Abstract
EPEC is an attaching and effacing diarrheal pathogen that carries a large pathogenicity island, locus for enterocyte effacement (LEE). Recently, the pathogenicity island PAI O-122 was described among non-LEE effectors and found to be associated with diarrhea among atypical EPEC strains. It is unknown if incomplete PAI O-122 could be associated with diarrhea duration and severity. To identify these virulence determinants we analyzed 379 EPEC strains isolated from Peruvian children. EPEC was diagnosed by PCR(eae+, stx−) and classified as typical(t-EPEC) or atypical(a-EPEC). To characterize PAI O-122 we amplified three modules by PCR: Module 1(pagC), Module 2(senA, nleB and nleE) and Module 3(lifA/efa-1). To characterize the large ORF lifA/efa-1 we amplified the regions known as efa-N, efa-M and efa-C. Clinical information was obtained from the cohort study. A total of 379 EPEC strains were able to analyze PAI O-122 genes, 128 (10.4%) EPEC strains were isolated from 1235 diarrhea episodes and 251(9.2%) from 2734 healthy controls. t-EPEC strains were isolated from 14.8% (19/128) of children with diarrhea and 25/251(10.0%) from healthy controls. The most frequent PAI O-122 genes were nleE(37.7%), senA(34.6%) and nleB(37.5%), with similar prevalence among diarrhea and control samples. However, lifA/efa-1 was more common among diarrhea cases than healthy control cases (30.5% vs. 21.1%, p<0.05). The presence of complete PAI O-122 was associated with diarrhea episodes of higher severity among single pathogen infection (33.3% vs. 1.8%, p<0.05) mainly due to the presence of a complete lifA/efa-1 gene. In summary, the gene lifA/efa-1 is significantly associated with diarrheal episodes of higher severity, suggesting to be an important virulent factor.
Keywords: Pathogenicity Island O-122, enteropathogenic Escherichia coli, diarrhea severity, children, Peru
INTRODUCTION
Diarrhea is the third most common cause of death worldwide in children under the age of five, with the majority of cases occurring in developing countries (Liu et al., 2012). A series of pathogens have been associated with diarrhea, however, their relative importance varies both in relation to geographical distribution and severity of the illness (Lanata et al., 2013; Kotloff et al., 2013). In a recent multicenter diarrhea study (GEMS), EPEC has been associated with increased risk of death in young children (Donnenberg and Finlay, 2013; Kotloff et al., 2013).
Multiple genetic determinants encode virulent factors that underlie the ability of EPEC to cause illness. Among the main genetic determinants are 1) EAF-plasmid (EPEC adherence factor) which mediates localized adherence to intestinal cells (Kaper et al, 2004) and discriminates typical EPEC (t-EPEC) strains (eae+, bfpA+) from atypical EPEC (a-EPEC) strains (eae+, bfpA−); 2) the “locus of enterocyte effacement” pathogenicity island (PAI-LEE) located on the bacterial chromosome which encodes more than 40 proteins for the type III secretion system and effector proteins necessary for the attaching and effacing (A/E) lesion in the intestinal epithelial surface (Nougayrède et al., 2003); and 3) the non-LEE genes including the O-122 pathogenicity island (PAI O-122), which comprises a series of putative virulence genes such as pagC, senA, nleB, nleE and lifA/efa-1 (Karmali et al., 2003).
The lifA/efa-1 (lymphocyte inhibitory factor A/EHEC factor for adherence-1) gene encodes lymphostatin, a very large surface protein that inhibits proliferation of mitogen activated lymphocytes and the synthesis of pro-inflammatory cytokines (Klapproth et al., 2000). lifA/efa-1 has also been implicated in the attachment of a-EPEC strains to host cells (Badea at al., 2003) and is critical for intestinal colonization by Citrobacter rodentium, an A/E lesion-producing bacterial murine pathogen (Klapproth et al., 2005). However, its association with diarrhea is controversial (Afset et al., 2006). Vieira et al, have shown that typical and atypical EPEC strains may harbor a complete (pagC, senA, nleB, nleE and lifA/efa-1 all together) or incomplete PAI O-122; a strong association between the presence of a complete PAI O-122 and diarrhea was observed only in a-EPEC strains (Vieira et al., 2010). It is unknown if incomplete PAI O-122 could also be associated with diarrhea.
Thus, the aim of this study was to determine the prevalence of PAI O-122 genes, their distribution pattern among EPEC strains, and their correlations with clinical characteristics in strains isolated from Peruvian Children with and without diarrhea.
