Abstract
Several genes belonging to the MADS box transcription factor family have been shown to be involved in the transition from vegetative to reproductive growth. The Petunia hybrida MADS box gene UNSHAVEN (UNS) shares sequence similarity with the Arabidopsis thaliana flowering gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, is expressed in vegetative tissues, and is downregulated upon floral initiation and the formation of floral meristems. To understand the role of UNS in the flowering process, knockout mutants were identified and UNS was expressed ectopically in petunia and Arabidopsis. No phenotype was observed in petunia plants in which UNS was disrupted by transposon insertion, indicating that its function is redundant. Constitutive expression of UNS leads to an acceleration of flowering and to the unshaven floral phenotype, which is characterized by ectopic trichome formation on floral organs and conversion of petals into organs with leaf-like features. The same floral phenotype, accompanied by a delay in flowering, was obtained when a truncated version of UNS, lacking the MADS box domain, was introduced. We demonstrated that the truncated protein is not translocated to the nucleus. Using the overexpression approach with both the full-length and the nonfunctional truncated UNS protein, we could distinguish between phenotypic alterations because of a dominant-negative action of the protein and because of its native function in promoting floral transition.
INTRODUCTION
The role of MADS box proteins as key regulators in many steps of development and differentiation has been acknowledged over the last decade (for a review, see Ng and Yanofsky, 2001). The first discovered members of this family in plants, AGAMOUS (AG) and DEFICIENS, were identified as flower homeotic genes in the model species Arabidopsis thaliana and Antirrhinum majus, respectively (Sommer et al., 1990; Yanofsky et al., 1990).
The origin of this DNA binding superfamily of proteins predates the divergence of plants and animals: conserved MADS box domains also occur in yeast and mammalian cells, where they are involved in the conversion of external signals into developmental or metabolic responses (Shore and Sharrocks, 1995), indicating ancient regulatory functions in no way related to floral organogenesis.
Various studies in the model species Arabidopsis revealed that MADS box genes also play prominent roles within the complex genetic network that regulates the transition to flowering: the switch between vegetative and generative growth is determined by the activation of interacting groups of genes that are responding to a specific combination of environmental and endogenous factors. The MADS box gene FLOWERING LOCUS C (FLC), together with FRIGIDA, accounts for most of the differences between early and late flowering ecotypes in Arabidopsis, being a repressor of flowering regulated by vernalization (Burn et al., 1993; Lee et al., 1993; Clarke and Dean, 1994; Michaels and Amasino, 1999, 2001; Sheldon et al., 1999, 2000, 2002). Analysis of five FLC paralogs, MADS AFFECTING FLOWERING1 (MAF1) (also known as FLOWERING LOCUS M [FLM]) to MAF5, revealed a similar role in flowering time as floral repressors, at least for MAF1/FLM and MAF2 (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001). SHORT VEGETATIVE PHASE (SVP) is also a MADS box gene that, similarly to FLC, acts in a dosage-dependent manner as a repressor of flowering; its position within the regulatory network, however, has yet to be established (Hartmann et al., 2000). Another MADS box gene, AGAMOUS-LIKE 20 (AGL20), later renamed as SOC1 (for SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1), appeared to be an immediate target of the transcription factor CONSTANS, which acts in the long-day pathway as a promoter of flowering (Simon et al., 1996; Samach et al., 2000). Because SOC1 expression is also regulated by FLC and by the gibberellin-dependent signaling pathway, SOC1 itself has been identified as one of the floral pathway integrators, together with FLOWERING LOCUS T and LEAFY (Lee et al., 2000; Hepworth et al., 2002; Simpson and Dean, 2002; Moon et al., 2003). Like SOC1, AGL24 also acts as promoter of flowering upregulated by vernalization; unlike SOC1, however, its mRNA level is not affected by FLC (Yu et al., 2002; Michaels et al., 2003). Furthermore, genetic and expression data indicate that these two MADS box transcription factors positively regulate each other's transcription (Michaels et al., 2003). Recent studies on flowering signals in crucifers and rice (Oryza sativa) showed that the important role of the SOC1 and FLC MADS box proteins is at least partly conserved between these species. Putative SOC1 homologs were isolated, and analyses of flowering time and gene expression indicated a correlation between an increase in expression levels and the floral transition (Kim et al., 2003; Tadege et al., 2003). In addition, overexpression of the Arabidopsis FLC gene in rice delayed flowering; no FLC homolog has been identified in this species yet. These observations clearly show conservation of the regulatory role of MADS box proteins in the flowering induction pathway.
Flowering in Petunia hybrida plants is also affected by photoperiod: long-day (LD) conditions promote early flowering in both early and late varieties. Each variety retains its characteristic blooming time when grown under short-day (SD) conditions; under LD conditions, however, plants of both varieties flower earlier and at the same moment (Sink, 1984). Despite the fact that physiological studies on the effect of photoperiod, light, and temperature on growth and flowering of petunia plants date back to the late 1950s, the genetic basis of flowering time in petunia has not been unraveled yet.
Nevertheless, nonflowering petunia plants obtained by suppression of at least the MADS box gene PETUNIA FLOWERING GENE (PFG) demonstrate that MADS box proteins are required for the switch from vegetative to generative growth in this species (Immink et al., 1999).
MADS box proteins are composed of a conserved DNA binding domain, the MADS box, which is located at the N terminus; furthermore, most of the plant MADS box proteins have an additional conserved region that resembles the coiled-coil segment of keratin (therefore named K box; Ma et al., 1991) located in the middle of the protein and separated from the MADS box by the highly variable I region (for a review, see Riechmann and Meyerowitz, 1997). Several studies using both in vitro and in vivo systems have been conducted to identify the regions responsible for DNA binding, dimerization, transactivation, and interaction with coaccessory regulatory factors. Given the complexity of regulatory networks, homodimerization, heterodimerization, or even multimeric complex formation efficiently creates a large collection of transcription activation or repression complexes, originating from a relatively limited number of proteins. The inclusion of nonfunctional MADS box proteins allows the creation of dominant inhibitory complexes that are potentially useful in generating loss-of-function phenotypes.
MADS box proteins lacking the activation domain have indeed been successfully produced to act as dominant-negative isoforms. The role of human SERUM RESPONSE FACTOR (SRF) and MYOCYTE ENHANCER FACTOR2 proteins in differentiation pathways have been confirmed by this strategy (Belaguli et al., 1997; Ornatsky et al., 1997; Okamoto et al., 2000). Similar mutant phenotypes can be obtained when a transcription factor is altered in its DNA binding ability, while it still retains its dimerization capacity. A mutated form of the avian SRF, in which a triple point mutation was introduced in the MADS domain, competes successfully with binding by the endogenous SRF to its DNA target site (Johansen and Prywes, 1993; Croissant et al., 1996). Although much research has been done on dominant-negative mutations in MADS box proteins in vertebrates, only a few examples of loss-of-function phenotypes as a result of such mutations have been described in plants. A truncated version of the AG protein, lacking the C-terminal region, generates an ag mutant phenotype when ectopically expressed in Arabidopsis (Mizukami et al., 1996). In rice, the naturally occurring mutant leafy hull sterile1 is caused by a point mutation in the MADS box of the O. sativa MADS1 gene, which inhibits the binding of the transcription factor to its DNA target site (Jeon et al., 2000). Lastly, overexpression of the lily (Lilium longiflorum) AP3 homolog (LMADS1) lacking the MADS box region in Arabidopsis is sufficient to generate an ap3-like phenotype through interaction of the corresponding protein with the endogenous PISTILLATA protein (Tzeng and Yang, 2001).
In this study, we report the cloning and functional characterization of the petunia MADS box gene FLORAL BINDING PROTEIN20 (FBP20), homologous to the Arabidopsis SOC1 and renamed as UNSHAVEN (UNS). The phenotypes obtained by overexpressing the full-length gene revealed its function in promoting floral transition, and truncated versions induced dominant-negative effects caused by interactions with different dimerization partners. The lack of phenotype in the knockout mutant obtained by transposon insertion mutagenesis strongly suggests redundancy between the UNS gene and the closely related FBP21, FBP22, and FBP28 MADS box genes.
