Abstract
In order to survive, bacteria continually sense, and respond to, environmental fluctuations. Stringent control represents a key bacterial stress response to nutrient starvation1,2 that leads to a rapid and comprehensive reprogramming of metabolic and transcriptional patterns3. In general, transcription of genes for growth and proliferation are down-regulated, while those important for survival and virulence are favored4. Amino acid starvation is sensed by depletion of the aminoacyl-tRNA pools5, which results in accumulation of ribosomes stalled with non-aminoacylated (uncharged) tRNA in the ribosomal A-site6,7. RelA is recruited to stalled ribosomes, and activated to synthesize a hyperphosphorylated guanosine analog, (p)ppGpp8, which acts as a pleiotropic second messenger. However, structural information for how RelA recognizes stalled ribosomes and discriminates against aminoacylated tRNAs is missing. Here, we present the electron cryo-microscopy (cryo-EM) structure of RelA bound to the bacterial ribosome stalled with uncharged tRNA. The structure reveals that RelA utilizes a distinct binding site compared to the translational factors, with a multi-domain architecture that wraps around a highly distorted A-site tRNA. The TGS domain of RelA binds the CCA tail to orient the free 3’ hydroxyl group of the terminal adenosine towards a β-strand, such that an aminoacylated tRNA at this position would be sterically precluded. The structure supports a model where association of RelA with the ribosome suppresses auto-inhibition to activate synthesis of (p)ppGpp and initiate the stringent response. Since stringent control is responsible for the survival of pathogenic bacteria under stress conditions, and contributes to chronic infections and antibiotic tolerance, RelA represents a good target for the development of novel antibacterial therapeutics.
Stringent control is a pleiotropic response to the failure of amino acid availability to keep up with the demands of protein synthesis1. It is mediated by a hyperphosphorylated nucleotide ((p)ppGpp) 9,10. In E. coli, the synthesis of (p)ppGpp is catalyzed by RelA2, a multi-domain ATP:GTP(GDP) pyrophosphate transferase, and a prototypic member of the RelA/SpoT homolog (RSH) family11. The majority of multi-domain RSH proteins are stimulated to generate (p)ppGpp in a ribosome-dependent manner when an uncharged and cognate tRNA, which acts as a marker for nutrient deficiency, is located in the ribosomal A-site7,8. Discrimination against aminoacylated tRNA prevents undesired activation of stringent control during the normal translation cycle.
Using cryo-EM we have solved the structure of the E. coli ribosome, programmed so that uncharged tRNA(Phe) occupies the A-site, in complex with RelA at an overall resolution of 3.0 Å (Fig. 1, Extended Data Figs. 1-2, and Extended Data Table 1). We did not observe any class in which RelA was bound to the ribosome in the absence of A-site tRNA. Both RelA and the A-site tRNA remain flexible when bound to the ribosome, primarily due to binding intrinsically flexible rRNA elements (notably the L7/L12 stalk base and the A-site finger). Although there are only minor differences in conformations (Extended Data Fig. 1b), the heterogeneity was sufficient to result in RelA having less well-resolved density than the ribosome. To distinguish conformational states and improve the local map quality we utilized a recent modification of the 3D classification process12, in which ribosome projections were subtracted from each experimental particle leaving signal only for RelA prior to classification focused on each domain (Methods and Extended Data Fig. 1). This improved the density for the RelA domains (Extended Data Figs. 2-3) allowing models to be built (Extended Data Table 2).
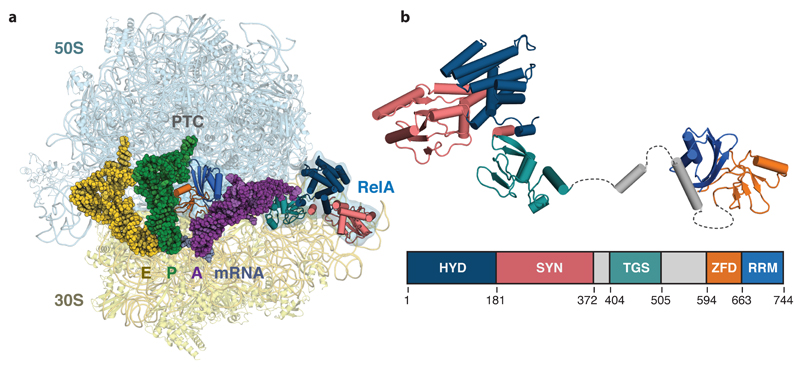
Figure 1. Structure of RelA bound to the ribosome.
a, Overall view of RelA in complex with a ribosome stalled with an uncharged tRNA in the A-site. Displayed are the 50S and 30S ribosomal subunits; E-, P- and A-site tRNAs; mRNA, and RelA coloured by domain. b, Structure of the ribosome-bound form of RelA oriented from N- to C-terminus with the domain organization below showing the boundaries of the hydrolase (HYD), synthetase (SYN), TGS, Zinc-finger (ZFD) and RNA recognition motif (RRM) domains. Unmodeled flexible elements that connect RelA domains are indicated with dashed lines.