MATERIALS AND METHODS
Patients and controls
Specimens analyzed in this study were obtained from a previous community-based, randomized, double blind placebo-controlled diarrhea trial, conducted from January 2008 through May 2011, in children from peri-urban areas in Lima, Peru. Since no difference in EPEC prevalence and severity with the study intervention (lactoferrin) was observed, we worked with all samples collected in the study (Ochoa et al., 2013). Diarrhea was defined as three or more liquid or semi-liquid stools within 24 hours or a single bloody semi-liquid stool within 24 hours. We have also included stool samples from healthy “control” children without diarrhea one week before and one week after the sample collection period. Samples were evaluated for common enteric pathogens (Shigella, Salmonella, Vibrio, Campylobacter, adenovirus, rotavirus, Giardia lamblia, Cryptosporidium and other parasite) by conventional methods (Ochoa et al., 2013). Five lactose positive colonies were isolated from MacConkey plates and tested by a multiplex real time PCR with specific primers to detect virulence factors associated with diarrheagenic E. coli (Guion et al., 2008).
Strains and DNA extraction
A total of 379 EPEC strains, stored at −70°C in Trypticase Soy Broth (TSB) containing 15% glycerol, were included in this study; one colony per sample was used. All EPEC strains were cultured on Mac Conkey agar plates for overnight grow at 37°C. A single colony from the plates was suspended in 50 uL of PCR water for DNA extraction by boiling method (Guion et al., 2008). Two microliters of DNA were used as a template in a 25 ul total PCR volume or stored at −20°C until use.
PCR analysis
PCR for determination of typical and atypical EPEC
To discriminate t-EPEC from a-EPEC strains, a duplex conventional PCR assay was performed, using primers for eae and bfpA genes (Table 1). PCR reaction mix (Promega Corp., Madison, WI) included: 1× GoTaq Flexi Buffer, 2mM MgCl2, 200 µM dNTPs, 0.5 µM of each primer, 0.75 U of GoTaq DNA Polymerase and 2 µl of DNA template adjusted to 25 µl per reaction with PCR water. The duplex conventional PCR reaction was programmed as follows: pre-denature at 94 °C for 5 min; 30 cycles at 94 °C for 20 s, 58 °C for 20 s, 72 °C for 30 s, and post-annealing at 72 °C for 5 min in a Applied Biosystems 2720 Thermal Cycler.
Table 1.
Primers and conditions used in this study.
| Gen | Orientation | Sequence (5′ -→ 3′) | Annealing temp (°C) |
No Of Cycles |
Product length (pb) |
Reference of primers |
|---|---|---|---|---|---|---|
| eae | F | ATGCTTAGTGCTGGTTTAGG | 58 | 30 | 248 | (Guion et al., 2008) |
| R | GCCTTCATCATTTCGCTTTC | |||||
| bfpA | F | GTCTGCGTCTGATTCCAATA | 58 | 30 | 460 | (Lacher et al., 2007) |
| R | TCAGCAGGAGTAATAGC | |||||
| pagC | F | ATGAGTGGTTCAAGACTGG | 58 | 30 | 521 | (Karmali et al., 2003) |
| R | CCAACTCCAACAGTAAATCC | |||||
| senA | F | GGATGGAACCATACCTGG | 60 | 30 | 551 | (Karmali et al., 2003) |
| R | CGCAATCAATTGCTAATGC | |||||
| nleB | F | GGTGTGCTGGTAGATGGA | 58 | 30 | 175 | (Afset et al., 2006) |
| R | CAGGGT ATGATTCTTGTT TATG | |||||
| nleE | F | CTA ATACTCAGGGCGTGTCC | 64 | 30 | 192 | (Afset et al., 2006) |
| R | ACCGTC TGGCTTTCTCGT TA | |||||
| lifA/efa-1 | F | AGAATGGAAGATCACACCAG | 56 | 30 | 310 | (Narimatsu et al., 2010) |
| R | ATAATGCCTTTCATCCACAC | |||||
| Efa-N | F | GGAATTCATGAGACTGCCAGAGAAAG | 60 | 30 | 1128 | (Badea et al., 2003) |
| R | CGTCGACTTAGCTGTATAGTTCTGCATGC | |||||
| Efa-M | F | GGAATTCACCACCGACCGTCTGGCG | 64 | 30 | 1215 | (Badea et al., 2003) |
