Abstract
22q11.2 deletion syndrome (22q11.2DS) is the most common chromosomal microdeletion disorder, estimated to result mainly from de novo non-homologous meiotic recombination events occurring in approximately 1 in every 1,000 fetuses. The first description in the English language of the constellation of findings now known to be due to this chromosomal difference was made in the 1960s in children with DiGeorge syndrome, who presented with the clinical triad of immunodeficiency, hypoparathyroidism and congenital heart disease. The syndrome is now known to have a heterogeneous presentation that includes multiple additional congenital anomalies and later-onset conditions, such as palatal, gastrointestinal and renal abnormalities, autoimmune disease, variable cognitive delays, behavioural phenotypes and psychiatric illness — all far extending the original description of DiGeorge syndrome. Management requires a multidisciplinary approach involving paediatrics, general medicine, surgery, psychiatry, psychology, interventional therapies (physical, occupational, speech, language and behavioural) and genetic counselling. Although common, lack of recognition of the condition and/or lack of familiarity with genetic testing methods, together with the wide variability of clinical presentation, delays diagnosis. Early diagnosis, preferably prenatally or neonatally, could improve outcomes, thus stressing the importance of universal screening. Equally important, 22q11.2DS has become a model for understanding rare and frequent congenital anomalies, medical conditions, psychiatric and developmental disorders, and may provide a platform to better understand these disorders while affording opportunities for translational strategies across the lifespan for both patients with 22q11.2DS and those with these associated features in the general population.
The eponymous description of DiGeorge syndrome — by the late Dr Angelo DiGeorge in 1965 — included infants with absence of the thymus (thymic aplasia) and parathyroid glands (hypoparathyroidism)1. Congenital heart disease (CHD), especially involving the outflow tract2, was later added to the list of symptoms, contributing to the theory that a mechanism leading to the perturbation of the third and fourth pharyngeal arches during embryonic development might be involved. Interestingly, a similar phenotype can be associated with maternal diabetes3,4, maternal retinoic acid exposure5, single-gene disorders due to mutations in chromo-domain helicase DNA-binding protein 7 (CHD7; known as CHARGE syndrome)6,7 or mutations in T-box 1 (TBX1)8,9 and other chromosomal deletions, including 10p13–14 (REF. 10) and 11q23-ter11.
Over time, multiple aetiologies for DiGeorge syndrome were identified, beginning with a cytogenetically apparent 22q11.2 deletion in the early 1980s12,13. In the early 1990s, fluorescence in situ hybridization (FISH) studies using probes within the commonly deleted region identified submicroscopic 22q11.2 deletions as the most frequent cause of DiGeorge syndrome14,15 (FIG. 1). This preceded recognition that several seemingly unrelated conditions with overlapping phenotypic features similarly resulted from a 22q11.2 deletion, including: velocardiofacial syndrome15, conotruncal anomaly face syndrome16,17, and subsets of patients with Opitz G/BBB18 and Cayler cardiofacial19 syndromes20. Together, these observations suggest that the previously described clinical diagnoses were actually one and the same condition with a common aetiology21.
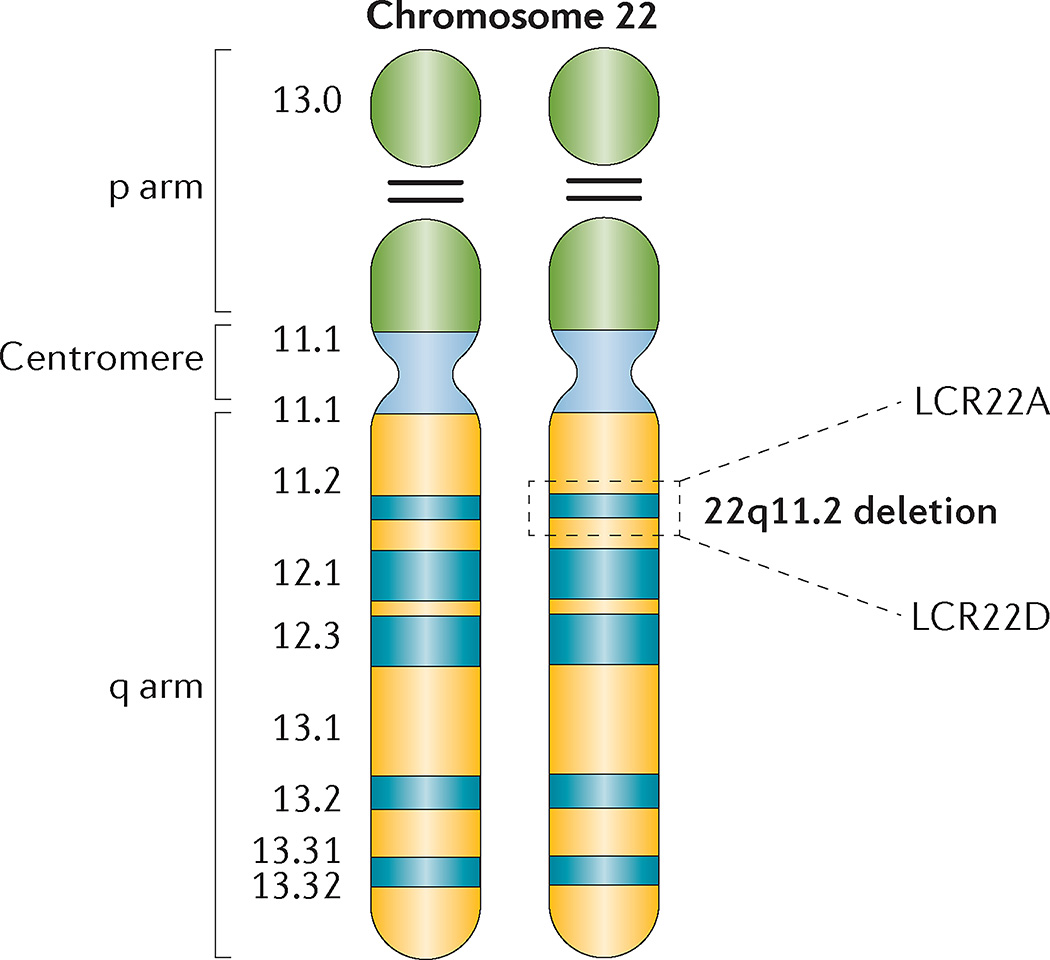
Figure 1. Chromosome 22 idiogram.
Cytogenetic representation of chromosome 22 showing the short (p) and long (q) arms along with the centromere, which functions to separate both arms. Chromosome 22 is an acrocentric chromosome, as indicated by the two horizontal lines in the p arm. The 22q11.2 deletion occurs on the long arm of one of the two chromosomes, depicted by dashed lines in the 22q11.2 band. The position of the two low copy repeats (LCRs) on 22q11.2 (LCR22A and LCR22D), which flank the typical 3-Mb deletion, are indicated.
Today, it is well established that 22q11.2 deletion syndrome (22q11.2DS) involves microdeletions (approximately 0.7–3 million base pairs in size), resulting in an heterogeneous clinical presentation, irrespective of deletion size, that can be associated with multi-organ dysfunction including cardiac and palatal abnormalities, immune and autoimmune differences, endocrine, genitourinary and gastrointestinal problems, and brain involvement as evinced by variable developmental delays, cognitive deficits and neuropsychiatric illnesses (such as anxiety disorders and schizophrenia). In fact, 22q11.2 deletion is the second-most common cause of CHD and developmental delays, and the most common cause of syndromic palatal anomalies. However, why the 22q11.2 region is particularly vulnerable to deletions remains under investigation. Furthermore, as a consequence of mechanistic understanding, the term DiGeorge syndrome is now reserved for those rare patients who share clinical symptoms with 22q11.2DS but do not harbour a 22q11.2 deletion. Otherwise, the broad phenotypic range of symptoms — including findings formerly associated with DiGeorge syndrome, velocardiofacial syndrome or conotruncal anomaly face syndrome — is referred to using the underlying cytogenetic nomenclature: 22q11.2DS22,23. In this Primer, we focus on our current understanding of the 22q11.2DS phenotype and its genetic underpinnings.
Epidemiology
22q11.2DS is common and is the most frequent chromosomal microdeletion syndrome. The prevalence of this disorder has been estimated to range from 1 per 3,000 to 1 per 6,000 live births, based on the diagnosis of infants with major birth defects and a few population screening studies conducted between the early 1990s and early 2000s using FISH technology24–28. Today, most (90–95%) newly identified patients with 22q11.2DS are found to have de novo deletions — that is, neither parent has the 22q11.2 deletion29. However, owing to improved survival and thus higher reproductive fitness of individuals with 22q11.2DS, the prevalence, especially of the inherited types, is expected to increase30,31. As 22q11.2DS is a haploinsufficient disorder, approximately half of the children of individuals with 22q11.2DS will have the deletion. Similarly, smaller, atypical nested deletions between the low copy repeats on chromosome 22 (LCR22B–LCR22D and LCR22C–LCR22D deletions; FIG. 2) — not typically detected by clinically available FISH probes and, therefore, not included in the population studies from the 1990s — are often familial and have reduced penetrance and/or a milder expression; thus, these patients are more likely to reproduce. As a consequence, the proportion of patients ascertained at The Children’s Hospital of Philadelphia with a LCR22B–LCR22D deletion inherited from an affected parent is higher (60%) than it is for patients with the typical LCR22A–LCR22D deletion (~10%)32.