RESULTS
Isolation and Sequence Analysis of UNS
A full-length cDNA clone, corresponding to the UNS (formerly known as FBP20) MADS box gene has been isolated from a cDNA library obtained from young petunia inflorescences (Immink et al., 2003). The UNS cDNA encodes a putative protein of 217 amino acids, which has a high sequence similarity with the Arabidopsis SOC1 protein (Samach et al., 2000) and other related sequences from tobacco (Nicotiana tabacum) (MADS1; Mandel et al., 1994), Sinapis alba (SaMADSA; Menzel et al., 1996), and tomato (Lycopersicon esculentum) (TDR3; Pnueli et al., 1991). UNS and SOC1 share 65% sequence identity, when the full-length proteins are compared, and this raises to 89% in the MADS box domain.
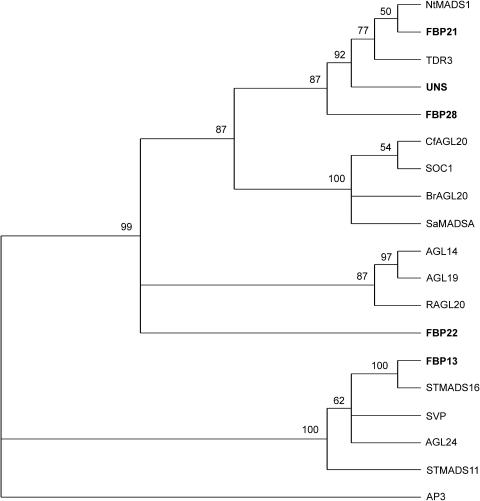
Figure 1 shows a phylogenetic tree generated using the MADS box, the I region, and the K box of UNS and related Arabidopsis protein sequences as present in the databases (Pařenicová et al., 2003). We also included the sequences from tobacco, S. alba, tomato, Brassica, Cardamine flexuosa (Kim et al., 2003), and rice (accession number AY332476), all related to UNS, and the potato (Solanum tuberosum) STMADS11 and STMADS16 (Carmona et al., 1998; Garcìa-Maroto et al., 2000), which are related to SVP together with FBP13 from petunia (Immink et al., 2003). The AP3 protein from Arabidopsis was used as an outgroup. Indeed, the phylogenetic analysis reveals that the Solanaceae UNS, NtMADS1, and TDR3 group together with the crucifers SOC1, BrAGL20, and SaMADSA, whereas FBP13 belongs to the SVP/AGL24 group together with the potato StMADS11 and StMADS16.
Figure 1.
Phylogenetic Tree of Predicted MADS Box Protein Sequences from Several Species.
Comparisons were made using the MADS box, the I region, and the K box of the proteins from the plant species Arabidopsis (SOC1, AGL14, AGL19, SVP, AGL24, and AP3), Brassica rapa (BrAGL20), Cardamine flexuosa (CfAGL20), petunia (FBP28, FBP21, UNS, FBP22, and FBP13, in bold in the figure), potato (STMADS16 and STMADS11), rice (RAGL20), S. alba (SaMADSA), tobacco (NtMADS1), and tomato (TDR3). Bootstrap values are indicated above each branch. The Arabidopsis AP3 protein was used as an outgroup.
Expression Analysis of UNS in Wild-Type Petunia
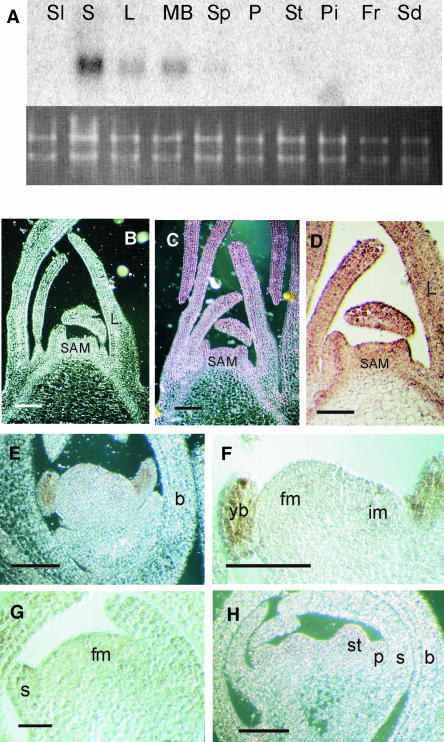
RNA gel blot analysis was performed to analyze the expression of UNS in wild-type petunia tissues (Figure 2A). An UNS-specific probe without the MADS box region was used to hybridize blots containing RNA isolated from seedlings, stems, leaves, mature bracts, sepals, petals, stamens, pistils, fruits, and developing seeds at 5 d after fertilization. UNS expression is confined to the green vegetative tissues and the first floral whorl. A more detailed picture of the UNS expression pattern was obtained by in situ hybridization of shoot apical meristem (SAM), inflorescence, and floral meristems (Figures 2B to 2H). UNS mRNA was detectable throughout the vegetative meristem, the young leaves, and at a relatively high level around the vascular bundles of the emerging first leaves (Figures 2C and 2D). The expression pattern of UNS is therefore similar to that of SOC1 in Arabidopsis plants that have been induced to flowering by 16-h LD conditions (Samach et al., 2000). UNS is highly expressed in the emerging young bracts and at a very low level in inflorescence meristems (Figures 2E and 2F). No hybridization signal above background was detectable in the flower meristem and in the emerging floral organ primordia (Figures 2G and 2H). mRNA levels rapidly declined with further development of the flower organs (Figure 2A).
Figure 2.
Expression of UNS in Wild-Type Tissues of Petunia.
(A) RNA gel blot using RNA from seedlings (Sl), stems (S), leaves (L), mature bracts (MB), sepals (Sp), petals (P), stamens (St), pistils (Pi), fruits (Fr), and seeds (Sd) of wild-type petunia hybridized with an UNS-specific probe. Below the blot is a picture of the gel before blotting, stained with ethidium bromide, illustrating equal loading of the samples.
(B) to (H) RNA in situ localization of UNS in SAMs and developing petunia inflorescences. Longitudinal sections were hybridized with a digoxygenin-labeled sense UNS probe (B) or antisense UNS RNA ([C] to [H]). (B), (C), (E), and (H) and (D), (F), and (G) were viewed using dark- and bright-field microscopy, respectively. L, leaf; b, bract; yb, young bract, fm, floral meristem; im, inflorescence meristem; s, sepal; p, petal; st, stamen. Bars = 200 μm in (B) to (F) and 500 μm in (G) and (H).
(B) to (D) SAMs with emerging leaves on their flanks. The UNS mRNA is equally distributed throughout the apex and the emerging young leaves. (D) is a higher magnification of (C).
(E) and (F) Inflorescence apex with developing floral meristem. (F) is a higher magnification of (E). The expression is strong in young bracts, but weak hybridization signals are also present in the floral and inflorescence meristems.
(G) Floral meristem with developing sepals.
(H) Young flower bud with petal and stamen primordia.
Knockout Mutant of UNS
To obtain an UNS loss-of-function mutant, we screened 5712 petunia W138 plants for transposon insertions in the open reading frame of UNS (Koes et al., 1995; Vandenbussche et al., 2003). One plant was found to be hemizygous for a dTph1 transposable element insertion in the MADS box coding region. The small transposable element of 284 nucleotides in length (Gerats et al., 1990) was inserted 104 nucleotides downstream of the ATG start codon in the first exon (see supplemental data online). The insertion was stabilized by crossing with a wild-type W115 line, which does not carry transposase activity. Homozygous mutant plants were obtained upon selfing and analyzed for phenotypical effects under both SD and LD conditions. Although carrying a null mutation in the UNS gene, the homozygous insertion plants did not show any clear phenotypic alteration or change in flowering time, suggesting the existence of functional redundancy with other genes.
