Abstract
In contrast with the class A heat stress transcription factors (HSFs) of plants, a considerable number of HSFs assigned to classes B and C have no evident function as transcription activators on their own. However, in the following article, we provide evidence that tomato (Lycopersicon peruvianum) HsfB1 represents a novel type of coactivator cooperating with class A HSFs (e.g., with tomato HsfA1). Provided the appropriate promoter architecture, the two HSFs assemble into an enhanceosome-like complex, resulting in strong synergistic activation of reporter gene expression. Moreover, HsfB1 also cooperates in a similar manner with other activators, for example, with the ASF1/2 enhancer binding proteins of the 35S promoter of Cauliflower mosaic virus or with yet unidentified activators controlling housekeeping gene expression. By these effects, HsfB1 may help to maintain and/or restore expression of certain viral or housekeeping genes during ongoing heat stress. The coactivator function of HsfB1 depends on a histone-like motif in its C-terminal domain with an indispensable Lys residue in the center (GRGKMMK). This motif is required for recruitment of the plant CREB binding protein (CBP) ortholog HAC1. HsfA1, HsfB1, and HAC1/CBP form ternary complexes in vitro and in vivo with markedly enhanced efficiency in promoter recognition and transcription activation in plant and mammalian (COS7) cells. Using small interfering RNA–mediated knock down of HAC1 expression in Arabidopsis thaliana mesophyll protoplasts, the crucial role for the coactivator function of HsfB1 was confirmed.
INTRODUCTION
All eukaryotic heat stress (HS)–inducible genes share conserved promoter elements with the palindromic consensus motif formed by a purine-rich head module (H) and a pyrimidine-rich tail module (T): 5′-AGAAnnTTCT-3′ (Pelham, 1982; Nover, 1987). They represent the recognition sites for the corresponding HS transcription factors, which have a modular structure with an N-terminal DNA binding domain characterized by a central helix-turn-helix motif, an adjacent domain with heptad hydrophobic repeats (HR-A/B region) involved in oligomerization, short peptide motifs essential for nuclear import (nuclear localization signals [NLS]) and export, and a C-terminal activation domain (Döring et al., 2000; Heerklotz et al., 2001).
A unique feature of plants is the multiplicity of heat stress transcription factors (HSFs) with 20 to 30 representatives, as derived from the Arabidopsis thaliana and rice (Oryza sativa) genome sequences and EST databank searches. Based on the structure of their HR-A/B regions, plant HSFs belong to three different classes. Only for class B HSFs, the compact structure of the HR-A/B region corresponds to the oligomerization domains of all nonplant HSFs, whereas class A and class C HSFs have extended HR-A/B regions because of insertions of 21 and 7 amino acid residues, respectively (Morimoto, 1998; Nover et al., 2001). Additional features for discrimination of the three classes reside in their C-terminal domains (CTDs). The CTDs of most class A HSFs are acidic with short peptide motifs containing central Trp or Phe residues (AHA motifs), which are essential for the activator function (Treuter et al., 1993; Döring et al., 2000). Similar motifs also have been identified as parts of the activator regions of mammalian and yeast (Saccharomyces cerevisiae) transcription factors (for references, see Nover and Scharf, 1997; Döring et al., 2000). In contrast with this, class B HSFs have no AHA motifs, and their CTDs are neutral or even positively charged. In reporter assays, class B HSFs act as weak activators or even as attenuators, if tested together with class A HSFs (Treuter et al., 1993; Czarnecka-Verner et al., 2000). According to the amino acid composition of their activation domains, transcription factors are generally classified as acidic, Gln rich or Pro rich (Regier et al., 1993; Tjian and Maniatis, 1994; Goodrich et al., 1996; Ptashne and Gann, 1997; Kadonaga, 1998). Following this concept, plant class A HSFs belong to the acidic activators, whereas class B HSFs do not correspond to any of these classes. Their CTDs are neither acidic nor Gln or Pro rich.
In support of the basic structural differences between class A and class B HSFs, only class A HSFs were found to replace the yeast Hsf1 in its survival functions in the corresponding hsf1 disruption strain (Boscheinen et al., 1997; Bharti et al., 2000). Moreover, cosuppression of tomato (Lycopersicon peruvianum) plants with small interfering RNA–mediated knock down of HsfA1 expression are unable to mount an HS response, and they are extremely heat sensitive. However, in mesophyll protoplasts, the wild-type level of thermotolerance could be restored by HsfA2 and other class A Hsfs but not by class B or class C HSFs (Mishra et al., 2002).
Probably intimately connected with the complexity of the plant HSF family are peculiarities of HSF-dependent promoters. In most cases, they are characterized by complex patterns of heat shock element (HSE) clusters frequently embedded in binding sites for other transcription factors. Although details are far from clear, the particular combinations of binding sites may be decisive for the expression patterns of HS-inducible genes (Nover, 1987, 1991). The existence of elaborate clusters of HSEs in promoter regions of HS-inducible genes of plants was recognized very early (Schöffl et al., 1984; Czarnecka et al., 1985; Nagao et al., 1985). On the basis of the Arabidopsis genome, we made a more detailed promoter analysis of genes encoding members of the Hsp20 (Scharf et al., 2001) and HSF families (Nover et al., 2001). It is striking that many HSE clusters contain defective head (h) or tail (t) modules, for example, because of a replacement of the invariant G residue in the head or C residue in the tail module (for explanations, see Figure 1A and Supplemental Table S1 online). It will be shown elsewhere (S.K. Baniwal and K. Bharti, unpublished data) that this pattern is important for the proper positioning and interaction of HSFs in the transcription complex.
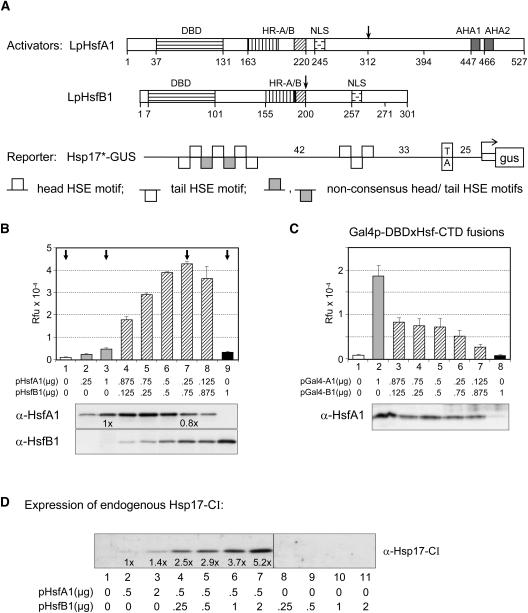
Figure 1.
Influence of Synergistic Interactions between HsfA1 and HsfB1 on Reporter Gene Activation in Tobacco Protoplasts.
(A) Block diagrams show basic structure of the two HSFs with their DNA binding domains (DBD), oligomerization domains (HR-A/B), NLS, and the two C-terminal activator motifs of HsfA1 (AHA1 and AHA2). The pHsp17*-GUS reporter contains a fragment of the soybean hsp17.3B-CI promoter (Schöffl et al., 1984) with a TATA proximal HTH trimer and a TATA distal THtHtHT cluster as potential HSF binding sites. Numbers indicate distance in base pairs.
(B) Expression of the pHsp17*-GUS reporter was tested in protoplasts transformed with the indicated amounts of plasmids encoding HsfA1 and HsfB1. The immunoblots at the bottom indicate the expression levels of both HSFs. Arrows mark the standard samples used for the subsequent figures with GUS reporter assays. Rfu, relative fluorescence units.
(C) Reporter assay with pGal4DBS-GUS and fusion activators Gal4DBDxHsfA1(amino acids 312 to 527) and Gal4DBDxHsfB1(amino acids 200 to 301). See arrows in (A) for the fusion points in HsfA1 and HsfB1, respectively. Expression levels of the HsfA1 fusion protein were detected by α-HsfA1, whereas the HsfB1 fusion protein could not be detected by our α-HsfB1 antiserum.
(D) Stimulation of chromosomal Hsp17-CI expression by coexpression of HsfA1 and HsfB1. The numbers, derived from densitometer scans, indicate the relative increase of Hsp17-CI expression as compared with sample 2 with HsfA1 alone.
To investigate the intriguing coexistence of class A and B HSFs in more detail, we tested HSF-dependent reporter constructs in tobacco (Nicotiana tabacum) protoplasts cotransformed with expression vectors encoding tomato HsfA1 and HsfB1. Depending on the promoter context, we observed strong synergistic effects, which can exceed the activator potential of HsfA1 alone by one order of magnitude. The coactivator function of HsfB1 depends on a histone-like motif in its C terminus (GRGKMMK) mediating the recruitment of histone acetyl transferase HAC1/CREB binding protein (CBP). Interestingly, the role of HsfB1 as coactivator also is observed with a given set of constitutively active promoters, providing the basis for the maintenance or even enhancement of transcription of certain housekeeping or viral genes during HS.
