Abstract
Plant cells develop various types of endoplasmic reticulum (ER)–derived structures with specific functions. ER body, an ER-derived compartment in Arabidopsis thaliana, is a spindle-shaped structure. The NAI1 gene regulates the development of ER bodies because mutation of NAI1 abolishes the formation of ER bodies. To better understand the role of NAI1, we cloned the NAI1 gene using a positional cloning strategy. The nai1-1 mutant had a single nucleotide change at an intron acceptor site of At2g22770 (NAI1 gene). Because of this mutation, aberrant splicing of NAI1 mRNA occurs in the nai1-1 mutant. NAI1 encodes a transcription factor that has a basic-helix-loop-helix (bHLH) domain. Transient expression of NAI1 induced ER bodies in the nai1-1 mutant. Two-dimensional electrophoresis and RT-PCR analyses showed that a putative lectin was depressed at both the mRNA and protein levels in nai1 mutants, as was a β-glucosidase (PYK10). Our results provide direct evidence that a bHLH protein plays a role in the formation of ER bodies.
INTRODUCTION
Endoplasmic reticulum (ER) is an extensive, morphologically continuous network of membrane tubes and flattened cisternae. Classically, the ER is subdivided into three compartments: rough ER, smooth ER, and the nuclear envelope (Baumann and Walz, 2001). In addition to these compartments, many ER-derived structures with specific functions have been identified in plant cells (Okita and Rogers, 1996; Staehelin, 1997; Chrispeels and Herman, 2000). The protein bodies in the endosperm of maize (Zea mays) and rice (Oryza sativa) are responsible for the accumulation of seed storage proteins (Herman and Larkins, 1999). Precursor-accumulating vesicles in maturing cotyledons of pumpkin (Cucurbita maxima) mediate the transport of seed storage protein to protein storage vacuoles directly from ER (Hara-Nishimura et al., 1998). KDEL (Lys-Asp-Glu-Leu)-tailed Cys proteinase-accumulating vesicles and ricinosomes are also ER-derived structures that are found in vegetative organs of black gram (Vigna mungo) and castor bean (Ricinus communis), respectively (Schmid et al., 1998; Toyooka et al., 2000).
In transgenic Arabidopsis thaliana expressing green fluorescent protein (GFP) with an ER-retention signal (GFP-HDEL, His-Asp-Glu-Leu), spindle-shaped GFP-fluorescent structures (∼10 μm long and ∼1 μm wide) have been visualized together with the ER networks (Haseloff et al., 1997; Ridge et al., 1999; Hawes et al., 2001; Hayashi et al., 2001). Electron microscopic studies show that the structures have a fibrous pattern inside, and they are surrounded by ribosomes (Hayashi et al., 2001). The presence of ribosomes on the surface of the structures indicates that they are directly derived from the ER. Therefore, we recently proposed to call them ER bodies (Hayashi et al., 2001). ER bodies develop in nontransgenic Arabidopsis, indicating that they are not artificial structures caused by overexpression of the transgene but, rather, accumulate some endogenous materials inside and have some specific role in plant cells (Matsushima et al., 2003a). Similar structures have been reported in the cells of various organs of Brassicaceae plants (Bonnett and Newcomb, 1965; Iversen, 1970; Behnke and Eschlbeck, 1978; Bones et al., 1989).
Transgenic Arabidopsis expressing GFP-HDEL (GFP-h) enabled us to study the unique distribution of ER bodies in living plants. A large number of ER bodies are observed in cotyledons, hypocotyls, and roots of young seedlings (Matsushima et al., 2002). Cotyledons decrease the number of ER bodies during senescence, whereas hypocotyls and roots maintain the ER bodies (Matsushima et al., 2002). In contrast with seedlings, rosette leaves of mature plants have few ER bodies and have GFP fluorescence only in ER networks (Matsushima et al., 2002). However, many spindle-shaped structures can be induced in rosette leaves by wounding them or treating them with exogenous methyl jasmonate (MeJA), a plant hormone that induces a defense response against wounding and chewing by insects (Matsushima et al., 2002). The induced structures have shapes and sizes similar to those of ER bodies of seedlings. We named them induced ER (i-ER) bodies because rosette leaves have no ER bodies under normal conditions (Matsushima et al., 2003b). These observations suggest that i-ER bodies play some roles in plant defense systems when wounding or insect feeding damages plants.
To study the molecular mechanisms responsible for the biogenesis of ER bodies, we previously screened Arabidopsis mutants from chemically mutagenized GFP-h plants and isolated a nai1 mutant in which fluorescent ER bodies were hardly detected (Matsushima et al., 2003a). The nai1 mutant shows no visual defects under normal conditions, other than the absence of ER bodies. However, GFP-h and nai1 seedlings exhibit different protein compositions. ER bodies are concentrated in a 1000g pellet (P1) fraction obtained from GFP-h seedlings (Hayashi et al., 2001), whereas no ER bodies were detected in the P1 fraction from nai1 (Matsushima et al., 2003a). A comparison of proteins in the two P1 fractions showed that a 65-kD protein (p65) is present in GFP-h seedlings but not in nai1 (Matsushima et al., 2003a). p65 is PYK10, a β-glucosidase with an ER-retention signal, KDEL. Immunofluorescent staining and immunoelectron microscopy confirmed that PYK10 is specifically localized in ER bodies (Matsushima et al., 2003a). The accumulation of PYK10 in wild-type seedlings is high; Coomassie blue staining can detect it in crude extracts of cotyledons, hypocotyls, and roots (Matsushima et al., 2003a). Therefore, PYK10 is a major component in ER bodies. The physiological role of PYK10 has not been determined. On the other hand, BGL1, a PYK10 homolog in Arabidopsis (70% identity), has been suggested to play a role in the defense against herbivores because it is induced after feeding by diamondback moth (Plutella xylostella) (Stotz et al., 2000). Therefore, we previously suggested that PYK10 also is involved in defense systems against herbivores and pathogens.
The nai1 mutant is an ideal tool to investigate the molecular basis of ER body biogenesis. In this study, we performed fine mapping of the NAI1 locus. The nai1 mutant had a single base pair change at the intron splicing acceptor site of the At2g22770 gene. A T-DNA insertion line containing an insertion in the second exon of the At2g22770 gene was allelic to the nai1 mutant with respect to the development of ER bodies. The At2g22770 gene encodes a 320–amino acid protein with a basic-helix-loop-helix motif. Therefore, NAI1 appears to act as a transcriptional factor. Two-dimensional electrophoresis and RT-PCR analyses revealed that PYK10 and a putative lectin were downregulated in nai1 mutants. NAI1 appears to regulate the expression of genes related to ER bodies and to play a key role in the formation of ER bodies.
RESULTS
Fine Mapping of the NAI1 Locus
We previously showed that the nai1 mutation segregated as a single recessive allele (Matsushima et al., 2003a). We crossed a nai1 mutant (Columbia background) with wild-type Arabidopsis (Landsberg erecta background) and identified 174 homozygous F2 plants. These plants were subsequently scored according to their genetic background with a series of molecular markers using the cleaved amplified polymorphic sequences mapping procedure (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) (Figure 1A). Our initial mapping of the NAI1 locus located it to the middle of chromosome 2, between PHYB and ERECTA. For high-resolution mapping, we generated six molecular markers, T9I22-5, T9I22-4, T9I22-7, T30L20-3, T30L20-4, and T20K9-1. The number of chromosomes that showed a Landsberg background represents the number of recombinations that occurred between the NAI1 locus and the position of each molecular marker because the genomic DNA of the nai1 mutant has a Columbia background. Fine mapping revealed that the NAI1 locus was located in the 69-kb region between T9I22-4 and T30L20-4 (Figure 1A). Fourteen putative open reading frames are predicted in this region. Sequence comparison of wild-type Columbia and the nai1 mutant revealed a single G-to-A transition in the At2g22770 gene. This G-to-A transition is consistent with the mode of action of EMS, the mutagen that was used to generate the mutagenized population from which the nai1 mutant was identified.
Figure 1.
Positional Cloning of the NAI1 Gene.
(A) Fine mapping of the NAI1 gene on chromosome 2. Names and positions of the molecular markers are indicated. T9I22, T30L20, and T20K9 are BAC clones. We analyzed 174 F2 progeny (348 chromosomes) having homozygous nai1 alleles. The numbers of recombinations that occurred between the NAI1 locus and the molecular markers are indicated. The NAI1 locus was mapped to a 69-kb region between two molecular markers (T9I22-4 and T30L20-4). This region contained 14 open reading frames (boxes). The nai1 mutant had a mutation in the At2g22770 gene (gray box).