Ectopic Expression of UNS and UNSΔMI Generate the Same Phenotype in Petunia Flowers but Have the Opposite Effect on Flowering Time
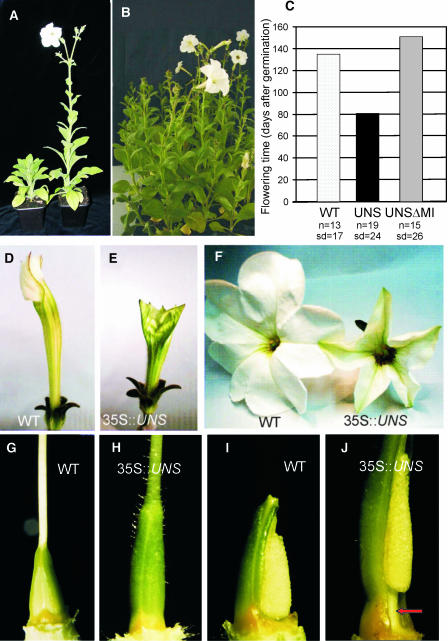
Transgenic petunia plants carrying the full-length UNS cDNA under the control of the constitutive 35S promoter of Cauliflower mosaic virus were generated by Agrobacterium tumefaciens–mediated transformation. Thirty-eight independent transformants were obtained, of which 14 showed transgene expression at variable levels, as determined by RNA gel blot analysis (data not shown). The progenies (T1-UNS) of the two primary transformants with the highest transgene expression produced after selfing were grown under SD conditions, and a clear reduction in flowering time was observed in both progenies. Transgenic petunia plants flowered much earlier compared with the wild type, as shown in Figure 3A. To quantify the effect of the transgene on flowering behavior, a third T1-UNS progeny was grown under SD, together with a wild-type population and a progeny from a primary transformant overexpressing a truncated version of UNS (T1-UNSΔMI) lacking the MADS box and the I region. As shown in Figure 3C, the T1-UNS population showed a clear reduction in flowering time with an average of 81 d after germination compared with the 135 d necessary for the wild type to flower. Surprisingly, overexpression of UNSΔMI caused an opposite effect on the flowering behavior of the transgenic petunia lines because the T1-UNSΔMI population flowered on average 2 weeks later than the wild type (Figure 3B). The delay in flowering time caused by overexpression of UNSΔMI was also observed in two independent transgenic populations (data not shown).
Figure 3.
Effect of UNS and UNSΔMI Ectopic Expression on Petunia Plants.
(A) Overexpression of UNS causes early flowering in transgenic petunia plants (right) compared with the wild type (left).
(B) Overexpression of UNSΔMI delays flowering in transgenic petunia plants (left) compared with the wild type (right).
(C) Flowering time of wild-type, 35S-UNS (UNS), and 35S-UNSΔMI (UNSΔMI) plants as measured by the number of days after germination until the first flower bud appears. Number of plants (n) and standard deviation (sd) are indicated below each bar. The significance of the differences in flowering time between the wild type and 35S-UNS and between the wild type and 35S-UNSΔMI plants were confirmed by Student's t test.
(D) to (J) Flowers and ovaries of wild-type and 35S-UNS petunia plants.
(D) and (E) Wild-type and transgenic (35S-UNS) petunia buds at the same stage of development. The transgenic bud is significantly shorter and greener than a bud of the wild type.
(F) Wild-type and 35S-UNS mature flowers. The reduced dimensions are maintained until full maturity of the transgenic flower, which does not open completely.
(G) to (J) Ovaries of wild-type ([G] and [I]) and 35S-UNS ([H] and [J]) plants. The transgenic ovaries are significantly longer than those from the wild type. The outer epidermis is covered with trichomes, and a stem-like structure develops at the basis of the septum (arrow in [J]). One carpel is removed from the ovaries in (I) and (J).
Because the truncation of UNS results in a nonfunctional protein, as shown in colocalization experiments (see below), the delay in flowering time is most likely because of a dominant-negative effect of the truncated MADS box protein.
Phenotypic alterations of the 35S-UNS transgenic plants were also apparent in flower organs where UNS transcript is normally not present. The overall architecture of the petunia inflorescence was not affected. Leaves, bracts, sepals, and stamens did not reveal obvious changes in their morphology, whereas modifications in petals and ovaries could be seen in all plants in which ectopic expression of UNS was observed.
As shown in Figure 3E, transgenic floral buds are significantly shorter and greener than wild-type buds (Figure 3D). The green hue of the petals in overexpression plants is maintained until full maturity of the flower. Transgenic flowers are reduced in size, and the corolla does not open completely (Figure 3F). Petals of wild-type and aberrant flowers were analyzed in detail by scanning electron microscopy (Figure 4). These analyses revealed that trichomes develop on both abaxial and adaxial sides of transgenic petals, equally distributed over the whole surface. By contrast, petals of wild-type plants have only a few trichomes on the abaxial side, predominantly along the midvein (Figures 4A and 4B). The shape of the petal cells was also affected: epidermal cells at both sides of the petals in overexpression lines were flatter than those of wild-type plants, which have conical shaped cells (Figures 4C, 4D, 4I, and 4J). Petals of overexpressor lines exhibit features that are typical of leaves and bracts of wild-type plants (Figures 4E and 4F), with flat epidermal cells and many trichomes on both abaxial and adaxial sides.
Figure 4.
Scanning Electron Microscopy of Wild-Type Petal and Bract and 35S-UNS Petals.
(A) to (D) Abaxial and adaxial side of a wild-type petal. A few trichomes are visible along the midvein at the abaxial side only (A). A higher magnification shows the conical shaped cells that are present on both sides ([B] to [D]).
(E) and (F) Adaxial side of a wild-type bract, which is covered with trichomes (E) and stomata (F).
(G) to (J) Abaxial and adaxial sides of a 35S-UNS line. Numerous trichomes are growing on both sides of the petals, and the cells are flatter compared with the wild type ([I] and [J]). (I) and (J) are higher magnifications of (G) and (H), respectively.
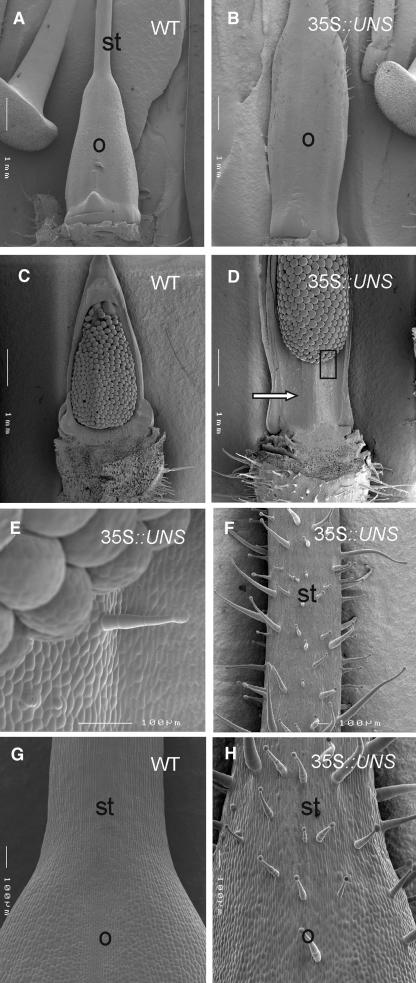
Pistils of the overexpression lines also exhibited a change in morphology. The ovary was elongated compared with wild-type ovaries (Figures 3G to 3J and 5A to 5D), and a stem-like structure developed at the basis of the septum inside the ovary (arrows in Figures 3J and 5D). Like on transgenic petals, trichomes developed ectopically on the surface of the transgenic ovaries and over the whole style to the basis of the stigma, whereas trichomes are completely absent on wild-type pistils (Figures 5F to 5H). Even inside the ovary, trichomes developed occasionally on the surface of the septum (Figure 5E). Because the presence of many trichomes on different organs was one of the most striking features of the transgenic plants, we renamed FBP20 to UNS. When UNS lacking the MADS box and the I region (UNSΔMI) was ectopically expressed in petunia plants, the same phenotypic alterations were observed in the flowers, as seen in transgenic plants overexpressing the full-length cDNA. Greenish petals covered with trichomes and elongated hairy pistils are features common to all transgenic plants, whereas the severity of the phenotype correlated with the expression level of the transgene (data not shown).
Figure 5.
Scanning Electron Microscopy of Pistils of Wild-Type and 35S-UNS Lines.