RESULTS
Synergistic Activation of Reporter Gene Expression by HsfA1 and HsfB1
A characteristic feature of the tomato HS response is a transient increase in HsfB1 level during the early phase. This led us to speculate about the role of a changing mixture of HSFs on the pattern of gene expression during the ongoing HS response (Scharf et al., 1990; Treuter et al., 1993). To investigate the role of HsfB1 as a functional partner of HsfA1, we tested different combinations of both in reporter assays performed in tobacco protoplasts in the presence of a ß-glucuronidase (GUS) reporter plasmid harboring a natural promoter fragment derived from the soybean (Glycine max) Hsp17.3B-CI gene (Figure 1A). The promoter fragment in the reporter pHsp17*-GUS contains two HSF binding sites, namely a TATA proximal head-tail-head (HTH) trimer and a TATA distal cluster of seven HSE modules (THtHtHT) with two defective tail modules (t) separating the central head module from the two peripheral TH and HT dimers (for sequence definitions, see Supplemental Table S1 online).
The GUS activity in sample 1 (Figure 1B) reflects the very low expression level resulting from the endogenous HSFs of the protoplasts. Compared with this, transformation with either 1 μg of HsfA1 encoding plasmid (sample 3) or 1 μg of HsfB1 encoding plasmid (sample 9) caused a 10- and 3-fold increase of GUS activities. These results were similar to those reported earlier when investigating the transactivation potential of both HSFs alone (Treuter et al., 1993; Döring et al., 2000). Most interesting was the outcome with protoplast samples expressing mixtures of decreasing amounts of HsfA1 and increasing amounts of HsfB1 (Figure 1B, samples 4 to 8). Compared with sample 3, the GUS expression levels increased approximately eightfold in sample 7 (i.e., in protoplasts transformed with a mixture of 0.25 μg of HsfA1 and 0.75 μg of HsfB1 encoding plasmids). The immunoblots at the bottom of Figure 1B serve as expression controls for the two HSFs.
Evidently, two effects of HsfB1 contribute to the outcome of this experiment: (1) HsfB1 acts as coactivator of HsfA1, resulting in an increased GUS expression level, and (2) the expression of the HsfA1 cassette connected with the 35S promoter of Cauliflower mosaic virus (CaMV35S) is enhanced. Despite the reduction in the amount of the HsfA1 encoding plasmid from 1 μg (sample 3) to 0.25 μg (sample 7), the level of HsfA1 was reduced only by 20% (see sample 2 with 0.25 μg of HsfA1 expression plasmid). More detailed explanations on the latter effect will be given in the context of Figure 2.
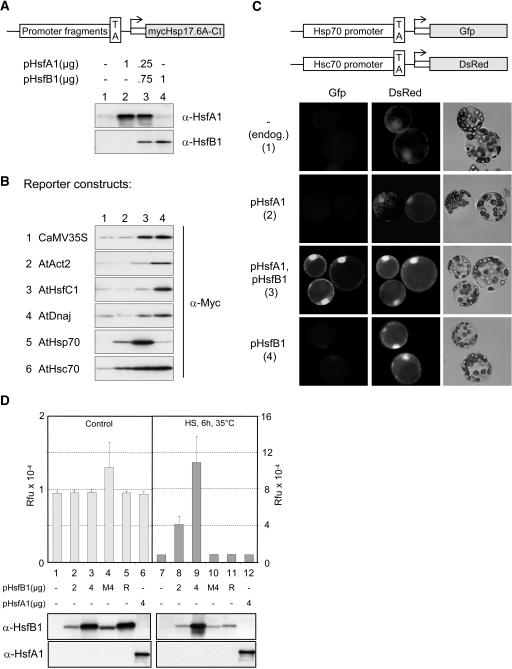
Figure 2.
Activation of Viral and Housekeeping Promoters by HsfB1 in a Transient Reporter Assay.
(A) Block diagram of the mycHsp17.6A-CI reporter constructs used in (B) and expression controls of the four standard samples (see arrows in Figure 1B).
(B) Immunoblot analyses of the expression levels obtained with reporter constructs harboring promoter/leader fragments of the indicated genes (for details, see Supplemental Table S1 online).
(C) Single cell reporter assay in tobacco protoplasts cotransformed with the indicated HSF expression plasmids and the two reporters with promoter/leader fragments derived from the AtHsp70 and AtHsc70 encoding genes (see block diagrams at top).
(D) Expression of a chromosomal CaMV35S-GUS construct in mesophyll protoplasts of tobacco transformed with the indicated plasmids encoding wild-type and mutant forms of HsfB1 and HsfA1, respectively. M4, DNA binding mutant of HsfB1; R, mutant of HsfB1 with K275R. Four micrograms of expression plasmid was used for M4 and R (for the detail of mutants, see Figure 3 and Methods). Protoplast samples represented in the left part were kept under control conditions; protoplasts in the right part were heat stressed for 6 h at 35°C before harvesting. Note the differences of scales. Immunoblots at the bottom reflect expression levels of HsfB1 and HsfA1.
The synergistic interactions between HsfA1 and HsfB1 depend on the promoter context (i.e., the proper positioning of both HSFs with respect to each other). Two special aspects are worth being mentioned. (1) In the soybean Hsp17.3B promoter fragment (Figure 1A), the TATA distal THtHtHT cluster was necessary and sufficient for the effect, whereas the TATA proximal HTH trimer was dispensible (data not shown). (2) Many other reporter constructs containing promoter fragments of Arabidopsis small heat stress protein (sHsp) encoding genes, for example, of Hsp17.4-CI, Hsp17.8-CI, Hsp17.6-CII, Hsp23.6-M, and Hsp25.3-P encoding genes (for identification, see Scharf et al., 2001), were tested under comparable conditions. All showed medium or strong effects of synergistic activation by HsfA1 and HsfB1.
However, there was no synergistic effect observed upon coexpression of the corresponding Gal4DBDxHsfCTD fusion proteins tested with the pGal4DBS-GUS. In contrast with the situation with the pHsp17*-GUS reporter, expression of the pGal4DBS-GUS reporter was very high with the HsfA1 fusion activator alone (Figure 1C, sample 2), and similar to results reported for a comparable test situation by Czarnecka-Verner et al. (2000), it was markedly reduced in the presence of increasing amounts of HsfB1 fusion protein (samples 3 to 7). The high affinity promoter of the Gal4DBS reporter is excellent for testing the activator potential of HSFs (Döring et al., 2000), but it is inappropriate for detection of the more subtle effects forming the basis of synergism.
To confirm that the synergistic gene activation by HsfA1 and HsfB1 was not restricted to plasmid-borne reporters, we analyzed the expression of endogenous sHsp encoding genes of tobacco protoplasts using specific antibodies against Hsp17-CI. As can be seen in Figure 1D, the mild stimulation of Hsp17-CI expression caused by HsfA1 alone (lanes 1 to 3) could be strongly enhanced by increasing levels of HsfB1 (lanes 4 to 7), whereas HsfB1 alone had no effect (lanes 8 to 11). The response of the chromatin-embedded sHsp encoding genes was basically similar to that observed in experiments with the pHsp17*-GUS reporter (Figure 1B).
Function of HsfB1 for Maintenance of Housekeeping Gene Transcription
The synergistic effect of HsfB1 evidently was not restricted to cooperation with HsfA1 on HS promoters, but it also extended to constructs harboring CaMV35S promoter (Figure 1B). Closer inspection of the 35S promoter demonstrates that the well-known enhancer region may have two functions. On the one hand, it represents the binding sites for ASF-1 and ASF-2 transcription factors, which are responsible for the strong constitutive activity of the promoter in many tissues (Katagiri et al., 1989; Lam and Chua, 1989; Lam, 1994; Niggeweg et al., 2000). On the other hand, it contains several potential recognition sites for an HSF dimer, which are interspersed with the ASF-1/2 binding sites (see details in Supplemental Table S1 online). Hence, we assumed that the threefold to fourfold stimulation of the plasmid-encoded HsfA1 expression in the presence of HsfB1 (Figure 1B) represents the synergistic interaction between the transcription factors binding to the 35S enhancer and HsfB1.
To provide more insight into this effect and to support the hypothesis that HsfB1 functions as general coactivator for the transcription of defined viral and housekeeping genes, we performed immunoblot analyses with a Myc-tagged Hsp17.6-CI as reporter gene, which was under the control of different 5′ upstream regions of constitutively expressed genes (see block diagram in Figure 2A). All reporters were tested with the standard combination marked by arrows in Figure 1B. The corresponding plasmids used for transformation and the immunoblot controls for expression of HsfA1 and HsfB1 are shown in Figure 2A (samples 1 to 4).
First, we used the 35S promoter construct (Figure 2B, construct 1) to investigate the influence of HsfA1 and HsfB1 on the expression level of Myc-Hsp17.6. As expected from the predicted interaction with the enhancer binding proteins, there was a strong stimulation of Hsp17.6 expression in the presence of HsfB1 (lanes 3 and 4), but presence of HsfA1 alone had no effect (lane 2). HS did not influence the outcome of the experiment, and we confirmed by RT-PCR that the stimulation resulted from a marked increase of the mRNA levels encoding the Hsp17.6 reporter protein (data not shown).