The structure reveals that RelA forms a highly extended conformation on the ribosome to cradle the uncharged tRNA in a distorted conformation in the A-site (Fig. 1). RelA has an N-terminal region formed by hydrolase (HYD), synthetase (SYN), and TGS domains that are located at the acceptor end of the A-site tRNA, and a C-terminal region formed by a zinc-finger domain (ZFD) and an RNA recognition motif (RRM) that run parallel to the anticodon arm of the tRNA. These five domains are connected by flexible and helical elements in a serpentine configuration that wind between the ribosome and the A-site tRNA (Fig. 1, 3 and Extended data Fig. 4). In this conformation, RelA inhibits accommodation of the acceptor arm of the uncharged tRNA into the peptidyl transferase center (PTC).
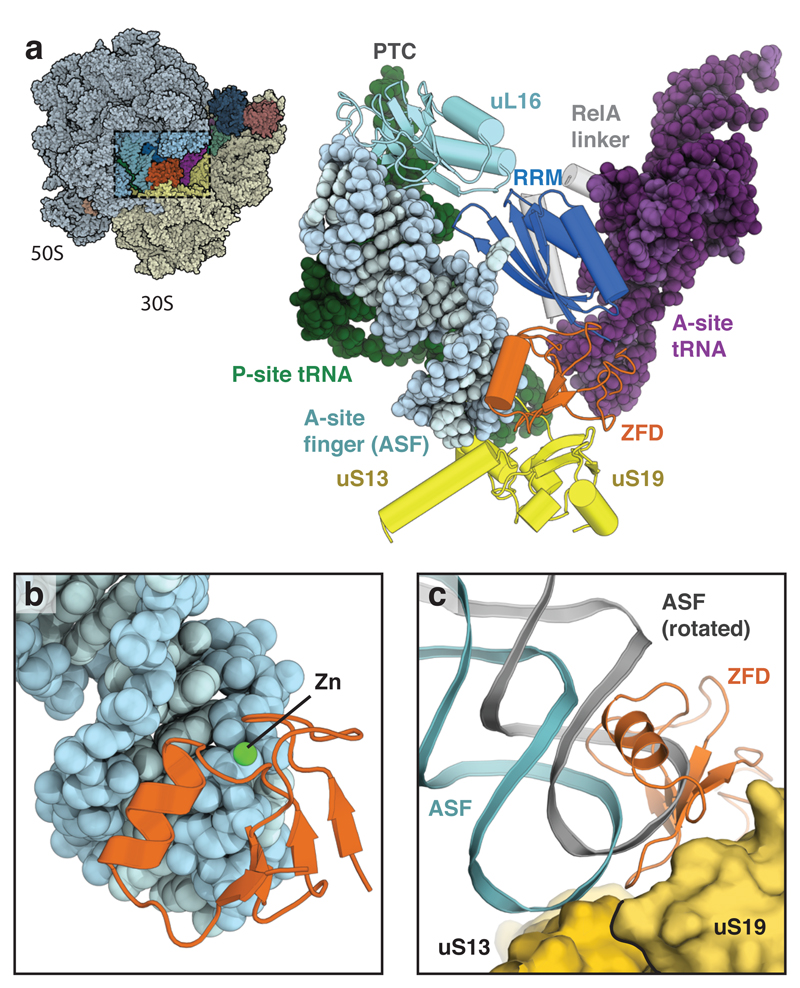
Figure 3. Interactions between RelA and the ribosome.
a, Overview (left) and details (right) of the interaction between the ZFD (orange) and RRM (blue) of RelA and the ribosomal ASF (light blue) that spans the intersubunit interface between the P-site (green) and A-site (purple) tRNAs. RelA acts as an additional intersubunit bridge by binding uL16 (cyan) in the large subunit and uS19 (yellow) in the small subunit. b, The α-helix of the ZFD binds in the major groove of the ASF, with the zinc-binding site interacting with the phosphate backbone. c, By binding to uS19, the ZFD occupies the position adopted by the ASF in the rotated ribosome (grey).