(A) and (B) Ovaries of the wild type and 35S-UNS. The ovary (o) of the transgenic line is elongated compared with the wild-type ovary and covered with trichomes. st, style.
(C) to (E) Ovaries of the wild type and 35S-UNS with removed ovary wall. The box in (D) is enlarged in (E) and shows a trichome that develops on the surface of the elongated septum inside the ovary (arrow in [D]).
(F) to (H) Stylar tissues of wild-type and 35S-UNS plants. Numerous trichomes develop at the stylar surface of 35S-UNS, whereas wild-type pistils do not produce trichomes. o, ovary; st, style.
Expression Analyses of Differentially Expressed MADS Box Genes in 35S-UNS and 35S-UNSΔMI Transgenic Plants
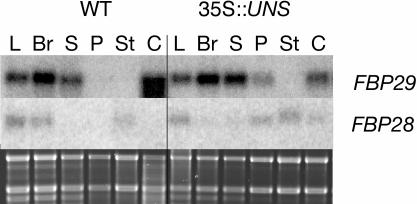
To assess the identity of the modified floral organs in the 35S-UNS plants, we checked the expression of two different petunia MADS box genes: FBP28 and FBP29. These genes are both expressed in leaves and bracts of a wild-type petunia plant, although at different levels, whereas their patterns of expression differ in the floral organs (Figure 6). FBP29 mRNA is detectable in sepals and carpels, but it is excluded from whorls 2 and 3. FBP28 mRNA, instead, is present only in stamens and absent from the other three floral whorls. In 35S-UNS plants, the expression of FBP29 is extended to whorl 2 organs, where FBP28 also is upregulated. A decrease in FBP29 expression and the appearance of FBP28 mRNA in transgenic carpels is also in line with the altered morphology of these organs. These observations together suggest that petals and carpels of the overexpression plants have partially lost their identity as floral organs and have acquired hallmarks of vegetative organs of the plant.
Figure 6.
RNA Gel Blot Analysis of Wild-Type and 35S-UNS Petunia Plants.
RNA gel blot using RNA from leaves (L), bracts (Br), sepals (S), petals (P), stamens (St), and carpels (C) of wild-type (left) and 35S-UNS (right) petunia plants. The blot was hybridized with an FBP29- or FBP28-specific probe. Below the blots is a picture of the gel before blotting, stained with ethidium bromide, illustrating the amount of RNA of the samples.
UNS and UNSΔMI Interacting Partners Assessed by a Yeast Two-Hybrid System
The ability of the wild-type and truncated UNS proteins to dimerize with corresponding or different MADS box partners was assessed by the yeast cyto-trap system (Immink et al., 2003). As shown in Table 1, UNS and UNSΔMI share most of the dimerization partners, including the SEPALLATA-like proteins FBP2 and FBP5 as well as FBP13, the putative petunia homolog of the Arabidopsis AGL24 protein, which functions as a dosage-dependent promoter of flowering and a positive regulator of SOC1 (Michaels et al., 2003).
Table 1.
Yeast Cyto-Trap Assay Using Different Protein Combinations
| UNS | UNSΔMI | |
|---|---|---|
| FBP13 | + | + |
| UNS | − | + |
| PFG | − | + |
| FBP2 | + | + |
| FBP5 | + | + |
| FBP9 | + | + |
| FBP23 | + | + |
Both UNS and UNSΔMI were tested against 22 known petunia MADS box proteins (Immink et al., 2003); only the positive combinations are reported here.
Unlike its wild-type counterpart, UNSΔMI is able to interact with the full-length UNS protein and with PFG, whose cosuppression causes a nonflowering phenotype in petunia plants (Immink et al., 1999).
Colocalization of UNS, UNSΔMI, and Interacting Partners in Petunia Protoplasts
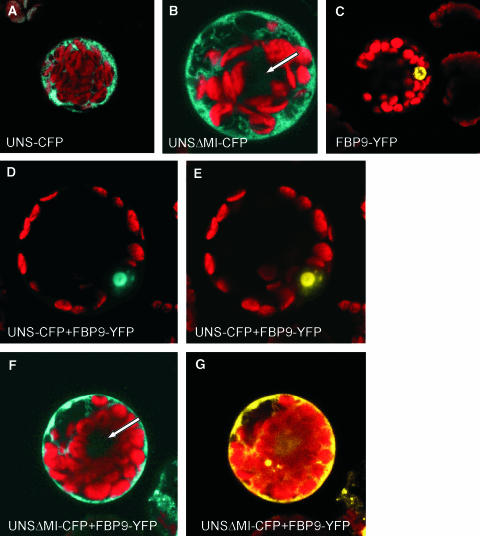
Surprisingly, both UNS and UNSΔMI resulted in similar phenotypic alterations in the flower when expressed ectopically. This suggests that the mutation is because of a dominant-negative effect rather than transcriptional activation by UNS. To provide additional evidence for this hypothesis, we analyzed the localization of different MADS box fusion proteins in petunia leaf protoplasts by confocal laser scanning microscopy. Fusion constructs containing the yellow fluorescent protein (YFP) and either the full-length UNS or the truncated UNSΔMI were expressed under the constitutive 35S promoter of Cauliflower mosaic virus. Similarly, a fusion between the cyan fluorescent protein (CFP) and the MADS box protein FBP9 was overexpressed in petunia W115 leaf protoplasts. FBP9 is a dimerization partner of UNS as determined by yeast two-hybrid experiments (Table 1).
The fluorescence because of YFP expression was localized in the cytoplasm when either UNS or UNSΔMI alone was expressed (Figures 7A and 7B). The cytoplasmic localization of the chimeric transcription factor constructs points to the absence of a heterodimerization partner and the inability to form homodimers (Table 1). In contrast with UNS, FBP9 can form homodimers (Immink et al., 2002), and the complex is transferred to the nucleus where the yellow fluorescence of the FBP9:YFP construct is localized (Figure 7C). The use of two variants of the green fluorescent protein with different emission spectra enables a simultaneous localization of the two different MADS box protein constructs. The combinations tested, FBP9/UNS and FBP9/UNSΔMI, gave rise to different fluorescence patterns. When both full-length proteins are present, the yellow and cyan fluorescence colocalize in the nucleus (Figures 7D and 7E). By contrast, both FBP9 and UNSΔMI remain located in the cytoplasm despite the ability to form heterodimers between FBP9 and UNSΔMI (Figures 7F and 7G). Deletion of the MADS box domain from one of the dimerizing proteins, and therefore the absence of one nuclear localization signal, thus seems to be sufficient to prevent the whole complex from being translocated into the nucleus. As a consequence, the FBP9/UNSΔMI heterodimer cannot function as a transcription factor. In this specific combination, UNSΔMI thus acts as a dominant-negative form toward FBP9, preventing the complex to reach its DNA targets inside the nucleus. This suggests that the aberrant phenotype in the flower is because of the elimination of active UNS dimerization complexes.
Figure 7.
Confocal Laser Scanning Microscopy of Petunia Leaf Protoplasts Expressing Different Fusion Proteins.
Chlorophyll autofluorescence is shown in red.
(A) and (B) The fusion proteins between CFP and UNS full-length or UNSΔMI are localized in the cytoplasm. The arrow in (B) indicates the position of the nucleus.
(C) The fusion protein FBP9-YFP is translocated to the nucleus.
(D) and (E) Protoplast imaged with CFP (D) and YFP (E) filters, respectively. UNS-CFP and FBP9-YFP are colocalized in the nucleus.
(F) and (G) Single protoplast expressing UNSΔMI-CFP and FBP9-YFP imaged with CFP (F) and YFP (G) filters, respectively. Both signals are localized in the cytoplasm. The arrow in (F) indicates the position of the nucleus.