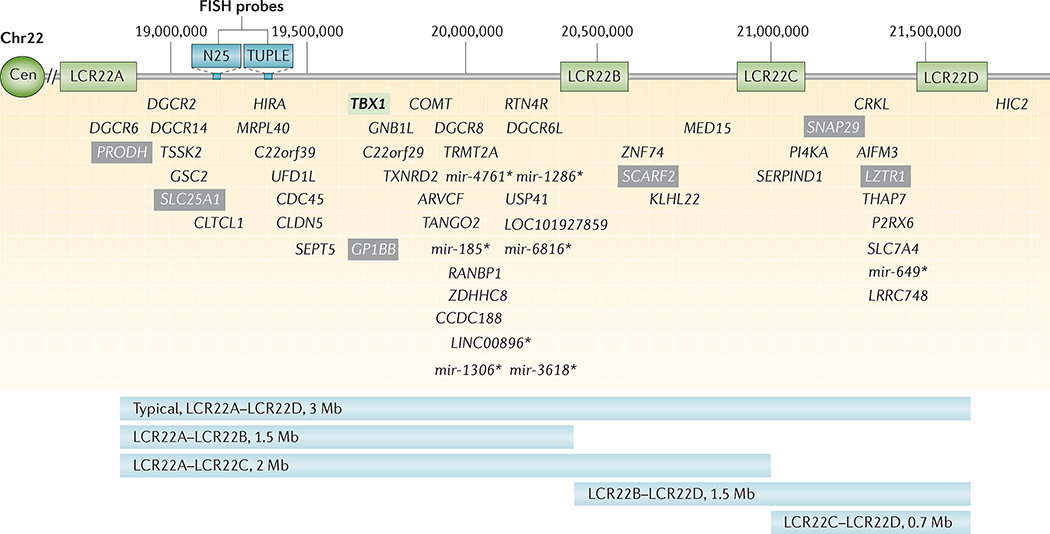
Figure 2. Low copy repeats and genes within the 22q11.2 deletion.
Schematic representation of the 3-Mb 22q11.2 region that is commonly deleted in 22q11.2 deletion syndrome, including the four low copy repeats (LCR22s) that span this region (LCR22A, LCR22B, LCR22C and LCR22D). Common commercial probes for fluorescence in situ hybridization (FISH) are indicated (N25 and TUPLE). The protein-coding and selected non-coding (*) genes are indicated with respect to their relative position along chromosome 22 (Chr22). T-box 1 (TBX1; green box) is highlighted as the most widely studied gene within the 22q11.2 region. Mutations in this gene have resulted in conotruncal cardiac anomalies in animal models and humans. Known human disease-causing genes that map to the region are indicated in grey boxes. These include proline dehydrogenase 1 (PRODH; associated with type I hyperprolinaemia), solute carrier family 25 member 1 (SLC25A1; encoding the tricarboxylate transport protein and is associated with combined D-2- and L-2-hydroxyglutaric aciduria), platelet glycoprotein Ib β-polypeptide (GP1BB; associated with Bernard–Soulier syndrome), scavenger receptor class F member 2 (SCARF2; associated with Van den Ende–Gupta syndrome), synaptosomal-associated protein 29 kDa (SNAP29; associated with cerebral dysgenesis, neuropathy, ichthyosis and palmoplantar keratoderma (CEDNIK) syndrome), and leucine-zipper-like transcription regulator 1 (LZTR1; associated with schwannomatosis 2). Further details on the location of non-coding RNAs and pseudogenes in the 22q11.2 region may be found in Guna et al.89. Common 22q11.2 deletions are shown, with the typical 3-Mb deletion flanked by LCR22A and LCR22D (LCR22A– LCR22D) on top and the nested deletions, with their respective deletion sizes indicated below. Each of the deletions portrayed is flanked by a particular LCR22. Those rare deletions not mediated by LCRs are not shown. AIF3M, apoptosis-inducing factor mitochondrion-associated 3; ARVCF, armadillo repeat gene deleted in velocardiofacial syndrome; CDC45, cell division cycle 45; Cen, centromere; CLDN5, claudin 5; CLTCL1, clathrin heavy chain-like 1; COMT, catechol-O-methyltransferase; CRKL, v-crk avian sarcoma virus CT10 oncogene homologue-like; DGCR, DiGeorge syndrome critical region; GNB1L, guanine nucleotide-binding protein (G protein), β-polypeptide 1-like; GSC2, goosecoid homeobox 2; HIC2, hypermethylated in cancer 2; HIRA, histone cell cycle regulator; KLHL22, kelch-like family member 22; LINC00896, long intergenic non-protein-coding RNA 896; LOC101927859, serine/arginine repetitive matrix protein 2-like; CCDC188, coiled-coil domain-containing 188; LRRC74B, leucine-rich repeat-containing 74B; MED15, mediator complex subunit 15; mir, microRNA; MRPL40, mitochondrial ribosomal protein L40; P2RX6, purinergic receptor P2X ligand-gated ion channel 6; PI4KA, phosphatidylinositol 4-kinase catalytic-α; RANBP1, Ran-binding protein 1; RTN4R, reticulon 4 receptor; SEPT7, septin 7; SERPIND1, serpin peptidase inhibitor clade D (heparin co-factor) member 1; TANGO2, transport and golgi organization 2 homologue; THAP7, THAP domain-containing 7; TRMT2A, tRNA methyltransferase 2 homologue A; TSSK2, testis-specific serine kinase 2; TXNRD2, thioredoxin reductase 2; UFD1L, ubiquitin fusion degradation 1-like; USP41, ubiquitin-specific peptidase 41; ZDHHC8, zinc-finger DHHC-type-containing 8; ZNF74, zinc-finger protein 74.
Two multicentre prenatal studies published in 2012 and 2015 reported 22q11.2 deletions in 1 per 347 and 1 per 992 fetuses, respectively, using invasive prenatal testing33,34. Both studies included analyses of fetuses with or without abnormal ultrasonographic findings. The prevalence of the 22q11.2 deletion was approximately 1 in 100 for fetuses with major structural anomalies such as CHD, and was 1 per ~1,000 in seemingly anatomically normal fetuses (in both studies)33,34. The true live birth incidence remains to be defined by global newborn screening35,36. Notably, a small subset of neonates with 22q11.2 deletions have already come to attention through newborn screening for severe combined immunodeficiencies. 22q11.2 deletions have been identified in 1 per 169 children referred for clinical laboratory testing based on a suspicion of developmental disabilities37. Conversely, the 22q11.2 deletion is not found in samples of seemingly healthy individuals37 as penetrance to develop clinical symptoms, frequently involving multiple organ systems, is high38.
Both sexes and all racial and ethnic groups are affected39. However, the deletion is slightly more often maternal in origin40 and non-white patients may be diagnosed less often, perhaps owing to less recognizable craniofacial features in these populations41,42.
In the general population, 22q11.2 deletion is among the most common detectable cause of several conditions. A large proportion of patients with CHD have 22q11.2 deletions: 52% of those with interrupted aortic arch type B, 34% of those with truncus arteriosus, 16% of those with tetralogy of Fallot43 and ~5–10% of those with ventricular septal defects44. Other conditions associated with 22q11.2 deletions include velopharyngeal insufficiency (12.5–30% of patients), cleft palate (10% of patients; cleft lip with or without cleft palate in 1–2% of patients)45–47, developmental disabilities (2–3% of patients)48 and schizophrenia (0.5–1% of patients)49,50. By contrast, the prevalence of the 22q11.2 deletion in other conditions with overlapping symptoms (for example, hypoparathyroidism) is as yet unknown. Ascertainment strategies will have a role in defining prevalence in these conditions. 22q11.2 deletions will be less likely identified in samples that deliberately or inadvertently exclude individuals with multiple medical or psychiatric features, or intellectual disabilities (that is, the very features that are common in 22q11.2DS), or in samples enriched for older individuals (given the premature mortality of the syndrome)51.
Premature mortality, although lower than suggested by early reports of DiGeorge syndrome, remains profound at all ages. About 4% of all infants with 22q11.2DS succumb, with mortality figures exceeding those for infants with similar malformations31,41. Cardiac defects, hypocalcaemia and airway malacia (in which cartilage defects lead to collapsibility of the airway) are risk factors for early death, with median age at death of 3–4 months31,41. In adults, premature death occurs at median age in the 40s; causes are multiple, including sudden unexplained death51, but are not necessarily related to cardiac defects or psychotic illness51. Larger studies are needed to more fully clarify mortality risks prospectively at all ages31,41,51.
Mechanisms/pathophysiology
22q11.2 deletion
The 22q11.2 region is one of the most structurally complex areas of the genome primarily due to several large blocks of LCRs or segmental duplications52–54 (FIG. 2). These LCRs are >96% identical, thereby making the locus vulnerable to meiotic error55,56. The two largest repeats, LCR22A and LCR22D, flank the typical 3-Mb 22q11.2 region that is hemizygous in ~85% of patients52,53. The 22q11.2 deletion results from nonallelic homologous recombination between LCR22A and LCR22D (FIG. 3). The same mechanism leads to the proximal (centromeric) nested 1.5-Mb (LCR22A–LCR22B) or 2-Mb (LCR22A–LCR22C) deletions (FIG. 2). Patients with these nested deletions have major phenotypic features in common with patients with the typical LCR22A–LCR22D deletion52–54. However, the frequency of the nested proximal deletions accounts for only 5–10% of all 22q11.2 deletions52,53,57. Likewise, distal nested (LCR22B–LCR22D and LCR22C–LCR22D) deletions lead to overlapping phenotypic features, including conotruncal cardiac anomalies, palatal defects and developmental differences, but the clinical features are less penetrant than those of the typical LCR22A–LCR22D deletion. Moreover, these deletions (LCR22B–LCR22D and LCR22C–LCR22D) are more frequently inherited32,58.