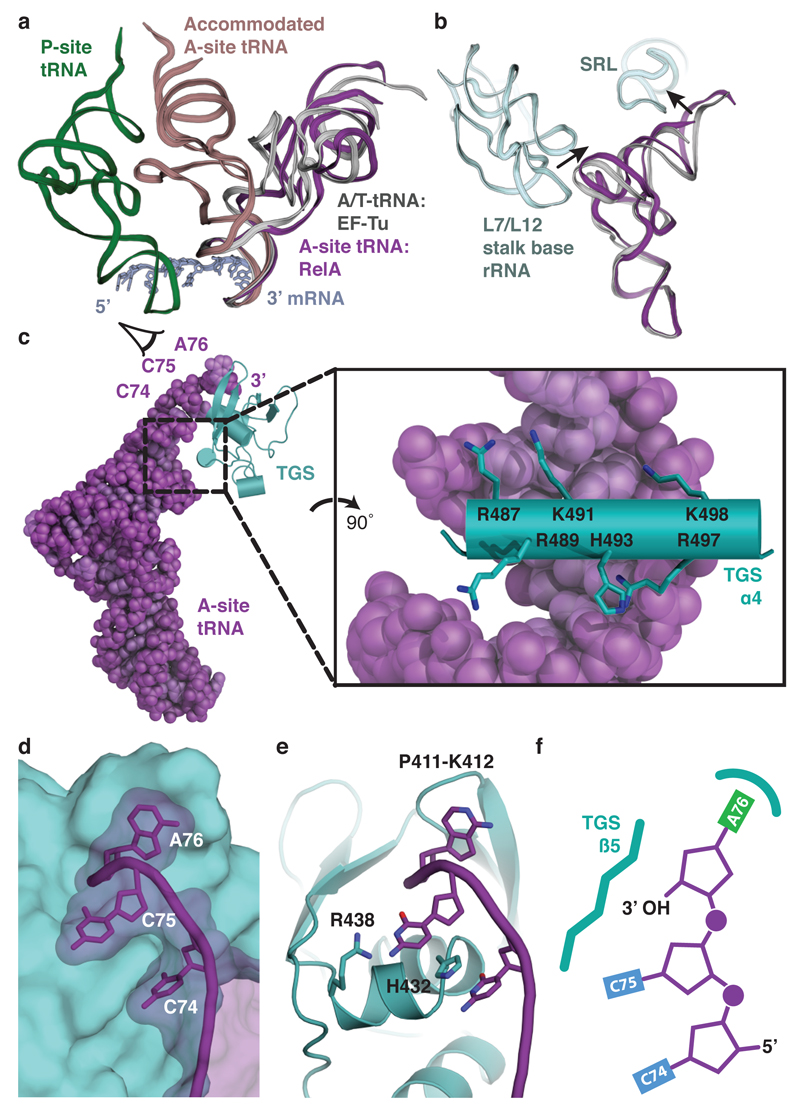
As was previously noted13, the overall conformation of the A-site tRNA in the presence of RelA resembles the A/T state adopted by pre-accommodated aminoacyl-tRNA in complex with EF-Tu14 (Fig. 2a). However, our high-resolution map reveals that the interactions between the tRNA and the ribosomal large subunit are very different, with the tRNA contacting both the RNA component of the L7/L12 stalk base (H42-44) and the sarcin-ricin loop (SRL; H95) (Fig. 2b). A stacking interaction between nucleotides C56 of the tRNA elbow and A1067 of the L7/L12 stalk base is reminiscent of how the L1 stalk (H76-78) recognizes E-site tRNA15. The antibiotic thiostrepton binds in the vicinity of A1067 and may prevent this interaction, explaining its ability to inhibit (p)ppGpp formation8. The contacts with rRNA, and also with RelA, distort the tRNA compared to the A/T state (Fig. 2a,b and Extended Data Fig. 5). Starting at base-pair 27:43 after the aligned anticodon stem-loops (ASLs), a 6° rotation away from the ribosome is coordinated with an outward movement of the L7/L12 stalk base. A further 11° rotation of the acceptor stem, starting at base-pair 7:66, allows the tRNA to contact the SRL, which is bound by EF-Tu in the decoding complex14.
Figure 2. Molecular basis for the recognition of uncharged A-site tRNA.
a, Comparison of A-site tRNAs. In the presence of RelA, A-site tRNA (purple) structurally resembles the A/T state adopted by pre-accommodated aminoacyl-tRNA in complex with EF-Tu (light grey). RelA prevents accommodation of tRNA into the canonical A-site position (brown). P-site tRNA (green) and mRNA (dark grey) are shown for reference. b, Conformational differences between RelA-bound A-site tRNA (purple) and A/T-tRNA (light grey) result from an outward position of the L7/L12 stalk base and movement of the tRNA to contact the SRL (both light blue). c, The TGS domain (teal) binds the acceptor end of the A-site tRNA (purple) through a positively-charged helix that interacts with the phosphate backbone (expanded view). d, When viewed from the back as indicated in c, the 5’ CCA (nucleotides 74-76) of the A-site tRNA wraps around the surface of the TGS domain. e, The conformation of the CCA is maintained by interactions with invariant residues of the TGS domain. f, The free 3’OH of the terminal adenine is positioned to face the β4-strand of the TGS domain to sterically preclude the binding of aminoacylated tRNAs.