Ectopic Expression of UNSΔMI in Arabidopsis Phenocopies the Floral Abnormalities Observed in the Petunia Overexpressors and Generates an Additional apetala1-Like Phenotype
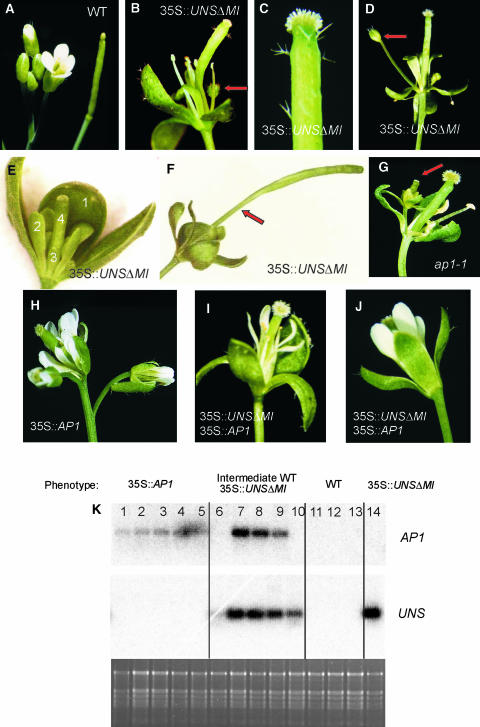
We transformed Arabidopsis plants with the 35S-UNSΔMI petunia construct to monitor the dominant-negative effects on flower morphology in a heterologous system. Twenty-nine out of 44 putative primary transformants showed variable degrees of phenotypic alterations. They all had small green petals, being almost absent in the four lines with the most severe aberrations (Figures 8B to 8G). In addition, these lines produced elongated unshaven carpels (Figures 8B and 8C). The long stem at the basis of the effected ovary (arrow in Figure 8F) and stellate trichomes arising from its surface are characteristics that are very similar to the ones produced in petunia by the same construct (cf. Figures 3 and 8).
Figure 8.
Flower Morphology of 35S-UNSΔMI in Wild-Type and 35S-AP1 Arabidopsis Lines.
(A) Wild-type Arabidopsis inflorescence (left) and silique (right).
(B) to (F) Flowers of 35S-UNSΔMI transformants. Stellate trichomes on the abaxial side of the sepals and on the pistil are visible in (B) and (C). The arrows in (B) and (D) point to the secondary buds arising at the axil of the first whorl organs. Petals are completely absent or largely reduced in size and are greenish.
(E) A young 35S-UNSΔMI flower with small green organs in whorl 2. The four flower whorls are numbered.
(F) Flower of 35S-UNSΔMI after fertilization. The arrow points to the elongated stem at the base of the silique.
(G) ap1-1 flower. The arrow indicates the axillary bud arising at the axil of the first whorl organs.
(H) to (J) Progeny plants from a cross between 35S-AP1 and 35S-UNSΔMI.
(H) Terminal flower of a 35S-AP1 plant.
(I) and (J) Flowers from two lines carrying both 35S-AP1 and 35S-UNSΔMI constructs and showing a phenotype intermediate between wild-type and UNSΔMI overexpression. The sepals are still bract-like, and petal formation is partly (I) or completely (J) restored.
(K) Expression of AP1 and UNSΔMI in leaves of Arabidopsis plants derived from a cross between a 35S-AP1 and a 35S-UNSΔMI line. The top blot was hybridized with an AP1 probe and the bottom one with an UNS probe. The phenotype is indicated at the top of the gel. Below the blots is a picture of the gel before blotting and stained with ethidium bromide.
Surprisingly, the Arabidopsis transformants exhibited extra features not seen in the transgenic petunia lines, which closely resembled abnormalities described for apetala1 (ap1) mutants (Irish and Sussex, 1990; Bowman et al., 1993). As shown in Figures 8B and 8D to 8F, first whorl organs tend to be bract-like, being elongated and pointed and with stellate trichomes on the abaxial surface. Another trait that is typical for all strong ap1 alleles is the presence of extra flowers arising in the axils of the first whorl organs of the primary flowers (Figure 8G). Flowers with this indeterminate branched structure that replaces the single flower of a wild-type plant are depicted in Figures 8B and 8D.
RNA gel blot analysis was performed to check whether the expression of the AP1 gene was affected in the transgenic plants overexpressing UNSΔMI. No reduction in the level of AP1 mRNA was detected in any of the transgenic plants when compared with wild-type plants (data not shown). This demonstrates that the ap1-like phenotype of the UNSΔMI overexpressing lines is not caused by a transcriptional silencing of the AP1 gene. However, in line with the mode of action in petunia, UNSΔMI may also act as a dominant-negative factor toward the AP1 protein, preventing the translocation of the heterodimer complex to the nucleus in Arabidopsis. Remarkably, transgenic Arabidopsis plants overexpressing the full-length UNS gene did not produce any floral morphological aberrations. Probably, this is because of different dimerization abilities for the full-length and truncated UNS proteins in Arabidopsis. To verify this hypothesis, we tested the heterologous interactions of the petunia UNS and UNSΔMI proteins with the Arabidopsis AP1 MADS box protein in the yeast two-hybrid system. These analyses revealed that only the petunia UNS lacking the MADS box and the I region dimerizes with the AP1 protein, whereas the full-length protein failed to interact with the Arabidopsis protein in any of the conditions tested (data not shown). This may explain the ap1-like phenotype as observed specifically in the UNSΔMI overexpression lines and the absence of this phenotype in the 35S-UNS lines.
In addition to the effect on flower morphology in Arabidopsis, the effect of UNS and UNSΔMI overexpression on flowering time was analyzed. 35S-UNS transgenic Arabidopsis plants showed a minor acceleration in flowering time, in terms of decreased rosette leaf numbers as well as days to flowering (see Supplemental Table 1 online), suggesting a similarity in function between the petunia gene and its Arabidopsis homolog. The truncated protein, however, did not cause any delay in flowering as it did in petunia, but similar to the full-length protein, accelerated the transition to flowering in Arabidopsis a little (see Supplemental Table 2 online). This suggests that different partners might interact with UNSΔMI in the heterologous species. Possible candidates are floral repressors, such as SVP (Hartmann et al., 2000), which are either not present or do not interact with UNS in petunia.
AP1 Complements the ap1-Like Phenotype in 35S-UNSΔMI Transgenic Arabidopsis Plants
To investigate whether the ap1-like phenotype, obtained by overexpression of the UNSΔMI construct, is caused by suppression of the AP1 function, crosses were made between 35S-AP1 plants and Arabidopsis plants carrying the UNSΔMI construct. The progeny was analyzed both at the molecular and phenotypic levels. Five out of the 45 progeny plants analyzed showed partial complementation in their flowers: wild-type petals were formed, and the secondary buds in the axils of the sepals were absent (Figures 8I and 8J). Among the other plants in the progeny, we observed some wild-type plants and plants displaying either an AP1 (Figure 8H) or an UNSΔMI overexpression phenotype. A subset of the individuals belonging to the four phenotypic classes was checked at the molecular level by means of RNA gel blot analysis (Figure 8K). The gel blot was probed either with the AP1 or the UNS gene. All wild-type plants lacked ectopic expression of both transgenes, and overexpression of AP1 or UNSΔMI was detected in the plants displaying the corresponding phenotype. The chimeric phenotype with both wild-type and 35S-UNSΔMI characteristics, as shown in Figures 8I and 8J, was either because of a complementation by high expression of the AP1 transgene (lanes 7, 8, and 9) or because of a relatively low expression of UNSΔMI (lanes 6 and 10). This suggests that the AP1 function is affected in the 35S-UNSΔMI overexpressor lines.
DISCUSSION
MADS box genes have been recruited during plant evolution as master regulatory genes that control developmental processes, such as inflorescence and flower formation. Although floral homeotic functions have been the first ones to be assigned to members of this family of transcription factors, the number of these genes that are found to be involved in processes not related to floral organ identity is rapidly increasing (Gu et al., 1998; Zhang and Forde, 1998; Liljegren et al., 2000). An intricate network of pathways that respond to environmental stimuli as well as endogenous signals tightly regulates the transition from vegetative to reproductive development. Arabidopsis is the model plant in which this complex regulation has been most extensively studied (for review, see Mouradov et al., 2002; Ratcliffe and Riechmann, 2002). Genetic analysis of Arabidopsis mutants and natural ecotypes have shown that several MADS box genes are involved in the regulation of flowering time.