| R | CGTCGACTTAGGAGAAATCCAGCAGACC | |||||
| Efa-C | F | GGAATTCATGGATACAACGGACAGG | 60 | 30 | 1200 | (Badea et al., 2003) |
| R | CGTCGACTTAGTTAAAAAGGTTGTCACC |
PCR for PAI O-122 characterization
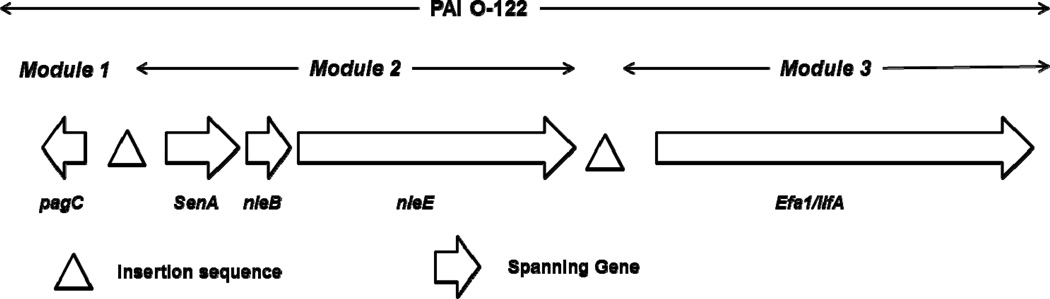
To characterize the PAI O-122, we searched five genes distributed in three modules (Konczy et al., 2008): Module 1 (pagC), Module 2 (senA, nleB and nleE) and Module 3 (lifA/efa-1) (Fig. 1), using primers and specific annealing temperatures for each gene as previously described (Table 1). The complete (all five PAI O-122 genes present) or Incomplete PAI O-122 (at least one but not all five PAI O-122 genes present) was consider for analyze.
Fig 1.
Genetic organization of the five genes (pagC, senA, nleB, nleE, lifA/efa-1) analyzed located in pathogenicity island O-122 (PAI O-122).
To characterize a large open reading frame (ORF) spanning of 9,669 bp, which is designate lifA/efa-1 (module 3), we used a set of primers to amplify three regions: 5’ known as efa-N; central, efa-M and 3’, efa-C (9). The PCR amplification of each region was performed with the same reagents used for PAI O-122, under the following conditions: 94 °C per 2 min., 30 cycles of 94°C per 15 s., a specific annealing temperature for each amplicon (table 1) per 20 s, and 72°C per 90 s. and final extension of 72°C per 5 min. The PCR products were analyzed by agarose electrophoresis (Invitrogen, Carlsbad, CA) using 12 uL of the amplicon. For the determination of typical and atypical EPEC and PAI O-122 characterization we use a 2% gel, and 1% for lifA/efa-1 characterization. The gels were stained with ethidium bromide then visualized and analyzed under ultraviolet light (Bio-Rad transilluminator). The product length was determined using GeneRuler 100bp plus DNA Ladder (Thermo Scientific, USA).
Clinical data
Clinical information about diarrheal episodes was obtained from the clinical trial. Children were followed for 6 months with daily home visits. Diarrhea duration was recorded until the last day an unformed stool was produced. Diarrhea duration was classified as acute (< 7 days) and prolonged (≥ 7 days) (Moore et al., 2010). We used a Modified Ruuska-Vesikari Score (MRV) to determine the severity of the EPEC episodes, as previously described (Ochoa et al., 2009). The scored included: duration of diarrhea in days (0–3 points), maximum number of stools per day during the episode (1–3), presence of vomiting (0–1), maximum number of emesis per day during the episode (0–3), fever (0–1), dehydration (0–1), and treatment (0–2). The maximum possible score was 19; a score > 8 was considered moderate/severe.
Statistical analysis
Differences between isolation rates, clinical characteristics, and presence of the PAI-O122 gene among EPEC strains isolated from diarrhea and control samples were evaluated by chi-square or Fisher exact tests. All statistical analyses were performed using free Epi Info version 7. The significance level was set at a p value of < 0.05.
RESULTS
EPEC prevalence
One hundred twenty eight (10.4%) EPEC strains isolated from 1235 diarrhea episodes and 251 (9.2%) EPEC strains isolated from 2734 healthy controls were analyzed. The presence of t-EPEC (bfpA+) was found in 19/128 strains isolated from children with diarrhea (14.8%) and 25/251 from healthy controls (10.0%). a-EPEC was found in 109/128 (85.2%) of diarrhea samples and 226/251 (90.0%) of healthy controls. Additionally, all EPEC strains have been checked for Stx1 and Stx2 to exclude Shiga toxin producing E. coli (STEC).
