Domesticated rice (Oryza sativa L.) accompanied the dawn of Asian civilization1 and has become one of world’s staple crops. From archaeological and genetic evidence various contradictory scenarios for the origin of different varieties of cultivated rice have been proposed, the most recent based on a single domestication2,3. By examining the footprints of selection in the genomes of different cultivated rice types, we show that there were three independent domestications in different parts of Asia. We identify wild populations in southern China and the Yangtze valley as the source of japonica gene-pool, and populations in Indochina and the Brahmaputra valley as the source of indica gene-pool. We reveal a hitherto unrecognized origin for the aus variety in central India or Bangladesh. We also conclude that aromatic rice is a result of a hybridization between japonica and aus, and that the tropical and temperate versions of japonica are later adaptations of one crop. Our conclusions are in accord with archaeological evidence that suggests widespread origins of rice cultivation1,4. We therefore anticipate that our results will stimulate a more productive collaboration between genetic and archaeological studies of rice domestication, and guide utilization of genetic resources in breeding programmes aimed at crop improvement.
Although genome sequences for Oryza sativa subsp. indica and subsp. japonica have been available for over a decade5,6, the phylogenetic, geographic, and temporal origins of these domesticated groups remain unresolved. The earliest archaeological evidence for human exploitation of rice dates to 9,000–10,000 years ago in India (Ganges plain) and China (middle and lower Yangtze valley) and suggests that there were independent origins of rice cultivation in these regions4. However, much of this work depends on the classification of archaeobotanical remains, which is problematic at the subspecies level. Moreover, large regions of southern and southeast Asia remain archaeologically unexplored1. Genetic studies of extant rice varieties aimed at resolving the questions of the origin of cultivated groups have resulted in diverse and often contradictory hypotheses. Genetic support for independent origins of domesticated rice7–11 includes the estimated divergence time for the two subspecies, indica and japonica, which at 200,000–440,000 years ago7,8,11 is much earlier than the beginning of agriculture in southeast Asia. Based on ecology, genetics and culinary properties, indica and japonica are subdivided into indica and aus, and temperate japonica, tropical japonica and aromatic, respectively12,13. Within long-grained indica, the aus group consists of drought-tolerant, early-maturing cultivars. Increasing evidence suggests that indica and aus could have separate origins that are obscured by their morphological similarities12–14, but a unique wild sister-population of aus has not been identified so far. Within the japonica subspecies, typically giving sticky rice after cooking, the temperate and tropical japonica groups are adapted to distinct climatic conditions, while aromatic includes rice with specific flavours popular in Pakistan and northern India (basmati) and Iran (sadri).
Other genetic studies have concluded that all cultivated Asian rice derives from a single domestication2,3,15–17, but with disparate explanations for the differentiation between indica and japonica. Proposed scenarios include an indica-like domesticate giving rise to japonica16, a proto-domesticate differentiating into indica and japonica under different climatic conditions17, or japonica being domesticated first and indica emerging from crosses between japonica and wild rice after cultivation spread into new areas3.
The last domestication scheme is derived from analysis of the largest dataset of genetic variations so far assembled, comprising ~8 million single nucleotide polymorphisms (SNPs) collected from genome-wide sequence data of 1,083 O. sativa and 446 wild rice accessions3. The relationship between wild and cultivated rice was inferred from comparison of a neighbour-joining (NJ) tree constructed from the genome-wide data with a tree constructed from what the authors refer to as 'domestication sweeps' – regions of depleted variation in the cultivated rice genomes relative to wild populations, assumed to result from selection during domestication. While indica and japonica form independent clusters on the first tree, pointing at differences on the genome-wide scale, both subspecies cluster together on the latter tree, evoking the hypothesis of hybrid origin. However, if as the authors conclude, indica was derived from crosses between japonica and wild rice, the genomic regions in the domestication sweeps would be passed from japonica to indica and should result in a phylogeny where the indica group is nested within japonica. Contrary to this expectation, japonica was nested within indica in the NJ tree constructed from the domestication sweeps3.
