ap2-1 pollen was used to fertilize blr-4 stigma. Seeds were collected from F2 blr-4 plants whose genotype was verified by PCR. Double mutants were observed in F3 plants. For ag-1 blr-4 lug-1 triple and ag-1 blr-4 double mutants, pollen from an ag-1 lug-1/++ plant was used to fertilize blr-4 stigma. Individual F2 blr-4 plants were identified by PCR. Those blr-4 plants that segregated ag-1 or ag-1 lug-1 in F3 were analyzed further.
To examine the AG:GUS reporter expression in blr-4, wild-type plants harboring the KB9 construct was crossed into blr-4 stigma. F2 plants homozygous for blr-4 were identified, and their flowers were stained for GUS activity. GUS staining, tissue fixation, section, and visualization were based on a previously described protocol (Blazquez et al., 1997).
Phylogenetic Footprinting
AG intronic sequences from the 29 Brassicaceae and 12 non-Brassicaceae species were published by Hong et al. (2003). These sequences were analyzed with sliding window with a window size of 24 bp at the steps of one character, which led to the identification of several highly conserved motifs in the AG intron, including a segment of the BBS region. Manual adjustment of the BBS1-3 alignment led to the result shown in Figure 7.
Microscopic Analyses
Scanning electron microscopy samples were collected, fixed, and coated as previously described (Bowman et al., 1989, 1991). Samples were examined on an AMRAY 1000A scanning electron microscope (AMRAY, Bedford, MA). Images were captured on a Polaroid camera. Whole-mount floral photomicrographs were taken through a Zeiss Stemi SV6 dissecting microscope (Zeiss, Thronwood, NY). In situ hybridization images were photographed under a Nikon Eclipse E600W microscope (Nikon, Melville, NY) with Nomarski optics equipped with a digital still camera.
Map-Based Cloning
A mapping population was constructed by crossing blr-4 lug-2 (Ler) into wild-type (Col). Eighty-nine F2 blr-4 lug-2 double mutants were assayed with various PCR-based markers on the top of chromosome 5. BLR was mapped to the BAC clone T7H20 within a region between 14K and 60K. Only one gene in this region (At5g02030) encodes a putative transcription factor. An 8.99-kb genomic DNA, covering the At5g02030 and 3.4-kb/2.4-kb upstream/downstream sequences, was introduced into blr-4 mutants. Six out of seventeen T1 transgenic blr-4 plants showed wild-type phenotypes in both phyllotaxy and floral morphology. Cosegregation of the transgene with the rescued phenotype was confirmed in the progeny of these six rescued plants.
Analyses of BLR Transcripts
A BLR full-length cDNA clone was isolated from the Weigel Ler flower cDNA library. In situ experiments were based on a protocol from http://carnegiedpb.stanford.edu/research/barton/in_situ_protocol.html. For generating antisense BLR probe, a NsiI DNA fragment containing full-length BLR cDNA was inserted into the PstI site of pBluescript II SK+ vector to create the pRSL(+) clone. XbaI was used to linearize pRSL(+), and an 819-bp antisense RNA corresponding the 3′ end of the BLR gene (from XbaI to NsiI) was transcribed using the T7 promoter and the DIG RNA labeling kit (Roche, Indianapolis, IN). The probe was used directly without hydrolysis.
Testing BLR Binding to AG cis-Elements
Full-length BLR cDNA was excised from pRSL(+) using BamHI/EcoRI and inserted into the pGEX-3X (Novagen, Madison, WI) vector to generate a GST-BLR fusion. BugBuster protein extraction reagent (Novagen) and protein refolding kit (Novagen) were used to purify GST-BLR from inclusion bodies. Various AG intronic sequences were PCR amplified and PCR labeled with [α-32P]dATP. Two micrograms of protein extracted from inclusion bodies (containing ∼20 ng of GST-BLR) was mixed with 1500 cpm probe in 1× gel shift binding buffer (Promega, Madison, WI). The mixture was incubated on ice for 30 min and ran on a 6% DNA retardation gel (Invitrogen, Carlsbad, CA) in 0.5× TBE buffer (Tris-borate/EDTA) for 80 min at 160 V. When large DNA fragments were used as probes (Figure 6B), 0.0002 units of DNase I were added into each binding reaction and incubated for 15 min at room temperature before the reactions were loaded onto the 6% DNA retardation gel.