Frequency and characterization of PAI O-122
The most frequent PAI O-122 genes were nleE (37.7%), senA (34.6%) and nleB (37.5%), with the same prevalence among diarrhea and control samples. However, lifA/efa-1 was more frequently found among EPEC strains isolated from diarrhea than healthy control samples (30.5% and 21.1%) respectively (p=0.031) (Table 2), using a single set of primers as marker of a large lifA/efa-1 gene (Narimatsu, et al., 2010). In general, there was no difference in the presence of PAI O-122 genes between t-EPEC and a-EPEC strains.
Table 2.
Prevalence of PAI O-122 genes in EPEC strains isolated from children with diarrhea and healthy controls
| t-EPEC | a-EPEC | All EPEC strains | |||||
|---|---|---|---|---|---|---|---|
| Genes O-122 | Diarrhea N=19 n (%) |
Control N=25 n (%) |
Diarrhea N=109 n (%) |
Control N=226 n (%) |
Diarrhea N=128 n (%) |
Control N=251 n (%) |
Total N=379 n (%) |
| PagC | 3 (15.8) | 2 (8.0) | 23 (21.1) | 40 (17.7) | 26 (20.3) | 42 (16.7) | 68 (17.9) |
| SenA | 8 (42.1) | 5 (20.0) | 37 (33.9) | 81 (35.8) | 45 (35.2) | 86 (34.3) | 131 (34.6) |
| NleB | 9 (47.4) | 5 (20.0) | 41 (37.6) | 87 (38.5) | 50 (39.1) | 92 (36.7) | 142 (37.5) |
| NleE | 8 (42.1) | 5 (20.0) | 40 (36.7) | 90 (39.8) | 48 (37.5) | 95 (37.8) | 143 (37.7) |
| lifA/efa-1 | 8 (42.1) | 4 (16.0) | 31 (28.4) | 49 (21.7) | 39 (30.5)a | 53 (21.1) | 92 (24.3) |
| Efa-N | 6 (31.6) | 3 (12.0) | 21 (19.3) | 42 (18.6) | 27 (21.1) | 45 (17.9) | 72 (19.0) |
| Efa-M | 6 (31.6) | 4 (16.0) | 27 (24.8) | 45 (19.9) | 33 (25.8) | 49 (19.5) | 82 (21.6) |
| Efa-C | 6 (31.6) | 5 (20.0) | 26 (23.9) | 42 (18.6) | 32 (25.0) | 47 (18.7) | 79 (20.8) |
| lifA/efa-1 complete b | 6 (31.6) | 3 (12.0) | 19 (17.4) | 36 (15.9) | 25 (19.5) | 39 (15.5) | 64 (16.9) |
| PAI O-122 complete c | 1 (5.3) | 0 (0.0) | 10 (9.2) | 24 (10.6) | 11 (8.6) | 24 (9.6) | 35 (9.2) |
, p= 0.031 (fisher exact), for the comparison between diarrhea and control strains.
, EPEC strains with all three regions present (efa-N, efa-M and efa-C)
, EPEC strains with all five genes present (pagC, senA, nleB, nleE, lifA/efa-1)
t-EPEC, typical enteropathogenic E coli; a-EPEC, atypical enteropathogenic E coli.
The presence of incomplete PAI O-122 (at least one but not all five PAI O-122 genes present) was similar among EPEC strains isolated from diarrhea and healthy control samples (44.5% and 39.4%), as well as among t-EPEC and a-EPEC strains (40.9% and 41.2%). However, among t-EPEC strains incomplete PAI O-122 was significantly associated with diarrhea episodes 11/19(57.9%) in comparison to healthy controls 7/25 (28.0%) (p=0.045) (data not shown).
Analyzing PAI O-122 in a modular way, Module 1 was present in 68(17.9%) strains; complete Module 2 was present in 124(32.7%); and Module 3 was present in 92(24.3%) (Table 3). The most common profiles were: only Module 2 present in 50(13.2%) strains; all three modules together in 35(9.2%) strains; and Module 2 and Module 3 together in 32(8.4%) strains. The presence of Module 2 and Module 3 together was significantly associated with EPEC strains isolated from children with diarrhea than healthy controls (12.5% vs. 6.4%, p=0.036) Mainly due to t-EPEC than a-EPEC strains (31.6% vs 9.2 %, p=0.02) (Table 3).
Table 3.