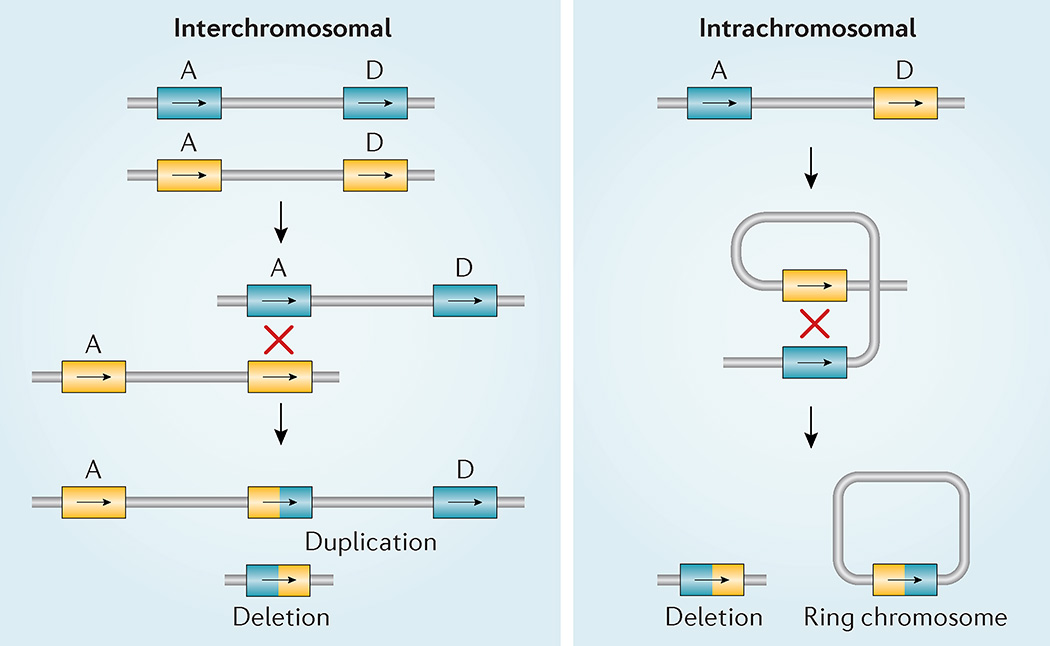
Figure 3. 22q11.2 non-allelic homologous recombination.
Diagram of two different types of meiotic non-allelic homologous recombination events that can occur between low copy repeats on chromosome 22 (LCR22s). Rearrangements between LCR22A and LCR22D are indicated (A and D) on each allele (blue versus yellow). Interchromosomal events (left) occur between paralogous LCR22s (A and D) in two different alleles owing to >99% sequence identity of direct repeats (‘X’ shows the crossover of the two chromosomes). The hybrid LCR22 is shown as half yellow and half blue. This process results in a duplication or deletion of intervening genes in resulting gametes. Intrachromosomal recombination events (right) result from crossing over (indicated by ‘X’) within one allele, resulting in a deletion (left) or a ring chromosome (right); the ring chromosome is not viable.
Much remains to be learned about the underlying mechanisms of vulnerability to non-allelic homologous recombination and the de novo occurrence of the 22q11.2 deletion40. The completeness and accuracy of the human genome reference assembly remains a major challenge to these research efforts59,60.
Furthermore, 22q11.2 hemizygosity alone cannot explain the genetic mechanism of the highly variable phenotypic expression of 22q11.2DS. Proposed mechanisms, in addition to the combined effect of this multi-gene deletion61–65 (FIG. 2) and stochastic phenomena, include the sensitivity of individual genes within the 22q11.2 region to gene dosage66,67, variants in genes on the intact 22q11.2 (REF. 68) and additional ‘modifying’ variants outside the 22q11.2 region, involving both protein-coding genes and regulatory mechanisms67,69–73. Researchers are currently evaluating common and rare single-nucleotide variants as well as copy number variations in genome-wide assays to explain these findings. Parental age and parental origin of de novo deletions seem to have no discernible phenotypic effect40,74. However, inherited deletions may result in a more-severe cognitive phenotype, perhaps related to a combination of socioeconomic factors and heritable components contributed by the unaffected parent75.
Developmental aspects of 22q11.2DS
The mouse is the main organism used for investigating developmental aspects of the syndrome, as its developmental anatomy is similar to that of a human and it is possible to generate individual gene mutations as well as multi-gene deletions63. Conditional mutagenesis studies have also examined the timing and/or tissue-specific requirements of certain genes76 and gene dosage requirements using an allelic series approach66,77. Importantly, as some phenotypes only become apparent at later ages63, conditional mutants allow bypass of the early embryonic lethality that is observed in some constitutive null mutants. Mouse models have also contributed greatly to understanding expression in developing and adult brain tissue61,63,67,78–82.
Much of the pathology related to typical congenital physical features associated with 22q11.2DS can be ascribed to problems with the morphogenesis and subsequent abnormal function of pharyngeal arch system derivatives, including the craniofacial structures, the thymus, the parathyroid glands, the aortic arch and the cardiac outflow tract (FIG. 4). These structures receive contributions from all three classic germ layers of the embryo — the endoderm, mesoderm and ectoderm — together with neural crest cells derived from the closing neural tube.
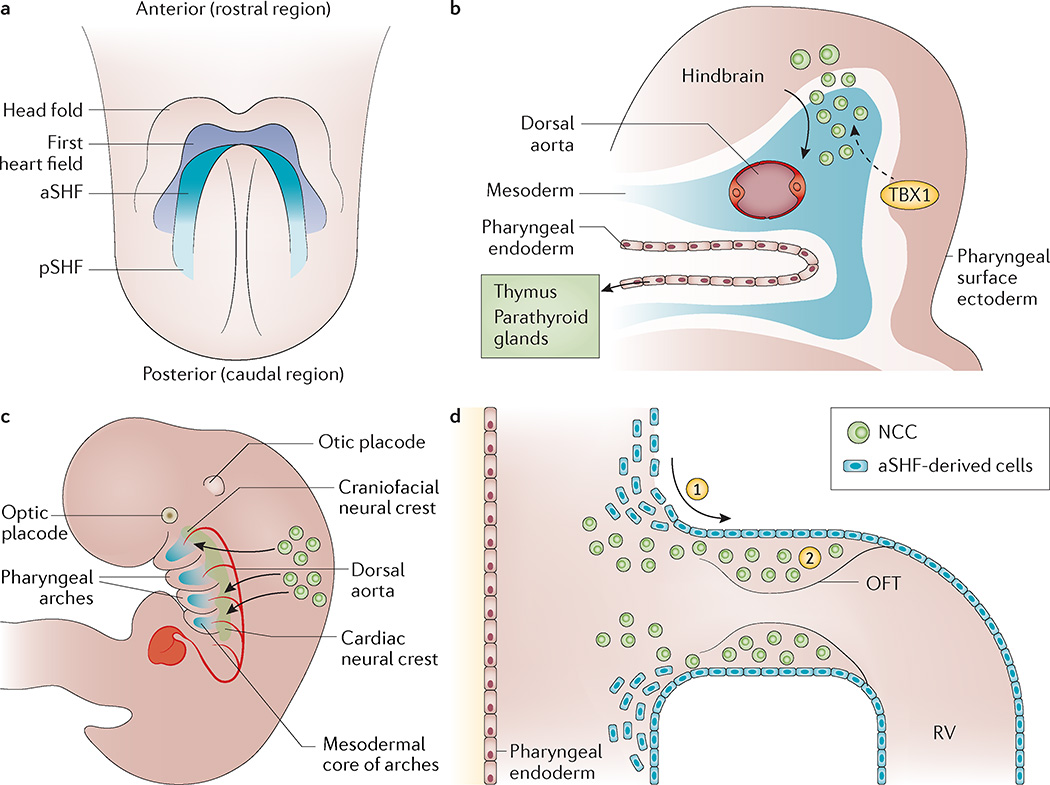
Figure 4. Development of the cardiovascular and pharyngeal structures affected in 22q11.2 deletion syndrome.
a | The schematic ventral view of an embryonic day 7.5 (E7.5) mouse embryo shows the relationship of the cardiac crescent to the head folds and also depicts the distinct cellular fields termed the first heart field and the second heart field (SHF). Within the second heart field, the anterior segment (aSHF) contributes to the outflow tract (OFT) and the right ventricle (RV) of the heart, whereas the posterior segment (pSHF) contributes to the inflow of the heart. b | Cardiac neural crest cells (NCCs) delaminate from the hindbrain and migrate ventrolaterally to populate the pharyngeal arches. The T-box transcription factor TBX1 is required within the pharyngeal surface ectoderm to regulate as yet unknown signalling pathways, which pattern the cardiac NCCs (dashed arrow). The pharyngeal endoderm and cardiac NCCs interact in the formation of the thymus and parathyroid glands. c | Lateral view of an E10.5 mouse embryo. The cardiac neural crest arises from the neural tube at the level between the otic placode and somite three, and migrates ventrolaterally to populate the pharyngeal arches, interacting with the core mesoderm and ultimately contributing the smooth muscle cells to the remodelling arch arteries. The caudal stream enters the OFT. d | Schematic presentation of the cell lineage that contributes to the OFT of the heart at approximately E10.5. (1) Cells derived from the aSHF enter the OFT where they contribute to the myocardium and endocardium. (2) These cells interact with cardiac NCCs that migrate in from the pharyngeal arch region. Signals to the cardiac neural crest are also received from the pharyngeal epithelium. Disruption to these cellular contributions or interactions can result in a common arterial trunk, alignment defects or ventricular septation defects.