Second, we tested the general validity of our hypothesis that HsfB1 acts as a coactivator for maintenance of housekeeping gene expression using many other reporters harboring promoter/leader fragments of the Arabidopsis Actin2, HsfC1, DnaJ, Hsc70, and Hsp70 encoding genes (Figure 2B, constructs 2 to 6). All reporters contain the complete 5′ upstream regions, including a 5′ untranslated region and a TATA box fused immediately upstream of the ATG start codon of the mycHsp17.6 expression cassette. They were all tested in the standard conditions as shown in Figure 2A. With the remarkable exception of the Hsp70 reporter (construct 5), the results were basically similar to those obtained with the 35S reporter (Figure 2B, construct 1). HsfB1 alone stimulated the Myc-Hsp17.6 expression approximately fourfold (lane 4), and HsfA1 had no effect (lane 1 versus 2) or even reduced the positive influence of HsfB1 (lane 3). This is particularly pronounced for reporters 2 and 3. In fact, closer inspection of the Hsp17.6 expression patterns show characteristic differences between the five reporters with promoter fragments derived from non-HS promoters (constructs 1 to 4 and 6). This indicates that the regulatory fingerprint of each construct is context dependent involving additional factors present in the tobacco protoplasts. Unfortunately, the lack of knowledge about the functional anatomy of these promoters precludes further discussions on this interesting point. At any rate, we identified potential HsfB1 binding sites in all of them (for sequence details, see Supplemental Table S1 online).
A striking example for the role of the promoter context stems from the comparison of results obtained with reporters constructed with the Hsp70 and Hsc70 promoter fragments (Figure 2B, constructs 5 and 6). The latter, representing the constitutively expressed gene, showed a response that was similar to the other constitutively expressed genes (constructs 1 to 4). However, the Hsp70 encoding gene in Arabidopsis tissues is expressed almost exclusively under HS conditions (Lin et al., 2001). In agreement with this, expression of the corresponding reporter construct was stimulated in the presence of HsfA1 but much more by the combination of HsfA1 and HsfB1. In this case, HsfB1 alone had no effect. The results were similar to those with the pHsp17*-GUS (Figure 1B).
Using this Hsc70/Hsp70 pair of reporters in a single cell reporter assay, we wanted to illustrate the interesting interplay of the two HSFs as part of a dynamically changing transcription machinery in the course of the HS response. To this aim, we fused the promoter/leader fragments of the Hsc70 and Hsp70 encoding genes to the autofluorescent proteins DsRed and green fluorescent protein, respectively. As shown before, both reporters were coexpressed in tobacco protoplasts in the absence or presence of HsfA1 and/or HsfB1 (Figure 2C). The DsRed fluorescence reflecting expression of the constitutively active construct was visible in all samples, albeit enhanced in the two protoplast samples expressing HsfB1 (samples 3 and 4). In contrast with this, the green fluorescence was very low except in protoplasts coexpressing HsfA1 and HsfB1 (sample 3). This situation reflects the early phase of a HS response in tomato cells, when the activation of HsfA1 as master regulator mediates the transient increase of the HsfB1 level (Scharf et al., 1990; Mishra et al., 2002) and both HSFs together become part of a HS enhanceosome.
Similar to the situation with the HS promoters, we wanted to know whether the stimulation of the 35S promoter in the presence of HsfB1 also could be observed on a chromatin embedded transgene. To this aim, we used sterile tobacco plants harboring a CaMV35S-GUS transgene and tested GUS expression in mesophyll protoplasts transformed with the indicated expression plasmids encoding HsfA1 and HsfB1 (Figure 2D). At control temperatures, no effects were seen on the high level of constitutive GUS expression, neither in the presence of HsfA1 nor of HsfB1 (samples 1 to 6). However, this situation changed when protoplasts were shifted 16 h after transformation for 6 h to 35°C before harvesting. In this case, GUS expression levels were indeed markedly increased in the presence of HsfB1 (samples 8 and 9) but not in the presence of the two inactive mutant forms (HsfB1 M4 and R; for details, see Methods) nor in the presence of HsfA1 (sample 12). Because we never observed a comparable HS effect in other reporter assays with HsfB1, it is tempting to speculate that HS-induced changes of chromatin structure and/or depletion of other activators binding to the 35S enhancer improve the accessibility of HsfB1 binding sites. As a result, this leads to a 4- to 10-fold increase of GUS expression in the presence of HsfB1 (mark the two different scales of GUS activity in Figure 2D).
Functional Requirements of HSFs for Synergism
To investigate the structural prerequisites of HsfA1 and HsfB1 for synergistic gene activation, we tested mutant forms of both HsfA1 and HsfB1 (Figure 3). Similar to the standard samples in Figure 1B, GUS reporter activities were measured with the two HSFs alone (Figure 3A, samples 2 and 3) and with the combination of HsfA1 plus HsfB1 (Figure 3A, sample 4 and samples in Figures 3B to 3D). The reference sample 1 in Figure 3A represents the background GUS activity obtained with the endogenous HSFs of tobacco protoplasts. In Figure 3B, mutants of HsfA1 were combined with wild-type HsfB1, whereas in Figure 3C, mutants of HsfB1 were combined with wild-type HsfA1. The same type of mutant HSFs were used in both parts, namely, HSFs defective in DNA binding (sample 1), deletion forms lacking the oligomerization domain (sample 2), mutant HSFs defective in NLS function (sample 3), and finally, HSFs with deletions in their CTDs (sample 4). In all cases, the synergistic effect observed with the wild-type HsfA1 and HsfB1 (Figure 3A, sample 4) was abolished irrespective of which partner was mutated and independent of the type of mutation. The signals of the immunoblot analyses reflect the expression levels of the corresponding HSFs. As expected from the stimulatory effect of HsfB1 on the activity of the 35S promoter (Figure 1B), the HsfA1 levels were lower in most samples with inactive HsfB1 (e.g., samples 1 to 4 in Figure 3C and samples 6 to 9 in Figure 3D), but this does not affect the conclusions drawn from this experiment.
Figure 3.
Structural Requirements of HsfA1 and HsfB1 for Synergistic Activation of the pHsp17*-GUS Reporter.
Immunoblots (IB) in (A) to (D) reflect the expression levels of the HSFs. Mutant forms of HsfB1 in the following figures are abbreviated as follows: M4, DNA binding mutant (C1); Δ, mutant with deletion of amino acids 272 to 279 (D9); R, mutant with 272-GRGK>GKGR.
(A) GUS activities measured with the four standard samples (arrows in Figure 1B). Sample 4 transformed with HsfA1 and HsfB1 represents the reference for all samples in (B) to (D).
(B) and (C) GUS activities of samples coexpressing the indicated mutant forms of HsfA1 and wild-type HsfB1 (B) or, vice versa, the wild type of HsfA1 with mutant forms of HsfB1. Block diagrams of HsfA1 and HsfB1 (C) were shown in Figure 1A. For further details about the mutants, see Methods and Supplemental Table S1 online.
(D) Functional dissection of the CTD (amino acids 271 to 301) of HsfB1. Amino acid residues that changed in the mutant forms of HsfB1 are highlighted by underlined boldface letters. All GUS activities were measured by coexpressing HsfA1 and the indicated mutant forms of HsfB1.
The peculiarities of the class B HSFs with respect to their CTDs have already been mentioned in the Introduction. To get further information on this, we created several deletions and point mutations in HsfB1 and tested them in the standard combination with wild-type HsfA1 (Figure 3D). The C-terminal amino acid sequence of HsfB1 (amino acids 271 to 301) and its derivatives are indicated along with the GUS activities and the signals of the corresponding immunoblots. Reference for samples 2 to 12 is the GUS activity in sample 1 of Figure 3D with coexpression of wild-type forms of HsfA1 and HsfB1.
The results can be summarized as follows. (1) In contrast with the essential role of aromatic and large hydrophobic amino acid residues for the activator function of class A HSFs (Döring et al., 2000), the two aromatic residues (W286 and Y282) were not important for the activity of HsfB1 (Figure 3D, samples 2 and 3). (2) Deletion of the C-terminal 7 or 15 amino acid residues had no effect (samples 4 and 5), but deletion of 27 amino acid residues destroyed the activity (HsfB1ΔC274, sample 6). Evidently, an essential element for the coactivator function of HsfB1 resides in the 12 amino acid residues between positions 274 and 286. (3) Decisive insights into the central motif of the HsfB1 CTD came from analysis of a mutant designed to introduce a unique SalI site by exchanging GK274/275>VD. Surprisingly, this mutant form of HsfB1 was inactive (sample 7). The same was true for the K275>Q mutant (sample 8), for HsfB1 with an internal deletion of amino acid residues 272 to 279 (sample 9), and for the mutant form with replacement of the GRGK motif by LWTT (sample 10). Most intriguing was the complete lack of activity of the HsfB1 mutant form with a mutual exchange of the two basic residues in the GRGK motif (i.e., HsfB1-GKGR; sample 11). In contrast with this, change of the adjacent Lys residue (K278>R) had no effect (sample 12).
Evidently, the Lys residue 275 embedded in the 272-GRGKMMK motif is indispensible for the function of HsfB1 as coactivator of HsfA1. It cannot be replaced by the adjacent Lys residue (K278) or by a Lys residue introduced in position 273 (i.e., in the mutant form with the GKGR motif).