PAI O-122 genes profiles harbored by EPEC strainsa
| PAI O-122 genes | All EPEC strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Module 1 | Module 2 | Module 3 | Diarrhea (N=128) | Control (N=251) | Total | ||||
| PagC | SenA | nleB | nleE | lifA/efa-1 | t-EPEC (N=19) n (%) |
a-EPEC (N=109) n (%) |
t-EPEC (N=25) n (%) |
a-EPEC (N=226) n (%) |
(N=379) n (%) |
| − | + | + | + | − | 1 (5.3) | 13 (11.9) | 1 (0.9) | 35 (15.5) | 50 (13.2) |
| + | + | + | + | + | 1 (5.3) | 10 (9.2) | 0 | 24 (10.6) | 35 (9.2) |
| − | + | + | + | + | 6 (31.6) b | 10 (9.2) | 4 (3.7) | 12 (5.3) | 32 (8.4) |
| − | − | − | − | + | 1 (5.3) | 6 (5.5) | 0 | 11 (4.9) | 18 (4.7) |
| + | − | − | − | − | 2 (10.5) | 5 (4.6) | 2 (1.8) | 7 (3.1) | 16 (4.2) |
| + | + | + | + | − | 0 | 3 (2.8) | 0 | 4 (1.8) | 7 (1.8) |
| − | − | + | + | − | 0 | 1 (0.9) | 0 | 6 (2.7) | 7 (1.8) |
| − | + | − | − | − | 0 | 1 (0.9) | 0 | 5 (2.2) | 6 (1.6) |
| − | − | − | + | − | 0 | 1 (0.9) | 0 | 5 (2.2) | 6 (1.6) |
| + | − | + | + | − | 0 | 1 (0.9) | 0 | 3 (1.3) | 4 (1.1) |
| + | − | − | − | + | 0 | 2 (1.8) | 0 | 0 | 2 (0.5) |
| − | − | + | − | − | 1 (5.3) | 0 | 0 | 1 (0.4) | 2 (0.5) |
| + | + | + | − | + | 0 | 0 | 0 | 1 (0.4) | 1 (0.3) |
| + | − | + | + | + | 0 | 1 (0.9) | 0 | 0 | 1 (0.3) |
| + | − | + | − | + | 0 | 1 (0.9) | 0 | 0 | 1 (0.3) |
| + | − | + | − | − | 0 | 0 | 0 | 1 (0.4) | 1 (0.3) |
| − | − | + | − | + | 0 | 1 (0.9) | 0 | 0 | 1 (0.3) |
| − | − | − | + | + | 0 | 0 | 0 | 1 (0.4) | 1 (0.3) |
| − | − | − | − | − | 7 (36.8) | 53 (48.6) | 18 (16.5) | 110 (48.7) | 188 (49.6) |
, The virulence profile was based on the presence (+) or absence (−) of genes by conventional PCR among 379 strains tested for all genes.
, P=0.02, for the comparison between t-EPEC and a-EPEC strains.
Characterization of Module 3 (lifA/efa-1)
The frequencies of efa-N, efa-M and efa-C were 19.0%, 21.6% and 20.8%, respectively. The complete Module 3 (all three regions present) was found in 16.9% (64/379) of strains (Table 2). The frequency of a complete Module 3, using three set of primers to amplify three different regions of a large lifA/efa-1 gene (Badea, et al., 2003), was similar among EPEC strains isolated from children with diarrhea and healthy controls, as well as among t-EPEC and a-EPEC strains (Table 2).
Association of PAI 0–122 with diarrhea duration and severity
For this analysis we have included only the diarrhea episodes with EPEC as a single pathogen infection (n=64). We have excluded episodes with co-infections with viruses, other bacteria and parasites, 50% of EPEC diarrhea episodes had a co-infection (64/136). The presence of complete PAI O-122 was associated with diarrhea episodes of higher severity (33.3% and 1.8%, p<0.05) mainly due to the presence of lifA/efa-1 gene (55.6% and 18.2%, p<0.05) (Table 4), suggesting to be an important virulent factor. There was no difference among t-EPEC and a-EPEC strains isolated with diarrhea duration and severity (data not shown).
Table 4.
Association of PAI O-122 genes with diarrhea duration and severity among episodes with EPEC as single pathogen infection (excluding co-infections) a.