AGL24 and SOC1, among others, act as dosage-dependent promoters of flowering and regulate each other's expression level (Lee et al., 2000; Samach et al., 2000; Yu et al., 2002; Michaels et al., 2003). Furthermore, SOC1 represents an integration point at which inductive signals from different pathways converge, preceding the upregulation of floral meristem identity genes (Lee et al., 2000; Simpson and Dean, 2002; Moon et al., 2003). SOC1 homologs have also been identified from different species, and functional analysis on a few of them confirmed their role as promoters of flowering (Bonhomme et al., 2000; Kim et al., 2003; Tadege et al., 2003). Here, we describe the identification and functional analysis of a petunia MADS box gene, UNS, which is specifically expressed in vegetative and green tissues and shares sequence similarity with the Arabidopsis SOC1 gene. The ectopic expression of this gene accelerates flowering time in petunia plants and induces vegetative features in flowers. Opposite effects on flower induction and similar floral phenotypic alterations in plants overexpressing a nonfunctional UNS protein reveal the function of the wild-type UNS in promoting flowering and the dominant-negative effects of the transgene in flower organs.
The Petunia UNS Gene Belongs to the SOC1 Clade and Acts as a Promoter of Flowering
The petunia UNS gene shares a high degree of sequence similarity with the Arabidopsis SOC1 gene and its mustard (S. alba) homolog, SaMADSA. SOC1 is a promoter of flowering in response to environmental and endogenous stimuli, such as a long photoperiod treatment, vernalization, and gibberellin inductive signals, and its expression has been observed in most Arabidopsis organs at different levels (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000). Although SOC1 mRNA could be detected in the SAM during vegetative development, its level increases dramatically during floral induction, it expands into the developing leaves, and it persists in the inflorescence meristem, but it is absent in the floral meristem (Borner et al., 2000; Samach et al., 2000). UNS mRNA is also present in the SAM, but it has been detected in emerging leaves as well long before the plant is committed to flowering. Nevertheless, a function as floral activator can be assigned to the petunia UNS, too, because of the drastic reduction of the vegetative phase in plants where the gene is overexpressed. A strong indication that the acceleration of flowering in 35S-UNS plants is indeed a gain-of-function phenotype and not a dominant-negative effect of the transgene comes from the opposite flowering behavior induced by ectopic expression of UNSΔMI. UNSΔMI encodes a truncated UNS protein lacking the MADS box and the I region and still retains its ability to dimerize with other MADS box proteins but cannot function as a transcription regulator because the lack of a nuclear localization signal prevents the protein complex from being transported to the nucleus. The fact that overexpression of UNSΔMI delays flowering suggests that the truncated protein acts antagonistically to wild-type UNS, most likely through dimerization with the corresponding partners, thus preventing them to reach their natural targets within the nucleus. Furthermore, unlike the full-length protein, UNSΔMI can also interact with other promoters of floral transition, such as PFG, thereby hindering them in their natural function. This is also in line with a delay or prevention of flowering observed in PFG suppression plants (Immink et al., 1999).
One of the dimerization partners of UNS is FBP13, which is most close in amino acid sequence to the Arabidopsis AGL24 protein. Remarkably, genetic analyses in Arabidopsis showed cross talk in the flowering pathway between AGL24 and SOC1 (Yu et al., 2002; Michaels et al., 2003). Based on the identified physical interaction between UNS and FBP13 and the conservation of the function of this kind of MADS box proteins in flowering, this suggests heterodimerization between SOC1 and AGL24 as well. Recently, this interaction was indeed identified by yeast two-hybrid analysis (G.C. Angenent and R.G.H. Immink, unpublished results). Most likely, these interactions occur in planta as well, and UNS (SOC1) and FBP13 (AGL24) act as heterodimers at the same level in the floral inductive pathway.
Unlike in Arabidopsis, however, UNS in petunia probably shares its function as promoter of flowering with one or more redundant genes. The lack of phenotype observed in uns knockout mutants strongly indicates this, and similarity in sequence, expression, and interaction patterns point to FBP21, FBP22, and/or FBP28 as possible candidates for functional redundancy (Immink et al., 2003).
UNS Confers Leaf-Like Features to Floral Organs in a Dominant-Negative Manner
Although one should be careful in drawing conclusions from ectopic expression studies, it can be particularly informative when homeotic or phase changes appear in overexpressor lines and when a gene is functionally redundant. It must be taken into account that aberrations induced by a transgene can be the result of a different mode of action of the transgenic protein, when expressed in a cellular environment different from its original one. This is the case for UNS, but it is most likely applicable to many other overexpression lines as well. All phenotypic alterations that were induced by ectopic expression of the full-length UNS protein in floral organs were also apparent in transgenic plants in which the truncated nonfunctional UNSΔMI protein was expressed. The leaf-like characteristics, conferred to floral organs by both proteins, cannot be because of direct activation of a set of genes by the transcription factors because UNS lacking the complete DNA binding domain and the I region simply cannot perform the function of the wild-type protein. Similar changes in flower morphology have also been reported for the overexpression of the potato MADS box gene STMADS16 in tobacco and SOC1 and AGL24 in Arabidopsis (Borner et al., 2000; Garcìa-Maroto et al., 2000; Michaels et al., 2003). The greenish sepaloid petals and elongated pistils observed in 35S-SOC1 and 35S-AGL24 Arabidopsis (Borner et al., 2000; Michaels et al., 2003) were also present in Arabidopsis and petunia plants overexpressing UNSΔMI, therefore suggesting a dominant-negative effect of SOC1 and AGL24 ectopic expression in Arabidopsis flowers. Likewise, tobacco transgenic flowers had leaf-like sepals and elongated ovaries with a ridged shape and trichomes developing all over the stylar surface. STMADS16 belongs to the AGL24 clade together with petunia FBP13, and a similar dominant-negative effect of the potato MADS box gene is likely to occur in transgenic tobacco flowers.
As stated before, the UNS protein lacking the MADS domain and the I region cannot act as a transcription factor; evidence for this phenomenon came from the localization experiments in petunia protoplasts. It has been shown with Arabidopsis and petunia MADS box proteins that dimer formation is necessary for the translocation into the nucleus; when only one of the two interacting proteins is present, the localization of either one is exclusively cytoplasmic (McGonigle et al., 1996; Immink et al., 2002). Dimerization as a prerequisite to target a transcription factor complex to the nucleus appears to be a general mechanism that has been shown to occur also with the minichromosomal maintenance complex in yeast and the Extradenticle/Homotorax homeodomain complex in Drosophila melanogaster (Abu-Shaar et al., 1999; Pasion and Forsburg, 1999). Similarly in petunia, the UNS protein is targeted to the nucleus only when the dimerization partner FBP9 is present because the UNS protein itself is not able to form a homodimer. Conversely, UNSΔMI still retains the ability to dimerize with FBP9, but it prevents the complex from being targeted to the nucleus, most likely because of the lack of the nuclear localization signal. The fact that dimerization still occurs is proven by the cytoplasmic localization of FBP9. This protein is able to homodimerize, and when present alone in a protoplast, it is only detectable in the nucleus. The affinity for heterodimer formation with UNSΔMI is apparently higher than for homodimer formation; UNSΔMI/FBP9 complexes are assembled rather than FBP9/FBP9 homodimers. The hypothesis of a dominant-negative interaction of UNSΔMI is in line with the phenotype that the heterologous protein generated in Arabidopsis. Besides features similar to the ones observed in petunia transformants, such as elongated hairy pistils and green leaf-like petals, additional ap1-like characteristics were prominent in transgenic Arabidopsis plants (i.e., bract-like sepals and new flower buds arising at the axils of the petals). Because these two features are typical for the Arabidopsis ap1 mutant, we tested the ability of UNSΔMI to dimerize with AP1 protein. The positive results obtained with the yeast two-hybrid system confirmed our assumption of a dominant-negative action of the petunia protein in the Arabidopsis floral whorls. This was also supported by the partial complementation that occurred in plants overexpressing both AP1 and UNSΔMI.
