Abstract
The Woronin body is a peroxisome-derived dense-core vesicle that is specific to several genera of filamentous ascomycetes, where it has been shown to seal septal pores in response to cellular damage. The Hexagonal peroxisome (Hex1) protein was recently identified as a major constituent of the Woronin body and shown to be responsible for self-assembly of the dense core of this organelle. Using a mutation in the Magnaporthe grisea HEX1 ortholog, we define a dual and essential function for Woronin bodies during the pathogenic phase of the rice blast fungus. We show that the Woronin body is initially required for proper development and function of appressoria (infection structures) and subsequently necessary for survival of infectious fungal hyphae during invasive growth and host colonization. Fungal mycelia lacking HEX1 function were unable to survive nitrogen starvation in vitro, suggesting that in planta growth defects are a consequence of the mutant's inability to cope with nutritional stress. Thus, Woronin body function provides the blast fungus with an important defense against the antagonistic and nutrient-limiting environment encountered within the host plant.
INTRODUCTION
Magnaporthe grisea is an ascomycetous fungus that causes devastating blast disease in graminaceous hosts such as rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), and millet (Pennisetum americanum). Establishment of excellent molecular genetics for M. grisea makes the rice blast pathosystem a good model for the investigation of fungus–host interactions (Valent, 1990). The disease cycle during a blast infection comprises the following sequence of developmental events: germination of the conidium; sensing of host surface toughness and hydrophobicity by the tip of the germ tube; elaboration of a specialized infection structure called an appressorium; cytoplasmic streaming into the appressorium; build-up of enormous turgor within the appressorium; and forcible entry into the host epidermis through production of an infection peg and subsequent invasive growth in the host tissue. Considerable progress has been made in identifying gene functions necessary for the initial establishment of the fungus on the host (for comprehensive reviews, see Dean, 1997; Tucker and Talbot, 2001; Talbot, 2003), but little is known about the later steps of infection related to invasiveness and in planta fungal growth.
The morphological and physiological transitions during the life cycle of phytopathogenic fungi appear to be induced by the environment encountered during each stage of pathogenesis: the plant surface, the intrinsic plant micro-environment, and the interface between plant and its surroundings. Signals that affect the later stages of in planta fungal development have received limited attention. The cellular environment within the host plant represents a challenge to an invading fungus, which must evade or eliminate constitutive and induced toxic molecules produced by the host (Hammond-Kosack and Jones, 1996; Osbourn 1996). Oxidative burst and resultant accumulation of reactive oxygen species is one of the first plant responses upon pathogen detection (Doke, 1983), and peroxide produced during the oxidative burst contributes to restriction of pathogen growth (Hammond-Kosack and Jones, 1996).
Several studies have suggested that lack of nutrients is one of the signals that controls expression of pathogenicity factors in various fungal pathogens of plants (Snoeijers et al., 2000) and humans (Lengeler et al., 2000). Starvation stress has also been implicated as a key influence on fungal gene expression during growth of M. grisea within the host plants (Talbot et al., 1997). It has been proposed that a subset of signaling pathways that regulate fungal pathogenicity have been co-opted from those involved in nutrient sensing and subsequent fungal response (Alspaugh et al., 1997; Pellier et al., 2003).
Woronin bodies are septal pore–associated organelles that are unique to the filamentous ascomycetes (Woronin, 1864; Trinci and Collinge, 1974; Markham and Collinge, 1987; Jedd and Chua, 2000; Momany et al., 2002). The major Woronin body structural protein, Hexagonal peroxisome (Hex1p), self-assembles to form the vesicle's dense core (Jedd and Chua, 2000). Deletion of the HEX1 gene in Neurospora crassa results in cells that lack Woronin bodies, and mutant hyphae bleed cytoplasm through septal pores after cellular damage (Jedd and Chua, 2000; Tenney et al., 2000). In addition, a dense core is required for Woronin body function because a mutant Hex1p specifically disrupted in self-assembly produces a soluble Woronin body core, which is nonfunctional (Yuan et al., 2003). Hex1p also uses a consensus peroxisome targeting signal-1 (PTS-1) for vesicular localization, confirming that Woronin bodies are related to peroxisomes (Jedd and Chua, 2000). Together, these data show that Woronin bodies are specialized peroxisomes that function to seal the septal pores in response to cellular damage. However, it is also possible that Woronin bodies execute development functions associated with the multicellular growth characteristic of filamentous ascomycetes.
Here, we describe important cellular functions of Woronin bodies during the pathogenicity phase of M. grisea. We have investigated the role of Hex1 protein during the rice blast infection cycle and found that it functions as a virulence determinant based on molecular characterization of a mutant with lesion in the HEX1 locus. We show that loss of Hex1p in M. grisea leads to morphological and functional defects in appressoria and also delays host penetration and subsequently disrupts invasive hyphal growth in planta. The host environment and starvation stress, particularly for nitrogen, were found to be important regulators of HEX1 function. Further analyses revealed that Woronin bodies provide M. grisea with a distinct advantage in the colonization of plant tissues and to tide over nutrient-limiting conditions.
RESULTS
Isolation of HEX1-Deficient Mutant of M. grisea
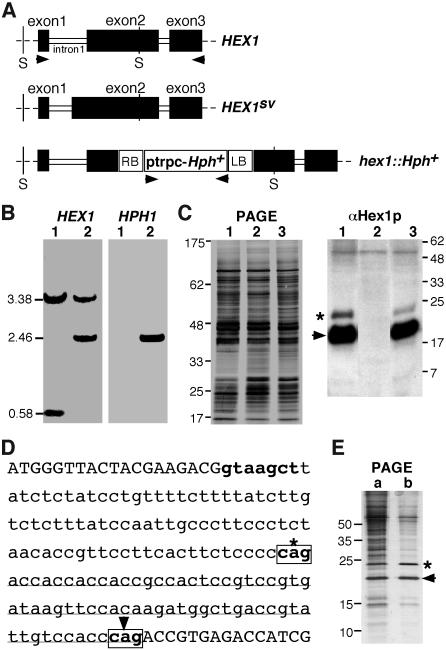
In an Agrobacterium tumefaciens T-DNA–mediated insertional mutagenesis screen designed to enrich for M. grisea mutants defective in pathogenicity, we identified an insertion mutant named TMV6 (see Methods for details) that produced highly misshapen appressoria. However, this mutation produced no change in mycelial or conidial architecture in TMV6. Identification and nucleotide sequence analysis of DNA sequences flanking the T-DNA insertion site in TMV6 showed that thisT-DNA disrupted MVP1, the HEX1 ortholog in M. grisea (GenBank accession number AY044846; hereafter referred to as HEX1). Figure 1A is a schematic representation of the T-DNA insertion at the HEX1 locus (hex1∷Hph) compared with the wild-type HEX1 locus. A splice variant arising from the HEX1 gene has been documented (Tenney et al., 2000) and is also schematically depicted in Figure 1A for comparison. DNA gel blot hybridization (Figure 1B, lanes 1 and 2, using HEX1 or HPH1 as probes) and protein gel blot analysis using αHex1p antibodies (Figure 1C; see Methods for details) confirmed that the single-copy insertion of HPH in TMV6 disrupted HEX1. As shown in Figure 1C, no protein product of the HEX1 gene was detected in the TMV6 strain. It is important to note that the aforementioned T-DNA insertion prevents expression of both forms of Hex1p: the longer 203–amino acid (Figure 1C, asterisk) protein as well as the more abundant 181–amino acid version (Figure 1C, arrowhead). We confirmed the two distinct splice variants arising from the wild-type HEX1 by identifying and sequencing the respective full-length cDNA clones to precisely establish the splice donor and acceptor sites (Figure 1D). Further and final confirmation was obtained by sucrose density gradient fractionation, partial purification of the Woronin-body core fraction, SDS-PAGE analysis, and subsequent Edman degradation of the two protein variants encoded by the HEX1 locus (Figure 1E). Under these conditions, the two forms of Hex1p (Figure 1E, b, asterisk and arrowhead) were found to be near equal in abundance, suggesting that the αHex1p antisera is less efficient in immunorecognition of the larger Hex1p variant (Figure 1C, asterisk). Thus, we conclude that an alternate splicing event in the first intron of HEX1 leads to two different mRNA species that are translated to give rise to two distinct Hex1p variants. Based on these data, the single-copy insertion in TMV6 represents a complete loss-of-function mutation in HEX1, and hereafter we refer to it as a hex1Δ strain.