(B) A schematic representation of the exon and intron organization of At2g22770 and deduced protein structure. The translation start codon (ATG) is designated nucleotide +1, and the stop codon (TAA) is +1464. At2g22770 contains three exons and two introns (bars). White boxes indicate untranslated regions in exons, and gray boxes indicate translated regions in exons. Two distinct transcription start sites (arrows) were determined based on the capping structure of mRNA (see Methods). The nai1 mutant had a single base pair change (G to A) at an intron splicing acceptor site (+867). At2g22770 encodes a 320–amino acid protein that contains an acidic region (12 to 24), two Ser-rich regions (64 to 87 and 212 to 233) and a basic-helix-loop-helix domain (125 to 177). Basic, helix, and loop regions are indicted by yellow, blue, and gray boxes, respectively.
(C) Alignment of amino acid sequences of the bHLH domain (125 to 177) of At2g22770 protein and other bHLH proteins, PIF3 (340 to 392, Arabidopsis), AtMYC2 (445 to 497, Arabidopsis), AN1 (468 to 520, Petunia x hybrida), DEL (436 to 488, Antirrhinum majus), PG1 (455 to 507, Phaseolus vulgaris), c-myc (351 to 406, Homo sapiens), and MAX (20 to 74, H. sapiens). These bHLH proteins are thought to function as transcription factors. The Glu residue indicated by an arrow is critical for the recognition of the E-box sequence. Identical residues are shown in black, and similar residues are shown in gray.
The At2g22770 gene contains three exons and two introns (DNA in Figure 1B). Determination of the 5′ untranslated region revealed that the At2g22770 gene has two distinct transcription start sites (arrows in Figure 1B). The single G-to-A transition in the nai1 mutant is located at the acceptor site of the first intron. The At2g22770 gene encodes a 320–amino acid protein that contains a basic-helix-loop-helix (bHLH) domain (protein in Figure 1B). This sequence similarity is clearly in alignment with sequences from a variety of organisms (Figure 1C). The bHLH proteins are a superfamily of transcription factors that bind as dimers to specific DNA target sites (Ferre-D'Amare and Burley, 1995; Heim et al., 2003; Toledo-Ortiz et al., 2003). The bHLH domain is comprised of ∼50 amino acids with two functionally distinct regions. The basic region, located at the N-terminal end of the domain, is involved in DNA binding (yellow box in Figure 1C). The HLH region functions as a dimerization domain and is constituted mainly of hydrophobic residues that form two amphipathic α-helices separated by a loop region of variable sequence and length (blue and gray boxes in Figure 1C). Outside the bHLH domain, the At2g22770 protein has little sequence similarity to other proteins with proven functions. The At2g22770 protein has one acidic region and two Ser-rich regions (protein in Figure 1B). The acidic region has seven acidic residues out of 13 residues. The Ser-rich regions have 14 and 12 Ser residues out of 24 and 22 residues, respectively. At2g22770 is most similar to the Arabidopsis At2g22750 and At2g22760 genes, both of which encode bHLH proteins.
Aberrant Splicing of At2g22770 mRNA in the nai1 Mutant
The point mutation (G to A) in the nai1 mutant disrupted the splicing acceptor site of the first intron. Therefore, the correct splicing of At2g22770 mRNA was probably prevented in nai1 plants. To examine this possibility, we performed RT-PCR analyses with gene-specific primers that cover the first intron. Using these primers, the amplified cDNA fragment should be 517 bp if correct splicing occurs. The amplified cDNA fragment was separated in 1.0% agarose gel and stained with ethidium bromide (Figure 2A). The band intensity of the At2g22770 cDNA fragment from nai1 was almost the same as that from wild-type Columbia (Col) and GFP-h plants. The negative control that omitted reverse transcriptase gave no signal (GFP-hRT−). Actin as an internal control yielded constant signals. Next, the amplified cDNA fragments were subjected to capillary electrophoresis to analyze them more precisely (Figure 2B). The signals of the Col and GFP-h cDNA fragments were similar in strength and occurred at the same peak position, whereas the signal of nai1 cDNA fragment was weaker and its peak had a shoulder. The peak and shoulder positions of the nai1 cDNA fragment were ∼9 and 16 base pairs shorter, respectively, than the peak position of Col and GFP-h cDNA fragments. These results indicated that the amplified cDNA from the nai1 mutant is a mixture made up of more than one fragment.
Figure 2.
Aberrant Splicing of At2g22770 mRNA in the nai1 Mutant.
(A) Analyses of At2g22770 expression in wild-type Col, GFP-h, and nai1. Actin was used as an internal control. The lane GFP-hRT− is the negative control in which reverse transcriptase was omitted from the reaction mixture.
(B) Electropherogram of the amplified cDNA fragment of At2g22770 RNA resolved by capillary electrophoresis. The 35-cycle products in (A) were analyzed. The amplified cDNA fragment of nai1 had a smaller size than the amplified cDNA fragments of Col (blue) and GFP-h (black). The peak of nai1 (red) had a shoulder. The peak and shoulder positions were 9 or 16 base pairs smaller, respectively, compared with the peak positions of Col and GFP-h.
(C) Effect of the single nucleotide mutation in nai1 on the translation of predicted proteins. Sequencing of the amplified cDNA fragment revealed that the G-to-A transition at the intron acceptor site resulted in aberrant splicing. Asterisks indicate the mutated nucleotide. Two distinct splicing patterns were detected in nai1 (nai1 cDNA1 and cDNA2). Boxed sequences in the genome are introns that are spliced out in the mature mRNA.
Sequencing of the nai1 cDNA fragment revealed two distinct sequences (nai1 cDNA1 and nai1 cDNA2 in Figure 2C). Figure 2C compares the genomic DNA and cDNA sequences and the predicted protein sequences. In the case of nai1 cDNA1, an AG sequence that is eight to nine bases after the point mutation site (indicated by an asterisk) was used as an intron splicing site. This led to a deletion of three amino acids after the mutation site, but it did not introduce a frame shift. In the case of nai1 cDNA2, an AG sequence that is 15 to 16 bases after the point mutation site (indicated by an asterisk) was used as an intron splicing site. This led to a frameshift of the coding sequence that, in turn, led to the creation of a premature stop codon. The lengths of nai1 cDNA1 and cDNA2 corresponded to the shoulder and peak positions in Figure 2B, respectively.
Characterization of a Mutant with a T-DNA Insertion in the At2g22770 Gene
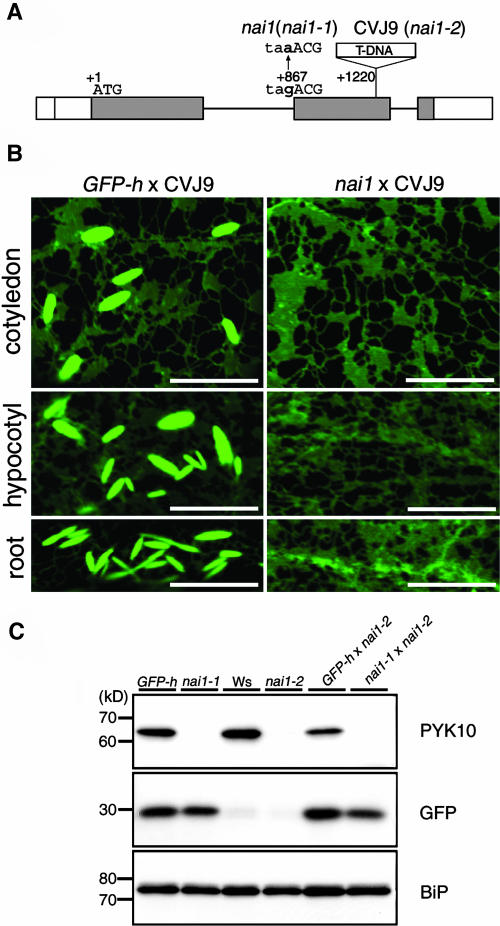
Next, we characterized a mutant (CVJ9, Wassilewskija [Ws] background) containing a T-DNA insertion in the At2g22770 gene. CVJ9 had a T-DNA insertion in the second exon of the At2g22770 gene (Figure 3A). To determine allelism, GFP-h and nai1 were crossed with CVJ9. The F1 progenies of GFP-h × CVJ9 exhibited fluorescent ER bodies in cotyledons, hypocotyls, and roots, being representative of the wild-type phenotype (Figure 3B), whereas no ER bodies were detected in the F1 seedlings of nai1 × CVJ9 (Figure 3B). These results indicated that nai1 and CVJ9 are allelic. Hereafter, we refer to nai1 and CVJ9 as nai1-1 and nai1-2, respectively.
Figure 3.
Allelism Test between nai1 and T-DNA Insertion Line.
(A) Schematic representation of the T-DNA insertion site in CVJ9 line.
(B) Fluorescent images of F1 progeny of a cross between GFP-h × CVJ9 and nai1 × CVJ9. No ER bodies were detected in the F1 seedlings of nai1 × CVJ9. Bars = 10 μm.
(C) Absence of PYK10 in nai1-1, nai1-2, or F1 progeny of nai1-1 × nai1-2. Extracts prepared from 7-d-old GFP-h, nai1-1, Ws, nai1-2, and F1 progeny of GFP-h × nai1-2 and F1 progeny of nai1-1 × nai1-2 were subjected to immunoblot analyses with anti-PYK10, anti-GFP, and anti-BiP antibodies. Molecular masses are given at the left in kD.