In the present study, we first tested the hypothesis that domestication alleles were introgressed from japonica to the progenitor of indica by comparing genomic regions under selection in both cultivated groups. We investigated the tree built from the sweep regions by Huang et al.3 that led the authors to the incongruent conclusion of the derived nature of the indica group. This tree was inferred from 55 ‘major domestication sweeps’ identified by comparing the nucleotide diversity present in all cultivated rice against the entire dataset of wild populations (Oryza rufipogon Griff.). This treatment makes an a priori assumption that all cultivated rice forms a single taxonomic group and that regions under selection coincide in indica and japonica rice (and in any other O. sativa subgroup). However, there is little coincidence between the domestication sweeps detected by Huang et al. for indica and japonica separately (Supplementary Tables 10 and 11 in ref. 3). In fact, the identified domestication sweeps of japonica and indica overlap in only 15 genomic regions, implying that the majority of the 55 major domestication sweeps are under differential effect of selection in these two groups. Inclusion of genomic regions under differential selection across the tree is a likely cause of artefactual results in the analysis by Huang et al.3. When we constructed a NJ tree from only those 15 overlapping sweep regions, we found that cultivated rice fell into three distinct clusters, one comprising indica accessions, one of japonica, and one of aus (Supplementary figure 1), again contrary to the proposed scheme in which indica-type rice is derived from japonica.
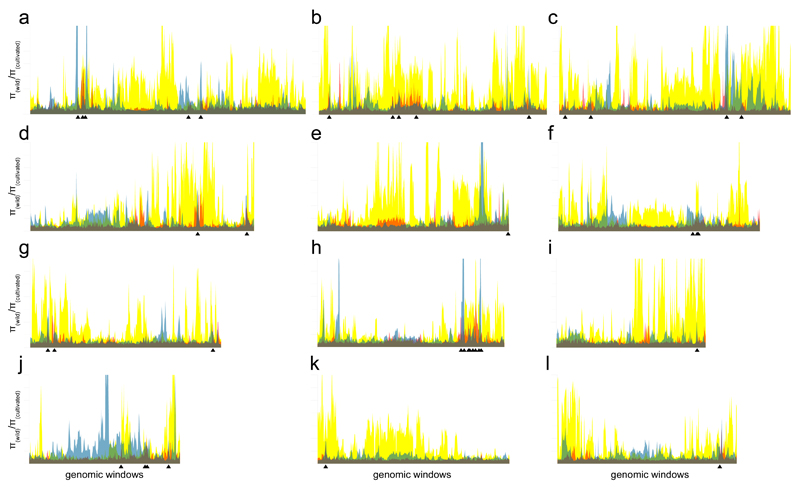
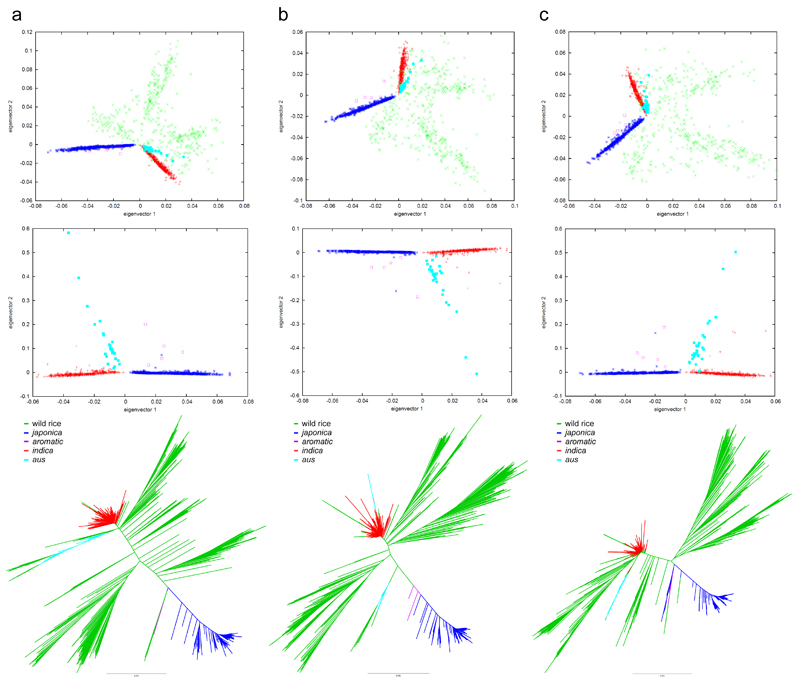
Guided by the results of the first test and by recent genetic studies13,14, we reanalyzed the genomic dataset3 to reveal what impact selection has left in the genomes of the three groups of cultivated rice – indica, japonica and aus. Selection profiles differ among all three groups with regards to both the position and magnitude of the selective sweeps, with the japonica genome most affected by a bottleneck and the indica genome least affected (Fig. 1; Supplementary Notes). The group-specific profiles do not necessarily imply independent domestications, because some of the selective pressures that resulted in the regions of low diversity could have operated after domestication, following the separation of indica, japonica and aus. However, if all cultivated rice is derived from a single domestication then selection occurring during that event would be expected to have left a number of selective sweeps that are identical in all three groups. We therefore focused on those parts of the rice genome that are under selection in indica, aus and japonica and identified co-located low-diversity genomic regions (CLDGRs) where the nucleotide diversity for each of these three groups is lower than it is in wild rice. We carried out three separate analyses applying π(wild)/π(domesticated) thresholds 3 (nucleotide diversity in each of the three domesticated groups >3 times lower than that of wild rice), 4 and 5. At a threshold of 3 we found 109 CLDGRs containing 421,000 SNP positions, at 4 there are 38 CLDGRs (127,000 SNPs), and only 17 CLDGRs (51,000 SNPs) remain at threshold 5. We examined the diversity of these three datasets by principal component analysis (PCA) and NJ clustering. The analysis for each of the three threshold values gave similar results, discriminating three statistically strong groups of