Once EF-Tu has dissociated from the ribosome, it is not known whether the A-site tRNA conformation could still fluctuate or the higher affinity of aminoacylated (compared to uncharged) tRNA for the A site in the PTC16,17 would stabilize the accommodated form of tRNA. However, with an uncharged tRNA, the fluctuations in its conformation could bring the acceptor end into contact with the RelA TGS domain and stabilize a distorted form in a manner that discriminates against aminoacylated tRNAs (Fig. 2c). This small domain has a β-grasp fold similar to that found in the ubiquitin family. In RelA, an additional pair of C-terminal α-helices, the second of which spans across the axis of the acceptor stem, extend the fold (TGS α3 and α4, see Extended Data Fig. 6). Highly conserved basic residues (Arg487, Lys491, His493, Arg497, and Lys498) at each end of this helix form electrostatic interactions with the tRNA phosphate backbone (Fig. 2c and Extended Data Fig. 7). The absence of base-specific contacts allows RelA to recognize all tRNAs equally. The 3’ CCA of the A-site tRNA extends ~14 Å around the outside of the TGS domain (Fig. 2d) and is maintained by a series of interactions; C74 stacks with His432, C75 can form hydrogen bonds with Arg438, and crucially A76 stacks beneath Pro411 and Lys412 (Fig. 2e and Extended Data Fig. 3a-c). This positions the free 3’ hydroxyl group of the terminal adenosine towards the β5-strand of the TGS domain (Fig. 2e, f). An aminoacylated tRNA at this position would be sterically precluded (Extended Data Fig. 3b). This agrees with data that shows a free 3’ hydroxyl group is a prerequisite of RelA activation18.
As well as binding the A-site tRNA, the TGS domain contacts the small subunit rRNA and the ribosomal proteins uS12 and uL14. Although ribosomes that lack, or have mutant, uL11 have impaired (p)ppGpp synthesis19,20, we do not observe a direct interaction between RelA and uL11.
The TGS domain together with the N-terminal hydrolase and synthetase domains, defines the minimal unit found in ribosome-dependent RSHs11. In RelA, the two C-terminal domains act in tandem to further anchor RelA to the ribosome by binding the ribosomal A-site finger (ASF; 23S rRNA H38) that spans the intersubunit space (Fig. 3a and Extended Data Fig. 7). Although the C-terminus was known to be involved in binding to the ribosome21,22, the nature of the interaction is unexpected and was not previously noted in the low-resolution reconstruction13. To our knowledge, this represents a novel binding site on the ribosome for an extrinsic factor.
RelA interacts with the tip of the ASF through an unpredicted, and unusual, CCHC-type zinc-finger domain (ZFD). This domain has a βββααβ topology (Extended Data Fig. 6), with a single zinc ion coordinated at the N-terminus of the α1-helix (Fig. 3b). The zinc-binding residues, Cys612 and His634, directly contribute to binding the ASF by interacting with the phosphate backbone of nucleotide U884 and help to orient the α1-helix in the major groove of the ASF. The ASF usually forms a dynamic bridge (B1a) with either uS13 or uS19 depending on the ratcheted state of the ribosome23. RelA stabilizes the bridge with uS13 by binding to the same interface of uS19 as occupied by the ASF in the rotated ribosome, which results in a better-defined ASF than in any previous ribosome structure (Fig. 3c).
The C-terminal domain is sandwiched between the upper part of the ASF and the A-site tRNA, and contacts the large subunit protein, uL16 (Fig. 3a). Although this domain had been predicted as an ACT fold11, the interaction between the face of its four-stranded antiparallel β-sheet and the ASF is more reminiscent of an RNA recognition motif (RRM), which shares the same topology (Extended Data Fig. 8). Furthermore, conjoint RRM/zinc-finger domains are common in eukaryotes24.
In contrast to the C-terminal domains, the hydrolase and catalytic synthetase domains form few contacts with the ribosome. The domains are extremely flexible, and occupy multiple positions between the SRL and the spur of the small subunit (18S rRNA h6). Using our focused classification approach we isolated a few well-populated states that were sufficiently resolved to orient a model based on a crystal structure of a homolog25 (Extended Data Fig. 1a). Although the synthetase domain can, in some classes, contact the tip of the spur (nucleotides 84-88), the general absence of interactions with the ribosome, suggests that RelA activation is indirect. Rather, as the isolated catalytic domain of RelA is constitutively active in a ribosome-independent manner22, our structure supports a model where association with the ribosome and uncharged A-site tRNA suppresses regulatory auto-inhibition that results from RelA homodimers or oligomers21,26, as previously suggested27. This contrasts with activation of the translational GTPases in which the SRL has a direct role in inducing catalysis28. Our structure should provide a framework for the design of experiments to differentiate among the various proposed mechanisms for RelA20,27,29.