We previously showed that PYK10 is a major component in ER bodies and that the accumulation of PYK10 is decreased in nai1-1 (Matsushima et al., 2003a). We examined whether this phenotype was also observed in nai1-2. Crude extracts of 7-d-old seedlings were subjected to immunoblot analyses (Figure 3C). PYK10 was hardly detected in nai1-1, nai1-2, or the F1 progeny of nai1-1 × nai1-2. GFP was used as a control of genetic crosses and was detected in seedlings with the transgene. Binding protein (BiP), a member of the Hsp70 family localized in ER, used as a loading control yielded almost the same signals among the analyzed plants. These results indicate that nai1-1 and nai1-2 are also allelic with respect to PYK10 accumulation.
Induction of ER Bodies by Transient Expression of At2g22770 Protein
To examine whether At2g22770 protein is responsible for the formation of ER bodies, we expressed At2g22770 protein transiently in nai1-1 cells by biolistic transformation. We introduced a plasmid (monomeric red fluorescent protein 1 [mRFP1] + At2g22770/pBI221) in 7-d-old nai1-1 seedlings to coexpress At2g22770 protein and mRFP1 (Campbell et al., 2002) under the control of 35S promoter (Figure 4A); mRFP1 was used to identify the bombarded cells. In these cells, many ER bodies were observed 48 to 53 h after bombardment. The percentage of bombarded cells that developed ER bodies was 83.8% (n = 74). As a negative control, we bombarded a plasmid (mRFP1/pBI221) from which only mRFP1 is expressed (Figure 4B). In this case, the percentage of bombarded cells that developed ER bodies was 1.4% (n = 74). These results indicate that expression of At2g22770 cDNA complemented the nai1 phenotype. Therefore, we concluded that At2g22770 is the NAI1 gene.
Figure 4.
Complementation of the nai1 Phenotype by Transiently Expressed At2g22770 Protein.
(A) Fluorescent images of nai1-1 epidermal cells that expressed At2g22770 protein and mRFP1. Plasmid (mRFP1 + At2g22770/pBI221) from which mRFP1 and At2g22770 protein were expressed under the control of the 35S promoter was bombarded into 7-d-old nai1-1 seedlings. Images were taken with a confocal laser scanning microscope 48 to 53 h postbombardment. Spindle-shaped ER bodies were observed in the bombarded cells. Bars = 20 μm.
(B) Fluorescent images of nai1-1 epidermal cells that expressed only mRFP1. Plasmid (mRFP1/pBI221) from which only mRFP1 was expressed under the control of the 35S promoter was bombarded. No ER bodies were detected in the bombarded cells. Bars = 20 μm.
Two-Dimensional Electrophoresis Analysis Identified Proteins Depressed in nai1-1 Mutant
Previously, we compared proteins of the P1 fraction of GFP-h and nai1-1 by one-dimensional SDS-PAGE and identified PYK10 as a component of ER bodies (Matsushima et al., 2003a). To further resolve proteins in P1 fractions, they were subjected to two-dimensional electrophoresis (Figure 5). Protein spots specific to GFP-h (spot numbers 1 to 4) were analyzed by matrix-assisted laser-desorption ionization time of flight mass spectrometry (Table 1), and the results confirmed by the other analyses. The N-terminal sequences of spot numbers 1, 2, and 3 were determined by Edman degradation to be DGPVCPPXNKLXRA, DGPVXPP, and DGPVXPPSNKLSRA, respectively (X, not determined). A peptide of spot number 4, having a mass of 1091.63, was identified to have the sequence IGVHVRPLSN by post source decay analysis. Spot numbers 1, 2, and 3 were PYK10. Spot number 4 was putative myrosinase binding protein (At3g16420).
Figure 5.
Two-Dimensional Electrophoresis of Proteins in the P1 Fraction from GFP-h and nai1-1.
Six-day-old seedlings of GFP-h and nai1-1 were homogenized and centrifuged at 1000g. The P1 fraction was separated by two-dimensional electrophoresis with denaturing isoelectric focusing (IEF) on immobilized pH gradients in the first dimension and SDS-PAGE in the second dimension. Proteins were detected by silver staining. Arrows indicate the protein spots that were specific to GFP-h. The protein spot numbers refer to the spot numbers listed in Table 1. Numbers on the x axis are pI, and numbers on the y axis are molecular mass (kD).
Table 1.
Identified Proteins Specific to the GFP-h P1 Fraction
| Spot No. | Identified Protein | Accession Number | Calculated Mass (kD) | Calculated pI | Sequence Coverage (%) |
|---|---|---|---|---|---|
| 1 | PYK10 | At3g09260 | 60.20 | 6.45 | 12 |
| 2 | PYK10 | At3g09260 | 60.20 | 6.45 | 36 |
| 3 | PYK10 | At3g09260 | 60.20 | 6.45 | 23 |
| 4 | Putative myrosinase binding protein | At3g16420 | 32.14 | 5.46 | 26 |
Proteins were identified with the MASCOT search engine (http://www.matrixscience.com/). Accession numbers, calculated molecular masses, and calculated pI values are cited from the MASCOT database.
Downregulation of Transcription of PYK10 and At3g16420 Genes in nai1 Mutants
We raised anti-At3g16420 antibodies that are specific to an internal amino acid sequence of At3g16420 protein. Immunoblot analysis of Arabidopsis extracts showed that At3g16420 protein was hardly detected in nai1-1, nai1-2, or the F1 progeny of nai1-1 × nai1-2 (Figure 6A). This result indicated that loss of NAI1 results in the absence of At3g16420 protein.
Figure 6.
Downregulation of PYK10 and At3g16420 mRNA in nai1-1 and nai1-2 Mutants.
(A) Absence of At3g16420 protein in nai1-1, nai1-2, or F1 progeny of nai1-1 × nai1-2. Immunoblot analysis was performed with anti-At3g16420 antibodies on the same extracts as Figure 3C. Molecular masses are given at the left in kD.
(B) The mRNA levels of PYK10 and At3g16420 genes were monitored by RT-PCR. Total RNA from 6-d-old whole seedlings of Col, GFP-h, nai1-1, Ws, and nai1-2 was subject to reverse transcription. Actin was used as an internal control. The lanes GFP-hRT− and WsRT− are the negative controls in which reverse transcriptase was omitted from the reaction mixture.
NAI1 is a putative transcription factor because it has a bHLH domain (Figure 1C). Therefore, it is possible that NAI1 regulates the transcription of the PYK10 and At3g16420 genes. The mRNA transcripts of these genes were examined by RT-PCR (Figure 6B). The level of PYK10 and At3g16420 RNAs in nai1-1 was reduced compared to their level in GFP-h, and the level of PYK10 and At3g16420 RNAs in nai1-2 was reduced compared to their level in Ws. The PYK10 and At3g16420 RNA levels in GFP-h and Col were about the same. No PYK10 RNA signal was observed in the negative controls, which did not have reverse transcriptase (GFP-hRT− and WsRT−). The level of Actin RNA, used as an internal control, was the same in all the strains. These results suggest that reduction of PYK10 and At3g16420 proteins in the nai1-1 and nai1-2 mutants is a consequence of lower levels of gene transcription.
nai1 Mutants Have Lower Activities of β-d-Glucosidase and β-d-Fucosidase than Wild-Type Plants
It was expected that PYK10 has β-d-glucosidase activity because PYK10 has high homology (40 to 70% identity) with experimentally confirmed β-d-glucosidases. We measured β-d-glucosidase activity in crude extracts of GFP-h, nai1-1, Ws, and nai1-2 using a fluorogenic substrate of 4-methylumbelliferyl (4-MU) β-d-glucopyranoside (Figure 7A). The β-d-glucosidase activity of nai1-1 was 19% of that in GFP-h, and the β-d-glucosidase activity of nai1-2 was 39% of that in Ws. To confirm the β-d-glucosidase activity of PYK10 more directly, the GFP-h and Ws extracts were incubated with anti-PYK10 antibodies before addition of substrate. Anti-PYK10 antibodies reduced the β-d-glucosidase activities of GFP-h and Ws to 27 and 36%, respectively, of the activities in the presence of preimmune serum (Figure 7B). These results indicated that PYK10 has β-d-glucosidase activity and that the reduced level of PYK10 in nai1 mutants leads to a sharp reduction of β-d-glucosidase activity in these mutants. Anti-PYK10 antibodies reduced the β-d-glucosidase activity of nai1-1 by only 2.7 pmol 4-MU/s/mg protein (Figure 7B). This result confirms that the inhibitory effect of the antibodies is mostly because of the specific binding with PYK10, not because of the nonspecific binding.
Figure 7.
Decreased β-d-Glucosidase and β-d-Fucosidase Activity in nai1-1 and nai1-2 Mutants.