cultivated rice (indica, japonica and aus; Anova p-values <0.001) and a fourth group (aromatic) with an ambiguous position (Fig. 2). FST estimates calculated from the CLDGRs supported the outcomes of the PCAs and NJ analyses (Supplementary Fig. 2). The implication is that even within the portion of the genome that was under selection in all three groups, distinct sequence types were selected during domestication, an observation that is incompatible with a period of shared domestication history between the indica, japonica and aus groups. These results contrast with the previous analysis of this dataset3 and we conclude that the tree topology in which the japonica group is nested within indica – which in itself is inconsistent with the authors’ interpretation that domestication-related genomic regions of indica are derived from japonica – is an analytical artefact caused by semi-random selection of genomic loci and by the generally higher diversity and shorter linkage disequilibrium (LD) decay of indica relative to japonica.
Figure 1.
Selection landscapes for the three groups of cultivated rice on each of the twelve rice chromosomes (a–l). The x-axis represents sliding windows of 2,000 (indica, japonica) or 4,000 (aus) SNPs, with sliding steps of 1,000 SNPs. The y-axis is scaled in intervals of 4 and represents the ratio of nucleotide diversity π(wild)/π(domesticated). Yellow, japonica; transparent blue, aus; transparent red, indica; green, overlaps of japonica and aus; orange, overlaps of japonica and indica; purple, overlaps of aus and indica; grey, overlaps of all three groups. Positions of 38 CLDGRs identified at π(wild)/π(domesticated)>4 are indicated with black arrowheads.
Figure 2.
Population structure analysis of low-variation regions in cultivated rice genomes. a, CLDGR threshold 3; b, CLDGR threshold 4; c, CLDGR threshold 5. Top panels, PCAs for all accessions; middle panels, PCAs without wild populations; bottom panels, unrooted NJ trees of concatenated CLDGRs. All shown eigenvectors discriminate the domesticated groups with statistical significance except: the first eigenvector fails to discriminate aromatic and japonica in all cases; the second eigenvector fails to discriminate aromatic and aus in all cases. Consequently, the aromatic accessions occupy the space between aus and japonica, indicating that aromatic could be an aus×japonica hybrid.
Phylogenetic analysis of sequences concatenated from different genomic regions can lead to incorrect conclusions because of incomplete lineage sorting, intergroup hybridizations or gene flow18 and such processes could also have contributed to misleading tree topologies in ref. 3 and in our analyses. To address this concern, we analyzed independently each of the CLDGRs detected at the threshold value of 4 (Supplementary Table 1) and produced 31 CLDGR-trees (Supplementary Fig. 3). The expectation from a single domestication – all five domesticated groups (indica, aus, aromatic, temperate japonica and tropical japonica) in a single cluster – held for only six of the CLDGR-trees, one of these including the PROG1 gene19 (Table 1, Supplementary Notes). The clusters observed in the remaining 25 CLDGR-trees gave strong evidence for separate domestications of indica, japonica and aus (Table 1). In 17 trees, temperate and tropical japonica appear in a single cluster (with or without aromatic), separate from the other cultivated groups. In the same number of trees, indica is separate from the other groups, and aus (with or without aromatic) branches separately on 16 CLDGR-trees.