In conclusion, our structural data reveal how RelA specifically recognizes ribosomes stalled under conditions of amino acid starvation to activate synthesis of (p)ppGpp and initiate the stringent response. By utilizing the ribosome as a signaling platform, RelA provides an immediate link between the status of translation and global adaptation to the environment. As the distribution of the RSH family is strictly limited to bacteria, and stringent control contributes to the virulence, persistence, and antibiotic tolerance of bacterial infections, the structure can provide a framework for the development of therapeutics that can selectively inactivate stringent control and re-sensitize resilient bacteria to antibiotics30.
Methods
RelA purification
The full-length E. coli RelA protein containing an in-frame N-terminal octahistidine tag and the recognition signal for tobacco etch virus (TEV) protease was recombinantly expressed in E. coli BL21 cells, as previously described13. In a modification to the reported procedure, the protein was expressed for 3 h at 37 °C after induction. Cells were collected by centrifugation at 4 °C and lysed by sonication in ice-cold lysis buffer (50 mM Tris pH 7.5, 1 M KCl, 1 mM MgCl2, 15 mM imidazole, 2 mM DTT) containing 1x protease inhibitor cocktail (Roche). After centrifugation, the supernatant of the lysate was applied to a 5 ml HisTrap HP column (GE Healthcare) equilibrated in lysis buffer. After copious washing, a linear gradient of lysis buffer supplemented with 450 mM imidazole was used to elute the histidine-tagged RelA protein. RelA was dialyzed against 2 l of buffer A (20 mM Tris pH 7.5, 0.6 M KCl, 1 mM MgCl2 and 2 mM DTT) at 4 °C for 2 h in the presence of TEV protease in a 1:200 mass ratio with RelA. A passage through a 5 ml HisTrap HP column equilibrated in buffer A was performed in a gradient of 0–450 mM imidazole to remove TEV protease and cleaved His tag. The early-eluting peak corresponding to digested RelA was used immediately to prepare complexes for freezing on cryo-EM grids to complete the procedure within a single day.
Sample preparation for electron microscopy
Ribosomes were purified from 50 g of E. coli MRE 600 cells grown to mid-log phase. Cells were disrupted by sonication in buffer R200 (20 mM Tris-HCl pH 7.5, 200 mM NH4Cl, 10 mM Mg(OAc)2, 6 mM β-mercaptoethanol, 0.1 mM benzamidine, and 0.1 mM PMSF). Cell debris was removed by two rounds of centrifugation at 30,000 × g for 30 min. Ribosomes were pelleted through a 20 ml 1.1 M sucrose cushion using a 45 Ti rotor (Beckman Coulter) at 205,000 × g for 18 h. This process was repeated twice in buffer R200, and then once in buffer R500 (as R200, but with 500 mM NH4Cl). Between pelleting steps, pellets were dissolved in the appropriate buffer by gentle shaking for 2 h on ice. The final pellet was dissolved in buffer R60 (20 mM Tris pH 7.5, 60 mM NH4Cl, 10 mM Mg(OAc)2, 6 mM β-mercaptoethanol).
To remove non-ribosomal material and isolated subunits, ~208 OD260 of sample were loaded on each of six 15-30% sucrose gradients in buffer R60 and centrifuged at 58,000 × g for 18 h with a SW28 rotor (Beckman Coulter). The peak fractions corresponding to 70S ribosomes were pooled and diluted to 180 ml with buffer G (5 mM Hepes pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(OAc)2, 6 mM β-mercaptoethanol), before a further pelleting step at 205,000 × g for 18 h. After washing with buffer G, the pellets were resuspended to a final concentration of ~6.8 μM in buffer G. Aliquots of 20 μl were flash cooled in liquid nitrogen and stored at −80 °C.
Aminoacylated fMet-tRNAfMet and uncharged tRNAPhe were produced as previously reported15. A modified version of Z4C mRNA15 was chemically synthesized (GE Dharmacon) to include six codons for Phe (UUC) after the initiator fMet codon (AUG).
Complexes were formed by incubating 100 nM ribosome with the step-wise addition of 1 μM mRNA in buffer G100 (5 mM K-HEPES pH 7.0, 100 mM KCl, 10 mM NH4Cl, 10 mM Mg(OAc)2, 6 mM β-mercaptoethanol), 200 nM fMet-tRNAfMet, and 800 nM tRNAPhe. The complexes were incubated for 5 minutes at 37 °C between each addition. Separately, 500 nM RelA was pre-incubated with 1 μM AMPCPP and 1 μM GDP for 10 minutes at 37 °C, and then added to the ribosome complex to a final volume of 100 μl. To stabilize the codon-anticodon interaction of the A-site tRNA, 1 μM paromomycin was added. After a further 5 minute incubation, the sample was cooled to 4 °C and used immediately to prepare grids for electron microscopy.