| Genes | Duration | Severity | Total | ||
|---|---|---|---|---|---|
| PAI O-122 | Acute <7 days (N=42) |
Prolonged ≥7 days (N=22) |
mild ≤8 (N=55) |
Moderate/severe >8 (N=9) |
(N=64) |
| PagC | 8 (19.0) | 4 (18.2) | 9 (16.4) | 3 (33.3) | 12 (18.8) |
| SenA | 14 (33.3) | 8 (36.4) | 17 (30.9) | 5 (55.6) | 22 (34.4) |
| nleB | 14 (33.3) | 8 (36.4) | 17 (30.9) | 5 (55.6) | 22 (34.4) |
| nleE | 15 (35.7) | 8 (36.4) | 18 (32.7) | 5 (55.6) | 23 (35.9) |
| lifA/efa-1 | 8 (19.0) | 7 (31.8) | 10 (18.2) | 5 (55.6) b | 15 (23.4) |
| efa-N | 4 (9.5) | 6 (27.3) | 6 (10.9) | 4 (44.4) b | 10 (15.6) |
| efa-M | 6 (14.3) | 6 (27.3) | 6 (10.9) | 6 (66.7) b | 12 (18.8) |
| efa-C | 5 (11.9) | 4 (18.2) | 5 (9.1) | 4 (44.4) b | 9 (14.1) |
| lifA/efa-1 complete | 4 (9.5) | 4 (18.2) | 4 (7.3) | 4 (44.4) b | 8 (12.5) |
| PAI O-122 complete | 1 (2.4) | 3 (13.6) | 1 (1.8) | 3 (33.3) b | 4 (6.3) |
, Including all EPEC strains as single pathogen (n=64) that were tested for all five genes (pagC, senA, nleB, nleE, lifA/efa-1).
, p<0.05 (fisher exact), for the comparison between mild and moderate/severe diarrhea episodes
Diarrhea duration: Acute, <7 days of diarrhea duration. Diarrhea severity: mild, ≤ 8 according Vesikari Score modified.
DISCUSION
Determinants of bacterial virulence are predominantly encoded by or associated with mobile genetic elements such as phages, plasmids, insertion elements, or transposons. A large number of these determinants are located within PAIs, and can be exchanged among different bacterial species, and assembled and stabilized by selective pressure, leading to new pathogenic variants (Schmidt and Hensel, 2004; Lee, 1996). Previous studies have described that PAI O-122 genes are frequently present in A/E E. coli strains (Morabito et al., 2003) and are associated with a higher virulence potential in non-O157:H7 STEC strains (Wickham et al., 2006)”. In this study we determined the frequency of PAI O-122 genes (pagC, senA, nleB nleE and lifA/efa-1) in EPEC strains isolated from children with and without diarrhea. Previous studies show that PAI O-122 pathogenicity island is a common attribute significantly associated with highly virulent EHEC and EPEC strains (Bugarel et al., 2011). The frequency of complete PAI O-122 (carrying pagC, senA, nleB nleE and lifA/efa-1) in EPEC strains isolated from diarrheal episodes was 8.6% and was more prevalent among a-EPEC strains than t-EPEC strains (10.1% and 2.3 % respectively), although this difference was not significant. These results differ from those reported by Vieira, where complete PAI O-122 was found in 32.5% of EPEC strains isolated from patients with diarrhea and was more prevalent in t-EPEC than in a-EPEC (48.9% versus 17.8%, respectively). Considering t-EPEC and a-EPEC strains together, the presence of complete PAI O-122 was similar among strains isolated from diarrhea (8.6%) and control (9.6%), different than the data reported by Vieira where EPEC and a-EPEC strains with complete PAI O-122 were significantly associated with diarrhea (32.5% and 22.4%) than control strains (10.5% and 6.5) respectively (Vieira et al., 2010).
Among strains with at least one of five PAI O-122 genes positive, t-EPEC strains were significantly associated with diarrhea episodes (63.2% in diarrhea vs. 28.0% in healthy control samples, p<0.05) (Table 2) These results suggest that the PAI-O122 genes will confer additional virulence to t-EPEC strains, which harbor EAF plasmid, to trigger clinical manifestations among human hosts.
Some genes tend to be related to each other and to be more retained as a set instead of being independent. For example, the presence of module 2 and 3 together are significantly more prevalent among t-EPEC than a-EPEC and with EPEC strains isolated from children with diarrhea than healthy controls (Table 3). The PAI O-122 modular patterns may reflect horizontal acquisition of one or more modules independently, or modular decay following transfer of a complete PAI O-122. The transference of modular components of PAI O-122 will be supported by the presence of Insertion Sequence-Associated (ISA) elements (or putative transposases) between the three modules, similar to that reported by Konczy in verotoxigenic-producing Escherichia coli (VTEC) (Konczy et al., 2008).
Previous reports using DNA microarray and PCR analysis in a case-control study among Norwegian children less than 5 years old, with and without diarrhea, found in a-EPEC strains that PAI O-122 genes (set/ent, nleB, and nleE) mainly efa1/lifA (lifA/efa-1) were statistically association with diarrhea (Vieira et al., 2010; Narimatsu et al., 2010). This result is different from what is reported in our study. Among a-EPEC strains, the frequency of PAI O-122 including lifA/efa-1 was similar in diarrhea and control cases. However, in t-EPEC strains, PAI O-122 genes, mainly lifA/efa-1, were more prevalent among children with diarrhea than healthy controls (data not shown); indicating that in our population, the t-EPEC will be more virulent.