(A) Assay was performed using a fluorogenic substrate, 4-MU β-d-glucopyranoside, by monitoring the increase of fluorescence. β-d-Glucosidase activity of nai1-1 decreased compared with that of GFP-h and β-d-glucosidase activity of nai1-2 decreased compared with that of Ws.
(B) Effect of anti-PYK10 antibodies (i) or its preimmune serum (p) on β-d-glucosidase activity. Extracts were incubated with anti-PYK10 antibodies or its preimmune serum before mixing with the substrate. The inhibitory effect of anti-PYK10 antibodies indicates that PYK10 has β-d-glucosidase activity.
(C) Assay was performed using a fluorogenic substrate, 4-MU β-d-fucoside, by monitoring the increase of fluorescence. β-d-Fucosidase activity of nai1-1 decreased compared with that of GFP-h, and β-d-fucosidase activity of nai1-2 decreased compared with that of Ws.
(D) Effect of anti-PYK10 antibodies (i) or its preimmune serum (p) on β-d-fucosidase activity. The inhibitory effect of anti-PYK10 antibodies indicates that PYK10 has β-d-fucosidase activity.
Each bar represents the mean value of three independent experiments, and error bars show standard deviations.
To identify other glycosidase activities of PYK10, we measured the hydrolytic activities of GFP-h and nai1-1 extracts against several fluorogenic (4-MU) substrates. A major difference in activities between GFP-h and nai1-1 was found for 4-MU β-d-fucoside (Figure 7C), but little difference was found for eight other substrates (4-MU α-d-glucoside, 4-MU α-l-fucoside, 4-MU β-l-fucoside, 4-MU β-d-lactoside, 4-MU β-d-cellobioside, 4-MU β-d-mannopyranoside, 4-MU 7-β-d-xyloside, and 4-MU β-d-galactopyranoside) (data not shown). The β-d-fucosidase activity in nai1-1 was 11% of that in GFP-h, and the β-d-fucosidase activity of nai1-2 was 37% of that in Ws. Anti-PYK10 antibodies reduced the β-d-fucosidase activity of GFP-h and Ws to 23 and 33%, respectively, of the activities in the presence of preimmune serum (Figure 7D). Anti-PYK10 antibodies also reduced the β-d-fucosidase activity of nai1-1 by only 6.3 pmol 4-MU/s/mg protein (Figure 7D). These results indicated that PYK10 has β-d-fucosidase activity in addition to β-d-glucosidase activity.
Irregular Shapes of i-ER Bodies in nai1-1 Rosette Leaves and Induction of NAI1 by MeJA Treatment
MeJA induces the formation of ER body–like structures (i-ER bodies) in rosette leaves that have no ER bodies under normal conditions (Matsushima et al., 2002). The shapes and sizes of ER bodies and i-ER bodies are so similar that the molecular mechanism(s) underlying the formation of these structures may be shared. We treated nai1-1 rosette leaves with 50 μM MeJA and studied the formation of i-ER bodies (Figure 8A). nai1-1 rosette leaves, like GFP-h leaves, developed many i-ER bodies. However, the i-ER bodies in nai1-1 had various shapes and sizes and included irregular-shaped and longer structures, whereas most i-ER bodies in GFP-h leaves had uniform shapes and sizes. Such irregular-shaped i-ER bodies (Figure 8A) were also observed in MeJA-treated GFP-h leaves occasionally (data not shown). Water had no effect on the ER network or the development of i-ER bodies.
Figure 8.
Irregular Shapes of i-ER Bodies in nai1-1 Rosette Leaves and Induction of the NAI1 Gene by MeJA Treatment.
(A) nai1-1 and GFP-h rosette leaves were treated with 50 μM MeJA or water. Epidermal cells of each rosette leaf were inspected with a confocal microscope 34 to 36 h later. Bars = 10 μm.
(B) Expression of NAI1, PYK10, and At3g16420 genes in water-treated, MeJA-treated, and MeJA plus ethylene–treated leaves was monitored by RT-PCR. Actin was used as an internal control. The lane MeJART− is a negative control in which reverse transcriptase was omitted in the reaction mixture of MeJA-treated leaves.
Figure 8B shows the expression patterns of NAI1, PYK10, and At3g16420 genes in water-treated, MeJA-treated, and MeJA plus ethylene–treated leaves. MeJA and ethylene has an antagonistic effect on the induction of i-ER bodies (Matsushima et al., 2002). MeJA treatment induced the expression of NAI1, PYK10, and At3g16420 genes, and this induction was suppressed in the presence of ethylene. The levels of Actin RNA, used as an internal control, were the same in all the treatments. These results showed that expression patterns of NAI1, PYK10, and At3g16420 genes is parallel to the induction pattern of i-ER bodies, suggesting that MeJA-induced NAI1 is needed for the correct formation of i-ER bodies.
DISCUSSION
NAI1 Is a bHLH Protein That Regulates the Formation of ER-Derived Structure
In this study, we performed map-based cloning of the NAI1 gene and showed that the nai1-1 mutant has a single nucleotide change in the At2g22770 gene. The hypothesis that At2g22770 is the NAI1 gene is supported by the aberrant splicing of At2g22770 RNA in the nai1-1 mutant (Figure 2), the results of an allelism test between nai1-1 and a T-DNA insertion allele (nai1-2) (Figure 3), and complementation of the nai1 phenotype by transient expression of At2g22770 protein (Figure 4). NAI1 is a bHLH protein that could function as a transcription factor.
bHLH proteins have been well characterized in nonplant eukaryotes, especially in mammalian systems. In these systems, bHLH proteins are important regulatory components in transcriptional networks controlling various processes, from cell proliferation to cell lineage establishments (Grandori et al., 2000; Massari and Murre, 2000). In plants, broad physiological roles of bHLH proteins have been reported, including roles in anthocyanin pigmentation (Goodrich et al., 1992; Spelt et al., 2000), phytochrome signal transduction (Ni et al., 1998; Fairchild et al., 2000), regulation of seed storage protein synthesis (Kawagoe and Murai, 1996), response to stress conditions (Abe et al., 1997; Chinnusamy et al., 2003), and organ development (Heisler et al., 2001). Physiological roles depend on the downstream genes regulated by each bHLH protein.
A transcription factor that is related to the formation of ER-derived structures has been described in maize. Opaque-2 (O2) is a basic Leu zipper-type transcription factor that regulates the expression of α-zein genes, whose product is a major component in protein bodies in maize endosperm (Hartings et al., 1989; Lending and Larkins, 1989; Schmidt et al., 1990, 1992). Therefore, α-zein synthesis is depressed in o2 mutants because of the absence of an upstream transcriptional activator (O2) (Hunter et al., 2002). As a result of the depressed amount of α-zein, protein bodies in o2 remain small (0.1 to 0.3 μm in diameter) compared with the size of wild-type protein bodies (1 to 2 μm in diameter) (Lending and Larkins, 1989; Geetha et al., 1991). We suggest that the absence of ER bodies in nai1 mutants is also attributable to the loss of a transcription factor (NAI1) that functions upstream of a major component (PYK10) (see below).
PYK10 and At3g16420 Genes Are Downstream of NAI1 Regulation
bHLH proteins are known to bind to a consensus hexanucleotide (CANNTG) called an E-box (Ferre-D'Amare and Burley, 1995). The basic region in the bHLH domain determines the DNA binding activity. In mammalian bHLH proteins, the analysis of crystal structures has shown that the Glu residue (arrow in Figure 1C) is critical because it contacts the first CA in the E-box (Ferre-D'Amare et al., 1993; Ma et al., 1994). This Glu residue is also conserved in NAI1 (Glu-137). Therefore, downstream genes of NAI1 may have E-box sequences in their promoter region. Because expression of PYK10 and At3g16420 genes were downregulated in nai1 mutants (Figure 6B), these genes must be downstream of NAI1. PYK10 has five E-box sequences in its promoter region (CAACTG at 805 to 810 bp, CATTTG at 881 to 886 bp, CATGTG at 977 to 982 bp, CAGCTG at 986 to 991 bp, and CATATG at 1350 to 1355 bp upstream of the putative first ATG). The PYK10 promoter region has been investigated through a promoter-deletion analysis using a β-glucuronidase reporter gene (Nitz et al., 2001). β-Glucuronidase activity was found to be 8 times higher when the promoter contained the 1457-bp region upstream of the putative start codon (construct C described in Nitz et al., 2001) than when it contained the 797-bp region upstream of the putative start codon (construct B described in Nitz et al., 2001). The former promoter contains the E-box sequences described above, whereas the latter does not. This result suggests that the E-box sequences in the PYK10 promoter have a strong effect on the expression of PYK10. It is possible that some or all E-box sequences of PYK10 are directly recognized by NAI1. The At3g16420 gene has three E-box sequences in the 2000-bp region upstream from the putative translation start site. The effectiveness of these E-box sequences has not been studied. Proteomic analysis of seed germination has shown that the relative abundance of PYK10 and At3g16420 proteins are most increased (>200-fold) just after germination (Gallardo et al., 2001). It is very likely that both genes are regulated by a common factor. MeJA induced expression of NAI1, PYK10, and At3g16420 genes, and ethylene suppressed the induction (Figure 8B). The common expression pattern of these genes is consistent with the idea that PYK10 and At3g16420 genes are regulated by a common factor, NAI1.