Electron microscopy
Aliquots of 3 μl of the RelA complex were incubated for 20 s on glow-discharged holey carbon grids (Quantifoil R2/2), on which a ~30 Å thick home-made amorphous carbon film had previously been deposited. The grids were blotted for 5 s in 100 % humidity at 4 °C before being flash cooled in liquid ethane using a Vitrobot MKII (FEI).
Grids were transferred to a Polara G2 microscope (FEI) operated at 300 kV. Images were recorded with the EPU automated data acquisition software on a Falcon III direct electron detector (FEI) at a calibrated magnification of 104,478 (yielding a pixel size of 1.34 Å). Images were collected with a total dose of 35 e-/Å2 and a defocus range of -1.8 to -3.0 μm. A bespoke system was used to intercept the videos from the detector at a speed of 30 frames for the 1.1 s exposures.
Image Processing
All micrographs that showed signs of astigmatism, substantial drift, or poor ice were discarded. The frames of the remaining micrographs were aligned using whole-image motion correction31 to reduce beam-induced blurring. Parameters of the contrast transfer function for each motion-corrected micrograph were obtained using Gctf32. The interactive semi-automatic swarm tool in the e2boxer.py program of EMAN233 was used to select particles from the images. We used reference-free two-dimensional class averaging in RELION34 to discard non-ribosomal particles and particles of isolated subunits. After two-dimensional classification, Euler angles for each particle were assigned using three-dimensional refinement in RELION34 using a 60 Å low-pass filtered reconstruction of EMDB-237313 as an initial reference.
This resulted in a consensus reconstruction in which density for tRNAs and RelA were apparent, but at lower occupancy than the ribosome. To enrich for particles containing RelA, we employed focused classification with signal subtraction (FCwSS)12 using a mask over the A-site tRNA and RelA. This yielded a class in which the A-site was fully occupied. The movements of particles within this class were corrected in RELION and the contributions of each frame weighted using a resolution-dependent radiation damage model35 to generate ‘polished particles’. Polished particles from different datasets were commingled and further classified and refined to yield a final dataset of 164,353 particles with a nominal resolution of 3.0 Å (Extended Data Table 1). Due to on-ribosome conformational heterogeneity, subsequent rounds of FCwSS were used to isolate different states of RelA. Prior to visualization, all density maps were corrected for the modulation transfer function (MTF), and then sharpened by applying a B-factor that was estimated using automated procedures36. Local resolution was quantified with ResMap37 (Extended Data Fig. 2).
Model building
The reconstruction was initially interpreted by docking the high-resolution crystal structure of the E. coli 70S ribosome (PDB accession code 4YBB38) into the map using Chimera39. Models for uL10, bL9, and bL31 were taken from the cryo-EM reconstruction of E. coli 70S in complex with EF-Tu (PDB accession code 5AFI40). Initial models for the tRNAs and mRNA were taken from the Thermus thermophilus 70S in complex with EF-Tu (PDB accession code 4V5L41). Density for paromomycin was clearly discernible within h44 of the small subunit, and fitted with a model. Unlike previous crystal structures42, we do not observe a second paromomycin binding site within the large subunit.
Homology models generated using I-TASSER43 were used to guide model building for E. coli RelA. A model for the hydrolase and synthetase domains was obtained from the crystal structure of the bifunctional catalytic domain from a Streptococcus equisimilis RSH25 (PDB accession code 1VJ7). A model for the RRM was generated using the structure of the RRM domain from a Chlorobium tepidum RSH (PDB accession code 3IBW). A model for the TGS domain was derived from the crystal structure of the TGS domain from a Clostridium leptum RSH (PDB accession code 3HVZ). The ZFD and connecting elements were built de novo (Extended Data Table 2). The fit of all models to the map was optimized using real space refinement in Coot44.
Model refinement and validation
Reciprocal space refinement was carried out in REFMAC v5.8 optimized for EM maps utilizing external restraints generated by ProSMART and LIBG44. FSCaverage was monitored during refinement and the final model was validated using MolProbity45 (Extended Data Table 1). Cross-validation against over-fitting was performed as previously described44,46.
Figures were generated using PyMOL47, Chimera39, or Coot/Raster3D48,49.