The truncated UNS protein can sequester its dimerization partners in the cytoplasm, preventing them from entering the nucleus and, thus, from functioning as transcriptional activators. Therefore, the phenotype observed in transgenic flowers is because of a dominant-negative action of UNSΔMI toward other MADS box proteins involved in the maintenance of specific floral organ characteristics. Because of the identical phenotypic alterations produced in petunia flowers by the wild-type UNS protein, we propose that a similar action toward the corresponding floral factors is accomplished by UNS in the flower domains. However, we cannot conclude at this stage whether UNS is acting directly on specific target genes in a dominant-negative way or whether it simply prevents other proteins from activating transcription by blocking their entrance into the nucleus. In the first hypothesis, UNS would bind target sites present in promoters, but without interacting with the correct transcription activator partners, it would prevent an active transcription complex to assemble at the same binding site. This competition between dominant-negative form and an active transcription factor complex has been shown to occur in mammals with the murine SRF factor (Belaguli et al., 1999). It has also been proposed as a possible mode of action for the negative regulator of floral transition SVP in Arabidopsis (Hartmann et al., 2000).
Based on the results described in this article, we suggest a role for UNS in promoting the transition from vegetative to reproductive development in a way that is similar to the Arabidopsis SOC1 and that most likely involves the interaction with other related MADS box proteins, such as FBP13 in petunia and AGL24 in Arabidopsis. We also provide evidence that the phenotypic alterations caused in the flowers by the ectopic expression of UNS do not reflect a gain-of-function phenotype but that they are the result of a dominant-negative effect of the transgene.
METHODS
Plant Material
Petunia hybrida lines W115 and W138, Arabidopsis thaliana ecotype Wassilewskija, and transgenic plants were grown under normal greenhouse conditions (22°C, 14 h light/10 h dark, LD). For SD treatments, 8 h light/16 h dark was applied.
Protein Sequence Analysis
Amino acid sequence comparisons were performed using the ClustalW multiple sequence alignment program (Thompson et al., 1994). Amino acid alignments, including the MADS box, the I region, and the K box, were used to obtain the phylogenetic tree with the Protdist and neighbor-joining programs of the PHYLIP 3.5c package (provided by J. Felsenstein, Department of Genetics, University of Washington, Seattle, WA). Bootstrap analysis was performed with Seqboot on 100 data sets, and branches with values of <50 were collapsed. Also included in the tree were sequences of MADS box genes from Arabidopsis, tobacco (Nicotiana tabacum), Sinapis alba, tomato (Lycopersicon esculentum), Brassica, Cardamine flexuosa, potato (Solanum tuberosum), and rice (Oryza sativa). AP3 from Arabidopsis was used as an outgroup.
Construction of Binary Vectors and Plant Transformation
The complete open reading frame (ORF) of UNS was generated by PCR using a 5′ primer on the ATG containing a HindIII site and a reverse primer designed after the STOP codon with a KpnI site. The PCR fragment was placed downstream of the double 35S promoter in the pGD120 plasmid (Immink et al., 2002), and the expression cassette was subsequently cloned into the pBIN19 binary vector (Bevan, 1984). The same cloning strategy was used to generate the AP1 overexpression vector.
The truncated ORF, UNSΔMI, lacking the MADS box and the I region, was also generated by PCR using the 5′ primer 5′-GGATCCATGCAAACTGAAAATCAAGCTGGG-3′, which introduces an ATG at bp231 of the original ORF and a BamHI recognition site, and the 3′ primer 5′-GGATCCGAAGAGGGATAGGTAGTCACC-3′, containing a BamHI site.
Transformation of petunia plants was performed as described before (Colombo et al., 1995). Transformation of Arabidopsis plants, ecotype Wassilewskija, was performed using the floral dip method as described by Clough and Bent (1998). C58C1 is the Agrobacterium tumefaciens strain used for the transformation.
RNA Gel Blot Analysis and in Situ Hybridization
Total RNA was isolated from different tissues of petunia and Arabidopsis plants according to Verwoerd et al. (1989). Ten micrograms of the total RNAs, denaturated with 1.5 M glyoxal, were fractionated on a 1.4% agarose gel and blotted onto Hybond N+ membrane. UNS, FBP28, FBP29, and AP1 gene-specific fragments were used as probes for hybridization. The probes were labeled by random oligonucleotide priming (Feinberg and Vogelstein, 1984), and blots were hybridized as described by Angenent et al. (1992).
In situ hybridizations were performed as described by Cañas et al. (1994). Digoxigenin-labeled RNA probes were synthesized by T7 polymerase-driven in vitro transcription from the PCR fragment containing the full-length UNS ORF according to the instructions of Boehringer Mannheim (Mannheim, Germany). The amplification product was obtained with the 5′ primer 5′- GTTCCTTGAAACATCTAAAAGG-3′ and the 3′ primer 5′-TAATACGACTCACTATAGGGATAGGTAGTCACCAATTAATTC-3′, containing the T7 polymerase promoter site.
Detection of UNS Knockout Mutants
A modified version of the PCR-based screening described by Koes et al. (1995) was adopted to identify dTph1 insertions in the UNS gene. The UNS-specific reverse primer 5′-TTCATATTGCTTGCCTCTAGGTG-3′ designed at the end of the MADS box and the dTph1 inverted repeat primer with an extra EcoRI site (5′-GAATTCGCTCCGCCCCTG-3′) were used in the PCR reactions on two different populations of 3840 plants each. The PCR products, blotted onto Hybond N+ membrane, were hybridized with the UNS cDNA.
Scanning Electron Microscopy
Samples were mounted on a stub, frozen in liquid nitrogen, coated, and observed as described by Angenent et al. (1995).
Protein–Protein Interaction
The full-length UNS and the truncated UNSΔMI were tested for dimerization with 22 known petunia proteins (Immink et al., 2003) using the Stratagene CytoTrap vector kit (catalog number 217438; La Jolla, CA). The assays were performed as described by Immink et al. (2003).
Construction of CFP/YFP Plasmids
C-terminal in-frame fusion proteins between FBP9, UNS, or UNSΔMI and CFP or YFP proteins were obtained by PCR amplification of the complete ORF of the MADS box genes and subsequent cloning in pECFP and pEYFP (catalog numbers 6075-1 and 6004-1, respectively; Clontech, Palo Alto, CA). MADS box gene–specific primers, introducing a SalI restriction site at the 5′-end and a BamHI site at the 3′-end, were used in the PCR reactions. The 3′ BamHI site replaces the endogenous stop codon. The obtained plasmids were digested with SalI and XbaI, and the chimerical genes were cloned in the expression vector pGD120 (Immink et al., 2002).
Protoplast Transfection
Protoplasts were prepared from W115 petunia leaves and transfected as described by Denecke et al. (1989). Before protoplasts isolation, leaves were surface sterilized by incubation in 1% bleach for 20 min. All measurements and imaging experiments were done after overnight incubation at 25°C in the dark.
Localization Studies
The localization of the fluorescent fusion proteins was analyzed by confocal scanning laser microscopy analyses (CSLM 510; Carl Zeiss, Jena, Germany). Protoplasts were excited by 458- and 514-nm argon laser lines controlled by an acousto-optical tunable filter. For CFP, an HFT458 dichroic mirror and BP470-500 emission filter were used, and for YFP, an HFT514 dichroic mirror and BP535-590 IR emission filter were used. Images were obtained with a 40× oil immersion objective. Protoplasts were scanned with step size of 101 μm (x axis) by 7.4 μm (y axis). The pinhole was set at 60 μm, corresponding to a theoretical thickness of ∼1 μm. Images were analyzed and adapted with Zeiss LSM510 software.
Supplementary Material
Acknowledgments
We thank Jos Mol for critical reading of the manuscript and helpful suggestions. Adriaan van Aelst is acknowledged for the scanning electron microscopy analysis. This project was in part financed by the European Union (BIO4 CT 972217) and the Ministry of Agriculture, Nature, and Food Quality.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is: Gerco C. Angenent (gerco.angenent@wur.nl).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019679.