Pharyngeal mesoderm progenitor cells give rise to craniofacial muscles and second heart field derivatives, including the cardiac outflow tract64,78. The facial bones and bony palate are variously derived from neural crest cells or the anterior mesoderm. The parathyroid glands and the thymus derive from tissue interactions between the pharyngeal endoderm and neural crest cells76,83. Defects in parathyroid gland development lead to hypocalcaemia, and defects in the developing thymus lead to immune deficiencies.
Congenital cardiac anomalies are related to defects in the arteries formed within the pharyngeal apparatus and in the cardiac outflow tract. As for the other affected tissues (the thymus, parathyroid glands and facial structures), there is a very close relationship between adjacent cell types. Mesoderm-derived endothelial cells and neural crest cells participate directly in the formation of the pharyngeal arch arteries. Signals from the pharyngeal ectoderm and a direct cellular contribution from the neural crest are required for remodelling into the mature aortic arch. Reciprocal interactions between the second heart field and neural crest cells are essential for outflow tract remodelling84–87. The most specific cardiovascular defects associated with 22q11.2DS are interrupted aortic arch type B (ascribed to aplasia of the left fourth pharyngeal artery)88 and tetralogy of Fallot (related to defects in the development of the pulmonary infundibulum).
DNA sequences in the 22q11.2 region
There are 90 known or predicted genes present in the typical 3-Mb 22q11.2 locus that are hemizygously deleted — including 46 protein-coding genes and seven microRNAs (miRNAs), ten non-coding RNAs (including one read-through transcript) and 27 pseudogenes (per genome build GRCh37 — a human reference sequence produced by the Genome Reference Consortium)89. A subset of genes, as provided in the University of California Santa Cruz (UCSC) gene prediction track, are shown in FIG. 2. The most studied gene of interest in the 22q11.2 deletion region is TBX1, encoding a T-box transcription factor. TBX1 was found to be a crucial gene in the LCR22A–LCR22B region using multiple mouse model approaches90–92. Heterozygous loss-of-function mutations of Tbx1 in the mouse result in partially penetrant cardiovascular, thymic and parathyroid defects that are reminiscent of congenital defects in 22q11.2DS91,92. Tbx1-null mice are embryonic lethal with a persistent truncus arteriosus, cleft palate and absence of the thymus and parathyroid glands. Conditional mutagenesis of Tbx1 in the mesoderm, pharyngeal surface ectoderm or endoderm each leads to an overlapping subset of the abnormalities mentioned above93,94, demonstrating the complexity of the tissue interactions that are required for morphogenesis of the pharyngeal derivatives95. Tbx1 expression has not been detected in neural crest cells, and neural crest cell-specific knockout mice have no discernible phenotype. However, neural crest cell patterning is affected in Tbx1-conditional mutants of both the surface ectoderm96 and the second heart field97. At the cellular level, mouse models have also been important in the detection of reduced proliferation and premature differentiation of progenitor cells expressing Tbx1 (REF. 98). Tbx1 has been implicated in brain microvascular development99 and may play some part in cognitive and behavioural deficits100.
Another gene of interest is DGCR8, encoding the DGCR8 microprocessor complex subunit (also known as Pasha), a double-stranded RNA-binding protein that mediates the biogenesis of miRNAs. This observation implicates an miRNA-related mechanism in 22q11.2DS81,82,101. miRNAs are small non-coding RNAs that regulate the expression of target genes by binding to specific sites in mRNAs for translational repression or degradation. In mouse models, heterozygosity of Dgcr8 results in neuronal deficits that are characteristic of 22q11.2DS, whereas inactivation of both alleles in neural crest cells101 results in heart defects101,102. Subtle alterations in miRNA expression levels can have profound effects on brain development and plasticity, especially involving synapses103,104. Recent studies propose that DGCR8 may play a part in modifying the expression of genes outside of the 22q11.2 deletion region that contribute to the neuropsychiatric and other phenotypes associated with 22q11.2DS63,72,101,105.
In addition to DGCR8-related changes in miRNAs, the high density of miRNAs in the 22q11.2 deletion region and the accumulative insight into their function indicate that these functional non-coding RNAs may themselves have a role in the variable expression of 22q11.2DS73,105,106. These effects on expression are likely to involve not only the central nervous system (CNS) but also the cardiovascular system and other aspects of embryonic development73,105,106.
Much evidence has been accumulated for a role of other individual 22q11.2 region protein-coding genes in major phenotypes of 22q11.2DS. These genes include v-crk avian sarcoma virus CT10 oncogene homologue-like (CRKL), encoding a cytoplasmic adaptor protein to growth factor signalling, which maps to the LCR22B–LCR22D region107 and acts in a dosage-sensitive manner108. Human and mouse model data indicate that haploinsufficiency of CRKL could be responsible for the aetiology of cardiac anomalies in individuals with nested distal deletions and seems to modulate natural killer cell function109. Another gene, synaptosomal-associated protein 29 kDa (SNAP29), encodes a soluble SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein that is predicted to mediate vesicle fusion at the endoplasmic reticulum or Golgi apparatus membranes, is highly expressed in myelinating glial cells, is required for lamellar body formation in the skin, and is indirectly required for β1-integrin endocytosis and cell migration. Mutations in this gene have been associated with cerebral dysgenesis, neuropathy, ichthyosis and palmoplantar keratoderma (CEDNIK), Kousseff and Opitz G/BBB syndromes68. Scavenger receptor expressed by endothelial cells 2 protein (SREC2), encoded by SCARF2, contains putative epidermal growth factor-like domains in its extracellular domain, along with numerous positively charged residues in its intracellular domain, indicating that it may be involved in intracellular signalling. Homozygous or compound heterozygous mutations of SCARF2 underlie Van den Ende–Gupta syndrome characterized by severe contractural arachnodactyly and distinctive facial dysmorphism, including triangular face, as well as skeletal anomalies110.
With respect to neuropsychiatric phenotypes, there are multiple gene candidates as the majority of genes in the 22q11.2 deletion region are expressed in the brain89. COMT encodes catechol-O-methyltransferase, one of several enzymes that degrade catecholamines, including dopamine. Its activity is of particular importance in brain regions with low expression of the presynaptic dopamine transporter, such as the prefrontal cortex. A polymorphism associated with a different level of enzymatic activity (that is, the COMT Val/Met functional polymorphism) has been explored in 22q11.2DS with respect to cognition and susceptibility to schizophrenia. However, multiple studies have found no association of the COMT functional Val/Met common allele with schizophrenia in adults with 22q11.2DS111–113. There may be some effects of this common variant on frontal lobe functioning and anatomy in 22q11.2DS111, although results for overall intellect are mixed114. PRODH, encoding the enzyme proline dehydrogenase, which breaks down proline, has also been studied, in part because pathogenetic mutations in PRODH are known to cause type I hyperprolinaemia, which in severe forms can cause seizures and intellectual disability. Approximately one-third of patients with 22q11.2DS have increased levels of proline115, and several studies, although not all, have shown significant associations between high proline levels and various brain outcome measures in 22q11.2DS116–119. However, studies of common variants in PRODH, as for those in COMT, in 22q11.2DS show contradictory results with respect to risks for intellectual disability or schizophrenia114. Zinc-finger DHHC-type-containing 8 (ZDHHC8), which encodes a palmitoyltransferase, has shown interesting results in studies of mutant mouse models, with effects on axonal growth and terminal arborization, and potential functional implications for synaptic connections and working memory62. Another 22q11.2 region candidate is RANBP1, encoding a binding protein for the small GTPase Ran. As a regulator of the Ran complex, this protein has multiple functions — including cilia formation and modulation of mitosis — that may contribute to the CNS and other phenotypes of 22q11.2DS117. Evidence for a role in neurogenesis places RANBP1 as a candidate for the cortical circuits implicated in disorders associated with 22q11.2DS, such as attention-deficit disorders, autism and schizophrenia79,117.
Together with the miRNA mechanism implicated by DGCR8, and downstream effects of dosage changes in individual genes such as TBX1, there is increasing evidence for effects of the 22q11.2 deletion on, and interaction with, signalling pathways and proteins encoded by multiple genes outside of the 22q11.2 deletion region. For example, mitogen-activated protein kinase 1 (Mapk1)120 and hypermethylated in cancer 2 (Hic2)121, are of interest for cardiovascular and other aspects of embryonic development in mice. Loss of Tbx1 may be partially rescued by hemizygosity of Trp53, implicating histone methylation as a mechanism and suggesting potential pharmacological strategies that could compensate for developmental defects associated with 22q11.2 deletions98. Defective cortical circuitry and some abnormalities of signalling, for instance, in Sonic Hedgehog and CXC chemokine receptor 4 (Cxcr4)– CXC chemokine ligand 12 (Cxcl12; also known as Sdf1) signalling, which are important in interneuron migration, have been detected in the brains of mouse deletion models that may involve a DGCR8-mediated miRNA mechanism and have relevance for schizophrenia in the general population79,122.
The plethora of signalling and other pathways that are affected by the hemizygosity of the 22q11.2 deletion region in model systems may be valuable to help to identify modifiers of the 22q11.2DS phenotype in humans63,79,114,122, perhaps particularly for genes outside of the 22q11.2 deletion region123,124. These studies also promise to elucidate mechanisms underlying the variable phenotypic expression related to other pathogenetic copy number variations and to identify genes involved in the mechanism of common complex conditions, such as congenital cardiac and palatal anomalies, schizophrenia, Parkinson disease and many others, emphasizing the importance of 22q11.2DS as a model for these diseases in the general population.