Physiological Role of At3g16420 Protein
At3g16420 protein consists of two repeated regions (At3g16420-N and At3g16420-C in Figure 9A). Each of these regions is homologous to the α-chain of jacalin, a carbohydrate binding protein (lectin) isolated from jackfruit (Artocarpus integrifolia) (Yang and Czapla, 1993). The α-chain of jacalin has been shown to bind IgA1 from human serum, specifically through the oligosaccharides of the IgA1 hinge region (Skea et al., 1988; Biewenga et al., 1989; Sankaranarayanan et al., 1996). Myrosinase binding protein 70p (MBP70p) from Brassica napus has three jacalin-homologous regions (repeats 1 to 3 in Figure 9A) (Geshi and Brandt, 1998). At3g16420 protein and MBP70p are homologous sharing the jacalin-homologous regions. These sequences are aligned in Figure 9B.
Figure 9.
Repeated Protein Structure of At3g16420 Protein Homologous to a Lectin.
(A) At3g16420 protein consists of two consecutive repeated regions (gray boxes of At3g16420-N and At3g16420-C, respectively). Both regions show high homology with α-chain of jacalin, a lectin isolated from jackfruit (Artocarpus integrifolia). Myrosinase binding protein 70p (MBP70p, Brassica napus) has three jacalin-homologous regions (gray boxes of repeats 1, 2, and 3, respectively).
(B) Alignment of amino acid sequences of the repeated regions of At3g16420 protein (At3g16420-N and At3g16420-C), MBP70p (70p-repeats 1, 2, and 3), and α-chain of jacalin (jacalin-α). Identical residues are shown in black, and similar residues are shown in gray.
MBPs are known to form large complexes (250 to 1000 kD) with myrosinase (Lenman et al., 1990; Falk et al., 1995; Eriksson et al., 2002). Myrosinase is a β-glucosidase responsible for the hydrolysis of glucosinolates, which are secondary metabolites found mainly in the Brassicaceae family (Bones and Rossiter, 1996; Rask et al., 2000). MBPs have lectin activity (Taipalensuu et al., 1997; Geshi and Brandt, 1998), so oligosaccharides of glycosylated myrosinase have been suggested to be targets of MBPs. PYK10 is also glycosylated with three high-mannose oligosaccharides (Matsushima et al., 2003a). Therefore, it is possible that At3g16420 protein forms complexes with glycosylated PYK10.
Immunogold labeling shows that MBP70p is specifically localized to the lumenal side of protein-body-like structures and vacuoles, although a cleavable ER signal peptide was not found at the N terminus of MBP70p (Geshi and Brandt, 1998). Some mammalian proteins are known to be secreted without cleavage of a signal peptide (Palmiter et al., 1978; Ye et al., 1988). MBP70p has been suggested to enter the secretary pathway by a mechanism that does not need a cleavable signal peptide (Geshi and Brandt, 1998). Hydropathy analysis shows that At3g16420 protein also has no cleavable signal peptides and no transmembrane domains (data not shown). Further studies are needed to determine the subcellular localization of At3g16420 protein.
Many plant lectins serve as natural plant protectants against herbivores (Chrispeels and Raikhel, 1991; Peumans and Van Damme, 1995a, 1995b). Lectins inhibit growth and increase mortality of insect larvae through binding to gut epithelial cells and/or to the peritrophic membrane (Gatehouse et al., 1984; Eisemann et al., 1994). Jacalin inhibits the growth of larval Southern corn rootworm (Diabrotica undecimpunctata howardi) when applied to artificial diets (Czapla and Lang, 1990). We previously suggested that the biological function of ER bodies and PYK10 is related to plant defense against herbivores and pathogens (Matsushima et al., 2002, 2003a, 2003b). It is possible that At3g16420 protein participates in the ER body–mediated defense systems.
PYK10 Exhibits Both β-d-Glucosidase and β-d-Fucosidase Activities
4-MU β-d-glucopyranoside is a general substrate for a broad range of β-d-glucosidases (Dharmawardhana et al., 1995; Esen and Blanchard, 2000; Kim et al., 2000). Therefore, the results in Figure 7B suggest that PYK10 is a major β-d-glucosidase in Arabidopsis roots. In addition to the β-d-glucosidase activity, we found PYK10 also exhibited β-d-fucosidase activity (Figures 7C and 7D). In the presence of anti-PYK10 antibodies, the β-d-fucosidase activity was reduced by 64 to 69 pmol 4-MU/s/mg protein (Figure 7D), whereas β-d-glucosidase activity was reduced by 24 to 26 pmol 4-MU/s/mg protein (Figure 7B). This indicates that PYK10 hydrolyzes the β-d-fucosidic linkage more effectively than the β-d-glucosidic linkage.
Fucose residues exist as α-l-fucose in oligosaccharides of cell walls and glycosylated proteins in plant cells (Staudacher et al., 1999; Reiter, 2002). α-l-Fucosidase hydrolyzes α-l-fucosidic linkages (Augur et al., 1993; Torre et al., 2002). However, little is known about the in vivo roles of β-d-fucosidase and β-d-fucose. Some β-d-glucosidases hydrolyze β-d-fucosidic linkages (Eksittikul and Chulavatnatol, 1988; Babcock and Esen, 1994; Srisomsap et al., 1996). The β-d-fucosidase activity of PYK10 may be mainly attributable to the close structural similarity of fucose and glucose.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col, Landsberg erecta, and Ws were used as wild-type plants. We also used GFP-h and nai1-1 plants. GFP-h is an Arabidopsis (ecotype Col) transformed with a Pro35S:SP-GFP-HDEL gene encoding SP-GFP-HDEL, which is composed of the signal peptide of pumpkin (Cucurbita maxima) 2S albumin, and GFP followed by an ER-retention signal, HDEL (Mitsuhashi et al., 2000; Hayashi et al., 2001). nai1-1 is an ER body–deficient mutant isolated from mutagenized GFP-h plants (Matsushima et al., 2003a). We also used a T-DNA insertion line (ecotype Ws), named nai1-2 (CVJ9), in which a T-DNA was inserted into At2g22770 gene. nai1-2 was isolated from the Versailles collection of T-DNA insertion mutants (Samson et al., 2002). Seeds of Arabidopsis were surface sterilized and then sown on soil or onto 0.5% (w/v) Gellan Gum (Wako, Tokyo, Japan) with MS medium (Wako) and 1.5% (w/v) sucrose and were grown at 22°C under continuous light conditions.
Mapping of the NAI1 Locus
To determine the map position of the NAI1 locus, nai1-1 and the wild-type Landsberg erecta plant were crossed. F2 seeds were obtained by self-fertilization of the F1 plants. Six- or seven-day-old seedlings of the F2 progenies were examined with a fluorescence microscope to select nai1-1 mutant plants. The genomic DNA of these nai1-1 mutant plants was individually isolated and analyzed using a combination of cleaved amplified polymorphic sequence markers and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) with data obtained from the Arabidopsis Information Resource (http://www.arabidopsis.org). Primers and enzymes used for molecular markers are as follows. T9I22-5: 5′-ATGCTAGAGCTATTCGGAGTGTG-3′ and 5′-AGATTATCCTCTTCGCCATCAAA-3′, SfcI; T9I22-4: 5′-GCTGGACTCGAAATGGTCACT-3′ and 5′-GTCACCACCACAACTACCAGATT-3′, BclI; T9I22-7: 5′-GCTAAGGGTGGCTTGAACAATG-3′ and 5′-CCCTGCAAAACAATAGACACAGA-3′; T30L20-3: 5′-TCTCAGCCGTTGAATCCTCTCAT-3′ and 5′-TTGTCATTCCCAGAGCATAAGTT-3′, AccI; T30L20-4: 5′-CTTTACTCTCCTCGCTCTCGTCT-3′ and 5′-CCCAAAACTCAAAATCGTTCCTT-3′, AluI; T20K9-1: 5′-TTCAGATGGAGCCTAAAGGTGT-3′ and 5′-TCCTCACCGCCAATAGCAAAA-3′, NdeI.