Figure 1.
Cloning of T-DNA Insertion Locus from the TMV6 Mutant.
(A) Schematic representation of the HEX1 locus in the wild-type strain and the TMV6 mutant in M. grisea. Closed bars indicate the coding regions. HEX1SV depicts a splice variant arising in intron 1. RB and LB represent the right and left border sequences, respectively, of T-DNA (open box) integrated in the mutant strain.
(B) DNA gel blot analysis to confirm the HEX1 disruption. SalI-digested genomic DNA from the wild type (1) or TMV6 mutant (2) was probed with HEX1 or HPH1. Arrowheads in (A) encompass the lengths of the respective probes used for DNA gel blotting. The estimated sizes of relevant SalI (abbreviated as S in [A]) fragments in kilobase pairs are indicated at the left.
(C) TMV6 lacks Hex1 protein. Silver-stained profile of the SDS-PAGE analysis of lysates prepared from wild-type (1), TMV6 (2), and TMV6 mutants rescued with a single-copy HEX1 gene (3). Protein gel blot analysis of the respective lysates using antibody against the N. crassa Hex1p to detect the M. grisea homolog (arrowhead). Asterisk indicates the variant Hex1p arising from HEX1SV shown in (A). The nonspecific protein band (Mr 50 kD) detected by antisera against Hex1p serves as an additional loading control. Molecular mass markers in kilodaltons are indicated at the right.
(D) Molecular basis for generation of an alternate splicing event in the HEX1 coding sequence. Nucleotide sequence representing the junction of exon 1 and exon 2 of HEX1 gene. Intron 1 is shown in lowercase letters. Splice donor and acceptor sites (boxed) are depicted in boldface letters. Asterisk indicates the splice acceptor that appends an additional 22 codons (underlined) to exon 2 in the splice variant mRNA. Arrowhead indicates the alternate splice acceptor that leads to a longer intron 1.
(E) Silver-stained profile of the SDS-PAGE analysis of lysates representing a purification intermediate (a) and a partially purified fraction of the Woronin body core (b). Asterisk and arrowhead denote the respective Hex1p variants as depicted in (C).
Growth Characteristics of hex1Δ Strain
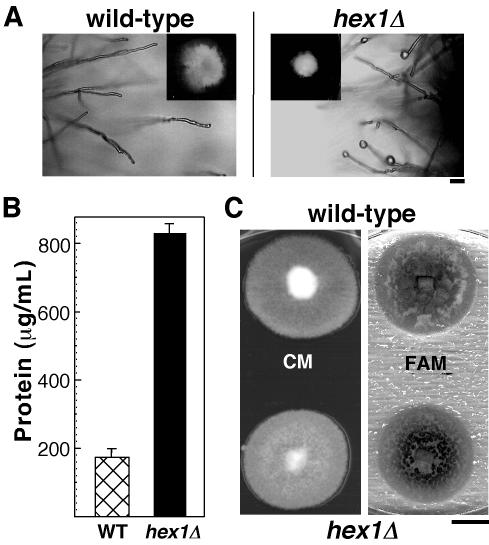
Earlier studies in N. crassa (Jedd and Chua, 2000; Tenney et al., 2000) showed that the Woronin body functions to seal the septal pore in response to cellular damage. We therefore analyzed the effects of similar cellular stress in the hex1Δ strain in M. grisea. The hex1Δ mutant showed poor and restricted growth on medium containing 2% sorbose (Figure 2A, inset), whereas the wild-type strain B157 showed a normal growth pattern under the same conditions. Moreover, mycelial tips of the hex1Δ strain displayed frequent cytoplasmic bleeding, characteristic of the lack of membrane resealing capability (Figure 2A). This observation was further supported by our finding that the hex1Δ mutant released more cytoplasm than the wild-type strain as judged by the amount of total protein quantified in the hypotonic exudates from the two strains (Figure 2B). Because Woronin bodies are derived from peroxisomes (Jedd and Chua, 2000), we checked whether there were any obvious defects in the hex1Δ mutant that were suggestive of deficiency in other peroxisome functions, such as β-oxidation of fatty acids. To this end, we assessed the growth of hex1Δ strain on medium supplemented with olive oil as the sole carbon source. Figure 2C shows that the mutant displayed comparable growth rate as the wild-type strain. However, a closer examination of the hex1Δ strain grown on fatty acid medium (FAM) revealed that branching and aerial hyphal growth were less profuse and restrictive in nature (data not shown). On complete medium with glucose as the carbon source, there was no significant difference in morphology or growth rate of hex1Δ as compared with the wild type (Figure 2C, CM). These results indicate that the major peroxisome function of metabolizing fatty acids seemed unaltered in the hex1Δ mutant, and the defects described above arise primarily as a consequence of the loss of Hex1p or Woronin bodies per se.
Figure 2.
Growth Characteristics and Analysis of hex1Δ Strain.
(A) Cytoplasmic bleeding in the hex1Δ mutant after growth for 3 d in the sorbose medium. Droplets of cytoplasm are seen at hyphal tips of the hex1Δ strain but not in wild-type hyphae. Insets show the colony morphologies on the sorbose medium. Bar = 20 μm.
(B) Hypotonic shock–induced release of cytoplasm in the wild type and the hex1Δ mutant. Values indicate the total amount of protein released per milliliter of culture filtrate by each strain. Mean values (± sd) of three independent quantitations are shown.
(C) Utilization of fatty acids is unaffected in the hex1Δ strain. The wild type and the hex1Δ mutant were grown on complete medium (CM) or FAM for 6 d. Bar = 1 cm.