The results presented so far indicate that the synergistic coactivation of transcription by HsfB1 with its GRGK motif in a positively charged CTD depends on the promoter architecture providing the basis for adjacent positioning with an acidic activator (e.g., with HsfA1). To check the general validity of this statement, we used the pHsp17*-GUS reporter and tested different combinations of acidic activators with wild-type HsfB1 (Table 1, samples 1 to 4) and, on the other hand, wild-type HsfA1 in combination with HsfB1 fusion proteins containing CTDs of the orthologs from tobacco, soybean, and Arabidopsis (Table 1, samples 5 to 7). With the exception of sample 7, all combinations resulted in high reporter gene expression and 2.5- to 12.5-fold synergism (details of calculation are given in Table 1). Evidently, HsfA1 can be substituted by tomato HsfA2 (sample 2) or by HSF fusion activators harboring the acidic activation domains of the yeast Gal4p (sample 3) or the viral VP16 (sample 4). On the other hand, the CTD of tomato HsfB1 can be replaced by those of tobacco or soybean HsfB1 (samples 5 and 6) but not by that of Arabidopsis HsfB1 (sample 7). Inspection of sequence details of the four types of HsfB1 may give a clue for an explanation of these interesting differences. The GRGKMMK motif of LpHsfB1 corresponds to GSRGKMMK in tobacco and GPRLKESK in soybean but to GSRMTETK in Arabidopsis (i.e., the decisive Lys residue is replaced by a Thr residue in the AtHsfB1). Interestingly, AtHsfB1 was tested previously in tobacco protoplasts in combination with AtHsfA4a as activator using the synthetic high-affinity pHSE9-GUS reporter (Czarnecka-Verner et al., 2000). In support of the failure of AtHsfB1 in the assay for synergism (Table 1, sample 7), the authors observed a strong inhibition of the AtHsfA4a activity, most likely caused by competition for DNA binding.
Table 1.
Synergism among Acidic Activators and Class B HSFs
| No. | Activator | GUS Activity (Rfu)a | Synergism (Fold Activity) |
|---|---|---|---|
| Group A: Acidic activators tested with LpHsfB1 | |||
| 1 | LpHsfA1 | 45.8 ± 0.9 | 7.8× |
| 2 | LpHsfA2 | 42.7 ± 1.0 | 5.0× |
| 3 | HsfA1xGal4AD | 44.3 ± 4.5 | 10.0× |
| 4 | HsfA1xVP16AD | 51.0 ± 7.3 | 2.5× |
| Group B: Class B HSFs tested with LpHsfA1 | |||
| 5 | HsfB1xNtB1CTD | 52.0 ± 2.5 | 12.5× |
| 6 | HsfB1xGmB1CTD | 64.4 ± 1.1 | 9.5× |
| 7 | HsfB1xAtB1CTD | 5.0 ± 0.7 | NDb |
To evaluate the synergistic activation of GUS expression, tobacco protoplasts were transformed and tested in the four standard combinations of reporter and activators (Figure 1B, arrows). Results are given only for combination 4 (i.e., 0.25 μg of acidic activator coexpressed with 0.75 μg of class B HSF). In group A, 0.75 μg of LpHsfB1 was cotransformed with 0.25 μg of acidic activator, for example, tomato HsfA1 and HsfA2 as well as the two indicated fusion proteins of tomato HsfA1 (amino acids 23 to 394; see arrow in Figure 1A) with the yeast Gal4 and the viral VP16 activation domains. In group B, 0.25 μg of LpHsfA1 was cotransformed with 0.75 μg of the indicated fusion proteins of LpHsfB1 (amino acids 1 to 198) with the CTDs of tobacco (Nt), soybean (Gm), and Arabidopsis (At) HsfB1. For details of the fusion constructs, see Supplemental Table S1 online. Calculation of synergism is based on the following formula (exemplified for GUS activities in samples 1, 2, and 6 in Figure 1B): GUS(6) − GUS(1)/GUS(2) − GUS(1) = 11-fold.
Rfu, relative fluorescence units.
ND, not determined.
Synergism Results from Corecruitment of HAC1/CBP
The GRGKMMK motif of HsfB1 is reminiscent of the highly conserved N-terminal motifs of histones (e.g., of histone H4) with the marked Lys residues acetylated by the mammalian p300/CBP (Kimura and Horikoshi, 1998): 1-SGRGKGGKGLGKGGAK. The intriguing role of the GRGK motif both in histones and HsfB1 led us to investigate the influence of CBP in reporter assays with coexpression of HsfA1 and HsfB1. In animal cells, CBP is a 300-kD global coactivator with histone acetyl transferase (HAT) activity, interacting with many transcription factors either bound to the N-terminal domain or to the CTD of the protein (Sterner and Berger, 2000; Bannister and Miska, 2000; Chan and LaThangue, 2001; Yuan and Giordano, 2002).
Five CBP-like proteins (HAC1 to HAC5) were identified recently in Arabidopsis (Bordoli et al., 2001; Yuan and Giordano, 2002). All contain the most conserved central parts with the three Zn finger (C/H1 to C/H3) and the embedded HAT domains (see block diagrams in Figure 4A). Because HAC1 (PCAT2) was shown to have HAT activity (Bordoli et al., 2001), we cloned its full-length cDNA by RT-PCR from Arabidopsis leaf RNA (see Methods). For reporter assays and pull-down experiments, we also created vectors for expression of the N-terminal domain (NTD) and CTD of HAC1, marked by arrows in Figure 4A.
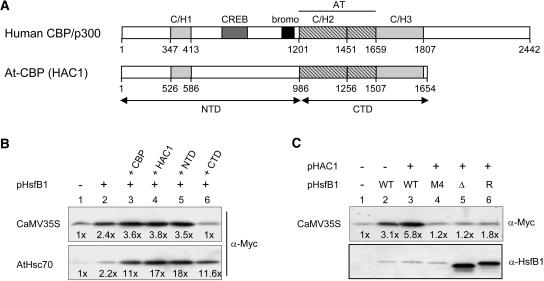
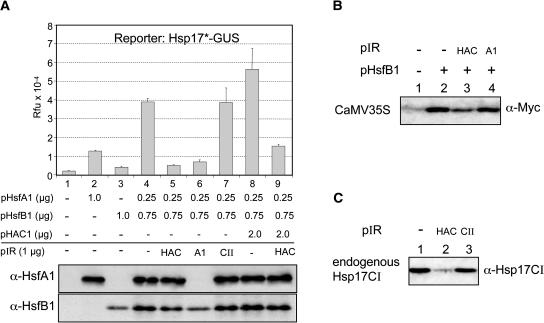
Figure 4.
Effect of Mammalian and Arabidopsis CBP/HAC1 on the Reporter Gene Expression in the Presence of HsfB1.
(A) Block diagrams represent the basic structures of mammalian CBP and the Arabidopsis ortholog HAC1 with conserved Zn-finger (C/H1, C/H2, and C/H3) and the HAT domains. Arrows mark the NTD and CTD fragments of HAC1 used in the assays.
(B) and (C) Effects of HsfB1 and CBP/HAC1 on constitutively active promoters. The two mycHsp17.6A reporters correspond to numbers 1 and 6 of Figure 2B. One microgram of HsfB1 encoding plasmid and 2 μg of HAC1/CBP constructs were used for transformation. Reporter gene expression was monitored with α-Myc. Numbers given at the bottom were derived from densitometer scans and indicate the fold stimulation of reporter gene expression as referred to for sample 1. For identification of the HsfB1 mutant forms in (C), see the legend of Figure 3.
From the results in Figures 1 and 2, we concluded that the synergistic effects of HsfB1 on reporter gene activation is not restricted to interaction with HsfA1 or related HSFs but results from the cooperation with acidic activators in general. If this conclusion is correct, then the positive influence of HAC1/CBP in the presence of HsfB1 also should be detectable with the constitutively active promoters analyzed in Figure 2B. To test this, we used the MycHsp17.6 reporter constructs with the CaMV35S and the AtHsc70 promoter fragments (Figures 4B and 4C). The stimulation by HAC1/CBP was clearly visible from the immunoblot detection of reporter gene expression and the numbers derived from corresponding densitometer scans. As shown by samples 2 to 6 in Figure 4C, the stimulation required the presence of wild-type HsfB1. The three mutant forms, defective in DNA binding (M4) or in the CTD (Δ and R), also were inactive in this context (see corresponding results in Figures 3C and 3D). Interestingly, the stimulatory effect of coexpression of HsfB1 with HAC1/CBP was much more pronounced with the AtHsc70 than with the CaMV35S reporter, and in the former case, even the CTD of HAC1 had a detectable positive influence. The differences indicate that the interaction of HAC1/CBP proteins may be markedly influenced by peculiarities of the promoter architecture and the activator proteins present at a given time. It is clearly visible that the extent of stimulation in the presence of HAC1/CBP was strongly influenced by the basal level of reporter expression in sample 1 (i.e., by the availability of the endogenous activators for the two promoters in the tobacco protoplasts).