Determination of the Transcriptional Start Site and 3′ Untranslated Region of the At2g22770 Gene
Transcriptional start site analysis based on the capping structure of mRNA was conducted using an Arabidopsis Cap Site cDNA dT kit (Nippon Gene, Toyama, Japan) according to the manufacturer's instructions. We used 5′-TCCGAAGAAGAAGACAGAGGAG-3′ and 5′-CCCACTTTCTGATCCATGCTTAC-3′ as gene-specific primers for initial PCR and nested PCR, respectively. The first PCR conditions were as follows: 95°C for 2 min; 40 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 45 s; and a final elongation step of 2 min at 72°C. The second PCR conditions were as follows: 95°C for 2 min; 30 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 30 s; and a final elongation step of 2 min at 72°C. The second PCR gave two products with different sizes. These products were subcloned into pT7Blue T-vector (Novagen, Madison, WI) and subjected to sequencing. The 3′ untranslated region of At2g22770 gene was determined by sequencing of an EST clone (AV527715) obtained from Kazusa DNA Research Institute (Kisarazu, Chiba, Japan).
Laser Scanning Confocal Microscopy
Fluorescent proteins were viewed with a laser scanning confocal microscope (LSM510; Carl Zeiss, Jena, Germany). For imaging GFP (Figure 3B), argon laser excitation lines of 488 nm were used with HFT 488 (Carl Zeiss) as a dichroic beam splitter and a 505/550-nm bandpass filter. For imaging GFP and mRFP1 (Figure 4), argon laser excitation lines of 488 nm for GFP and helium-neon laser lines of 543 nm for mRFP1 were used with line switching using the multitrack facility of the microscope. Fluorescence was detected using HFT 488/543 (Carl Zeiss) as a main dichroic beam splitter and NFT 545 (Carl Zeiss) as a secondary dichroic beam splitter. A 505/530-nm bandpass filter for GFP and a 560/615-nm bandpass filter for mRFP1 were used. The fluorescent images were analyzed with LSM 5 (Carl Zeiss) and Adobe Photoshop 5.5 (Adobe Systems, Tokyo, Japan).
Specific Antibodies
To prepare anti-At3g16420 antibodies, a peptide derived from At3g16420 protein, CKNGQPEQAPLRGTKG, was chemically synthesized with a peptide synthesizer (model 431A; Applied Biosystems, Tokyo, Japan). Cross-linking of the peptide with BSA and immunization of a rabbit were conducted as described previously (Matsushima et al., 2003a). We also used antibodies against each of GFP (Clontech, Palo Alto, CA), BiP (Hatano et al., 1997), and PYK10 [anti-PYK10(IM) antibody] that we had raised previously (Matsushima et al., 2003a).
Immunoblot Analysis
Extracts were prepared from 7-d-old GFP-h, nai1-1, Ws, nai1-2, F1 progeny of GFP-h × nai1-2, and F1 progeny of nai1-1 × nai1-2. A whole seedling was homogenized in 75 μL of extraction buffer (50 mM Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% [v/v] glycerol, and 5% [v/v] 2-mercaptoethnol). The extracts (8 μL) were subjected to SDS-PAGE. After SDS-PAGE, proteins were transferred electrophoretically to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Billerica, MA). The membrane was thoroughly dried for blocking and then incubated in Tris-buffered saline, pH 7.5, plus 0.05% (v/v) Tween 20 and antibodies for 1 h. Dilutions of antibodies were as follows: anti-PYK10(IM) 1:15,000 (v/v), anti-GFP 1:30,000, anti-BiP 1:30,000, and anti-At3g16420 (1:5000). Horseradish peroxidase–conjugated goat antibodies against rabbit IgG (Pierce, Rockford, IL) were diluted (1:5000) to be used as second antibodies. Immunodetection was performed with an ECL kit (Amersham Biosciences, Buckinghamshire, UK).
Plasmid Construction
To construct mRFP1/pBI221 (Figure 4B), a DNA fragment was produced by PCR amplification using mRFP1 in pRSETB (Campbell et al., 2002) as template and primers 5′-GCTCGAGATGCGGGGTTCTCATCATCATCA-3′ and 5′-CGAGCTCTTAGGCGCCGGTGGAGTGGCGGC-3′. The fragment produced from this amplification was inserted into XhoI and SacI sites of pBI221 (Clontech).
To construct At2g22770/pBI221, At2g22770 cDNA was produced by PCR using the EST clone as template and primers 5′-GTTCTCGAGATGGATGATTCAAGCTTTATG-3′ and 5′-TTGGATCCTTATTCAGCTAACGCAACTCTT-3′. The amplified fragment was then inserted into the XhoI-BamHI site of SP-GFP-2SC/pBI221 (Tamura et al., 2003).
To create mRFP1+At2g22770/pBI221 (Figure 4A), a HindIII-EcoRI (blunted with Klenow) fragment of mRFP1/pBI221 that contains the 35S promoter of Cauliflower mosaic virus, the sequence encoding mRFP1, and nopaline synthase terminator were inserted at the SphI site (blunted) of At2g22770/pBI221.
Biolistic Transformation
Seven-day-old nai1-1 seedlings were bombarded with 1-μm gold particles coated with mRFP1/pBI221 and mRFP1+At2g22770/pBI221 using a Helios gene gun system (Bio-Rad Laboratories, Hercules, CA) based on the manufacturer's protocol. The bombarded samples were kept in darkness at 22°C for 48 to 53 h and observed with the laser scanning confocal microscope.
Preparation of P1 Fraction
Six-day-old GFP-h and nai1-1 seedlings (1.3 g) were chopped with a razor blade in a Petri dish on ice in 3.9 mL of chopping buffer that contained 50 mM Hepes-NaOH, pH 7.5, and 400 mM sucrose. The homogenates were filtered through cheesecloth and centrifuged at 1000g at 4°C for 20 min. The pellet was resuspended with 400 μL of 20 mM sodium phosphate, pH 7.0, and sonicated using a sonicator (Sonifier 250D; Branson Japan, Atsugi, Japan) with an output of 1 for 30 s. The sonicated suspension was centrifuged at 8000g at 4°C for 5 min. The supernatant was used for two-dimensional electrophoresis.
Two-Dimensional Electrophoresis
The P1 fractions obtained from GFP-h and nai1-1 seedlings were subjected to two-dimensional electrophoresis. Isoelectricfocusing was conducted using dry polyacrylamide gel strips (Immobiline DryStrip, pH 3 to 10, 13 cm; Amersham Biosciences, Uppsala, Sweden) with an immobilized pH gradient from 3 to 10 at 20°C. Strips were rehydrated with 220 μL of reswelling buffer (7 M urea, 2 M thiourea, 2% [w/v] 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, 1.2% [v/v] Destreak reagent [Amersham Biosciences], 0.5% [v/v] immobilized pH gradient buffer, pH 3 to 10 [Amersham Biosciences], and 0.005% [w/v] bromophenol blue) and 30 μL of each P1 fraction for 1 h. Isoelectricfocusing was performed at 30 V for 11 h, 500 V for 3 h, 1000 V for 2 h, and 8000 V for 50,000 Vh in an Ettan IPGphorII isoelectric focusing system (Amersham Biosciences). Strips were equilibrated by SDS equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% [v/v] glycerol, and 2% [w/v] SDS) with 1% (w/v) DTT for 15 min and then with 2.5% (w/v) iodoacetamide for 15 min. Equilibrated gel strips were placed on a 12.5% (w/v) polyacrylamide gel, and SDS-PAGE was performed.
Mass Spectrometry
The gel was treated with deampholyte buffer (50% [v/v] methanol and 7% [v/v] acetic acid) for 12 h. Silver staining was performed by the method of Yan et al. (2000). Spots specific to the GFP-h P1 fraction were excised from the silver-stained gel, and destaining was performed by methods of Gharahdaghi et al. (1999). The sample was digested with 0.1 μg of trypsin (Promega, Tokyo, Japan) in 20 μL of 0.2 M ammonium bicarbonate, pH 8.0, for 12 h at 37°C. Peptides were extracted from gel slices by 20 μL of 5% (v/v) trifluoroacetic acid and 50% (v/v) acetonitrile for 30 min twice, and the solution of extracted peptides was dried using an evaporator. The dried sample was reconstituted by adding 3 μL of 0.1% trifluoroacetic acid and 50% acetonitrile and gently pipetting up and down to dissolve the extracted peptides. The peptides were purified and concentrated with reverse-phase media (C18 Zip-Tips; Millipore). The peptides and 150 ng of α-cyano-4-hydroxy-cinnamic acid (Bruker Daltonics, Bremen, Germany) were mixed and subjected to peptide mass fingerprinting analysis by matrix-assisted laser-desorption ionization time of flight mass spectrometry (REFLEX III; Bruker Daltonics). Proteins were identified by searching the MASCOT search engine (http://www.matrixscience.com/). Protein identity of spot number 4 was further confirmed from post source decay spectra. The searches with the post source decay data were conducted using the MASCOT search engine. Protein identities of spot numbers 1 to 3 were confirmed by the N-terminal amino acid sequence analysis as described previously (Matsushima et al., 2003a).