Appressorial Defects in hex1Δ Mutant
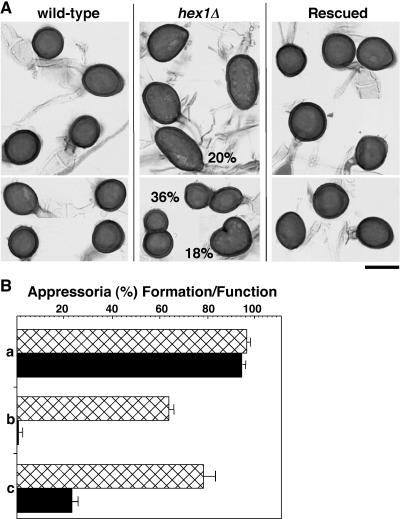
The hex1Δ strain showed no obvious anomalies in vegetative growth (Figure 2C) or in sexual mating. Asexual spores or conidia produced by the hex1Δ mutant were normal in morphology and germination; however, upon germination, these conidia produced aberrant appressoria. Figure 3A shows that hex1Δ appressoria exhibit a wide range of shapes, from oblong and elongated to kidney shaped and occasionally even bilobed. These distorted appressoria were not defective in melanization. However, results obtained using onion epidermis assays revealed that whereas there was no discernable difference in the ability of conidia from the wild type and hex1Δ mutant to germinate and form appressoria (Figure 3B, a), only a small percentage of hex1Δ appressoria (∼1.5%) could produce penetration pegs after 24 h (Figure 3B, b). The ability to form penetration pegs in the wild type was ∼62% at this time point. Forty-eight hours postinoculation, ∼22% appressoria of the hex1Δ mutant showed host infiltration (Figure 3B, c). By contrast, ∼78% of the wild-type appressoria achieved penetration and produced infectious hyphae within the host (Figure 3B, c) after 48 h. Thus, the majority of appressoria produced by the hex1Δ mutant were defective in morphology and showed diminished host penetration.
Figure 3.
Appressorium Formation and Host Penetration Defects in hex1Δ Strain.
(A) Morphological defects in appressoria of the hex1Δ mutant. Conidia from the wild type, hex1Δ mutant, and a hex1Δ strain complemented with a full-length HEX1 (rescued) were allowed to germinate on onion epidermal strips and resultant melanized appressoria stained with acid fuchsin and assessed for morphological aberrations. Overall occurrence of a particular defect is indicated as a percentage value for the mutant appressoria. Bar = 10 μm.
(B) hex1Δ appressoria are reduced in host penetration. (a) Equal number of conidia were assessed for their appressorium formation capabilities in the wild type (hatched bar) or the hex1Δ mutant (closed bar). (b) Appressoria from the wild type (hatched) or hex1Δ strain (closed) were tested for their potential to penetrate the host cells and produce penetration hyphae 24 h postinoculation. (c) Penetration and infectivity assessed 48 h postinoculation in the wild type (hatched) or hex1Δ strain (closed). Mean values (± sd) presented as percentage points were calculated from three independent assessments.
hex1Δ Mutant Is Reduced in Pathogenicity
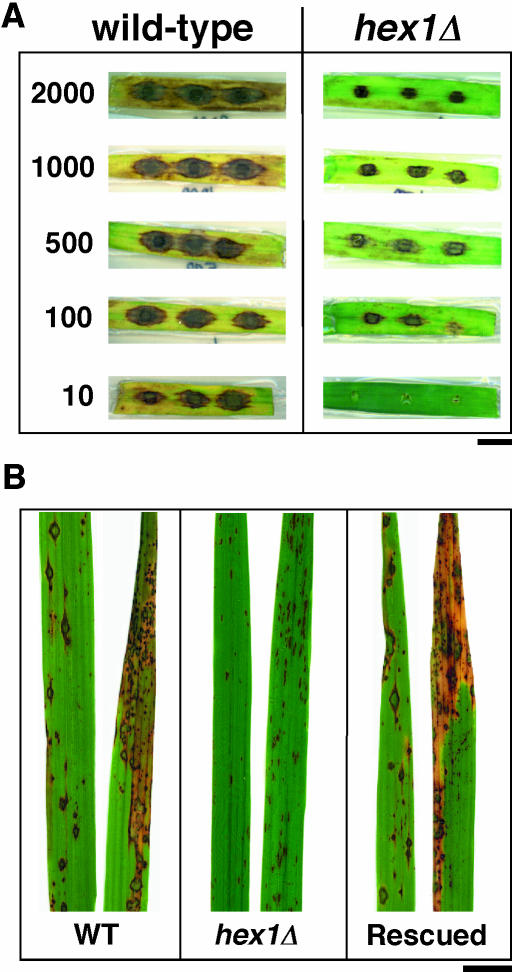
We tested the pathogenicity of the hex1Δ mutant on two different hosts. Barley leaves (variety Express) seeded with various dilutions of the wild-type conidial suspensions developed typical blast symptoms, showing spindle-shaped lesions with gray centers (Figure 4A, wild type). Under similar conditions, the hex1Δ mutant conidia failed to cause proper lesions on host leaf explants, with the difference being apparent at all dilutions tested (Figure 4A, hex1Δ). In another experiment, seedlings of the rice strain CO39 were sprayed with conidia of wild-type and hex1Δ mutant in parallel, and the resulting lesions were analyzed after 7 d. Figure 4B shows that whereas wild-type strain caused typical spindle-like, gray centered, and severe blast lesions that merged into one another on the inoculated rice leaves, the hex1Δ mutant failed to infect the host efficiently and showed marked decrease in virulence (Figure 4B). The lesions caused by the hex1Δ mutant were brown, nonconidiating, minute, and reminiscent of hypersensitive reactions elaborated normally by resistant host plants. Furthermore, the lesions caused by the mutant also failed to expand and coalesce (Figure 4B). Based on the marked reduction in lesion number, size, and quality, we conclude that the hex1Δ mutant shows a high reduction in its virulence capacity and is unable to infect and colonize the two different host species (barley and rice) tested.
Figure 4.
Infection Assays on Barley and Rice Leaves.
(A) Leaf explants from barley were inoculated with the indicated number of conidia from the wild type or hex1Δ strain, and disease symptoms assessed after 10 d. Bar = 1 cm.
(B) Seedlings from rice cv CO39 were spray inoculated individually with conidia from the wild type, hex1Δ mutant, or hex1Δ strain complemented by the introduction of HEX1 (rescued). Lesion formation on leaves was documented 10 d postinoculation. Bar = 1 cm.
Behavior of Mutant Infection Hyphae in Planta
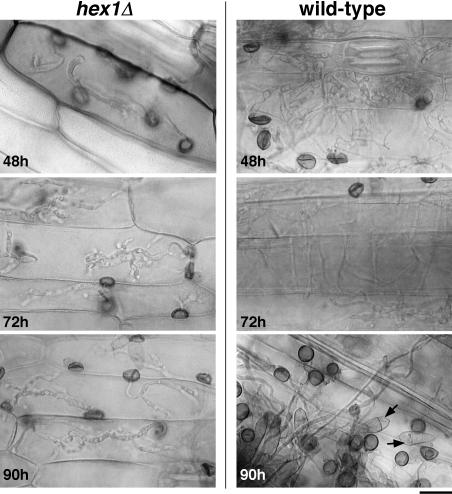
To understand the cause underlying the failure of mutant infection hyphae to colonize host plants, we performed a detailed microscopic analysis of the infection hyphae produced by the hex1Δ mutant and wild-type appressoria at different time periods after penetration (see Methods for further details). As shown in Figure 5 (hex1Δ, 48 h), the mutant infection hyphae were delayed in their in planta growth and colonization of the neighboring cells or host tissue layers, with all the primary infection hyphae being restricted to the cellular compartment they initially penetrated. The failure to advance the infection stage persisted 3 d postinoculation (Figure 5, hex1Δ, 90 h). By contrast, wild-type infection hyphae achieved cross-wall penetration and spread within 48 h postinoculation and produced secondary infectious hyphae and resultant conidia toward the end of the 90 h time point (Figure 5, wild type, 90 h). We therefore conclude that the hex1Δ mutant lesions are restricted in quality and quantity primarily because of the delayed and restricted in planta growth of the mutant strain and its inability to penetrate cross-walls within host plant tissue. As a result of these anomalies, the hex1Δ mutant is unable to expand the disease lesions and elaborate normal blast disease symptoms.