Physical Interaction of HAC1 with HsfA1 and HsfB1 in Vitro
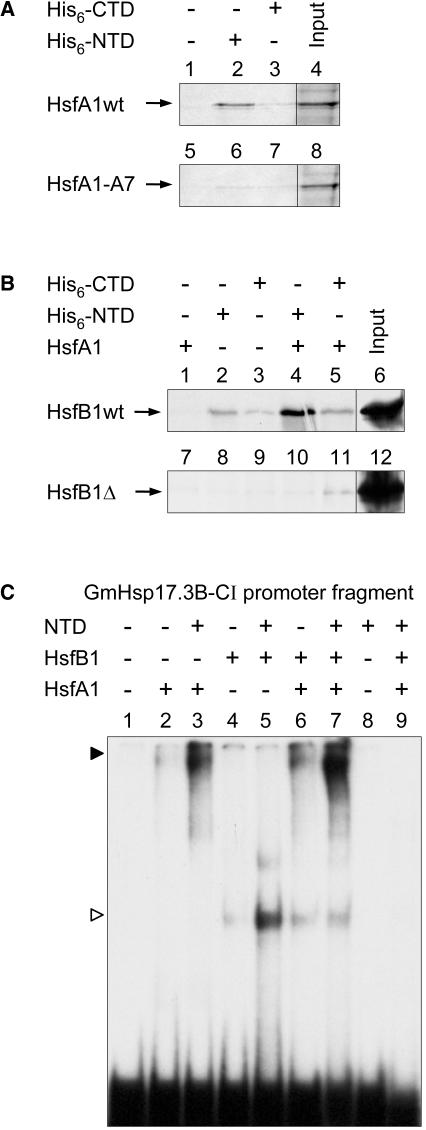
Unfortunately, the expression levels of HAC1/CBP in tobacco protoplasts were too low to allow immune detection of the proteins despite the fact that we expressed different tagged versions and used up to 4 μg of expression plasmids for transformation. To overcome these drawbacks and to provide direct evidence for the interaction of HsfA1 and HsfB1 with HAC1, we used two approaches (Figure 5). First, pull-down assays with recombinant His-tagged NTD and CTD of HAC1 and radioactively labeled HSFs were performed. For HsfA1, we observed a strong binding with nickel-nitrilotriacetic acid agarose (Ni-NTA) beads loaded with the NTD (Figure 5A, lane 2) but not with the beads alone (lane 1) and much weaker binding with beads loaded with the CTD (lane 3). In contrast with this, an inactive mutant of HsfA1 with permutated AHA motifs (HsfA1-A7; see Methods) showed strongly reduced or nondetectable binding (lanes 5 to 7). The input for both pull-down reactions is shown in lanes 4 and 8. Basically similar results were obtained with HsfB1 (Figure 5B), but in this case, interaction was found with both domains of HAC1 (Figure 5B, lanes 2 and 3). Interestingly, the pull-down of HsfB1 by the NTD, but not by the CTD, could be markedly enhanced by addition of recombinant HsfA1 (cf. Figure 5B, lanes 4 and 5). As expected, binding of the mutant form of HsfB1 lacking the GRGK motif (HsfB1Δ) was much weaker or undetectable (Figure 5B, lanes 7 to 11).
Figure 5.
In Vitro Interaction of HAC1 with HsfA1 and HsfB1.
(A) and (B) HsfA1, HsfA1-A7, HsfB1, and HsfB1-Δ were radioactively labeled by in vitro transcription/translation (see Methods). The labeled HSFs (input, see lanes 4 and 8 of [A] and lanes 6 and 12 of [B]) were used for pull-down reactions with Ni-NTA beads alone or in the presence of 20 μg of the His-tagged NTD or CTD of HAC1. In samples 1, 4, and 5 of (B), 100 ng of recombinant HsfA1 was added to the reaction mixture before incubation with the Ni-NTA beads.
(C) For the EMSA, 32P-labeled PCR fragment (118 bp) of the GmHsp17.3-CI promoter containing the active THtHtHT cluster (samples 1 to 8) or the inactive mutant cluster ththtHT (sample 9) were incubated with the indicated recombinant proteins (25 ng of NTD, 50 ng of HsfA1, and 30 ng of HsfB1), and the complexes were separated by PAGE (Lyck et al., 1997). The closed arrowhead points to the HsfA1, and the open arrowhead points to the HsfB1-specific bands.
For sequence of the radiolabeled probes used in (C) and (D), see Supplemental Table S1 online, numbers 66 and 67.
For the second approach, we used electrophoretic mobility shift assays (EMSA) with recombinant HsfA1 and HsfB1 and NTD of HAC1 (Figure 5C). The PCR-labeled 118-bp Gmhsp17.3B-CI promoter fragment used as probe contained either the well-characterized cluster motif THtHtHT (Figure 1A) highly responsive to the synergistic effects of HsfA1 and HsfB1 (lanes 1 to 8) or its inactive mutant derivative ththtHT as negative control (lane 9). The weak signal obtained with HsfA1 alone (lane 2) was markedly enhanced in the presence of the NTD (lane 3) or HsfB1 (lane 6), and the same type of enhancement was observed with HsfB1 in combination with NTD (lanes 4 and 5). However, by far the strongest signal was obtained in the presence of all three interacting proteins (lane 7). Because the HsfA1-specific band migrated very slowly in this gel, a supershift in the presence of HsfB1 and NTD could not be detected.
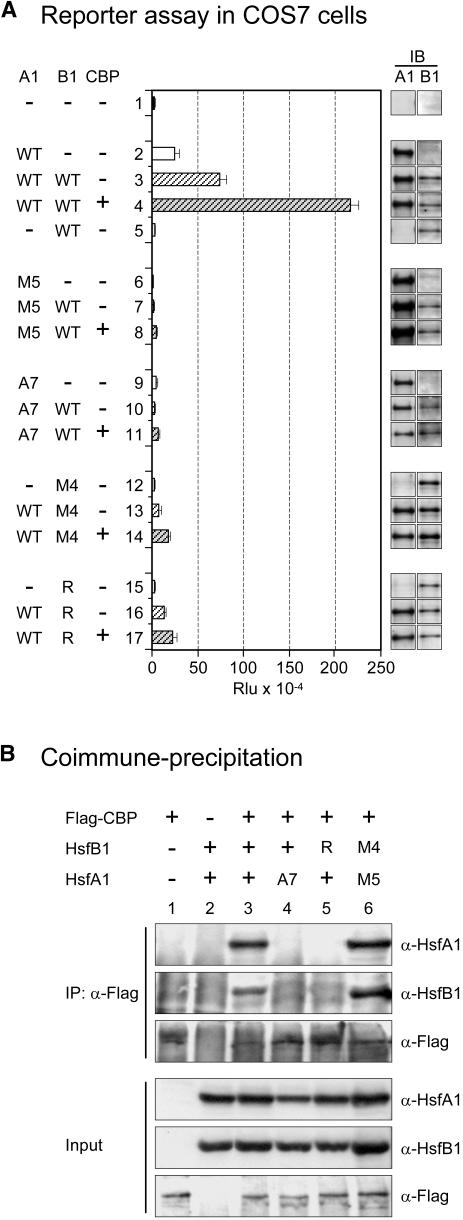
A Ternary Complex of CBP with HsfA1 and HsfB1 Detected in COS7 Cells
Based on our observation that the mammalian CBP could function in tobacco protoplasts similar to the plant HAC1 (Figure 4), we wanted to test the synergistic activation of reporter gene expression by tomato HsfA1 and HsfB1 and the corecruitment effect of CBP also in mammalian cells. To this aim, we used the pHsp17-luciferase (LUC) reporter shown previously to function highly selectively in mammalian cells (Heerklotz et al., 2001). COS7 cells were nicely responsive to the coexpression of HsfA1 with HsfB1 and/or CBP (Figure 6A, samples 2 to 4). Because of the lack of an endogenous class A HSF in mammalian cells, HsfB1 alone had no detectable effect (sample 5). Similar to the results in Figure 3, the mutants of either HsfA1 or HsfB1 abolished the effects (Figure 6A, samples 6 to 17), indicating that the structural prerequisites for corecruitment were similar or identical for mammalian CBP and Arabidopsis HAC1. The results in COS7 cells faithfully reflect the situation in tobacco protoplasts, but they are easier to evaluate because there is no influence of HsfB1 on the expression level of HsfA1 as observed in mesophyll protoplasts. Unlike the 35S promoter in plant cells, the cytomegalovirus promoter used for expression in animal cells does not respond to HsfB1.
Figure 6.
Interaction of Tomato HsfA1 and HsfB1 in COS7 Cells and Coimmunoprecipitation with CBP.
(A) Expression of the pHsp17-LUC reporter in the presence of the indicated forms of HsfA1 and/or HsfB1 and CBP. IB, immunoblot expression controls of the indicated HSFs. Rlu, relative luminescence units.
(B) Use of α-Flag for coimmunoprecipitation of Flag-CBP with HsfA1 and HsfB1. Four percent of the whole-cell extracts was used for the input controls.
See Methods for explanation of the mutant forms M5 (DNA binding mutant) and A7 (activation domain mutant) of HsfA1 as well as M4 (DNA binding mutant) and R (K275>R mutant) of HsfB1 used in (A) and (B).
The coexpression in COS7 cells also allowed the investigation of the physical interaction of CBP with HsfA1 and HsfB1 by coimmunoprecipitation using Flag-tagged CBP (Figure 6B). In agreement with our assumption of corecruitment, strong interactions of the three proteins were observed in the presence of all three partners in active form (lane 3) but not if one of the partner Hsfs was defective in its CTD (lanes 4 and 5). However, as expected for the coimmunoprecipitation situation, the two DNA binding mutant forms of HsfA1 and HsfB1 were equally effective in binding to CBP (lane 6). The results clearly demonstrate that the synergistic reporter gene activation by HsfA1 and HsfB1 is not dependent on other plant-specific proteins and that CBP can fully replace HAC1 at least in the two criteria applied in our tests (i.e., reporter gene activation and physical interaction of the three proteins), which was observed even without binding to DNA.