Table 1. Positioning of domesticated rice groups in the 31 CLDGR-trees*.
| Group | Sweep-trees in which the group forms a single cluster | |||
|---|---|---|---|---|
| Without aromatic | With aromatic | |||
| n | Tree numbers † | n | Tree numbers † | |
| indica | 17 | 1, 3, 7, 9, 11, 13, 15–17, 19, 21, 22, 24, 27–29, 31 | 0 | - |
| temperate + tropical japonica | 11 | 7–9, 15, 16, 21, 23, 24, 29–31 | 6 | 1, 6, 11, 17, 27, 28 |
| aus | 10 | 1–3, 11, 15–17, 27–29 | 6 | 7, 9, 21, 22, 24, 31 |
| aromatic | 7 | 8, 10, 15, 16, 20, 29, 30 | - | - |
| aus + indica | 3 | 6, 8, 30 | 1 | 23 |
| temperate japonica | 2 | 4, 22 | 1 | 3 |
| tropical japonica | 2 | 3, 22 | 0 | - |
| aus + temperate japonica + tropical japonica | 0 | - | 2 | 13, 19 |
| aus + indica + temperate japonica + tropical japonica | 2 | 10, 20 | 6‡ | 5, 12, 14, 18, 25, 26 |
| indica + temperate japonica + tropical japonica | 0 | - | 1 | 2 |
| aus + indica + tropical japonica | 0 | - | 1 | 4 |
Randomness of observed cluster frequencies was rejected at p<0.001 by chi-square test.
Tree numbers correspond to Supplementary Fig. 3.
All five domesticated groups (indica, aus, aromatic, temperate japonica and tropical japonica) in a single cluster, the expectation for a single domestication.
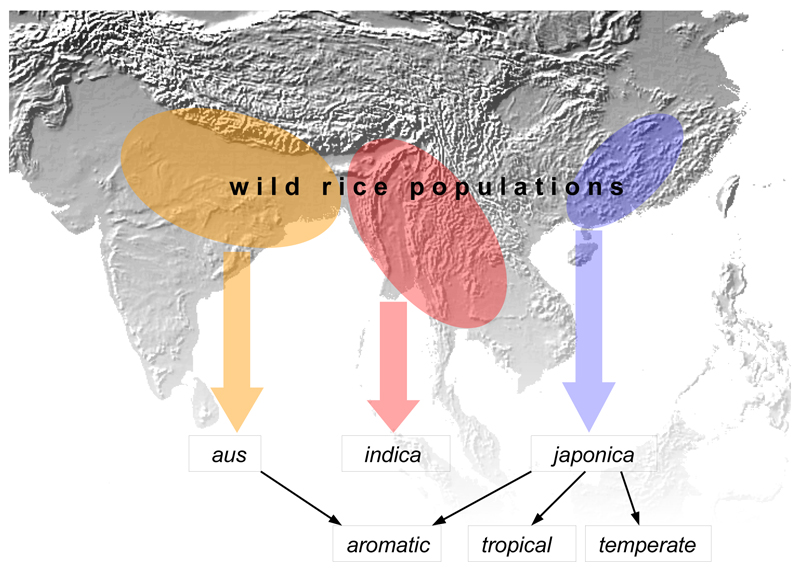
The distribution of wild rice accessions bearing sequences similar to the regions selected during domestication can provide an indication of the geographical origins of cultivation. In the 17 CLDGR-trees where japonica branches separately, most of the closely affiliated wild accessions are from southern China and Poyang Lake near the Yangtze river (Supplementary Fig. 4a), suggesting that japonica originated in this region, in agreement with previous genetic and archaeological studies4,3. The consistent placement of temperate and tropical japonica together implies that these two groups represent later adaptations of the original japonica crop to different climatic conditions. In the 16 trees with a separate aus branch, the related wild accessions originate from a broad region from central India to Bangladesh (Supplementary Fig. 4b), which we identify as the source of the aus gene-pool. The frequent coupling of aromatic with both japonica and aus in the CLDGR-trees provides additional evidence that aromatic is a hybrid between these two types. The closest wild accessions in the 17 CLDGR-trees with a separate indica branch are located in a second broad region from southern Indochina to the Brahmaputra valley (Supplementary Fig. 4c). For each of the remaining six CLDGR trees, in which all five domesticated groups appear as a single cluster, the most closely related sequences are widely distributed in the wild population (Supplementary Fig. 5). Such observation suggests that identical or similar alleles located in these genomic regions, potentially favourable for cultivation, could have been selected from the standing variation multiple times during separate domestication processes. This explanation reconciles the scenarios of multiple and entirely independent domestications with infrequent observations of alleles that are fixed in all cultivated rice (such as prog119 and sh420; Supplementary Notes). In conjunction with the existence of numerous CLDGRs bearing specific sequences with consistently distinct geographic origins in the three rice groups, we conclude that rice domestication was a multiregional process separately producing the indica, japonica and aus types of rice (Fig. 3).