Figure 5.
Infection Hyphae of the hex1Δ Mutant Are Unable to Spread in the Host Plant.
Conidia from hex1Δ strain (left) or the wild type (right) were inoculated on barley leaf explants and infection hyphae produced by the resultant appressoria visualized at the indicated time points postinoculation. Arrows indicate the in planta secondary conidiation cycle of wild-type fungal strain. Bar = 20 μm.
Subcellular Localization of Woronin Bodies and Hex1p
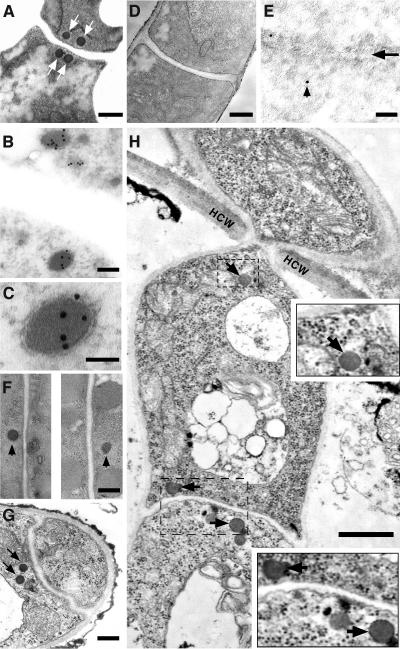
Thin-section electron microscopy was used to locate the dense-core Woronin bodies within various cell types in M. grisea. To determine the subcellular localization of Hex1 protein, we used immunoelectron microscopy (IEM) and the aforementioned αHex1p antibody. Woronin bodies were found to be present at the sites of septation (arrows in Figures 6A and 6B) in wild-type mycelia at prototypal numbers of two to three pairs per septum. As judged by IEM, Hex1p was found to localize specifically to the Woronin bodies and was seen uniformly distributed within the matrix of these organelles (Figures 6B and 6C). Similar electron microscopy (EM) and IEM analyses further revealed that Woronin bodies were absent in the hex1Δ mutant (Figures 6D and 6E). Ultrastructural analyses and subsequent comparisons with mycelial sections revealed that Woronin bodies were not as abundant in conidia (Figure 6F; see Discussion), and mostly a single Woronin body was observed adjacent to a conidial septum. However, the tips of the germ tubes showed the typical occurrence of a pair of Woronin bodies adjacent to the appressorial septa albeit only on the germ tube side (Figure 6G). In comparison with the prototypical number of Woronin bodies observed around the hyphal septa, the incidence of Woronin bodies in appressoria and penetration pegs was found to be extremely poor (data not shown; Figure 7C, wild type). We further attempted to identify Woronin bodies within in planta structures elaborated by M. grisea. In contrast with relatively low abundance of Woronin bodies in conidia, appressoria, and infection pegs (in comparison with mycelia), the primary and secondary infectious hyphae showed the typical incidence of Woronin bodies at the regions of septation and at the zone of host cross-wall invasions (Figure 6H, arrows and insets). This led us to conclude that the Hex1 protein in M. grisea localizes to the Woronin body matrix and that Woronin bodies (or the resident Hex1p) are present at sites that govern septation and/or invasive growth of M. grisea in host tissues.
Figure 6.
Subcellular Localization of Woronin Body and Hex1p by EM.
(A) Transmission electron micrograph showing a near-median section through the septal region of a mycelium in M. grisea. Arrows indicate the dense Woronin bodies on either side of the septum. Bar = 250 nm.
(B) and (C) Hex1p localizes to the core of the Woronin bodies (B). Vegetative mycelia were fixed and probed with affinity-purified αHex1p antisera followed by 10-nm gold particles. Bar = 100 nm. A magnified view of same is shown in (C), where the bar = 50 nm.
(D) Loss of Hex1p leads to complete loss of Woronin bodies. TEM section through a hex1Δ hypha depicting a septum. Bar = 250 nm.
(E) IEM analysis as detailed in (B) of a hex1Δ mycelium. Arrowhead indicates nonspecific staining in the septal region indicated with an arrow. Bar = 100 nm.
(F) Woronin bodies in conidiospores. TEM of conidium depicting solitary Woronin body (marked with arrowheads) associated with individual septa. Bar = 250 nm.
(G) TEM section through the tip of a germ tube and an appressorium initial. Arrows indicate a pair of Woronin bodies observed in the germ tube. Bar = 250 nm.
(H) TEM showing a section along the length of an intracellular infection hypha of wild-type M. grisea in host cells 48 h after penetration. Arrows indicate the Woronin bodies present therein. HCW, host cell wall; V, vacoule. Inset panels depict a 2× magnified view of the boxed region. Bar = 500 nm.
Figure 7.
Role and Regulation of Hex1p during M. grisea Pathogenesis.
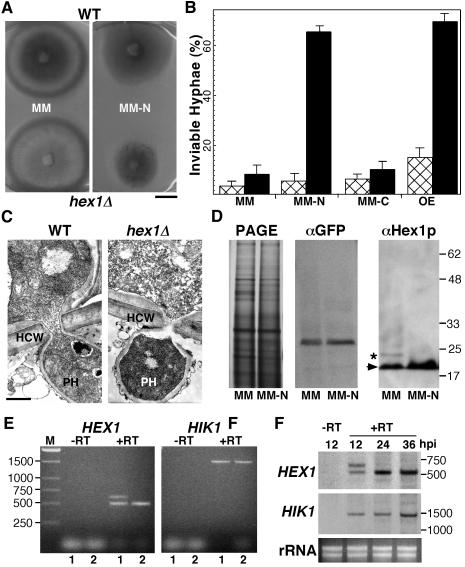
(A) Growth characteristics of hex1Δ mutant under nitrogen-deficient conditions. The wild type and the hex1Δ mutant were grown on minimal medium with abundant nitrogen (MM) or limiting nitrogen source (MM-N) for 1 week, and resultant colonies were photographed. Bar = 1 cm.
(B) Hex1p is necessary for survival in nitrogen-deficient environment and under in planta conditions. Wild-type strain (hatched bar) and the hex1Δ mutant (closed bar) were cultured on MM, MM-N, or MM-C, and inviable hyphae (positive for Phloxine B staining) were quantitated under respective conditions. OE refers to infection assays performed on onion epidermal strips with conidia from the respective strains and viability of the resultant penetration pegs and infectious hyphae estimated. Mean values (± sd) observed in three independent assessments are represented.