RT-PCR Analyses
Total RNA was isolated from 6-d-old Col, GFP-h, nai1-1, Ws, and nai1-2 seedlings using an RNeasy plant mini kit (Qiagen, Valencia, CA). One microgram of total RNA was treated with DNaseI (Invitrogen, Carlsbad, CA) and subjected to first-strand cDNA synthesis using SuperScriptII reverse transcriptase (Invitrogen) and oligo(dT)12-18 primer (Invitrogen). Negative controls were used in experiments in which the reverse transcriptase was omitted. PCR conditions were as follows: 94°C for 2 min; 25 to 40 cycles of 94°C for 30 s, 60°C (Actin, PYK10, and At3g16420 genes) or 55°C (At2g22770 [NAI1] gene) for 45 s, and 72°C for 1 min; and a final elongation step of 4 min at 72°C. The following gene-specific primers were used: for At2g22770 [NAI1] gene, 5′-AGACATCTTTGAACTTCTCCAACC-3′ and 5′-CAAGACGAAACTTCTCCAAGGAAC-3′; for Actin, 5′-AGAGATTCAGATGCCCAGAAGTCTTGTTCC-3′ and 5′-AACGATTCCTGGACCTGCCTCATCATACTC-3′ (Ratcliffe et al., 2003); for PYK10, 5′-ATCTCCTTATCTCTCACGCAGAAGCAG-3′ and 5′-ATCTTGGTCACTCACAAAGTAAAACATAGG-3′; for At3g16420 gene, 5′-TACTATCAAGAAGCAGAAGATGGCCCAAAA-3′ and 5′-CATCATCAGTTGGATAAAGGACGAACATGG-3′. The PCR products were separated by 1.0% agarose gel electrophoresis or analyzed by a capillary electrophoresis (Agilent 2100 Bioanalyzer; Agilent Technologies, Waldbronn, Germany).
Assay of β-d-Glucosidase and β-d-Fucosidase Activities
Total protein extracts were prepared from 17-d-old roots of GFP-h, nai1-1, Ws, and nai1-2. Six to eight roots were homogenized in 1 mL of 50 mM sodium phosphate, pH 7.0, and centrifuged at 8000g for 5 min. The supernatants were filtrated through a 5-μm filter (low-binding Durapore polyvinylidene difluoride membrane, Ultrafree-CL; Millipore). Protein concentration in the filtrates was determined using the advanced protein assay reagent (Cytoskeleton, Denver, CO). We used 4-MU β-d-glucopyranoside (Seikagaku-kogyo, Tokyo, Japan) and 4-MU β-d-fucoside (Sigma) as a fluorescent substrate of β-d-glucosidase and β-d-fucosidase, respectively. The filtrates containing 3-μg protein were incubated in 100 mM sodium phosphate, pH 7.0, for 10 min at 35°C (total 198 μL). Two microliters of 100 mM substrate was added to the solution and incubated at 35°C. The fluorescence intensity was measured using GENios (TECAN, Mannedorf, Switzerland) for kinetic analysis. The fluorescence was monitored under an excitation wavelength of 360 nm and an emission wavelength 465 nm. For an inhibitory assay of anti-PYK10 antibodies, the IgG fractions were purified from anti-PYK10(IM) antiserum and preimmune serum using a HiTrap protein A HP column (Amersham Biosciences). The filtrates containing 3-μg protein were incubated in 100 mM sodium phosphate, pH 7.0, together with 25-μg IgG at 35°C for 30 min (total 198 μL). The reaction was started by addition of 2 μL of a 100 mM solution of the substrates and monitored as described above.
Chemical Treatment
For MeJA treatment, rosette leaves of 16- to 17-d-old GFP-h and nai1-1 plants were floated on 50 μM MeJA solution and incubated at 22°C under continuous light conditions. As a negative control, water was used instead of MeJA. The rosette leaves were inspected with a laser scanning confocal microscope at 34 to 36 h after the treatments.
To study gene expressions, RT-PCR analyses were performed with chemically treated rosette leaves of 17-d-old GFP-h plants. For treatment of MeJA and ethylene simultaneously, the floating leaves were transferred to an airtight box containing 20 μL/L of ethylene gas. The procedures of chemical treatments were described above except that the treatment time was 12 h. The procedures of RT-PCR were the same as described above.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers: AC006340 (BAC clone T9I22), AC005617 (BAC clone T30L20), AC004786 (BAC clone T20K9), O80536 (PIF3, At1g09530), Q39204 (AtMYC2, At1g32640), AAG25928 (AN1), AAA32663 (DEL), T10861 (PG1), P01106 (c-myc), P25912 (MAX), CAB50792 (PYK10, At3g09260), Q96292 (Actin, At3g18780), 1UGXA (α-chain of jacalin), T08080 (MBP 70p), AV527715 (an EST clone of At2g22770 gene), and NP_850594 (Atg16420).
Acknowledgments
We are grateful to R.Y. Tsien (Howard Hughes Medical Institute Laboratories at the University of California, San Diego) for his gift of a plasmid (mRFP1 in pRSETB), to Kentro Tamura of Kyoto University for his construct of mRFP1/pBI221, and to Y. Makino and T. Mori of the National Institute for Basic Biology for their helpful support with peptide sequencing and synthesis, respectively. The T-DNA tagged line (CVJ9) was provided by the Institut National de la Recherche Agronomique (Versailles, France). This work has been supported by Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation, Grants-in-Aid for 21st Century Center of Excellence Research, Kyoto University (A14), and by a postdoctoral fellowship to R.M. from the Japan Society for the Promotion of Science (14001468).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ikuko Hara-Nishimura (ihnishi@gr.bot.kyoto-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021154.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augur, C., Benhamou, N., Darvill, A., and Albersheim, P. (1993). Purification, characterization, and cell wall localization of an α-fucosidase that inactivates a xyloglucan oligosaccarin. Plant J. 3, 415–426. [DOI] [PubMed] [Google Scholar]
- Babcock, G.D., and Esen, A. (1994). Substrate specificity of maize β-glucosidase. Plant Sci. 101, 31–39. [Google Scholar]
- Baumann, O., and Walz, B. (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205, 149–214. [DOI] [PubMed] [Google Scholar]
- Behnke, H.-D., and Eschlbeck, G. (1978). Dilated cisternae in Capparales—An attempt towards the characterization of a specific endoplasmic reticulum. Protoplasma 97, 351–363. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Biewenga, J., Hiemstra, P.S., Steneker, I., and Daha, M.R. (1989). Binding of human IgA1 and IgA1 fragments to jacalin. Mol. Immunol. 26, 275–281. [DOI] [PubMed] [Google Scholar]
- Bones, A.M., Evjen, K., and Iversen, T.-H. (1989). Characterization and distribution of dilated cisternae of the endoplasmic reticulum in intact plants, protoplasts, and calli of Brassicaceae. Isr. J. Bot. 38, 177–192. [Google Scholar]
- Bones, A.M., and Rossiter, J.T. (1996). The myrosinase-glucosinolate system, its organization and biochemistry. Physiol. Plant 97, 194–208. [Google Scholar]
- Bonnett, H.T., Jr., and Newcomb, E.H. (1965). Polyribosomes and cisternal accumulations in root cells of radish. J. Cell Biol. 27, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B., Hong, X., Agarwal, M., and Zhu, J.-K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels, M.J., and Herman, E.M. (2000). Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol. 123, 1227–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels, M.J., and Raikhel, N.V. (1991). Lectins, lectin genes, and their role in plant defense. Plant Cell 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czapla, T.H., and Lang, B.A. (1990). Effect of plant lectins on the larval development of European corn borer (Lepidoptera: Pyralidae) and southern corn rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 83, 2480–2485. [Google Scholar]
- Dharmawardhana, D.P., Ellis, B.E., and Carlson, J.E. (1995). A β-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 107, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisemann, C.H., Donaldoson, R.A., Pearson, R.D., Cadogan, L.C., Vuocolo, T., and Tellam, R.L. (1994). Larvicidal activity of lectins on Lucilia cuprina: Mechanism of action. Entomol. Exp. Appl. 72, 1–10. [Google Scholar]
- Eksittikul, T., and Chulavatnatol, M. (1988). Characterization of cyanogenic β-glucosidase (linamarase) from cassava (Manihot esculenta Crantz). Arch. Biochem. Biophys. 266, 263–269. [DOI] [PubMed] [Google Scholar]
- Eriksson, S., Andreasson, E., Ekbom, B., Graner, G., Pontoppidan, B., Taipalensuu, J., Zhang, J., Rask, L., and Meijer, J. (2002). Complex formation of myrosinase isoenzymes in oilseed rape seeds are dependent on the presence of myrosinase-binding proteins. Plant Physiol. 129, 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen, A., and Blanchard, D.J. (2000). A specific β-glucosidase-aggregating factor is responsible for the β-glucosidase null phenotype in maize. Plant Physiol. 122, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Falk, A., Taipalensuu, J., Ek, B., Lenman, M., and Rask, L. (1995). Characterization of rapeseed myrosinase-binding protein. Planta 195, 387–395. [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare, A.R., and Burley, S.K. (1995). DNA recognition by helix-loop-helix proteins. In Nucleic Acids and Molecular Biology, F. Eckstein and D.M.J. Lilley, eds (Berlin, Heidelberg: Springer-Verlag), pp. 285–298.