Figure 3.
Scheme for the origins of domesticated rice derived from phylogeographic analysis of 31 CLDGR-trees. Separate domestications gave the indica, japonica and aus types of rice, these domestications occurring in different parts of southeast and southern Asia, with subsequent hybridization between japonica and aus giving aromatic rice, and the temperate and tropical versions of japonica evolving as later adaptations. The scheme is consistent with archaeological evidence indicating independent origins of rice cultivation in India and China1,4, with genetic evidence of unique aus gene space14 and genome-wide relationships among the five cultivated groups13. (Background map obtained from public resources at http://www.ngdc.noaa.gov/mgg/).
Methods
The complete genotype dataset for 1,529 wild and cultivated rice accessions3 consisting of ~8 million SNPs, was downloaded from the Rice Haplotype Map Project database (http://www.ncgr.ac.cn/RiceHap3). The initial test of the hypothesis that domestication alleles were introgressed from japonica into the progenitor of indica was conducted on domestication sweeps co-located in the indica and japonica groups (Supplementary Tables 10 and 11 in ref. 3). Fifteen such sweeps were identified and the corresponding genomic windows extracted: 22.7–22.8 Mb from chromosome 1; 1.6–2.0 Mb, 6.3 Mb, 11.6–11.7 Mb from chromosome 2; 29.4–29.9 Mb from chromosome 3; 21.7–21.8 Mb, 23.3–23.5 Mb, 25.8–26.3 Mb, 27.5 Mb, 34.6–34.7 Mb from chromosome 4; 9.2–9.4 Mb, 20.7–20.8 Mb from chromosome 5; 28.5–28.6 Mb from chromosome 6; 23.9–24.0 Mb from chromosome 8; and 2.6–2.9 Mb from chromosome 12. All genomic coordinates refer to the rice reference genome IRGSP 4. The genotypes of 44 ‘intermedia’ accessions were excluded as suspected intergroup hybrids, because hybrid samples could distort tree inference21. The resulting matrix consisted of 1,485 rice accessions and c.86,000 SNPs, with 62.7% mean proportion of missing data. Genetic distances were calculated with PHYLIP22 using the F84 distance method and empirical base frequencies. An unrooted NJ tree was constructed with the same package and visualized in Geneious 6.1 (Biomatters, http://www.geneious.com).
The complete genotype dataset for each of the twelve rice chromosomes was converted into PHYLIP format and divided into four groups: indica (520 accessions), japonica (489 accessions), aus (30 accessions) and wild (446 accessions). For each group, nucleotide diversity π was calculated in VariScan 2.0.323, with sites containing missing data included. The NumNuc parameter was set to 2, the sliding window width to 2,000 (indica vs. wild, japonica vs. wild) or 4,000 bp (aus vs. wild, increased to compensate for higher variance due to the small sample size of aus), and the sliding window step was 1,000 bp. Optimum window width, in respect to the LD decay and the amount of informative data, may vary across the created groups as well as across the rice chromosomes. We did not account systematically for such possible differences and chose fixed window sizes of 2,000 and 4,000 SNPs instead, corresponding to average genomic windows of ~100 and ~200 kb similar to values used in other studies3,24. Selection landscapes for each of the three cultivated rice groups were identified by dividing the nucleotide diversity of the wild accessions by the nucleotide diversity of the cultivated groups in corresponding genomic windows. A permutation test was performed to estimate a random noise threshold. Two groups consisting of 30 and 446 randomly selected accessions were assembled using the complete genotype dataset and a new random pick for each of the twelve chromosomes. The nucleotide diversity ratios were calculated as above. The maximum π(446)/π(30) value recorded was 2.65, suggesting that the genomic regions of cultivated groups with higher diversity ratios represent true selective sweeps free from false positive signals. In order to analyze genomic parts with low diversity and limited recombination in the three cultivated groups yet sufficient diversity in the wild populations, we focused on genomic regions where the π(wild)/π(domesticated) values were >3 in all three cultivated groups, and we refer to these regions as CLDGRs.