(C) Transmission electron micrographs depicting near-median sections through the appressorial penetration pegs and primary infectious hypha of a wild-type strain and a hex1Δ mutant in a barley leaf explant. Bar = 500 nm for each panel. HCW, host cell wall; PH, primary infectious hypha.
(D) Nitrogen starvation stress positively regulates Hex1p. Protein gel blot analyses of lysates prepared from wild-type strain (expressing GFP-PTS1 constitutively) grown in MM or MM-N, using αGFP (as control) or αHex1p to detect the M. grisea Hex1p (arrowhead). Asterisk indicates the variant Hex1p arising from HEX1SV as shown in Figure 1A. Molecular mass markers in kilodaltons are indicated at the right and are applicable to the three panels. PAGE refers to the silver-stained profile of the respective protein samples used for the protein gel blot.
(E) Differential regulation of HEX1 transcripts. Ethidium bromide–stained profile of RT-PCR products amplified from total RNA extracted from wild-type strain grown under MM (1) or MM-N (2) conditions using HEX1- or HIK1-specific primers. Negative control (−RT) refers to the RNA sample being processed without a reverse transcriptase step before the PCR amplification.
(F) Differential regulation of HEX1 in planta. Total RNA extracted from barley leaf explants infected with wild-type conidia (∼104 per set) for the indicated hours postinoculation (hpi) was subjected to RT-PCR analyses as in (E) followed by DNA gel blot analysis using HEX1- or HIK1-specific probes to detect the respective amplification products. −RT refers to the negative control as discussed earlier. Ethidium bromide–stained profile of total RNA (rRNA) is depicted in the bottom panel. Molecular mass standards are represented in base pairs.
Rescue of the hex1Δ Mutant
We transformed plasmid pFGL118 (containing the full-length HEX1 gene) or pBarKS (control) into the hex1Δ strain. Among 15 bialaphos-resistant transformants screened by DNA gel blot analysis, we identified two strains that carried single-copy integration of the HEX1 gene at an ectopic site (data not shown). Hex1p expression in one of these strains was confirmed by protein gel blotting (Figure 1C, lane 3) and was found to be equivalent to wild-type levels. The complemented hex1Δ transformants were then analyzed for the rescue of the various morphological defects seen in the hex1Δ mutant. The appressorial defects seen in the hex1Δ strain were completely suppressed with the introduction of the wild-type HEX1 gene (Figure 3A, rescued), which also restored its ability to proliferate on sorbose-containing growth medium. By contrast, the vector control could not suppress these abnormalities (data not shown). The complemented strain was found to be as virulent as the wild-type isolate when spray inoculated on rice seedlings (Figure 4B, rescued). Thus, the appressorial, host penetration, and lesion development defects in the hex1Δ mutant could be completely restored by reintroduction of the wild-type HEX1 allele. These results show that the phenotypic and morphological changes observed in hex1Δ strain resulted from the disruption of Hex1p function in this mutant.
Regulation and Role of Hex1p in M. grisea
Our observation that hex1Δ infection hyphae are restricted in growth and incapable of advancing blast disease symptoms raised a possibility that these mutant infection hyphae were under some stress that presumably leads to cytolysis and premature death because of failure in septal plugging and membrane resealing.
Earlier studies have presented evidence that nutritional starvation, particularly nitrogen-limiting condition, is one of the key environmental factors that influences M. grisea growth in rice leaves (Lau and Hamer, 1996; Talbot et al., 1997). We therefore tested the growth characteristics of the hex1Δ mutant under conditions of nutrient deprivation (see Methods for details). Contrary to the wild-type strain, the hex1Δ mutant showed severely restricted growth on minimal medium that contained limiting amounts of nitrogen source (Figure 7A). However, under conditions of carbon limitation, hex1Δ mutant showed growth rates and proliferation comparable to the wild type (data not shown). To assess the viability of mutant hyphae, we stained the minimal medium (MM) and medium with reduced nitrogen (MM-N) or carbon (MM-C) grown cultures with Phloxine B, which accumulates in dead cells. As shown in Figure 7B, the mycelia produced by the hex1Δ strain exhibit an increased incidence of hyphal death (Phloxine B positive hyphae) as compared with the wild-type strain when tested under conditions of nitrogen starvation. The occurrence of hyphal death under nitrogen-abundant (and carbon limiting, MM-C) conditions was found to be equivalent between the wild type and the hex1Δ strain (Figure 7B). These results proved that Woronin body function is crucial to tide over starvation stress induced by nitrogen-deficient environment and that absence of Hex1p (or Woronin bodies) leads to increased cell death in hyphae under such stress conditions.
To assess the viability of primary infectious hyphae in planta, we stained onion epidermal strips infected with hex1Δ mutant and the wild type (48 h postinfection) with Phloxine B. The percentage of Phloxine B-positive infectious hyphae was found to be ∼4.5-fold higher in the hex1Δ mutant as compared with the wild-type strain (Figure 7B). We conclude that the in planta infection pegs and primary infection hyphae generated by the mutant appressoria in hex1Δ strain were inviable likely because of loss of a septal plugging mechanism that appears to be important also for the survival of the fungus under nitrogen-deficient conditions. We then performed EM examination to assess the subcellular differences (if any) between the primary infectious hyphal growth of the mutant and the wild-type strains in the host. As illustrated in Figure 7C, a striking difference was that wild-type penetration pegs showed an even apportioning of ribosomes during the infection phase, whereas the ribosomal distribution within the cytoplasm of mutant penetration pegs was found to be uneven. Taken together, these data indicated that Hex1p/Woronin body function is essential for survival of the blast fungus in the nitrogen-deficient milieu of the host plant.
We next investigated whether starvation stress, particularly for nitrogen, also regulates Hex1p directly. To address this question, we tested the levels of Hex1 protein in total cell lysates derived from a wild-type strain expressing green fluorescent protein fused to a PTS-1 tripeptide (GFP-SRL, under the constitutive P2 promoter; M. Ramos-Pamploña and N.I. Naqvi, unpublished results) grown in parallel under conditions of nitrogen abundance and nitrogen limitation. Equal amounts of total proteins as ascertained by spectrophotometric estimations were analyzed for these experiments. Protein gel blotting with αGFP antibody further confirmed this estimation (Figure 7D, αGFP). Figure 7D shows that the level of Hex1p is upregulated (∼2.4-fold, based on densitometric analysis using Bio-Rad Quantity 1 software) in cultures starved for nitrogen. Interestingly, the larger form of Hex1p was found to be absent under stress conditions imposed by such nutrient starvation.
We therefore investigated the role of transcription and splicing in such a differential expression of HEX1. As shown in Figure 7E, RT-PCR amplification using HEX1-specific primers revealed that the larger HEX1 mRNA (expected 609-bp cDNA) was absent in the wild-type strain cultured under conditions of nitrogen deficiency, whereas both the HEX1 splice variants (609- and 543-bp cDNAs) could be easily amplified when the RNA was derived from wild-type strain grown in the presence of nitrogen. The HIK1 (encoding a histidine kinase function) derived cDNA amplified from the above mentioned conditions served as a control for total RNA used in the RT-PCR reactions (Figure 7E, HIK1). Thus, under nutritional-starvation conditions, HEX1 expression is controlled at the level of mRNA splicing. We then proceeded to assess whether this splicing-dependent regulation of HEX1 occurred in vivo. Because the amount of fungal material in such an experiment was in meager amounts, we resorted to DNA gel blot analysis to detect the respective cDNA moeities upon RT-PCR amplification. Figure 7F depicts that whereas both the splice variants arising from HEX1 locus could be observed up to 12 h postinoculation on barley, the HEX1SV was entirely absent 24 h postinoculation. Moreover, HEX1 expression was upregulated (∼2.8-fold) after 24 h in the host. HIK1 mRNA levels remained largely unchanged during this period (Figure 7F). Taken together, this lead us to conclude that a unique mechanism dependent on mRNA splicing regulates enrichment of one of the variants of Hex1 protein during nutritional starvation stress as well as under growth environment encountered by M. grisea during its infection-related development in the host.