Diagnosis, screening and prevention
Clinical manifestations
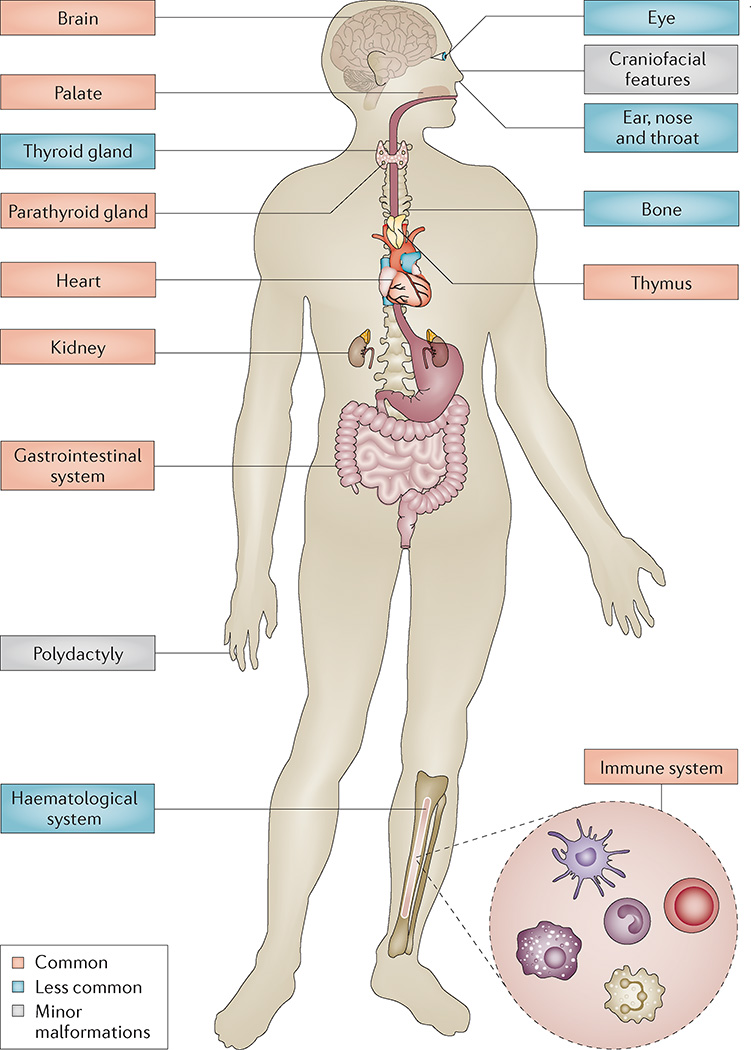
Clinical manifestations that urge for diagnostic testing vary by age. In infancy or childhood, typical symptoms include some combination of congenital heart defects, chronic infection, nasal regurgitation, hypernasal speech, hypocalcaemia, feeding difficulties, developmental and language delays, behavioural differences and learning disabilities20,22,39,125. Renal abnormalities, laryngo-tracheo-oesophageal abnormalities, hypothyroidism, intrauterine growth retardation, short stature, skeletal differences such as vertebral anomalies, club feet, polydactyly and scoliosis, thrombocytopenia, hearing loss, microcephaly, idiopathic seizures and hypotonia are less frequent (FIG. 5).
Figure 5. Organ and system involvement in 22q11.2 deletion syndrome.
22q11.2 deletion syndrome leads to significant morbidity (and some premature mortality), with frequent multi-organ system involvement, such as cardiac and palatal abnormalities, immune differences, endocrine and gastrointestinal problems, and later-onset conditions across the lifespan including variable cognitive deficits and psychiatric illness that is attributable to functional brain changes. Less-frequent manifestations, when present, contribute to substantial morbidity (examples include: idiopathic seizures; polymicrogyria; sclerocornea; coloboma; deafness; choanal atresia; laryngeal cleft or web; tracheo-oesophageal fistula; hypothyroidism or hyperthyroidism; juvenile rheumatoid arthritis; idiopathic thrombocytopenia; autoimmune haemolytic anaemia; craniosynostosis; scoliosis; intestinal malrotation; Hirschsprung disease; and imperforate anus). Minor malformations generally confer little indisposition but may enhance ascertainment. These generally include: mild dysmorphic craniofacial features, such as hooded eyelids, auricular anomalies, nasal differences including a dimple or crease, and asymmetric crying facies; and, cervical and thoracic vertebral anomalies or butterfly vertebrae, arachnodactyly, camptodactyly, 2–3 toe syndactyly and polydactyly (preaxial and postaxial of the hands and postaxial of the feet).
In adolescence and adulthood, behavioural abnormalities, in many instances indicative of (emerging) psychiatric illness, can lead to the diagnosis125, frequently (but not always) with a history of associated medical and developmental differences, such as hypocalcaemia or learning difficulties29. The presence of subtle but characteristic facial features can assist with identification at any age (FIG. 6). However, the opportunity for diagnosis can be missed when typical craniofacial and other typical congenital features such as cardiac or palatal defects are absent. This can be the case even when presenting with other high-prevalence conditions associated with 22q11.2DS, such as hypocalcaemia or psychotic illness. Major medical centres when less experienced with caring for patients with 22q11.2DS are not exempt from overlooking the 22q11.2DS diagnosis in part owing to the broad phenotypic variability22,126. Some adults are only diagnosed following the birth of an affected child29.
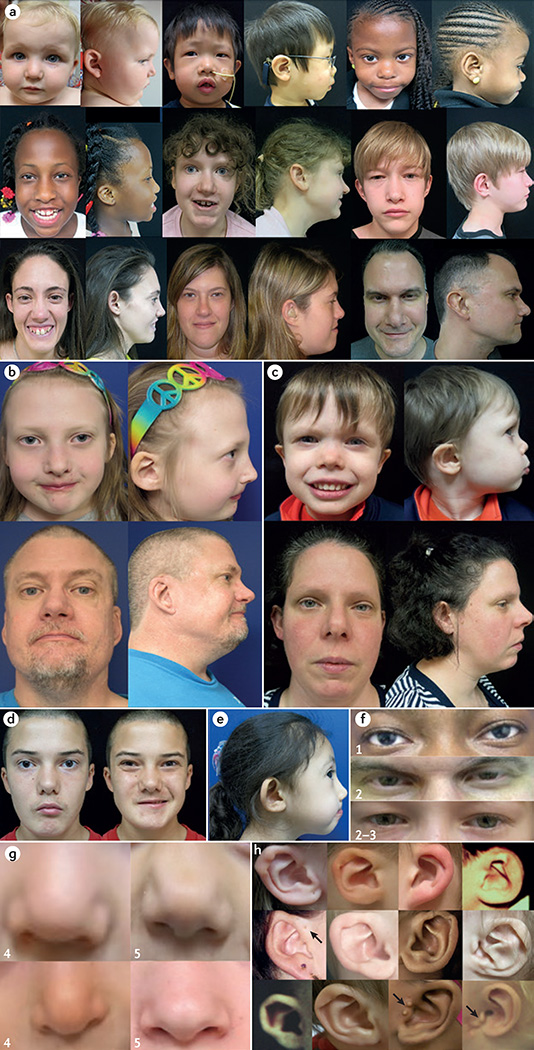
Figure 6. Craniofacial features associated with 22q11.2 deletion syndrome.
Patients with 22q11.2 deletion syndrome (22q11.2DS), shown here from infancy through to adulthood, demonstrate variability of associated craniofacial features — most with few recognizable dysmorphia (part a). A person with 22q11.2DS has a 50% recurrence risk with each pregnancy for this microdeletion syndrome, but some adults only come to attention following the diagnosis in a child with associated features, as in these unrelated nuclear families (daughter and father (part b) and son and mother (part c)). When viewed individually, some craniofacial features provide important clues to the diagnosis, for example, microstomia and asymmetric crying facies (part d), and malar flatness and micrognathia (part e). External eye findings (part f) may include upslanting palpebral fissures and hypertelorism (1), hooded eyelids and/or ptosis (2) and mild epicanthal folds (3). Nasal features (part g) may include a bulbous nasal tip with hypoplastic alae nasi (4) often with a nasal dimple or crease with or without a faint haemangioma (5). Auricular differences (part h) frequently include thick overfolded, squared-off and crumpled helices, microtic, cupped or posteriorly rotated ears, attached lobes and preauricular pits or tags (arrows).
The overall prevalence of considerable medical problems varies by age and ascertainment. In childhood, the triad of DiGeorge syndrome (although with highly variable degree of severity) is often ascertained: immunodeficiency (~75% of patients); congenital cardiac anomalies (~75% of patients); and hypocalcaemia due to hypoparathyroidism (~50% of patients). Other complications include palatal abnormalities (~75% of patients); manifest gastrointestinal, feeding and swallowing problems (~30% of patients); and genitourinary anomalies including renal agenesis (~30% of patients) (FIG. 5). Phenotypic expression is highly variable and ranges from severe life-threatening conditions to only a few less-severe associated features23,114. Additional complexities include considerable interfamilial and intrafamilial variability127, even between identical twins29. Diagnosis on clinical grounds of a child with mild features requires familiarity with the condition. Dual diagnosis of other unrelated conditions (for example, ‘café-au-lait’ spots due to neurofibromatosis, skeletal disproportion due to achondroplasia or Marfan syndrome, or deep palmar and plantar creases due to trisomy 8 mosaicism) is possible, especially when features appear unusual, as 22q11.2DS is common.