A common domestication history is expected to leave selective sweeps at identical genomic positions in the groups under investigation. However, decreased nucleotide diversity at a particular locus in two or more groups does not mean that the same alleles have been selected in those groups. In order to reveal whether the CLDGRs differ genetically among the three groups of cultivated rice, we performed an analysis of population structure. We identified the CLDGRs at thresholds 3, 4 and 5 and extracted those regions from the SNP data matrices (DOI: 10.5281/zenodo.20740). We conducted PCA using smartpca in EIGENSOFT5.0.225, excluding no outliers and inferring genetic distance from physical distance.Two PCAs were performed for each threshold, one on the full set of rice accessions and one on domesticated accessions only. FST values were calculated using the same software. The extracted CLDGRs were also subjected to a concatenation NJ analysis, using the HKY substitution model in Geneious 6.1. To account for the potentially confounding effects of incomplete lineage sorting, interpopulation gene flow and hybridization on phylogenetic inference from concatenated datasets, all 38 CLDGRs detected at the threshold value of 4 were also analyzed individually. In these analyses, the actual sequences of the indica, aus, aromatic, temperate japonica and tropical japonica accessions were replaced by the majority consensus sequences for each group. The use of consensus sequences for each group mitigates the problem of missing data which becomes more pronounced in datasets of only a few thousand sites; their use assumes that intragroup nucleotide variability within the CLDGRs is minimal. Unrooted NJ trees were constructed as described above. In cases where neighboring CLDGRs on the same chromosome yielded a similar tree topology, the respective alignments were concatenated and a new tree was constructed. In total, 31 trees were produced from the 38 CLDGRs. Clusters containing the consensus sequences of cultivated groups were identified and the locations of related wild accessions plotted on maps.
Supplementary Material
Acknowledgements
This work was supported by European Research Council grant 339941 awarded to TAB. Authors declare no competing financial interests. We thank Elham Karimi for sparking interest in the origin of aromatic rice. We also thank Jeffrey Ross-Ibarra for constructive comments on an earlier draft of this paper.
Footnotes
Author contributions
P.C. conceived the project and led the data analysis. H.C. contributed the geographical data analysis. C.F.C. and T.A.B. contributed conceptual development and data interpretation. P.C. and T.A.B. wrote the manuscript and all co-authors contributed manuscript editing.
References
- 1.Fuller DQ. Pathways to Asian civilizations: tracing the origins and spread of rice and rice cultures. Rice. 2011;4:78–92. [Google Scholar]
- 2.Molina J, et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci USA. 2011;108:8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–503. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross BL, Zhao Z. Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad Sci USA. 2014;111:6190–6197. doi: 10.1073/pnas.1308942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 7.Vitte C, Ishii T, Lamy F, Brar D, Panaud O. Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.) Mol Genet Genomics. 2004;272:504–511. doi: 10.1007/s00438-004-1069-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
- 9.Londo JP, Chiang Y-C, Hung K-H, Chiang T-Y, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C-C, et al. Independent domestication of Asian rice followed by gene flow from japonica to indica. Mol Biol Evol. 2012;29:1471–1479. doi: 10.1093/molbev/msr315. [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao K, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schatz MC, et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 2014;15:506. doi: 10.1186/s13059-014-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao L-Z, Innan H. Nonindependent domestication of the two rice subspecies, Oryza sativa ssp. indica and ssp. japonica, demonstrated by multilocus microsatellites. Genetics. 2008;179:965–976. doi: 10.1534/genetics.106.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu B-R, Zheng KL, Qian HR, Zhuang JY. Genetic differentiation of wild relatives of rice as assessed by RFLP analysis. Theor Appl Genet. 2002;106:101–106. doi: 10.1007/s00122-002-1013-2. [DOI] [PubMed] [Google Scholar]
- 17.Oka H-I, Morishima H. Phylogenetic differentiation of cultivated rice, XXIII. Potentiality of wild progenitors to evolve the indica and japonica types of rice cultivars. Euphytica. 1982;31:41–50. [Google Scholar]
- 18.Maddison WP. Gene trees in species trees. Syst Biol. 1997;46:523–536. [Google Scholar]
- 19.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 21.Allaby RG, Brown TA. AFLP data and the origins of domesticated crops. Genome. 2003;46:448–453. doi: 10.1139/g03-025. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6.Distributed by the author. Department of Genome Sciences, University of Washington; Seattle (WA): 2005. [Google Scholar]
- 23.Hutter S, Vilella AJ, Rozas AJ. Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinf. 2006;7:409. doi: 10.1186/1471-2105-7-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Z, et al. Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 2011;7:e1002100. doi: 10.1371/journal.pgen.1002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.