Our data demonstrate that the PAI O-122 gene lifA/efa-1 is statistically associated with EPEC strains isolated from diarrhea, when analyzing individual genes; however, when analyzing all PAI O-122 genes the only profile associated with diarrhea is the presence of module 3 (lifA/efa-1) plus module 2 (senA, nleB and nleE) without module 1 (pagC). It is important to highlight that this is only a descriptive analysis, since we are not analyzing all other virulence determinants of EPEC. This difference remains when analyzing the prevalence of lifA/efa-1 among t-EPEC (42.1% in diarrhea vs. 16.0% in controls) and among a-EPEC (28.4% in diarrhea vs. 21.7% in controls) (Table 2); however, these differences are not statistical significant; in the case of t-EPEC probably due to the small number of samples. Other PAI O-122 genes (pagC, senA, nleB nleE) were not associated with diarrheal disease. It is known that nleB encodes another putative effector involved in virulence of A/E pathogens; deletion of nleB gene in C. rodentium reduces colonization of mice and colonic hyperplasia (Kelly et al., 2006). However nleE, other non-LEE-encoded effector genes in C. rodentium, immediately downstream had no effect on host colonization or disease (Kelly et al., 2006).
Previous studies suggest that atypical EPEC might have an innate propensity to persist longer in the gut than other diarrheogenic E. coli strains, which are more transient in nature (Nguyen et al., 2006). However, among prolonged diarrhea samples infected with EPEC, as the only pathogen (excluding co-infections), we found that typical EPEC and atypical EPEC were isolated in similar proportions, 35.9% and 34.4% respectively (data not shown).
In order to determine the correlation between a specific virulence gene and the clinical outcome of the diarrheal episode (duration and severity) it is critical to exclude episodes with co-infections, since it will be difficult to attribute the clinical outcome to a specific pathogen. Therefore, in this study we have excluded all episodes in which EPEC was isolated together with other pathogens (viral, other bacteria and parasites) (Ochoa, et al., 2013). In this line it is need to take into account that in our setting it is very common to find mixed infections (Ochoa et al., 2009). Although previous studies have not detected a correlation of certain phylogroups with t-EPEC or a-EPEC strains (Afset et al., 2008), we suggest in the future to determine the phylogenetic background of the different t-EPEC and a-EPEC strains in order to detect a possible correlation of certain phylogroups and the presence of a complete or incomplete pathogenicity island O-122.
This study has some limitations. First, were not able to recover all strains in order to complete the analysis of all PAI O-122 genes. Second, we were not able to determine the location of PAI O-122 genes analyzed in EPEC genome.
In summary, this study correlates the presence of complete PAI O-122 among EPEC strains, isolated as single-pathogen in children, and with diarrhea episodes of higher severity, mainly due to the presence of the lifA/efa-1 gene, determined by a single set of primers for the entire gene. This could be used in the future for better diagnosis and management of severe diarrhea in children.
Acknowledgments
This work was partially funded by 1K01TW007405 (TJO). And by the Agencia Española de Cooperación Internacional para el Desarrollo (AECID), Spain, Programa de Cooperación Interuniversitaria e Investigación Científica con Iberoamérica (D/019499/08, D/024648/09, D/030509/10, and A1/035720/11) (J.R and T.J.O). JR has a fellowship from the program I3, of the ISCIII (grant number: CES11/012)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
We have no conflict of interest.