Effects of Expression Knock Down of HAC in Arabidopsis Protoplasts
To provide further support for the role of plant HAC for the synergistic stimulation of gene expression in the presence of HsfB1, we used an small interfering RNA approach (Figure 7). To this aim, a vector harboring inverted repeats (pIR) encoding the conserved HAT domain of HAC1 was used in a transient reporter assay in Arabidopsis mesophyll protoplasts. As controls, we included corresponding vectors with inverted repeats of tomato HsfA1 and Hsp17.4A-CII (for details, see Methods). The outcome and reproducibility of the reporter assays in Arabidopsis protoplasts (Figure 7A, samples 1 to 4; Figure 7B, lanes1 to 3) were similar to the results in tobacco protoplasts (Figure 1B).
Figure 7.
Endogenous HAC1 Is Required for Synergistic Gene Activation in Arabidopsis Protoplasts.
(A) Gus assay using pHsp17*-GUS as reporter and the standard combination of HsfA1, HsfB1, and HAC1 expression plasmids in Arabidopsis protoplasts. Expression of the endogenous HAC1 was knocked down using a vector harboring an inverted repeat (pIR) of a gene fragment encoding the HAC1 HAT domain (HAC, samples 5 and 9). HsfA1- (A1) and Hsp17-CII-specific (C) pIR were used as controls (samples 6 and 7, respectively). See Methods for details of the inverted repeat vectors.
(B) Immunoblot analysis to detect the expression of CaMV35S promoter in the absence or presence of pIR-HAC1 (samples 2 and 3) or pIR-HsfA1 (sample 4).
(C) Immunoblot analysis showing the expression of endogenous Hsp17-CI induced by 3-h HS at 35°C (sample 1) in presence of pIR-HAC1 (sample 2) and pIR-Hsp17-CII (sample 3).
Addition of pIR-HAC1 (sample 5) abolished the GUS expression resulting from the synergistic interaction between HsfA1 and HsfB1 without affecting the levels of the two HSFs in the protoplasts (see results of protein blot analysis in Figure 7B). GUS expression also was strongly reduced by cotransforming pIR-HsfA1 (sample 6), but in this case, the complete lack of HsfA1 expression was responsible for the outcome. The latter result also demonstrated that the knock down worked properly in this transient expression system. In contrast with this, no effect was observed in the presence of the pIR-Hsp17-CII vector as neutral control (sample 7). In samples 8 and 9, the specific inhibitory effect of pIR-HAC1 was demonstrated in protoplasts expressing plasmid-borne HAC1 in addition to the unknown mixture of endogenous HAC proteins. Unfortunately, our only information about the expression of endogenous HAC proteins stems from RT-PCR analyses showing that HAC1-4 mRNAs are well expressed in leaves, stem, flower, and seedlings of Arabidopsis (Bordoli et al., 2001). Because the pIR-HAC1 was constructed with a gene fragment encoding the conserved HAT domain of HAC1, we assume that expression of other HAC proteins is affected as well. RT-PCR analysis with total cell RNA from sample 5 indicated a reduction of HAC1-specific RNA to <20% of that found in sample 4 (data not shown).
As expected, the successful reduction of HAC expression in Arabidopsis protoplasts transformed with pIR-HAC1 also led to an inhibition of the plasmid-borne CaMV35S-MycHsp17.CI expression cassette (Figure 7B) as well as of the HS-induced expression of the chromosomal Hsp17-CI genes (Figure 7C). Expression levels were reduced by ∼50% (Figure 7B) and 70% (Figure 7C) compared with the controls with pIR-HsfA1 and pIR-Hsp17-CII, respectively.
DISCUSSION
Function of Class B HSFs: Coactivators versus Repressors
The seemingly controversial results about the role of class B HSFs (Treuter et al., 1993; Czarnecka-Verner et al., 2000) are now clarified by our observations about a strong synergistic enhancement of reporter gene expression by coexpression of tomato HsfA1 and HsfB1. Although the divergent structure of their oligomerization domains precludes direct physical interaction of both HSFs, they cooperate in transcription activation, if the promoter architecture allows a proper positioning of both HSFs on the DNA. This cooperation of the two HSFs provides a clue for an understanding of the role of the imperfect HSE clusters frequently observed in plant HS genes (for a review, see Nover, 1987).
Actually, the stimulating effect of HsfB1 is not restricted to cooperation with HsfA1 only but also can be observed with similar class A HSFs from tomato or Arabidopsis or even by appropriate HSF fusion activators harboring activation domains from the yeast Gal4 or the viral VP16 transcription factors (Table 1). Moreover, related HSFs (e.g., tobacco and soybean HsfB1) have comparable coactivator functions in combination with HsfA1. However, we also have evidence for plant-specific peculiarities that need further clarification by testing homologous combinations of class A and class B/C HSFs. Thus, the Arabidopsis class B HSFs did not show any positive effects, neither in combination with tomato HsfA1 nor with any of the Arabidopsis class A HSFs (data not shown). Clearly, the coactivator function of class B HSFs requires a certain promoter architecture and structural peculiarities of the CTDs as prerequisites for interaction with HAC1/CBP. HSFs lacking the crucial Lys residue in the histone motif (e.g., AtHsfB1) are inactive in synergism and may even interfere with the activator function of class A HSFs as observed earlier by Czarnecka-Verner et al. (2000). They compete for the DNA binding sites without being able to help in the recruitment of HAC1/CBP. Interestingly, similar repressor effects also were evident from the tests with mutant forms of LpHsfB1 (Figures 3C and 3D) or with the Gal4DBDxHsfB1 fusion protein (Figure 1C).
It is important to state that assays with plasmid-borne reporter constructs harboring complex promoter fragments derived from HS genes are essential to elaborate details of HSF interaction. However, they must be complemented by corresponding tests using the endogenous, chromatin-embedded genes as reporters. Whenever possible, we combined results from both types of assays, and the results were basically comparable (Figures 1, 2, and 7). On the other hand, the intriguing details of the strong stimulation of the CaMV35S-GUS transgene in the presence of HsfB1 only under HS conditions (Figure 2D) emphasize important differences between plasmid-borne and chromatin-embedded reporters, which deserve further attention.
HsfB1 as Maintenance Factor for Housekeeping Gene Transcription
Most intriguing are the stimulatory activities of tomato HsfB1 on the activity of CaMV35S and on several housekeeping promoters. Tomato HsfB1 may represent the long searched, HS-induced factor essential for maintenance and/or restoration of housekeeping or viral gene transcription during ongoing HS. It is important to notice that, depending on the promoter architecture, even HsfA1 may act as repressor because it competes with HsfB1 for DNA binding (Figure 2B, constructs 1 to 4). Unfortunately, our lack of knowledge about the fine structure of most housekeeping promoters and the transcription factors required for their constitutive activity precludes a more detailed description of these effects. However, closer inspection of the 5′ upstream regions used for the reporter constructs represented in Figure 2B showed that all of them contain a multiplicity of potential HSF binding sites, in particular dimer binding sites for HsfB1 or similar factors (see Supplemental Table S1 online).
The Histone-Like Motif of HsfB1 and the Role of HAC1/CBP
Our results demonstrate that the basis for the stimulatory effects of HsfB1 on HS-inducible as well as on housekeeping or viral promoters is similar in all cases. HsfB1 binding in close vicinity to an acidic activator leads to cooperative recruitment of components of the transcriptional machinery. Evidently, the GRGKMMK motif in the C terminus of HsfB1 plays a central role for its coactivator function because it serves as binding site for the plant CBP orthologs HAC1 to HAC5 recently identified by sequence comparison in the Arabidopsis genome (Bordoli et al., 2001; Yuan and Giordano, 2002). We have provided evidence that AtHAC1 and mammalian CBP exert a strong and specific stimulatory effect on gene activation in plant cells (Figure 4). Moreover, the physical and functional interaction of tomato HsfA1 and HsfB1 with mammalian CBP also could be confirmed by experiments with COS7 cells using reporter gene activation and coimmunoprecipitation (Figure 6). It is remarkable that the evolutionary conservation between CBP and HAC1 does not only include the residues in the CTD required for HAT activity but also the binding sites for transcription factors as divergent as the adenovirus E1A protein (Bordoli et al., 2001) and tomato HsfA1 and HsfB1 (Figures 5 and 6). Using pull-down assays, we have shown that AHA motifs of class A HSFs are essential for interaction with the components of the transcription machinery, but we never detected any components interacting with HsfB1 (Kotak et al., 2004). HAC1/CBP is the first interaction partner for both types of HSFs, and there is a mutual enhancement of HAC1/CBP and the two HSFs in the assembly of the ternary complex (Figures 5 and 6). Despite the differences in the recognition motifs (AHA motifs for HsfA1 versus GRGK motif for HsfB1), it is tempting to speculate that both HSFs interact with the C/H1 region, which is the only domain conserved in the NTD of HAC1 and CBP.
Despite several attempts (in vitro acetylation and/or immune detection with anti-acetyl Lys antibody), we could not find any evidence for acetylation of the GRGK motif in HsfB1 (data not shown). In support of this, the GRGQ mutant was completely inactive (Figure 3D, sample 8), despite the fact that Q was reported to mimic the role of acetyl-Lys residues (Zhang et al., 1998). Furthermore, the NTD of HAC1 lacking the HAT domain was fully functional in the reporter (Figure 4) as well as in the electrophoretic mobility shift and pull-down assays (Figure 5). Taken together, the results strongly indicate that HAC1/CBP exerts a scaffold function (Nakashima et al., 1999; Chan and LaThangue, 2001) helping to form a ternary complex with both HSFs and to enhance affinity for DNA binding.