Extended Data
Extended Data Figure 1. In silico 3D classification scheme.
a) All particles were subjected to 2D classification, from which non-ribosomal particles were discarded, prior to 3D refinement. To isolate particles containing A-site tRNA and RelA, 3D classification focused on occupancy of the ribosomal A-site was performed. Refinement of these 183,615 particles resulted in a reconstruction with a nominal resolution of 2.9 Å. A second round of 3D classification isolated 164,353 well-aligned particles. Conformational heterogeneity of the ribosome was resolved by 3D classification without alignment, which identified two dominant classes in which the body of the small subunit occupies different positions (indicated with an arrow). Class 1 was used as the reference for model building, refinement, and interpretation. To resolve additional conformational heterogeneity of RelA, focused classification with signal subtraction (FCwSS) was performed on each domain, with the hydrolase (HYD) and synthetase (SYN) domains treated as a single unit. For the RRM, ZFD and TGS domains a single class was isolated in which the density was better resolved than in the reference class. The overall resolution of the reconstructions are reported according to the FSC=0.143 criterion. Multiple conformations of the HYD and SYN domains were identified, with the four best resolved classes shown. Together these account for 42% of the particles. b, The two main classes for the TGS domain provide an example of the small conformational differences that were isolated using FCwSS.
Extended Data Figure 2. Quality of maps and models.
a, Fourier shell correlation (FSC) curve for the EM map. b, The unfiltered and unsharpened density map, in both surface and slice view, coloured by local resolution. c, Fit of models to maps. FSC curves calculated between the refined model and the final map (black), with the self- and cross-validated correlations in blue and magenta, respectively. Information beyond 3.0 Å was not used during refinement and preserved for validation. d-e, Examples of high-resolution features of the map. d, Density for selected rRNA modifications and paromomycin. e, Density for the codon:anticodon interaction in the A-site. e, Unfiltered and unsharpened map of RelA bound to the bacterial ribosome, showing the ribosome-binding RelA domains colored by local resolution according to the FCwSS maps (see Extended Data Fig. 1). The regions amplified in panels f-g are highlighted. f, The RelA ZFD and RRM colored by local resolution. g, The TGS domain colored by local resolution.
Extended Data Figure 3. Examples of RelA density.
a, Density for the interaction between the 3’ CCA of the A/T-tRNA and the TGS domain. b, A modeled tRNAAla demonstrates that even the smallest aminoacyl groups would clash with RelA. The sphere size of the atoms of the aminoacyl group corresponds to their Van der Waals radii. c, C74 stacks with His432 and C75 can potentially interact with Arg438 of the TGS domain. d, Density for helix α4 of the TGS domain. e, Density for TGS α3. f, Density for the interaction between the ZFD and uS19, showing distinctive density for two consecutive histidine residues. g, Example of side chain density used for the de novo building of the ZFD.
Extended Data Figure 4. RelA domains are connected by flexible linkers.
Two related views showing the density that connects the RelA HYD, SYN and TGS domains with the ZFD/RRM. The linker runs between the A/T-tRNA and the ribosome, but remains flexible as suggested by the weak and broken density. The figure shows the unfiltered, unsharpened density map for class 4 (Extended Data Fig. 1) with the large subunit removed for clarity.
Extended Data Figure 5. The conformation of uncharged A-site tRNA in the presence of RelA is distinct from aminoacylated A/T-tRNA in the presence of EF-Tu.
a, The ASLs of A-site tRNA (purple) and A/T-tRNA (grey) superpose until base-pair 27:43. At this point, the A-site tRNA is distorted so that the tRNA elbow regions are separated by a 6° rotation. b, A second 11° rotation occurs at base-pair 7:66 of the acceptor stem so that the A-site tRNA in the presence of RelA is closer to the ribosomal SRL.
Extended Data Figure 6. RelA topology diagram.
Secondary structure elements for RelA residues 404-740 are numbered separately for each domain. Unbuilt sections are shown as dashed lines. Topologies were extracted using Pro-origami50.
Extended Data Figure 7. RelA binds RNA through electropositive surfaces.
a, The ZFD and RRM of RelA act together to recognize the ASF of the LSU rRNA. b, As in a, but with the ZFD and RRM shown in surface representation colored by electrostatic potential. c, The RelA TGS domain binds the acceptor arm of the A/T-tRNA. d, As in c, but with the TGS domain in surface representation colored by electrostatic potential. Electrostatic potentials were calculated using APBS51, where k is Boltzmann’s constant, T is the temperature of the calculation (310 K) and ec is the charge of an electron.
Extended Data Figure 8. RelA contains an RNA recognition motif (RRM).
a, The RRM from RelA binds the ASF (nucleotides 894-899 shown) through the face of the β-sheet. b, RRMs recognize a wide variety of RNA molecules, but share a common fold and a similar protein-RNA interface, for example in the interaction between PRP24 and U6 small nuclear RNA (PDB accession code 4N0T).
Extended Data Table 1. Refinement and model statistics.
The RelA HYD/SYN domain (residues 15-353) was modeled as a rigid-body fitted homology model and excluded from refinement.
Extended Data Table 2. Modeled residues.
Supplementary Material
Acknowledgements
We thank A. Kelley for providing tRNAs, A. Xu and J. Murray for their contributions to the early stages of this project, C. G. Savva for help in data collection, J. Grimmett and T. Darling for computing support, and X. Bai, G. Murshudov, S. H. W. Scheres, and S. Tan for discussions. The work was supported by grants to V.R. from the UK Medical Research Council (MC_U105184332), the Wellcome Trust (WT096570), the Agouron Institute, and the Jeantet Foundation.