REFERENCES
- 1.Afset JE, Bruant G, Brousseau R, Harel J, Anderssen E, Bevanger L, Bergh K. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J. Clin. Microbiol. 2006;44:3703–3711. doi: 10.1128/JCM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afset JE, Anderssen E, Bruant G, Harel J, Wieler L, Bergh K. Phylogenetic Backgrounds and Virulence Profiles of Atypical Enteropathogenic Escherichia coli Strains from a Case-Control Study Using Multilocus Sequence Typing and DNA Microarray Analysis. J. Clin. Microbiol. 2008;46:2280–2290. doi: 10.1128/JCM.01752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badea L, Doughty S, Nicholls L, Sloan J, Robins-Browne RM, Hartland EL. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 2003;34:205–215. doi: 10.1016/s0882-4010(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 4.Bugarel M, Martin A, Fach P, Beutin L. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 2011;21:11–142. doi: 10.1186/1471-2180-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg MS, Finlay BB. Combating enteropathogenic Escherichia coli (EPEC) infections: the way forward. Trends Microbiol. 2013;21:317–319. doi: 10.1016/j.tim.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J. Clin. Microbiol. 2008;46:1752–1757. doi: 10.1128/JCM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. Association of genomic O island 122 of Escherichia coli EDL 933 with verocyto- toxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 2003;41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 9.Kelly M, Hart E, Mundy R, Marchès O, Wiles S, Badea L, Luck S, Tauschek M, Frankel G, Robins-Browne RM, Hartland EL. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect. Immun. 2006;74:2328–2337. doi: 10.1128/IAI.74.4.2328-2337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klapproth JM, Scaletsky IC, McNamara BP, Lai LC, Malstrom C, James SP, Donnenberg MS. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 2000;68:2148–2155. doi: 10.1128/iai.68.4.2148-2155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klapproth JM, Sasaki M, Sherman M, Babbin B, Donnenberg MS, Fernandes PJ, Scaletsky IC, Kalman D, Nusrat A, Williams IR. Citrobacter rodentium lifA/efa1 is essential for colonic colonization and crypt cell hyperplasia in vivo. Infect. Immun. 2005;73:1441–1451. doi: 10.1128/IAI.73.3.1441-1451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konczy P, Ziebell K, Mascarenhas M, Choi A, Michaud C, Kropinski AM, Whittam TS, Wickham M, Finlay B, Karmali MA. Genomic O island 122, locus for enterocyte effacement, and the evolution of virulent verocytotoxin-producing Escherichia coli. J. Bacteriol. 2008;190:5832–5840. doi: 10.1128/JB.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;20:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 14.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 2007;189:342–350. doi: 10.1128/JB.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Aryee MJ, Black RE Child Health Epidemiology Reference Group of the World Health Organization and UNICEF. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One. 2013;4:e72788. doi: 10.1371/journal.pone.0072788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CA. Pathogenicity islands and the evolution of bacterial pathogens. Infect. Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 17.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, Rudan I, Campbell H, Cibulskis R, Li M, Mathers C, Black RE Child Health Epidemiology Reference Group of WHO and UNICEF. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;9:2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 18.Moore SR, Lima NL, Soares AM, Oriá RB, Pinkerton RC, Barrett LJ, Guerrant RL, Lima AA. Prolonged episodes of acute diarrhea reduce growth and increase risk of persistent diarrhea in children. Gastroenterology. 2010;139:1156–1164. doi: 10.1053/j.gastro.2010.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morabito S, Tozzoli R, Oswald E, Caprioli A. A mosaic pathogenicity island made up of the locus of enterocyte effacement and a pathogenicity island of Escherichia coli O157:H7 is frequently present in attaching and effacing E. coli. Infect Immun. 2003;71(6):3343–3348. doi: 10.1128/IAI.71.6.3343-3348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narimatsu H, Ogata K, Makino Y, Ito K. Distribution of non-locus of enterocyte effacement pathogenic island-related genes in Escherichia coli carrying eae from patients with diarrhea and healthy individuals in Japan. J. Clin. Microbiol. 2010;48:4107–4114. doi: 10.1128/JCM.00677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 2006;12:597–603. doi: 10.3201/eid1204.051112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nougayrède JP, Fernandes PJ, Donnenberg MS. Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 2003;5:359–372. doi: 10.1046/j.1462-5822.2003.00281.x. [DOI] [PubMed] [Google Scholar]
- 23.Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Gil AI, Contreras C, Molina M, Amemiya I, Verastegui H, Hall ER, Cleary TG, Lanata CF. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from Periurban areas in Lima, Peru. Clin Infect Dis. 2009;49(11):1694–1702. doi: 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochoa TJ, Chea-Woo E, Baiocchi N, Pecho I, Campos M, Prada A, Valdiviezo G, Lluque A, Lai D, Cleary TG. Randomized double-blind controlled trial of bovine lactoferrin for prevention of diarrhea in children. J. Pediatr. 2013;162:349–356. doi: 10.1016/j.jpeds.2012.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vieira MA, Salvador FA, Silva RM, Irino K, Vaz TM, Rockstroh AC, Guth BE, Gomes TA. Prevalence and characteristics of the O122 pathogenicity island in typical and atypical enteropathogenic Escherichia coli strains. J. Clin. Microbiol. 2010;48:1452–1455. doi: 10.1128/JCM.01944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickham ME, Lupp C, Mascarenhas M, Vazquez A, Coombes BK, Brown NF, Coburn BA, Deng W, Puente JL, Karmali MA, Finlay BB. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J Infect Dis. 2006;194:819–827. doi: 10.1086/506620. [DOI] [PubMed] [Google Scholar]