DISCUSSION
Occurrence and Distribution of Woronin Bodies in M. grisea
The Woronin body is a dense-core vesicle unique to filamentous fungi belonging to the class Ascomycetes (Woronin, 1864; Trinci and Collinge, 1974; Markham and Collinge, 1987). The peroxisomal origin of the Woronin body and its major function of maintaining cellular integrity after cell lysis have been well established (Jedd and Chua, 2000; Tenney et al., 2000). Ultrastructural analyses have confirmed the presence of Woronin bodies during the vegetative phase and infection stage of several fungal pathogens (McKeen, 1971; Bourett and Howard, 1989; Wharton et al., 2001). We have shown here that Woronin bodies in the rice blast fungus M. grisea are present adjacent to septa in mycelia, germ tubes, and infection hyphae. Woronin bodies are also found at the invasion zone in fungal infection hyphae within the host plant, but these organelles were relatively less abundant in conidia, appressoria, and penetration pegs. Unoccluded septal pores and a low frequency of Woronin bodies associated with conidial septa have been previously documented inM. grisea (Money and Howard, 1996). Our current hypothesis is that septal plugging is not required in conidia (to allow bulk cytoplasmic streaming via septal pore) and mature appressoria (to allow release of turgor and host penetration), and these cell types therefore contain relatively few Woronin bodies. Our analysis of Woronin body distribution in the aforementioned cell types represents an estimate based on typical number of Woronin bodies (two to three pairs) associated with a mycelial septum. Comprehensive serial sectioning as detailed by Momany et al. (2002) is necessary, however, to determine the exact number of Woronin bodies in these infection-specific fungal cell types.
Function of Woronin Bodies in M. grisea
Using a loss-of-function mutation in the HEX1 locus, we have uncovered an important and essential role for Woronin bodies in the pathogenesis cycle of the rice blast fungus M. grisea. During the vegetative growth phase, a hex1Δ mutant showed normal mycelial growth, conidiation, and mating behavior. The mutant mycelia differentiated conidia similar to the wild type, but the resultant appressoria showed severe morphological defects and a delayed host penetration response. The phenotypic and cytological analysis of hex1Δ mutant revealed major defects in infection-related morphogenic events: first, only 22% of the total appressoria were capable of gaining entry into the host tissue, and entry was delayed. Second, the infectious hyphae produced were incapable of invasive growth and remained confined to the site of entry into the host. Such mutant infectious hyphae lacking HEX1 function did not survive the host environment. As a result of these anomalies, the hex1Δ strain shows highly diminished virulence toward rice and barley. By complementation analysis of hex1Δ with a wild-type HEX1 gene, we ascertained that the phenotypic defects observed in the mutant were a direct consequence of inactivation of this gene. Furthermore, introduction of the mutation/disruption (as reported in hex1Δ mutant here) by homology-dependent recombination in a different wild-type M. grisea strain Guy11 lead to identical phenotypic defects associated with appressorial morphology and reduced virulence (data not shown).
Several reports on Woronin bodies from other fungi have established presence of these organelles in nonseptal regions, such as the tips of the germlings or at the cell periphery (Markham and Collinge, 1987; Lim et al., 2001; Momany et al., 2002). We have identified Woronin bodies at the tips of secondary infectious hyphae that are involved in penetrating the cross-walls within the host tissue. Such Woronin bodies showed no association with the hyphal septum. Further work is needed to determine the exact function of Woronin bodies in subcellular compartments other than medial septa. An inability to cope with nitrogen starvation stress upon loss of Woronin bodies turned out to be an important and novel finding from our characterization of the hex1Δ mutant. Surprisingly, limiting the amount of carbon source did not elicit the same effect in the mutant. This aspect of Woronin body function will form the basis of our future investigations as well.
Regulation of Hex1p in M. grisea
Using IEM, we have demonstrated that Hex1p is present in the lumen of M. grisea Woronin bodies and that strains lacking Hex1p are unable to occlude septal pores in response to cellular damage. Our data further suggest that the major peroxisomal function of β-oxidation of fatty acids is unaltered in Hex1-deficient cells, and this is consistent with similar results obtained in N. crassa (Jedd and Chua, 2000). Thus, Woronin bodies and peroxisomes probably execute distinct functions in filamentous asomycetes. Our studies on regulation of Hex1p revealed that in planta environment as well as stress conditions, such as nitrogen deprivation, upregulate the production of Hex1p. This adds HEX1 to a growing list of fungal genes induced during infection of the host and during nitrogen starvation (Snoeijers et al., 2000). During the course of this study, we confirmed the existence of a splice variant of the HEX1 coding sequence resulting in a Hex1 protein of 203 amino acid residues and a shorter form of Hex1p (181 amino acid residues). Interestingly, only the higher mobility form of Hex1 polypeptide accumulates during the stimulatory conditions described above. Our data supports the possibility that such a differential regulation of HEX1 is controlled at the level of transcription and splicing (Figures 1 and 7).
The regulatory proteins Nut1, Npr1, and Npr2 have been reported to govern nitrogen metabolism and/or pathogenicity in M. grisea (Froeliger and Carpenter, 1996; Lau and Hamer, 1996). Molecular identities of NPR1 and NPR2 remain elusive (Lau and Hamer, 1996); however, NUT1 has been shown to encode a transcription factor that regulates nitrogen source utilization (Froeliger and Carpenter, 1996; Talbot et al., 1997). The upstream regulatory region of HEX1 does not contain any GATA motif–related consensus sequence (TATCTM; M = C/A) that has been predicted and subsequently shown to bind Nut1-like transcription factors (Froeliger and Carpenter, 1996; Soanes et al., 2002). Thus, the increased expression of Hex1p under nitrogen starvation stress may be controlled by some other factor(s) specific to later stages of infection.
Future investigations will define the exact role of Woronin body/Hex1p under starvation stress condition and during the rice blast infection cycle. Such studies should reveal novel mechanism(s) governing fungal development during critical stages of disease establishment and invasiveness. Our findings could also be extrapolated to other fungal pathogens that harbor Woronin bodies to help understand the role of these highly specialized vesicles in protecting synctitial fungi from cellular damage within their plant hosts.