The synergistic activation of gene expression by coordinate binding of HsfB1 with different acidic activators and the recruitment of HAC1/CBP is a general effect observed in tobacco and Arabidopsis protoplasts and in COS7 cells irrespective of plasmid-borne or chromosomal reporter genes used for detection (Figures 1, 2, 6, and 7). The important role of HAC1/CBP in this respect is strongly supported by the demonstration of a ternary complex with HsfA1 and HsfB1 (Figures 5 and 6), by the enhancement of DNA binding (Figure 5), and above all by the results of the HAC1/CBP expression knock down in Arabidopsis protoplasts (Figure 7). Although further details remain to be elaborated, the major reason for quantitative differences in the outcome of assays in different experimental systems very likely stems from the unknown levels of endogenous factors (e.g., other HSFs and/or HAC1/CBP) contributing to the background activities. Because of the lack or low level of interfering components, the COS7 cells represent a valuable tool to study certain details of this plant-specific phenomenon (Figure 6).
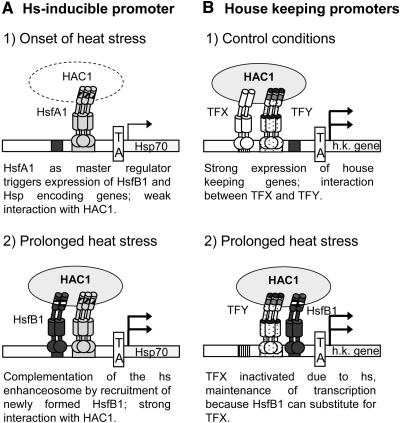
Function of HsfB1 in an Enhanceosome-Like Complex
The role of HsfB1 and related HSFs from other plants can be summarized in a model presented in Figure 8. HS gene transcription is triggered by activation of the constitutively expressed class A HSFs (1 in Figure 8A). In tomato, the dominant HSF for this event is the master regulator HsfA1 (Mishra et al., 2002). HsfB1, newly formed in response to the HS, joins the transcription complex with HsfA1, and together, they evidently provide the specific interface for recruitment of HAC1. The complementation of the HS enhanceosome results in a marked increase of transcription (2 in Figure 8A). Observations of transcriptional synergy in the context of enhanceosomes were originally described for mammalian cells, for example, for the interferon-β (Thanos and Maniatis, 1995; Merika et al., 1998) and the T cell receptor-α genes (Giese et al., 1995). Although many details remain to be elaborated for plants, the interaction of HsfA1 with HsfB1 to form a ternary complex with HAC1 required for synergistic transcription activation of appropriate promoters is reminiscent of essential aspects of the enhanceosome function in mammalian cells (Carey, 1998).
Figure 8.
Model for Synergistic Interactions of HsfB1 with Acidic Activators (HsfA1 and TFY) and Corecruitment of HAC1.
See text for further explanations. h.k., housekeeping gene.
As depicted in Figure 8B, the situation may be similar for constitutively active promoters (35S promoter and promoters of housekeeping genes). The activity under HS conditions can be strongly stimulated by cooperation with HsfB1 (2 in Figure 8B) replacing an unknown transcription factor inactivated during the HS response. By this mechanism, HsfB1 may help to maintain transcription of defined housekeeping genes.
The model with the newly detected role of HsfB1, although valid for many class A HSFs or other acidic activators in combination with HsfB1 and related HSFs from other plants (Table 1), cannot be applied in general, at least not in this simplified form. It is an interesting speculation that among the representatives of class B and class C HSFs, repressor HSFs are found (Czarnecka-Verner et al., 2000) together with coactivators and that the latter may form specific combinations with different HSFs or other acidic activators. The regulatory outcome could vary depending on the partners, the promoter architecture, and potential coregulator(s) involved. HAC1 is evidently only one possible coregulator for corecruitment. Further test systems and techniques, such as microarray analyses and chromatin immunoprecipitation, will help to clarify these questions.
METHODS
General Materials and Methods
Standard procedures were used for gene technology work (Ausubel et al., 1993; Sambrook and Russell, 2001). For cloning, PCR fragments were amplified with Taq plus precision system (Stratagene, La Jolla, CA) and purified by QIAquick gel extraction kit (Qiagen, Valencia, CA). GUS reporter assays and analysis of protein expression in tobacco protoplasts (Nicotiana plumbaginifolia) were described previously (Scharf et al., 1998; Döring et al., 2000). For single cell reporter assays with green fluorescent protein and DsRed as reporters, tobacco protoplasts were processed as described by Heerklotz et al. (2001). Transgenic tobacco plants (Nicotiana tabacum) harboring a CaMV35S-GUS cassette were obtained as a gift from Günther Kahl (Goethe-Universität, Frankfurt/Main).
In all transient expression assays with GUS or LUC reporters, three samples were transformed and analyzed independently. Activities are given as relative fluorescence units and relative luminescence units, respectively. Error bars in Figures 1, 2, 6, and 7 indicate standard deviations of the mean value. For the immunoblot analyses with the MycHsp17-CI reporter, three samples were pooled. One representative experiment was always used for presentation, but each experiment was repeated at least three times.
Genomic DNA from Arabidopsis thaliana cell suspension culture was prepared with the cetyl-trimethyl-ammonium bromide buffer (Ausubel et al., 1993). Total RNA from Arabidopsis leaves was isolated by the guanidinium thiocyanate method (Ausubel et al., 1993). cDNA was prepared using Moloney murine leukemia virus reverse transcriptase according to the manufacturer's protocol (MBI Fermentas, Vilnius, Lithuania). Plasmids and details of primers and cloning procedures are summarized in Supplemental Table S1 online.
Rabbit antisera against tomato (Lycopersicon peruvianum) HsfA1 and HsfB1 were described previously (Lyck et al., 1997; Mishra et al., 2002). Myc and Flag antisera were obtained from Babco (Denver, PA) and Sigma (St. Louis, MO), respectively. Secondary antibodies against rabbit/mouse immunoglobulins conjugated with horseradish peroxidase were obtained from Sigma.
Activator Constructs for Plant/Animal Cells
Plant expression and reporter constructs are based on the pRT and pBT series of vectors (Töpfer et al., 1988; Döring et al., 2000), whereas animal expression and reporter constructs are based on pcDNA3 and pluc vectors (Heerklotz et al., 2001). MycHsp17.6A-CI reporter constructs are based on pRT-mycHsp17.6A-CI (Kirschner et al., 2000). His-tagged fusion constructs were created in the pJC series of vectors (Clos and Brandau, 1994). For in vitro transcription/translation, pcDNA3 vectors were used for wild-type HsfA1and HsfA1-A7 mutant, whereas wild-type HsfB1 and HsfB1-Δ were cloned into pβstop vector, containing the β-globin translation enhancer. For construction of inverted repeat vectors (pIR) of HAC1, HsfA1, and Hsp17-CII, pJawohl vector (kind gift from Imre Somssich, MPI Züchtungsforschung, Köln, Germany) and Gateway technology (Invitrogen, Carlsbad, CA) were used.
Important HsfA1 and HsfB1 mutants are defined as follows (for further details, see Supplemental Table S1 online). For HsfA1: HsfA1-M5(R93>D), HsfA1ΔHRA/B (deletion of amino acids 164 to 238), HsfA1mutNLS (KR253/4>NS), HsfA1ΔC394 (deletion of C-terminal amino acids 395 to 527), and HsfA1-A7 (heptaA mutant form of HsfA1 with 450-IDWQSGLL, 468-DPFWEKFL- >450-IDAQSGAA, 468-DPAAEKAA-); for HsfB1: HsfB1-M4 (KH54/5>EL), HsfB1ΔHRA/B (deletion of amino acids 145 to 213), HsfB1ΔNLS (deletion of amino acids 214 to 268), HsfB1ΔC198 (deletion of C-terminal amino acids 199 to 301), HsfB1-Δ (deletion of amino acids 272 to 279 with GRGK motif), and HsfB1-R (HsfB1 with 272-GRGK>GKGR).
Culture, Transfection of COS7 Cells, Luciferase Assays, and Coimmunoprecipitation
COS7 cells were maintained in nutrient mixture Dulbecco's modified eagle medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (Life Technologies, Cleveland, OH). Transfections were done with polyfect (Qiagen) according to the manufacturer's protocol. Luciferase assays were performed as described previously (Heerklotz et al., 2001). Conditions for coimmunoprecipitation with Flag-CBP were described by Gingras et al. (1999).
Purification of Recombinant Proteins and Pull-Down Assays
His-tagged HsfA1, HsfB1, NTD-HAC1, and CTD-HAC1 were purified by binding to Ni-NTA sepharose beads followed by elution with 250 mM imidazole. For pull-down assays, HsfA1, HsfA1-A7, HsfB1, and HsfB1-Δ were in vitro transcribed and translated in the presence of 35S-Met using the TNT-coupled reticulocyte lysate (Promega, Madison, WI). Pull-down assays were performed as described previously (Kaufmann et al., 2000).