Cardiovascular abnormalities
Cardiovascular abnormalities become evident in the prenatal or neonatal period and are often the initial manifestation that leads to diagnosis23. Most abnormalities are conotruncal heart defects — defined as malformations of the outflow tract — and include tetralogy of Fallot (with or without pulmonary atresia), truncus arteriosus, interrupted aortic arch type B (between the left carotid and the left subclavian arteries) and ventricular septal defect. Anomalies of the aortic arch and/or of the pulmonary arteries may occur as isolated entities (~40%) or in association with conotruncal defects (~60%)44, contributing to the relative specificity of the cardiovascular patterns of this syndrome86–88. Aortic arch anomalies most frequently include a right-sided or a double aortic arch with or without aberrant subclavian arteries sometimes resulting in a vascular ring (~13%)128. Pulmonary artery anomalies include diffuse hypoplasia and discontinuity with or without major aorto-pulmonary collateral arteries. Approximately 10% of paediatric patients have aortic root dilation with unclear clinical significance129. Other types of CHD are rarer in patients with 22q11.2DS39. Cardiovascular defects that are less obvious, such as a vascular ring, may not be diagnosed until the patient is older and presents with respiratory symptomatology128. Similarly, a ductal-dependent lesion, for instance, an interrupted aortic arch type B, might escape neonatal detection in the absence of an audible murmur, pass neonatal screening with pulse oximetry in the presence of a patent ductus arteriosus and might only present in extremis, which increases overall morbidity and at times mortality. Overall, CHD represents the main cause of mortality (~87%)29,31 in children with 22q11.2DS.
Immunodeficiency
Immunodeficiency affects up to 75% of paediatric patients with 22q11.2DS owing to thymic hypoplasia and impaired T cell production7,23,29,130. However, the condition is heterogeneous, ranging from patients with normal thymic development and normal T cell production to a small subset of patients with absent T cell production22,29,131,132. Manifestations include chronic infections130,131,133, impaired humoral (antibody) immune response resulting in poor response to vaccines134,135, IgA deficiency136, allergy and asthma5,137,138. Autoimmune diseases such as juvenile rheumatoid arthritis139, idiopathic thrombocytopenia140, haemolytic anaemia141 and thyroid disease142 are collectively common7,22,29,130–136,138,139.
Testing is required to define the immune dysfunction. Assessment of thymic function is best performed by analysing peripheral blood T cells by flow cytometry. Imaging of the thymus is not accurate as the size, absence or presence of thymic tissue does not predict individual immune function, although when absent, it may lead to diagnostic studies both prenatally and postnatally143. Current recommendations support an early assessment of T cells using a flow cytometry panel that includes total T cells (CD3), naive T cells (usually CD4/CD45RA), memory T cells (usually CD4 or CD45RO), B cells (CD19) and natural killer cells (CD3−CD56+CD16+)36,92,134,144. The absolute counts of T cells should be analysed rather than percentages. T cell lymphocytopenia can also be detected in newborn screens for severe combined immunodeficiencies.
Palatal abnormalities
Only 11% of paediatric patients have overt cleft palate, of whom 1–2% have cleft lip or cleft palate, and even fewer have Pierre Robin sequence20,29,39,145,146. However, ~65% of patients have milder but often medically actionable manifestations such as: occult submucosal cleft palate, bifid uvula and velopharyngeal dysfunction, making the palatal diagnosis more challenging in the prenatal or early-neonatal period. Initial signs may only include a history of polyhydramnios (an excessive amount of amniotic fluid surrounding the fetus) or nasal regurgitation (that is, secretions, food or liquids coming through the nose with feeding and spitting up in young children or reflux of thin liquids into the nasal cavity in older individuals). Later, symptoms include abnormal nasal resonance and nasal emissions (air escape through the nasal passage with speech). Compensatory articulation errors are often present, which can worsen speech intelligibility in children with velopharyngeal dysfunction. In older children and adults, recurrent and chronic sinus infections may be a sign of nasopharyngeal reflux resultant from repeated contamination of the nasal cavity. Persistent otorrhoea with myringotomy tubes in place may also be due to nasopharyngeal reflux20,22,29,39,145,147–150. Additional craniofacial features that might facilitate diagnosis include asymmetric crying faces (found in 14% of patients with 22q11.2DS), auricular (ear) anomalies with or without hemifacial microsomia20, craniosynostosis and characteristic facial features, such as hooded eyelids and a nasal dimple151 (FIG. 6).
Endocrine abnormalities
Hypoparathyroidism, with hypocalcaemia as a consequence, is found in >50–65% of patients with 22q11.2DS152,153. Symptoms of hypocalcaemia can include tetany, seizures, feeding difficulty, stridor and fatigue. Transient neonatal hypocalcaemia or new-onset hypocalcaemia often occurs or recurs during times of stress (such as during illness, perioperatively or during adolescence and pregnancy)22,152–154. Additional endocrine manifestations that may also be features of 22q11.2DS include: hypothyroidism in children and ~20% of adults, and hyperthyroidism in children142 and ~5% of adults; and growth hormone deficiency, intrauterine growth retardation (~4% of patients) and short stature (~15% of patients)22,126,152–157.
Gastrointestinal abnormalities
Considerable gastrointestinal abnormalities are found in ~30% of patients and can result in substantial feeding and swallowing problems that might necessitate tube-feeding158. In early childhood, gastrointestinal complications can present as gastro-oesophageal reflux disease, oesophageal dysmotility, nasopharyngeal reflux, vomiting and constipation158. Serious complications include oesophagitis, aspiration, failure to thrive, malnutrition, feeding refusal and respiratory symptomatology, such as choking and recurrent pneumonia145,158. Importantly, CHD may be erroneously blamed for feeding difficulties. Other possible causes that should be taken into account include endocrine (aberrant calcium and/or thyroid hormone levels), CNS (hypotonia, polymicrogyria and/or cerebellar), respiratory (congestion, increased work of breathing, vascular ring and/or laryngeal anomaly) and gross motor (posture, oral motor coordination and tongue retraction) dysfunctions158. Rare but important complications of 22q11.2DS include intestinal malrotation or non-rotation, imperforate anus, Hirschsprung disease and oesophageal atresia or tracheo-oesophageal fistula159.
Genitourinary abnormalities
Approximately one-third of patients have genitourinary abnormalities20,22,23,27,29,39,153,160 including: bilateral or unilateral renal agenesis25,161,162, dysplastic or cystic kidneys161,162, duplicated collecting system161,162, hydronephrosis161,162, cryptorchidism18, hypospadias18, absent uterus163 or inguinal hernia164.
Other
Other important somatic associations include major malformations such as: congenital diaphragmatic hernia (~1%), laryngeal anomalies (clefts or webs), eye abnormalities (sclerocornea, retinal coloboma or microphthalmia), choanal atresia, polymicrogyria and neural tube defects22,165,166. Preaxial and postaxial polydactyly, camptodactyly, arachnodactyly and radial ray defects may be observed in the upper extremities, whereas 2–3 syndactyly, overlapping toes, hammer toes, postaxial polydactyly and club foot may be observed as lower extremity differences167. Anomalies are also observed in the cervical spine (platybasia, fusion and/or block, anomalous dens, C2 swoosh, increased motion) and thoracic vertebrae168 (butterfly vertebrae), eyelids (ptosis and hooded eyelids), eye (hypertelorism, tortuous retinal vessels, posterior embryotoxon and upslanting palpebral fissures) and umbilical hernia (especially in non-white patients)151. Conditions that require ongoing surveillance include scoliosis (~30%); sensorineural and conductive hearing loss or cochlear abnormities; and rarely malignancies that include hepatoblastoma, Wilms tumour, renal cell carcinoma, thyroid carcinoma, leukaemia, neuroblastoma and melanoma22,23,29,41,169. Treatable later-occurring conditions include unprovoked seizures (~15%), early-onset Parkinson disease and psychiatric illnesses (~60%)126,153,170.
Developmental delays
Developmental and educational concerns are frequently reported in association with 22q11.2DS. Gross and fine motor difficulties171, and expressive language delays and speech problems dominate in infants and toddlers172. Children with 22q11.2DS often demonstrate a significant delay in language onset; an early study indicated that ~70% of children did not speak or used only a few words or signs at 24 months of age or older172. Speech deficits should be discriminated from language disorders as the former often improves after velopharyngeal corrective surgery, whereas language disorders may occur independently of palatal findings172. Intelligence in children and adolescents follows a normal distribution that is comparable to the general population173,174. However, mean IQ is only ~70, with about two-thirds of individuals falling in the IQ range of 55–85, compared with the reference IQ range of 85–115 (mean: 100) in the typically developing population. Thus, learning difficulties are very common in preschool and primary school, especially within the domains of mathematics175,176 and language comprehension177. More-severe levels of intellectual disability are uncommon178. However, paediatric patients with secondary insults (for example, following cardiac arrest, prolonged hypocalcaemia or neonatal seizures) or primary brain malformations (for example, polymicrogyria) can have a poorer cognitive prognosis68,179. Several studies indicate that cognitive development varies with divergent trajectories125,180 and that the level of intellectual ability is not necessarily stable across the lifespan of the patient. Although IQ is generally considered to be a more or less stable trait in the typically developing youth, an average decline of 7 full-scale IQ points is observed in individuals with 22q11.2DS between 8 years and 24 years of age181.