METHODS
Fungal Strains and Growth Conditions
Magnaporthe grisea wild-type strain B157 was a kind gift from the Directorate of Rice Research (Hyderabad, India). B157 and the hex1Δ strain were cultured on prune agar medium (per liter: 40 mL of prune juice, 5 g of lactose, 1 g of yeast extract, and 20 g of agar) or complete medium (CM; 0.6% yeast extract, 0.6% casein hydrolysate, and 1% sucrose) at 28°C. FAM contained 1.6% yeast nitrogen base without amino acids, 1% ammonium nitrate, and 1% olive oil, pH 6.0, adjusted with sodium phosphate. Conidiation was induced in the prune agar medium–grown cultures by incubation under constant fluorescent light for 4 d. Mycelia collected from 2-d-old CM-grown cultures were used for the isolation of DNA and spheroplasts. Genetic crosses and random ascospore isolation methods were as described previously (Xu and Hamer 1996; Balhadère et al., 1999). The composition of MM, MM-N (used for nitrogen starvation), and MM-C were as reported earlier (Talbot et al., 1993). Typically, the fungal strains were cultured for a week on solid MM or MM-N or MM-C media to assess the growth and colony characteristics. Mycelia used for total protein extractions was obtained by growing the relevant strains in liquid CM, MM, or MM-N for 3 d at 28°C.
Appressorial Assays and Pathogenicity Tests
Conidia harvested from 10-d-old mycelial cultures were filtered through two layers of Miracloth (Calbiochem, San Diego, CA) and resuspended to 105 conidia per milliliter in sterile distilled water. Droplets of conidial suspensions (20 to 50 μL) were placed on plastic cover slips and incubated under humid conditions at room temperature. Microscopic observations were made after 3, 6, 12, 24, and 36 h.
Host penetration by the wild-type or mutant appressoria were assayed on epidermal strips from onion (a nonhost) usually 24 and 48 h postinoculation or on leaf explants from host barley, usually 24, 48, 72, and 90 h after conidial application (Chida and Sisler, 1987; Balhadère et al., 1999). Appressoria in these assays were stained by acid fuchsin as described (Balhadère et al., 1999). For pathogenicity tests, established protocols (Naqvi et al., 1995; Xu et al., 1997) were followed with slight modifications. Rice (Oryza sativa cv CO39) or barley (Hordeum vulgare cv Express) were used for these assays.
Nucleic Acid and Protein-Related Methodologies
Standard protocols (Sambrook et al., 1989) were followed for molecular manipulations and gel blot analyses of DNA and RNA. Fungal DNA was obtained using the potassium acetate method standardized for rice genomic DNA extraction (Naqvi et al., 1995). Plasmid DNA was isolated using the Qiagen plasmid preparation kits (Qiagen, Valencia, CA) and nucleotide sequencing performed using the ABI Prism big dye terminator method (PE-Applied Biosystems, Foster City, CA). Homology searches of DNA/protein sequences were performed with the BLAST program (Altschul et al., 1997). GeneWise-based predictions were performed using the public domain database of the European Bioinformatics Institute (http://www.ebi.ac.uk/Wise2/index.html). Total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA), and cDNA synthesis and subsequent PCR amplification was conducted using the Superscript kit (Life Technologies, Rockville, MD). The following primers were used for cDNA synthesis and subsequent amplification: OligodT (5′-TTTTTTTTTTTTTTTTTTTT-3′), Hex1F (5′-ATGGGTTACTACGAAGACG-3′), and Hik1F (5′-ATCTTCGACACATTCCAGC AG-3′; GenBank accession number AB041647). The HIK1 gene encodes a two-component histidine kinase function that acts as a virulence factor in M. grisea (N. Naqvi, unpublished results). For every cDNA reaction set, one RNA sample was processed without reverse transcriptase to provide a negative control in subsequent PCR reactions.
Protein purification, partial purification of Woronin body core, and immunoblotting procedures were as detailed previously (Jedd and Chua, 2000). The affinity-purified αHex1p antibodies specific to the N. crassa Hex1p that were shown to cross-react with the Hex1 protein from M. grisea (Jedd and Chua, 2000) were used at a 1:1000 dilution. Anti-GFP antibodies were purchased from Amersham Biosciences (Piscataway, NJ) and used at 1:1000 dilution for protein gel blot analysis. Protein concentrations were determined according to Bradford (1976) using a commercial kit (Bio-Rad Laboratories, Hercules, CA). The enhanced chemiluminescent method (ECL kit; Amersham Biosciences, Freiburg, Germany) was used for developing DNA and protein gel blots.
Isolation of the HEX1-Minus Mutant
Agrobacterium tumefaciens T-DNA–mediated random insertional mutagenesis was performed in M. grisea (L. Hao, S. Soundararajan, and N.I. Naqvi, unpublished results) using hygromycin resistance (encoded by hygromycin phosphotransferase gene HPH) as a selectable marker. T-DNA insert copy number was analyzed by DNA gel blot hybridization using standard procedures (Sambrook et al., 1989). Mutant strain(s) of interest were purified through monoconidial isolations and by random-ascospore analysis or tetrad dissection after a genetic cross. TMT201 was obtained in the above screen as a single-copy insertional mutant that produced aberrant appressoria and showed diminished host penetration. A monoconidial isolate from TMT201 was subjected to two rounds of purification and then used in a backcross with Mat1-1 strain (field isolate, MAT1-1, rice and barley pathogen). Results obtained for progeny testing from seven complete tetrads from this cross showed that hygromycin resistance segregated 4:4 in these tetrads. Progeny from three complete tetrads was tested for hygromycin resistance and defects in appressorium morphology and function (host penetration) in cosegregation analyses. Comparison of χ2 distribution (0, 0.1, and 0.1, respectively) at the expected 1:1 ratio supported single gene inheritance for the three phenotypes tested. TMV3 and TMV6 were two such hygromycin-resistant F1 progeny that produced aberrant appressoria and showed reduced pathogenicity and were considered for future characterizations. In parallel, a random ascospore analysis was performed on progeny obtained from a TMT201 × Mat1-1 cross to determine the inheritance of related mutant phenotypic defects. χ2 analysis here further confirmed the results obtained through tetrad analysis.
Flanking DNA sequences (right and left T-DNA border flanks) for the insertion in TMV6 were identified using a standard thermal asymmetric interlaced PCR method (Liu et al., 1995) and subsequently confirmed by nucleotide sequencing.
Rescue of the hex1Δ Mutant
The M. grisea HEX1 coding region and its 2-kb upstream sequence were amplified with primers NIN359 (5′-GAGAGTGAGGATCCCCGTTGCAGACCAGACCA-3′) and NIN59 (5′-GAGAGTGAGAATTCCTCAAGGCATAGAGTATC-3′) and cloned between the BamHI and EcoRI sites in pBarKS (Pall and Brunelli, 1993) to obtain pFGL118. Nucleotide sequencing was used to confirm the PCR-amplified fragments. Resistance to bialaphos or ammonium gluphosinate (Cluzeau Labo, Saint Foy La Grande, France) was used as a fungal selectable marker during hex1Δ transfection with NotI-linearized pFGL118 (or pBarKS as control), and DNA gel blot analysis was performed to confirm successful single-copy genomic integration.