- Ferre-D'Amare, A.R., Prendergast, G.C., Ziff, E.B., and Burley, S.K. (1993). Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45. [DOI] [PubMed] [Google Scholar]
- Gallardo, K., Job, C., Groot, S.P.C., Puype, M., Demol, H., Vandekerckhove, J., and Job, D. (2001). Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatehouse, A.M.R., Dewey, F.M., Dove, J., Fenton, K.A., and Pusztai, A. (1984). Effect of seed lectins from Phaseolus vulgaris on the development of larvae of Callosobruchus maculates: Mechanism of toxicity. J. Sci. Food Agric. 35, 373–380. [Google Scholar]
- Geetha, K.B., Lending, C.R., Lopes, M.A., Wallace, J.C., and Larkins, B.A. (1991). opaque-2 modifiers increase γ-zein synthesis and alter its spatial distribution in maize endosperm. Plant Cell 3, 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshi, N., and Brandt, A. (1998). Two jasmonate-inducible myrosinase-binding proteins from Brassica napus L. seedlings with homology to jacalin. Planta 204, 295–304. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi, F., Weinberg, C.R., Meagher, D.A., Imai, B.S., and Mische, S.M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Carpenter, R., and Coen, E.S. (1992). A common gene regulates pigmentation pattern in diverse plant species. Cell 68, 955–964. [DOI] [PubMed] [Google Scholar]
- Grandori, C., Cowley, S.M., James, L.P., and Eisenman, R.N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Shimada, T., Hatano, K., Takeuchi, Y., and Nishimura, M. (1998). Transport of storage proteins to protein-storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings, H., Maddaloni, M., Lazzaroni, N., Di Fonzo, N., Motto, M., Salamini, F., and Thompson, R. (1989). The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 8, 2795–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano, K., Shimada, T., Hiraiwa, N., Nishimura, M., and Hara-Nishimura, I. (1997). A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol. 38, 344–351. [DOI] [PubMed] [Google Scholar]
- Hawes, C., Saint-Jore, C., Martin, B., and Zheng, H.-Q. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP. Trends Plant Sci. 6, 245–246. [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N.K., Nishimura, M., and Hara-Nishimura, I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894–899. [DOI] [PubMed] [Google Scholar]
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G.B., Atkinson, A., Bylstra, Y.H., Walsh, R., and Smyth, D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Herman, E.M., and Larkins, B.A. (1999). Protein storage bodies and vacuoles. Plant Cell 11, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, B.G., Beatty, M.K., Singletary, G.W., Hamaker, B.R., Dikes, B.P., Larkins, B.A., and Jung, R. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14, 2591–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen, T.-H. (1970). The morphology, occurrence, and distribution of dilated cisternae of the endoplasmic reticulum in tissues of plants of the Cruciferae. Protoplasma 71, 467–477. [Google Scholar]
- Kawagoe, Y., and Murai, N. (1996). A novel basic region/helix-loop-helix protein binds to a G-box motif CACGTG of the bean seed storage protein β-phaseolin gene. Plant Sci. 116, 47–57. [Google Scholar]
- Kim, Y.-W., Kang, K.-S., Kim, S.-Y., and Kim, I.-S. (2000). Formation of fibrillar multimers of oat β-glucosidase isoenzymes is mediated by the As-Glu1 monomer. J. Mol. Biol. 303, 831–842. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Lending, C.R., and Larkins, B.A. (1989). Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenman, M., Rödin, J., Josefsson, L.-G., and Rask, L. (1990). Immunological characterization of rapeseed myrosinase. Eur. J. Biochem. 194, 747–753. [DOI] [PubMed] [Google Scholar]
- Ma, P.C.M., Rould, M.A., Weintraub, H., and Pabo, C.O. (1994). Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell 77, 451–459. [DOI] [PubMed] [Google Scholar]
- Massari, M.E., and Murre, C. (2000). Helix-loop-helix proteins: Regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R., Hayashi, Y., Kondo, M., Shimada, T., Nishimura, M., and Hara-Nishimura, I. (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130, 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R., Hayashi, Y., Yamada, K., Shimada, T., Nishimura, M., and Hara-Nishimura, I. (2003. b). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44, 661–666. [DOI] [PubMed] [Google Scholar]
- Matsushima, R., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2003. a). A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER retention signal in Arabidopsis. Plant J. 33, 493–502. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi, N., Shimada, T., Mano, S., Nishimura, M., and Hara-Nishimura, I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41, 993–1001. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Nitz, I., Berkefeld, H., Puzio, P.S., and Grundler, F.M.W. (2001). Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana. Plant Sci. 161, 337–346. [DOI] [PubMed] [Google Scholar]
- Okita, T.W., and Rogers, J.C. (1996). Compartmentation of proteins in the endomembrane system of plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 327–350. [DOI] [PubMed] [Google Scholar]
- Palmiter, R.D., Gagnon, J., and Walsh, K.A. (1978). Ovalbumin: A secreted protein without a transient hydrophobic leader sequence. Proc. Natl. Acad. Sci. USA 75, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans, W.J., and Van Damme, E.J.M. (1995. a). Lectins as plant defense proteins. Plant Physiol. 109, 347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans, W.J., and Van Damme, E.J.M. (1995. b). The role of lectins in plant defence. Histochem. J. 27, 253–271. [DOI] [PubMed] [Google Scholar]
- Rask, L., Andreasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, W.-D. (2002). Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5, 536–542. [DOI] [PubMed] [Google Scholar]
- Ridge, R.W., Uozumi, Y., Plazinski, J., Hurley, U.A., and Williamson, R.E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40, 1253–1261. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan, R., Sekar, K., Banerjee, R., Sharma, V., Surolia, A., and Vijayan, M. (1996). A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a β-prism fold. Nat. Struct. Biol. 3, 596–603. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Simpson, D., Kalousek, F., and Gietl, C. (1998). A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta 206, 466–475. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Burr, F.A., Aukerman, M.J., and Burr, B. (1990). Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87, 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R.J., Ketudat, M., Aukerman, M.J., and Hoschek, G. (1992). Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4, 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skea, D.L., Christopoulous, P., Plaut, A.G., and Underdown, B.J. (1988). Studies on the specificity of the IgA-binding lectin, jacalin. Mol. Immunol. 25, 1–6. [DOI] [PubMed] [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J.N.M., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisomsap, C., Svasti, J., Surarit, R., Champattanachai, V., Sawangareetrakul, P., Boonpuan, K., Subhasitanont, P., and Chokchaichamnankit, D. (1996). Isolation and characterization of an enzyme with β-glucosidase and β-fucosidase activities form Dabergia cochinchinesis Pierre. J. Biochem. 119, 585–590. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A. (1997). The plant ER: A dynamic organelle composed of a large number of discrete functional domains. Plant J. 11, 1151–1165. [DOI] [PubMed] [Google Scholar]
- Staudacher, E., Altmann, F., Wilson, I.B.H., and Marz, L. (1999). Fucose in N-glycans: From plant to man. Biochim. Biophys. Acta 1473, 216–236. [DOI] [PubMed] [Google Scholar]
- Stotz, H.U., Pittendrigh, B.R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., and Mitchell-Olds, T. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against cotton worm but not diamondback moth. Plant Physiol. 124, 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipalensuu, J., Eriksson, S., and Rask, L. (1997). The myrosinase-binding protein from Brassica napus seeds possesses lectin activity and has a highly similar vegetatively expressed wound-inducible counterpart. Eur. J. Biochem. 250, 680–688. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Shimada, T., Ono, E., Tanaka, Y., Nagatani, A., Higashi, S., Watanabe, M., Nishimura, M., and Hara-Nishimura, I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35, 545–555. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre, F.D.L., Sampedro, J., Zarra, I., and Revilla, G. (2002). AtFXG1, an Arabidopsis gene encoding α-L-fucosidase active against fucosylated xyloglucan oligosaccharides. Plant Physiol. 128, 247–255. [PMC free article] [PubMed] [Google Scholar]
- Toyooka, K., Okamoto, T., and Minamikawa, T. (2000). Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J. Cell Biol. 148, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J.X., Wait, R., Berkelman, T., Harry, R.A., Westbrook, J.A., Wheeler, C.H., and Dunn, M.J. (2000). A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21, 3666–3672. [DOI] [PubMed] [Google Scholar]
- Yang, H., and Czapla, T.H. (1993). Isolation and characterization of cDNA clones encoding jacalin isolectins. J. Biol. Chem. 268, 5905–5910. [PubMed] [Google Scholar]
- Ye, R.D., Wun, T.-C., and Sadler, J.E. (1988). Mammalian protein secretion without signal peptide removal. Biosynthesis of plasminogen activator inhibitor-2 in U-937 cells. J. Biol. Chem. 263, 4869–4875. [PubMed] [Google Scholar]