Psychiatric disorders
Individuals with 22q11.2DS are at an increased risk for developing several psychiatric disorders; the prevalence of anxiety, attention-deficit and autism spectrum disorders is increased in children with 22q11.2DS182. Anxiety disorders are also profoundly increased in adults with 22q11.2DS183. Conversely, bipolar disorder does not seem to be increased, and it remains unclear if major depressive disorder shows greater prevalence than in the general population182,183. As for any phenotypic feature of 22q11.2DS, prevalence is likely to be influenced by ascertainment bias and the use of diverse assessment methods. Approximately 25% of individuals with 22q11.2DS are diagnosed with schizophrenia112,184, and, in turn, the 22q11.2 deletion can be found in ~1 per 100–200 individuals with schizophrenia49, making 22q11.2 deletion the strongest known molecular genetic risk factor for schizophrenia. The manifestation of schizophrenia in patients with 22q11.2DS does not differ from other types of schizophrenia with respect to prodromal stage, age of onset (although a prospective study of youths is needed), core signs and symptoms, treatment response and cognitive profile apart from an overall lower IQ74,112,114,185–189. In addition to standard categorical classifications, dimensional assessment of psychiatric and cognitive symptoms190 can be helpful in understanding the individual’s profile of strengths and weaknesses. Education about associated psychiatric conditions that arise early in life (including autism spectrum, attention-deficit and anxiety disorders) as well as in adolescence and later (for example, psychotic illnesses and anxiety and mood disorders), and their early signs and symptoms, can assist the family and facilitate prompt access to psychiatric assessment, diagnosis and treatment22,126.
Reproduction
With the current standard of clinical care, survival of patients with 22q11.2DS to reproductive age is the norm29. Reproductive fitness for women with 22q11.2DS, in the absence of schizophrenia or intellectual disability, is similar to that of their unaffected sisters and general population expectations30. Disproportionately reduced reproductive fitness of men with 22q11.2DS might contribute to an observation of excess maternal transmission of 22q11.2 deletions30. However, reproductive health and decision making present challenges for patients, their families and clinicians191. The ability to act as an independent parent may be affected by the complex phenotypic expression of the syndrome, including psychiatric and neurodevelopmental features22,29. In addition, features of the syndrome may be important considerations for pregnancy planning. For example, the use of necessary but teratogenic medications (such as anticonvulsants, for example, valproate or phenytoin) can increase the risk for birth defects, and the presence of CHD in the expectant mother can increase the risk for pregnancy complications. As for all women with CHD considering pregnancy, recommendations suggest stratifying the patient’s risk by integrating the specific defect with the presence of additional risk factors (such as a history of arrhythmias, the need for systemic anticoagulation and smoking, among other factors)191–193.
Diagnostic tests
Historically, the most frequent diagnostic test used to identify a 22q11.2 deletion involved FISH, using a probe mapping to the LCR22A–LCR22B region14,15(FIG. 2). Thus, nested deletions excluding the LCR22A–LCR22B region (that is, LCR22B–LCR22D and LCR22C–LCR22D deletions) can be detected using customized FISH probes from the LCR22B–LCR22D region or more easily using clinically available whole-region methodologies, such as multiplex ligation-dependent probe amplification194,195 or single-nucleotide polymorphism microarrays. As genome-wide microarrays detect pathogenic copy number variants on all chromosomes, this methodology has the added benefit of not introducing bias by requiring pre-selection of a specific genomic region. This may be especially beneficial for those patients with few clinical features or multiple atypical features, with less experienced health care providers58,108.
Screening
Prenatal genetic counselling for couples in which one partner has the 22q11.2 deletion with an a priori 50% recurrence risk includes discussions about non-invasive screening (including fetal ultrasonography and echocardiography), definitive 22q11.2 deletion studies through chorionic villus sampling or amniocentesis, use of donor gametes, pre-implantation genetic diagnosis using in vitro fertilization and/or adoption. All parents of children with 22q11.2DS should be tested to identify mildly affected individuals and those with low-level somatic mosaicism. As germline mosaicism196 confers a recurrence risk above that of the general population for those couples in which neither parent has the 22q11.2 deletion, these couples may choose non-invasive methodologies, such as non-invasive prenatal screening and fetal imaging, or definitive diagnostic testing in subsequent pregnancies. Screening for 22q11.2 deletions can be included when invasive testing is already being considered for another reason, such as advanced maternal age. However, in many instances, parental anxiety is enough to support these investigations.
Currently, screening for the 22q11.2 deletion in individuals from the general population is considered when anatomic abnormalities, for example CHD, are identified on fetal ultrasonography, or when a fetus is considered high risk following non-invasive prenatal screening197. Invasive prenatal studies, using newly available methods such as single-nucleotide polymorphism microarrays, can diagnose a fetus with 22q11.2DS with or without prenatally recognizable features. Moreover, some parents are identified with 22q11.2DS only following ascertainment in their fetus29 or through non-invasive prenatal screening in previously undiagnosed mothers. Long available, fetal ultrasonography can reveal several 22q11.2DS-related findings, including CHD, cleft palate, renal anomalies, polyhydramnios, polydactyly, diaphragmatic hernia, club feet, tracheo-oesophageal fistula and neural tube defects. Should such findings lead to prenatal diagnostic testing to rule out aneuploidy (for example, trisomy 13, 18 and 21), the addition of 22q11.2 deletion studies as an adjunct to standard cytogenetics should be considered29,198. Early diagnosis may facilitate the ability to prevent neonatal seizures due to hypocalcaemia179.
Management
Management of 22q11.2DS requires an individualized, multidisciplinary and coordinated care plan that takes into account the associated features of the patient. An increasing emphasis of many health care systems on acute, readily managed, cross-sectional, ‘one-size-fits-all’ care in silos can be problematic. A broad flexible perspective, embracing multi-system issues that are often chronic and require longitudinal coordinated management, in the context of a frequently changing clinical picture and in the context of social and/or learning difficulties is required to manage patients with 22q11.2DS.
The International 22q11.2DS Consortium was formed in 2006 with a goal of developing management parameters (BOX 1). Guidelines were proposed for children and adults with 22q11.2DS22,126. The consortium currently operates under the umbrella of the 22q11.2 Society, a professional body that was established in 2013 to support basic science and clinical collaborations and to maintain access to contemporary guidance for health care providers.
Box 1. General principles to manage 22q11.2 deletion syndrome.
Early diagnosis and effective treatment of each condition improves general outcome
Attention to the multi-system nature of 22q11.2 deletion syndrome is key
Goals to promote an optimal quality of life and functioning for the individual patient
Appreciation of the level of the individual’s neurocognitive abilities when providing advice, regardless of chronological age
Family members and caregivers to be included, as much as possible, in developing a health care plan for patients of all ages as allies in maximizing the understanding of the illness and recommended treatments and follow-up
Relatives and social service agents can be instrumental in implementing health care recommendations for extended family members, in particular children, when the 22q11.2 deletion is familial
Times of transition (for example, from paediatric to adult care) require special attention to prevent gaps in care
The effect of treatment for one condition may positively or negatively impact another condition
Patients benefit from a single invested health care manager or team who will coordinate care and facilitate communication between providers
Cardiovascular system
CHD is typically diagnosed prenatally or during the first days to months of life199–202. The diagnostic tools and surgical treatment are, in general, the same as for any child with CHD199,200. However, 22q11.2DS is associated with longer hospital stays, greater resource use and more medications at discharge201,202. Specific perioperative care should be focused on the prevention of hypocalcaemia, immunological depression, vasomotor instability, bronchospasm and airway bleeding199,201,202. In addition to standard antimicrobial prophylaxis, an antifungal agent may be considered199. The results of surgery are usually good, and operative mortality is comparable to that of other patients199–202. However, overall mortality for those with 22q11.2DS exceeds that for non-syndromic individuals with similar cardiac defects31.
Lifetime surveillance is mandatory for patients with major CHD, with many patients requiring cardiac catheterization and a subgroup interventional procedures and/or repeat cardiac surgery126,203,204. Specific treatments relate to each cardiac defect in agreement with international protocols203. Residual valve lesions and outflow obstruction, ventricular function, arrhythmias, heart failure, aortic root dilatation and bacterial endocarditis should be monitored126,129,203,204. Cardiac components of premature and sudden death should be prevented, as much as possible, in all patients51,126,129,203,204.
Women with 22q11.2DS and CHD seeking advice about pregnancy should be evaluated using standard risk assessments developed for all women with CHD, given the risks for maternal, fetal and neonatal complications and mortality192,193. Although severe forms of systemic ventricular dysfunction, cyanosis and/or pulmonary hypertension pose excessive risks, in most other situations, pregnancies may be managed safely from a cardiac perspective, given that access to specialized high-quality care during pregnancy and delivery is guaranteed192,193. However, general risks relating to pregnancies in which the fetus has a 22q11.2 deletion, such as small size and prematurity, remain increased157.
Immune system and autoimmune conditions
Management of immune deficiency ranges from a thymus transplant (rare) to no intervention66. Assessment of newborns allows for the identification of infants with no CD45RA T cells who will require a thymus transplant or a matched T cell transplant. Early assessment also provides guidance for live viral vaccine administration. In the setting of very low T cells, patients will require protective isolation and live viral vaccines should not be given. Patients with no CD45RA T cells require prophylaxis against pneumocystis. T cell counts should be reassessed at approximately 1 year of age, before administration of the measles, mumps and rubella vaccines and the Varivax vaccine. Data support a CD4 T cell count of >500 cells per µl as being the lower limit for safe vaccine administration135,146,205–207. A small percentage of patients will develop antibody deficiencies208, but the patient characteristics that predispose to this condition are not yet clear; periodic monitoring seems prudent. Patients with hypogammaglobulinaemia can be treated with immunoglobulin replacement.