Electron and Light Microscopy
At intervals after fungal inoculation, pieces of leaf sheath tissue (∼4 × 4 mm) were excised from beneath the inoculation sites and placed immediately in glutaraldehyde (2.5%, v/v) in 0.01 M phosphate buffer, pH 6.8. Tissue was fixed for 6 h, including a 15-min vacuum infiltration, washed in buffer (3 × 10 min), and postfixed in osmium tetraoxide (1%,w/v) for 3 h. After washing in buffer (3 × 10 min), tissue was dehydrated in a graded ethanol series, rinsed with propylene oxide, and embedded in Spurr's epoxy resin within flat molds. Ultrathin sections through infection sites (identified by light microscopy) were cut on a Jung Reichert microtome (Leica Mikroskopie and Systeme, Wetzlar, Germany), collected on formvar-coated nickel grids, stained with uranyl acetate and lead citrate, and examined using a JEM1010 transmission electron microscope (Jeol, Tokyo, Japan) at 100 kV. Details pertaining to EM observation of Woronin bodies have been published (Jedd and Chua, 2000). Estimation of Woronin body numbers was performed on ∼15 to 20 sections for each cell type. Light microscopic examination was performed using a Leica DMLB microscope with appropriate filters. Images were captured with an Optronics DEI-750T cooled CCD camera (Muskogee, OK) and Leica Qwin software. Images were processed with Adobe Photoshop 5.5 (Mountain View, CA) and assembled using Canvas 5.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY044846 and AB041647.
Acknowledgments
We thank A. Suresh for blast infection assays and Y. Chan, Q. Lin, and H. Shio for excellent EM support. We are grateful to H. Bohnert andM-H. Lebrun for their help in setting up the barley infection assays. We thank members of Fungal Genomics, Cell Division, and Cell Dynamics Laboratories at Temasek Life Sciences Laboratory for helpful discussions, suggestions, and/or critical comments on the manuscript. Continued support and encouragement from M. Balasubramanian and S. Naqvi are gratefully acknowledged. This work was supported by intramural research funds from Temasek Life Sciences Laboratory to N.I.N. and by grants from the National Science Foundation to G.J. and N.H.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Naweed I. Naqvi (naweed@tll.org.sg).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020677.
References
- Alspaugh, J.A., Perfect, J.R., and Heitman, J. (1997). Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11, 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Shaffer, A.A., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhadère, P., Foster, A., and Talbot, N.J. (1999). Identification of pathogenicity mutants of the rice blast fungus Magnaporthe grisea by insertional mutagenesis. Mol. Plant-Microbe Interact. 12, 129–142. [Google Scholar]
- Bourett, T.M., and Howard, R.J. (1989). In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68, 329–342. [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chida, T., and Sisler, H.D. (1987). Restoration of appressorial penetration ability by melanin precursors in Pyricularia oryzae treated with anti-penetrants and in melanin-deficient mutants. J. Pestic. Sci. 12, 49–55. [Google Scholar]
- Dean, R.A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. [DOI] [PubMed] [Google Scholar]
- Doke, N. (1983). Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with the incompatible race of Phytophthora infestans and to hyphal cell wall components. Physiol. Plant Pathol. 35, 211–234. [Google Scholar]
- Froeliger, E.H., and Carpenter, B.E. (1996). NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol. Gen. Genet. 251, 647–656. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd, G., and Chua, N.H. (2000). A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2, 226–231. [DOI] [PubMed] [Google Scholar]
- Lau, G.W., and Hamer, J.E. (1996). Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell 8, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler, K.B., Davidson, R.C., D'souza, C., Harashima, T., Shen, W.C., Wang, P., Pan, X., Waugh, M., and Heitman, J. (2000). Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, D., Hains, P., Walsh, B., Bergquist, P., and Nevalainen, H. (2001). Proteins associated with the cell envelope of Trichoderma reesei: A proteomic approach. Proteomics 1, 899–909. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Markham, P., and Collinge, A.J. (1987). Woronin bodies in filamentous fungi. FEMS Microbiol. Rev. 46, 1–11. [Google Scholar]
- McKeen, W.E. (1971). Woronin bodies in Erysiphe graminis DC. Can. J. Microbiol. 17, 1557–1560. [DOI] [PubMed] [Google Scholar]
- Momany, M., Richardson, E., Van Sickle, C., and Jedd, G. (2002). Mapping Woronin body function in Aspergillus nidulans. Mycologia 94, 260–266. [PubMed] [Google Scholar]
- Money, N.P., and Howard, R.J. (1996). Confirmation of a link between fungal pigmentation, turgor pressure and pathogenicity using a new method of turgor measurement. Fungal Genet. Biol. 20, 217–227. [Google Scholar]
- Naqvi, N.I., Bonman, J.M., Mackill, D.J., Nelson, R.J., and Chattoo, B.B. (1995). Identification of RAPD markers linked to a major gene for blast resistance in rice. Mol. Breed. 1, 341–348. [Google Scholar]
- Osbourn, A.E. (1996). Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 8, 1821–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall, M.L., and Brunelli, J.P. (1993). A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl. 40, 59–62. [Google Scholar]
- Pellier, A.L., Lauge, R., Veneault-Fourrey, C., and Langlin, T. (2003). CLNR1, the AREA/NIT2-like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol. Microbiol. 48, 639–655. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Snoeijers, S.S., Perez-Garcia, A., Joosten, M.H., and De Wit, P.J. (2000). The effect of nitrogen on disease development and gene expression in bacterial and fungal pathogens. Eur. J. Plant Pathol. 106, 493–506. [Google Scholar]
- Soanes, D.M., Kershaw, M.J., Cooley, R.N., and Talbot, N.J. (2002). Regulation of the MPG1 hydrophobin gene in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15, 1253–1267. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J., Ebbole, D.J., and Hamer, J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J., McCafferty, H.R.K., Ma, M., Moore, K., and Hamer, J.E. (1997). Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol. Mol. Plant Pathol. 50, 179–195. [Google Scholar]
- Tenney, K., Hunt, I., Sweigard, J., Pounder, J.I., McClain, C., Bowman, E.J., and Bowman, B.J. (2000). Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31, 205–217. [DOI] [PubMed] [Google Scholar]
- Trinci, A.P., and Collinge, A.J. (1974). Occlusion of the septal pores of damaged hyphae of Neurospora crassa by hexagonal crystals. Protoplasma 80, 57–67. [DOI] [PubMed] [Google Scholar]
- Tucker, S.L., and Talbot, N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39, 385–417. [DOI] [PubMed] [Google Scholar]
- Valent, B. (1990). Rice blast as a model system for plant pathology. Phytopathology 80, 33–36. [Google Scholar]
- Wharton, P.S., Julian, A.M., and O'Connell, R.J. (2001). Ultrastructure of the infection of Sorghum bicolor by Colletotrichum sublineolum. Phytopathology 91, 149–158. [DOI] [PubMed] [Google Scholar]
- Woronin, M. (1864). Zur Entwicklungsgeschichte der Ascobolus pulcherrimus Cr. und einiger Pezizen. Abh. Senkenb. Naturforsch. 5, 333–344. [Google Scholar]
- Xu, J.R., and Hamer, J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Xu, J.R., Urban, M., Sweigard, J., and Hamer, J.E. (1997). The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol. Plant-Microbe Interact. 10, 187–194. [Google Scholar]
- Yuan, P., Jedd, G., Kumaran, D., Swaminathan, S., Shio, H., Hewitt, D., Chua, N.H., and Swaminathan, K. (2003). A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 10, 264–270. [DOI] [PubMed] [Google Scholar]