References
- Abu-Shaar, M., Ryoo, H.D., and Mann, R.S. (1999). Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent, G.C., Busscher, M., Franken, J., Mol, J.N.M., and Van Tunen, A.J. (1992). Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent, G.C., Franken, J., Busscher, M., Van Dijken, A., Van Went, J., Dons, H.J.M., and Van Tunen, A.J. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaguli, N.S., Schildmeyer, L.A., and Schwartz, R.J. (1997). Organization and myogenic restricted expression of the murine serum response factor gene. J. Biol. Chem. 272, 18222–18231. [DOI] [PubMed] [Google Scholar]
- Belaguli, N.S., Zhou, W., Trinh, T.H.T., Majesky, M.W., and Schwartz, R.J. (1999). Dominant negative murine serum response factor: Alternative splicing within the activation domain inhibits transactivation of serum response factor binding targets. Mol. Cell. Biol. 19, 4582–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M.W. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme, F., Kurz, B., Melzer, S., Bernier, G., and Jacqmard, A. (2000). Cytokinin and gibberellin activate SaMADS A, a gene apparently involved in regulation of the floral transition in Sinapis alba. Plant J. 24, 103–111. [DOI] [PubMed] [Google Scholar]
- Borner, R., Kampmann, G., Chandler, J., Gleißner, R., Wisman, E., Apel, K., and Melzer, S. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Alvarez, J., Weigel, D., Meyerowitz, E.M., and Smyth, D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743. [Google Scholar]
- Burn, J.B., Smyth, D.R., Peacock, W.J., and Dennis, E.S. (1993). Genes conferring late flowering in Arabidopsis thaliana. Genetica 90, 147–155. [Google Scholar]
- Cañas, L.A., Busscher, M., Angenent, G.C., Beltran, J.P., and Van Tunen, A.J. (1994). Nuclear localization of the petunia MADS box protein FBP1. Plant J. 6, 597–604. [Google Scholar]
- Carmona, M.J., Ortega, N., and Garcìa-Maroto, F. (1998). Isolation and characterization of a new vegetative MADS-box gene from Solanum tuberosum L. Planta 207, 181–188. [DOI] [PubMed] [Google Scholar]
- Clarke, J.H., and Dean, C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242, 81–89. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colombo, L., Franken, J., Koetje, E., Van Went, J., Dons, H.J.M., Angenent, G., and Van Tunen, A.J. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croissant, J.D., Kim, J., Eichele, G., Goering, L., Lough, J., Prywes, R., and Schwartz, R.J. (1996). Avian serum response factor expression restricted primarily to muscle cell lineages is required for α-actin gene transcription. Dev. Biol. 177, 250–264. [DOI] [PubMed] [Google Scholar]
- Denecke, J., Gosselé, V., Botterman, J., and Cornelissen, M. (1989). Quantitative analysis of transiently expressed genes in plant cells. Methods Mol. Cell. Biol. 1, 19–27. [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1984). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137, 266–267. [DOI] [PubMed] [Google Scholar]
- Garcìa-Maroto, F., Ortega, N., Lozano, R., and Carmona, M.J. (2000). Characterization of the potato MADS-box gene STMADS16 and expression analysis in tobacco transgenic plants. Plant Mol. Biol. 42, 499–513. [DOI] [PubMed] [Google Scholar]
- Gerats, A.G.M., Huits, H., Vrijlandt, E., Maraña, C., Souer, E., and Beld, M. (1990). Molecular characterization of a nonautonomous transposable element (dTph1) of petunia. Plant Cell 2, 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Q., Ferrándiz, C., Yanofsky, M.F., and Martienssen, R. (1998). The FRUITFUL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Hartmann, U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H., and Huijser, P. (2000). Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Valverde, F., Ravenscroft, D., Mouradov, A., and Coupland, G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink, R.G.H., Ferrario, S., Busscher-Lange, J., Kooiker, M., Busscher, M., and Angenent, G.C. (2003). Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genomics 268, 598–606. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Gadella, T.W., Jr., Ferrario, S., Busscher, M., and Angenent, G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99, 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink, R.G.H., Hannapel, D.J., Ferrario, S., Busscher, M., Franken, J., Campagne, M.M.L., and Angenent, G.C. (1999). A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126, 5117–5126. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Sussex, I.M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S., Chung, Y., Kim, S., Lee, Y.H., Cho, Y., and An, G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, F.-E., and Prywes, R. (1993). Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol. Cell. Biol. 13, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.W., Shin, J.H., Moon, J., Kim, M., Lee, J., Park, M.C., and Lee, I. (2003). The function of the flowering time gene AGL20 is conserved in Crucifers. Mol. Cells 16, 136–141. [PubMed] [Google Scholar]
- Koes, R., et al. (1995). Targeted gene inactivation in petunia by PCR-based selection of transposon insertion mutants. Proc. Natl. Acad. Sci. USA 92, 8149–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J.H., Kim, S.-G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Bleecker, A., and Amasino, R.M. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1991). AGL1–AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5, 484–495. [DOI] [PubMed] [Google Scholar]
- Mandel, T., Lutziger, I., and Kuhlemeier, C. (1994). A ubiquitously expressed MADS-box gene from Nicotiana tabacum. Plant Mol. Biol. 25, 319–321. [DOI] [PubMed] [Google Scholar]
- McGonigle, B., Bouhidel, K., and Irish, V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10, 1812–1821. [DOI] [PubMed] [Google Scholar]
- Menzel, G., Apel, K., and Melzer, S. (1996). Identification of two MADS box genes that are expressed in the apical meristem of the long-day plant Sinapis alba in transition to flowering. Plant J. 9, 399–408. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS-domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M., and Amasino, R.M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33, 867–874. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., Huang, H., Tudor, M., Hu, Y., and Ma, H. (1996). Functional domains of the floral regulator AGAMOUS: Characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell 8, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J., Suh, S.-S., Lee, H., Choi, K.-R., Hong, C.B., Paek, N.-C., Kim, S.-G., and Lee, I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M., and Yanofsky, M.F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2,186–195. [DOI] [PubMed] [Google Scholar]
- Okamoto, S., Krainc, D., Sherman, K., and Lipton, S.A. (2000). Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc. Natl. Acad. Sci. USA 97, 7561–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatsky, O.I., Andreucci, J.J., and McDermott, J.C. (1997). A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272, 33271–33278. [DOI] [PubMed] [Google Scholar]
- Pařenicová, L., De Folter, S., Kieffer, M., Horner, D.S., Favalli, C., Busscher, J., Cook, H.E., Ingram, R.M., Kater, M.M., Davies, B., Angenent, G.C., and Colombo, L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasion, S.G., and Forsburg, S. (1999). Nuclear localization of Schizosaccaromyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell 10, 4043–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli, L., Abu-Abeid, M., Zamir, D., Nacken, W., Schwarz-Sommer, Z., and Lifschitz, E. (1991). The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1, 255–266. [PubMed] [Google Scholar]
- Ratcliffe, O.J., Nadzan, G.C., Reuber, T.L., and Riechmann, J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., and Riechmann, J.L. (2002). Arabidopsis transcription factors and the regulation of flowering time: A genomic perspective. Curr. Issues Mol. Biol. 4, 77–91. [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101. [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of COSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Scortecci, K.C., Michaels, S.D., and Amasino, R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Conn, A.B., Dennis, E.S., and Peacock, W.J. (2002). Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14, 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Rouse, D.T., Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, P., and Sharrocks, A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Simon, R., Igeño, M.I., and Coupland, G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384, 59–62. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Sink, K.C., ed (1984). Monographs on Theoretical and Applied Genetics 9: Petunia. (Berlin: Springer-Verlag), pp. 211–215.
- Sommer, H., Beltran, J.P., Huijser, P., Pape, H., and Lonnig, W.-E. (1990). DEFICIENS, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege, M., Sheldon, C.C., Gelliwell, C.A., Upadhyaya, N.M., Dennis, E.S., and Peacock, W.J. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotech. J. 1, 361–369. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, T.-Y., and Yang, C.-H. (2001). A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol. 42, 1156–1168. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, M., Zethof, J., Souer, E., Koes, R., Tornielli, G.B., Pezzotti, M., Ferrario, S., Angenent, G.C., and Gerats, T. (2003). Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15, 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, I.L., Drews, G., Feldmann, K., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Yu, H., Xu, Y., Tan, E.L., and Kumar, P.P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 99, 16336–16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and Forde, B.G. (1998). An Arabidopsis MADS box gene that controls nutrient induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.