Footnotes
Author contributions
A.B., I.S.F, and V.R. designed the study. Y.G. purified ribosomes. I.S.F. prepared samples, optimized conditions and collected data. A.B. processed data and interpreted the cryo-EM reconstructions. A.B., I.S.F., and V.R. and wrote the manuscript. All authors discussed and commented on the final manuscript.
Author information
The maps have been deposited with the EMDB under accession codes 8107-8115. Coordinates have been deposited with the Protein Data Bank under the accession code 5IQR. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Sands MK, Roberts RB. The effects of a tryptophan-histidine deficiency in a mutant of Escherichia coli. Journal of Bacteriology. 1952;63:505–511. doi: 10.1128/jb.63.4.505-511.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stent GS, Brenner S. A genetic locus for the regulation of ribonucleic acid synthesis. Proc Natl Acad Sci USA. 1961;47:2005–2014. doi: 10.1073/pnas.47.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Micro. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends in Microbiology. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Fangman WL, Neidhardt FC. Protein and Ribonucleic Acid Synthesis in a Mutant of Escherichia coli with an Altered Aminoacyl Ribonucleic Acid Synthetase. J Biol Chem. 1964;239:1844–1847. [PubMed] [Google Scholar]
- 6.Haseltine WA, Block R. Synthesis of Guanosine Tetra- and Pentaphosphate Requires the Presence of a Codon-Specific, Uncharged Transfer Ribonucleic Acid in the Acceptor Site of Ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen FS, Lund E, Kjeldgaard NO. Codon specific, tRNA dependent in vitro synthesis of ppGpp and pppGpp. Nature New Biol. 1973;243:13–15. [PubMed] [Google Scholar]
- 8.Haseltine WA, Block R, Gilbert W, Weber K. MSI and MSII made on ribosome in idling step of protein synthesis. Nature. 1972;238:381–384. doi: 10.1038/238381a0. [DOI] [PubMed] [Google Scholar]
- 9.Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 10.Cashel M, Kalbacher B. The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. J Biol Chem. 1970;245:2309–2318. [PubMed] [Google Scholar]
- 11.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai X-C, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. elife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agirrezabala X, et al. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14:811–816. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeing TM, et al. The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selmer M. Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 17.Schilling-Bartetzko S, Franceschi F, Sternbach H, Nierhaus KH. Apparent association constants of tRNAs for the ribosomal A, P, and E sites. J Biol Chem. 1992;267:4693–4702. [PubMed] [Google Scholar]
- 18.Sprinzl M, Richter D. Free 3'-OH group of the terminal adenosine of the tRNA molecule is essential for the synthesis in vitro of guanosine tetraphosphate and pentaphosphate in a ribosomal system from Escherichia coli. Eur J Biochem. 1976;71:171–176. doi: 10.1111/j.1432-1033.1976.tb11103.x. [DOI] [PubMed] [Google Scholar]
- 19.Friesen JD, Fiil NP, Parker JM, Haseltine WA. A New Relaxed Mutant of Escherichia coli with an Altered 50S Ribosomal Subunit. Proc Natl Acad Sci USA. 1974;71:3465–3469. doi: 10.1073/pnas.71.9.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 21.Gropp M, Strausz Y, Gross M, Glaser G. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. Journal of Bacteriology. 2001;183:570–579. doi: 10.1128/JB.183.2.570-579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber G, et al. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 23.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 24.Maris C, Dominguez C, Allain FHT. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 25.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Ishiguro EE. Dimerization of the RelA protein of Escherichia coli. Biochem Cell Biol. 2001;79:729–736. doi: 10.1139/o01-144. [DOI] [PubMed] [Google Scholar]
- 27.English BP, et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci USA. 2011;108:E365–73. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 29.Li W, et al. Effects of amino acid starvation on RelA diffusive behavior in live Escherichia coli. Molecular Microbiology. 2016;99:571–585. doi: 10.1111/mmi.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen D, et al. Active Starvation Responses Mediate Antibiotic Tolerance in Biofilms and Nutrient-Limited Bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K. Gctf: Real-time CTF determination and correction. Journal of Structural Biology. 2015;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. Journal of Structural Biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal PB, Henderson R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2013;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22:336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Fischer N, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 41.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The Mechanism for Activation of GTP Hydrolysis on the Ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borovinskaya MA, et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLano WL. The PyMOL molecular graphics system. 2002 [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods in enzymology. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 50.Stivala A, Wybrow M, Wirth A, Whisstock JC, Stuckey PJ. Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics. 2011;27:3315–3316. doi: 10.1093/bioinformatics/btr575. [DOI] [PubMed] [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.