Sequences for the 17 Brassicaceae species shown in Figure 7 are under accession numbers AL161549/AL021711, AY253237, AY253251, AY253258, AY253255, AY253248, AY253260, AY253252, AY253243, AY253235, AY253250, AY253246, AY253239, AY253238, AY253244, AY253262, and AY253263.
Acknowledgments
We thank the TILLING facility and ABRC for blr-5, R. Martienssen for blr-2, D. Weigel for KB9, H. Sze for equipment, T. Mougel for microscopy assistance, M. Rivarola for help with in situ experiments, Y. Yan for assisting phylogenetic footprinting, and E. Baehrecke, S. Mount, and members of the Liu lab for comments on the manuscript. This work has been supported by USDA Grants 98-35304-6714 and 2001-35304-10926 and by National Science Foundation Grant IBN 0212847 to Z.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhongchi Liu (zl17@umail.umd.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021147.
References
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo, S., and Posakony, J.W. (2002). Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Laux, T. (2003). Apical meristems: The plant's fountain of youth. Bioessays 25, 961–970. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez, M.A., Soowal, L.N., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., Dagenais, N., and Weigel, D. (1999). Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 216, 260–264. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Busch, M.A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Groover, A.T., Fontana, J.R., and Martienssen, R.A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130, 3941–3950. [DOI] [PubMed] [Google Scholar]
- Chan, R.L., Gago, G.M., Palena, C.M., and Gonzalez, D.H. (1998). Homeoboxes in plant development. Biochim. Biophys. Acta 1442, 1–19. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, J., and Liu, Z. (2000). LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97, 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos, M.K., and Sieburth, L.E. (2000). Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12, 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002. [DOI] [PubMed] [Google Scholar]
- Franks, R.G., and Liu, Z. (2001). Floral homeotic gene regulation. Hortic. Rev. 27, 41–77. [Google Scholar]
- Franks, R.G., Wang, C., Levin, J.Z., and Liu, Z. (2002). SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129, 253–263. [DOI] [PubMed] [Google Scholar]
- Gehring, W.J., Affolter, M., and Burglin, T. (1994). Homeodomain proteins. Annu. Rev. Biochem. 63, 487–526. [DOI] [PubMed] [Google Scholar]
- Gusmaroli, G., Tonelli, C., and Mantovani, R. (2002). Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283, 41–48. [DOI] [PubMed] [Google Scholar]
- Hong, R.L., Hamaguchi, L., Busch, M.A., and Weigel, D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15, 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, J.Z., Fletcher, F.C., Chen, X., and Meyerowitz, E.M. (1998). A genetic screen for modifiers of UFO meristem activity identifies three novel FUSED FLORAL ORGANS genes required for early flower development in Arabidopsis. Genetics 149, 579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Liu, Z., and Meyerowitz, E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975–991. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., and Weigel, D. (2002). Building beauty: The genetic control of floral patterning. Dev. Cell 2, 135–142. [DOI] [PubMed] [Google Scholar]
- Martienssen, R.A. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., and Hake, S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17, 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27, 13–23. [DOI] [PubMed] [Google Scholar]
- Phillips, K., and Luisi, B. (2000). The virtuoso of versatility: POU proteins that flex to fit. J. Mol. Biol. 302, 1023–1039. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301, 334–336. [DOI] [PubMed] [Google Scholar]
- Ray, A., Robinson-Beers, K., Ray, S., Baker, S.C., Lang, J.D., Preuss, D., Milligan, S.B., and Gasser, C.S. (1994). Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl. Acad. Sci. USA 91, 5761–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83, 735–742. [DOI] [PubMed] [Google Scholar]
- Reiser, L., Sanchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42, 151–166. [PubMed] [Google Scholar]
- Roeder, A.H., Ferrandiz, C., and Yanofsky, M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99, 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M., and Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, J.C., VanderZwan, C., Xu, X., Swantek, D., Tracey, W.D., and Gergen, J.P. (2002). Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nat. Genet. 32, 206–210. [DOI] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–40. [DOI] [PubMed] [Google Scholar]