Electrophoretic Mobility Shift Assay
For details of the EMSAs, see Boscheinen et al. (1997) and Mishra et al. (2002). Probes were labeled with 32P by PCR with primers F, 5′-tacgccaagcttggatccgtcg-3′, and R, 5′-ccttatatagaggaagggtcttgcg-3′, for the Hsp17.3B-CI promoter fragment (for sequence of the probes, see Supplemental Table S1 online, numbers 66 and 67). Approximately 1 ng of 32P-labeled probes (105 cpm) was incubated for 30 min at 25°C with indicated combinations of recombinant proteins (Figure 6C). Twenty microliters of the EMSA buffer contained 20 mM Hepes, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT, 50 mM imidazole, 10% glycerol, 2% Ficoll, 2 μg of polydIdC, and 4 μg of acetylated BSA.
Supplementary Material
Acknowledgments
We are grateful to following people for providing expression plasmids for CBP (M. Rosenfeld, University of California, San Diego), for Flag-tagged CBP (E. Pfitzner, Georg-Speyer-Haus, Frankfurt), for TGA2.1 and TGA2.2 (C. Gatz, Göttingen), for fragments of HAC1 (R. Eckner, ETH, Zurich), for pβstop vector (M. Jantzen, University of California, Berkeley), and for transgenic tobacco plants (G. Kahl, Frankfurt/Main). We thank A. Starzinski-Powitz for help with animal cell culture and in vitro pull-down experiments. We thank Gisela Englich and Daniela Bublak for excellent technical assistance. We thank E. Pfitzner, Klaus Dieter-Scharf, Sanjeev Baniwal, Shravan K. Mishra, Christian Weber, and Markus Fauth for many helpful discussions and comments during the preparation of the manuscript. This work was supported by grants of the Deutsche Forschungsgemeinschaft to L.N.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lutz Nover (nover@cellbiology.uni-frankfurt.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019927.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds (1993). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Bannister, A.J., and Miska, E.A. (2000). Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 57, 1184–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti, K., Schmidt, E., Lyck, R., Bublak, D., and Scharf, K.-D. (2000). Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 22, 355–365. [DOI] [PubMed] [Google Scholar]
- Bordoli, L., Netsch, M., Lüthi, U., Lutz, W., and Eckner, R. (2001). Plant orthologs of p300/CBP: Conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res. 29, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheinen, O., Lyck, R., Queitsch, C., Treuter, E., Zimarino, V., and Scharf, K.-D. (1997). Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 255, 322–331. [DOI] [PubMed] [Google Scholar]
- Carey, M. (1998). The enhanceosome and transcriptional synergy. Cell 92, 5–8. [DOI] [PubMed] [Google Scholar]
- Chan, H.M., and La Thangue, N.B. (2001). p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373. [DOI] [PubMed] [Google Scholar]
- Clos, J., and Brandau, S. (1994). pJC20 and pJC40—Two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Protein Expr. Purif. 5, 133–137. [DOI] [PubMed] [Google Scholar]
- Czarnecka, E., Gurley, W.B., Nagao, R.T., Mosquera, L.A., and Key, J.L. (1985). DNA sequence and transcript mapping of a soybean gene encoding a small heat shock protein. Proc. Natl. Acad. Sci. USA 82, 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka-Verner, E., Yuan, C.X., Scharf, K.-D., Englich, G., and Gurley, W.B. (2000). Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol. Biol. 43, 459–471. [DOI] [PubMed] [Google Scholar]
- Döring, P., Treuter, E., Kistner, C., Lyck, R., Chen, A., and Nover, L. (2000). Role of AHA motifs for the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278. [PMC free article] [PubMed] [Google Scholar]
- Giese, K., Kingsley, C., Kirschner, J.R., and Grosschedl, R. (1995). Assembly and function of a TCR α enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9, 995–1008. [DOI] [PubMed] [Google Scholar]
- Gingras, S., Simard, J., Groner, B., and Pfitzner, E. (1999). p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res. 27, 2722–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J.A., Cutler, G., and Tjian, R. (1996). Contacts in context: Promoter specificity and macromolecular interactions in transcription. Cell 84, 825–830. [DOI] [PubMed] [Google Scholar]
- Heerklotz, D., Döring, P., Bonzelius, F., Winkelhaus, S., and Nover, L. (2001). The balance of nuclear import and export determines the intracellular distribution of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga, J.T. (1998). Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell 92, 307–313. [DOI] [PubMed] [Google Scholar]
- Katagiri, F., Lam, E., and Chua, N.H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340, 727–730. [DOI] [PubMed] [Google Scholar]
- Kaufmann, U., Zuppinger, C., Waibler, Z., Rudiger, M., Urbich, C., Martin, B., Jockusch, B.M., Eppenberger, H., and Starzinski-Powitz, A. (2000). The armadillo repeat region targets ARVCF to cadherin-based cellular junctions. J. Cell Sci. 113, 4121–4135. [DOI] [PubMed] [Google Scholar]
- Kimura, A., and Horikoshi, M. (1998). How do histone acetyltransferases select lysine residues in core histones? FEBS Lett. 431, 131–133. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., Winkelhaus, S., Thierfelder, J.M., and Nover, L. (2000). Transient expression and heat stress induced coaggregation of endogenous and heterologous small heat stress proteins in tobacco protoplasts. Plant J. 24, 397–411. [DOI] [PubMed] [Google Scholar]
- Kotak, S., Port, M., Ganguli, A., Bicker, F., and von Koskull-Döring, P. (2004). Characterization of C-terminal domains of Arabidopsis heat stress transcription factors and identification of a new signature combination of plant class A HSFs with AHA and NES motifs essential for activator function and intracellular localization. Plant J., in press. [DOI] [PubMed]
- Lam, E. (1994). Analysis of tissue-specific elements in the CaMV 35S promoter. In Plant Promoters and Transcription Factors, L. Nover, ed (Berlin, Germany: Springer), pp. 181–196. [DOI] [PubMed]
- Lam, E., and Chua, N.H. (1989). ASF-2: A factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B.L., Wang, J.S., Liu, H.C., Chen, R.W., Meyer, Y., Baraket, A., and Delseny, M. (2001). Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyck, R., Harmening, U., Höhfeld, I., Treuter, E., Scharf, K.-D., and Nover, L. (1997). Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 202, 117–125. [DOI] [PubMed] [Google Scholar]
- Merika, M., Williams, G.C., Collins, T., and Thanos, D. (1998). Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1, 277–287. [DOI] [PubMed] [Google Scholar]
- Mishra, S.K., Tripp, J., Winkelhaus, S., Tschiersch, B., Theres, K., Nover, L., and Scharf, K.-D. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16, 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, R.I. (1998). Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones and negative regulators. Genes Dev. 12, 3788–3796. [DOI] [PubMed] [Google Scholar]
- Nagao, R.T., Czarnecka, E., Gurley, W.B., Schöffl, F., and Key, J.L. (1985). Genes for low-molecular-weight heat shock proteins of soybeans: Sequence analysis of a multigene family. Mol. Cell. Biol. 5, 3417–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, K., Yanagisawa, M., Arakawa, H., Kimura, N., Hisatsune, T., Kawabata, M., Miyazono, K., and Taga, T. (1999). Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284, 479–482. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- Nover, L. (1987). Expression of heat shock genes in homologous and heterologous systems. Enzyme Microb. Technol. 9, 130–144. [Google Scholar]
- Nover, L. (1991). Heat Shock Response. (Boca Raton, FL: CRC Press).
- Nover, L., and Scharf, K.-D. (1997). Heat stress proteins and transcription factors. Cell. Mol. Life Sci. 53, 80–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover, L., Bharti, K., Döring, P., Mishra, S.K., Ganguli, A., and Scharf, K.-D. (2001). Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H.R.B. (1982). A regulatory upstream promoter element in the Drosophila hsp70 heat-shock gene. Cell 30, 517–528. [DOI] [PubMed] [Google Scholar]
- Ptashne, M., and Gann, A. (1997). Transcriptional activation by recruitment. Nature 386, 569–577. [DOI] [PubMed] [Google Scholar]
- Regier, J.L., Shen, F., and Triezenberg, S.J. (1993). Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90, 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scharf, K.-D., Heider, H., Höhfeld, I., Lyck, R., Schmidt, E., and Nover, L. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18, 2240–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, K.-D., Rose, S., Zott, W., Schöffl, F., and Nover, L. (1990). Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 9, 4495–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, K.-D., Siddique, M., and Vierling, E. (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins). Cell Stress Chaperones 6, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl, F., Raschke, E., and Nagao, R.T. (1984). The DNA sequence analysis of soybean heat shock genes and identification of possible regulatory promoter elements. EMBO J. 3, 2491–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D.E., and Berger, S.L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos, D., and Maniatis, T. (1995). Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Tjian, R., and Maniatis, T. (1994). Transcriptional activation—A complex puzzle with few easy pieces. Cell 77, 5–8. [DOI] [PubMed] [Google Scholar]
- Töpfer, R., Schell, J., and Steinbiss, H.H. (1988). Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res. 16, 8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter, E., Nover, L., Ohme, K., and Scharf, K.-D. (1993). Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol. Gen. Genet. 240, 113–125. [DOI] [PubMed] [Google Scholar]
- Yuan, L.W., and Giordano, A. (2002). Acetyltransferase machinery conserved in p300/CBP-family proteins. Oncogene 21, 2253–2260. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Bone, J.R., Edmondson, D.G., Turner, B.M., and Roth, S.Y. (1998). Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17, 3155–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.