Infections are often problematic across the lifespan of an individual with 22q11.2DS. Young children with 22q11.2DS have very frequent sinopulmonary infections, owing to immune deficiency, unfavourable anatomy and immune immaturity137. In addition, patients are at increased risk of allergies, and this may also contribute to sinopulmonary infections137. Infections seem to correlate with the humoral immune deficiency in adults209. The approach to recurrent infections in children and adults involves minimizing the anatomical contribution with ear tubes, sinus rinses, treatment of concomitant allergies and, in infants, feeding practices that discourage pooling of formula in the pharynx. Prophylactic antibiotics represent an approach that can provide some relief, and immunoglobulin replacement should be offered to those with demonstrated humoral defects. Patients with autoimmune diseases are typically managed using strategies that are appropriate for patients with autoimmune diseases without 22q11.2DS, but immune suppression should be minimized210.
Palatal disorders
Overt cleft palate and cleft lip or cleft palate in patients with 22q11.2DS are addressed routinely in very early childhood using treatments that are, in general, the same as for those of any child with clefts211. Velopharyngeal dysfunction can lead to maladaptive articulation patterns with unintelligible speech, resulting in frustration that affects psychosocial development. This often causes parental distress and an eagerness to proceed with surgical interventions. The goals of surgery, generally at approximately 4–6 years of age, include normal speech production, effective communication and improved quality of life (QOL). An individualized treatment plan to optimize speech outcomes while minimizing perioperative and postoperative complications requires an initial assessment of speech and language status.
Too little speech may preclude obtaining an adequate sample for surgical decision making, thus pre-surgical speech therapy with total communication strategies is recommended beginning in infancy. In addition, velopharyngeal imaging using nasendoscopy or multi-view videoflouroscopy is typically required to determine the pattern, motion and degree of velopharyngeal closure before making a surgical plan. Additional considerations include airway size and preoperative co-morbid conditions, including visibly enlarged tonsils or adenoids, obstructive sleep apnoea212, asthma, cervical spine instability, carotid displacement, and cardiac, endocrine, haematological and feeding problems; ascertainment of developmental, emotional and behavioural status is also important. Patients also benefit from perioperative calcium monitoring and preoperative and postoperative sleep studies150,212.
Endocrine system
Adequate treatment of hypocalcaemia and thyroid dysfunction is essential22,126,154. Special attention to calcium levels at times of biological stress (for example, peri-operatively or during puberty, pregnancy or delivery), and vigilance in the neonatal period to prevent seizures is crucial22,152,179. Growth hormone deficiency responds well to therapy154. Specific growth curves are available156.
Central nervous system
CNS involvement is common in 22q11.2DS. In infancy, neonatal seizures, developmental delays, language impairment and autism spectrum disorder each require their own management strategies. These include infant stimulation, total communication strategies beginning in infancy and specialized educational interventions. Neuromotor deficits, especially in the domains of balance and coordination, occur early and require remediation171,213,214. The diagnosis of autism spectrum disorder may be relevant to clarify limitations in social interaction and communication and the presence of repetitive behaviours215. Awareness of these and other developmental deficits is crucial to avoid situations in which the environmental expectations exceed the abilities of the child.
Standard pharmacological and non-pharmacological treatments for attention-deficit, anxiety and other mood disorders are effective22,126,216–218, as are standard treatments for major psychotic illnesses such as schizophrenia126. Attention to increased risk of seizures and movement disorders is important when considering medication choices, and adjunctive or prophylactic treatments may be necessary126,189. Standard treatments for epilepsy and Parkinson disease seem to be effective126. In light of the prominence of CNS expression, targeted pro-active awareness and management of these aspects are required for every individual with 22q11.2DS, starting early in life. The available treatment strategies, both pharmacological and non-pharmacological, for paediatric and later-onset neuropsychiatric disorders are effective and should be applied in accordance with general clinical practice guidelines for the associated conditions22,126.
Quality of life
The challenges of intellectual deficits, psychiatric illnesses and other somatic consequences of 22q11.2DS can have far-reaching effects on daily functioning and QOL of patients, which can result in high burden for the family. Providing adequate support to help optimize functioning and QOL of the patient is essential29. As the well-being of the affected individual is closely related to the well-being of the caregivers, it is also important to address caregivers’ stress and provide support when necessary across the lifespan219,220. As disability-related family problems increase with the age of the patient, a growing need for counselling, especially for aspects of parenting and discipline, and for treatment can be presumed221.
QOL among children with 22q11.2DS is often characterized by struggles in cognitive, social and emotional domains222 as compared with healthy peers and those with other chronic conditions, such as diabetes, asthma, CHD and cancer. Moreover, children and adolescents with 22q11.2DS and repaired CHD have more hospitalizations, worse exercise performance and lower reported QOL ratings than seemingly healthy peers201,202,223. Insofar as immunodeficiency contributes to infection and hospitalization, more-severe immune compromise would be associated with reduced QOL, however, no studies have yet been performed. The complexity of multiple affected systems in individuals with 22q11.2DS may have a compounding effect on QOL.
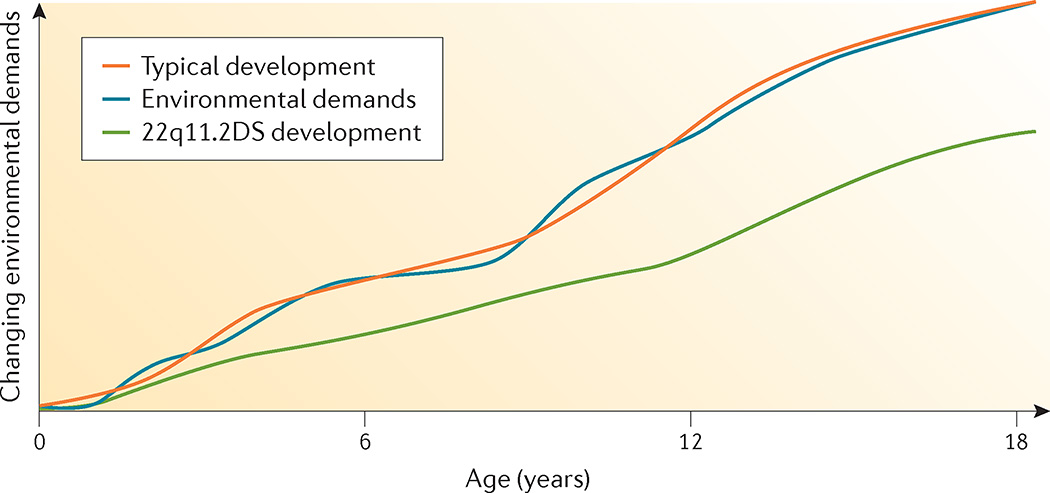
With respect to neurocognitive abilities, determining weaknesses as well as relative strengths and competencies can facilitate educational and vocational planning and optimize QOL and functional outcomes. Depending on overall cognitive capacities (ranging from average to intellectual disability), students will follow either typical school placements with educational support, often requiring an individualized educational plan, or special education with individualized educational plans. Given that the cognitive phenotype can change over time181, the intellectual abilities of all patients (regardless of age) should be followed and re-evaluated at regular intervals. Furthermore, continuous adaptation of expectations and learning environments is necessary to provide a balance between individual capacities and demands of the environment125 (FIG. 7). In this way, guidance can be implemented at home, school and work, whereas unnecessary stress can be averted. Importantly, cognitive decline during childhood may be a risk indicator for schizophrenia181, but may also represent a complicating factor in the struggle to maintain the balance between competencies and environmental demands.
Figure 7. Developmental trajectory.
As the child with 22q11.2 deletion syndrome (22q11.2DS) ages, the discrepancy between developmental level (based on chronological age) and environmental demands widens owing to associated neurocognitive and behavioural developmental deficits. Note that IQ decline observed in 22q11.2DS may not only be due to a relative but also to an absolute decline in cognitive abilities.
Functional impairment in adulthood is primarily mediated by more-severe intellectual disabilities and psychiatric phenotypes such as schizophrenia205. Functional impairment appears unrelated to a history of treated CHD, or mood or anxiety disorders205. Relative strengths in activities of daily living and employment are apparent29,205, and support may help to compensate for relative weaknesses in social and communication abilities29. Individuals tend to have deficits in executive functioning and mental arithmetic224, but relatively strong rote memory224,225, so may be well suited to perform structured tasks. Receptive and expressive communication may be more effective using indirect methods (that is, with a computer or smartphone), as compared with direct verbal communication. Hands-on training, demonstrations and written instructions may help205.
Adult patients living into middle-age present challenges for long-term care, mainly shouldered by their ageing parents, which may cause profound financial, physical and emotional stress29,219. Professional agencies, including social work and vocational training, can help47. Spouses, siblings and other relatives may also have important roles29,126. Prompt recognition and treatment of psychiatric disorders will maximize functioning and improve QOL, whereas the integration of medical, educational, behavioural and environmental treatment methods and approaches will be necessary to enhance QOL for individuals with 22q11.2DS and their families.
Outlook
Understanding the pathophysiology
The discovery that the 22q11.2 deletion is the genetic cause of what was previously called DiGeorge syndrome led to the identification of the main dosage-sensitive genes (for example, TBX1 and CRKL, among other genes) underlying the main developmental anomalies. However, all phenotypic features of the syndrome are not fully penetrant in any individual patient, and the clinical presentation is remarkably variable. This variability remains largely unexplained. It may be that the expression of certain features is more-directly related to individual gene dosage effects, as identified for TBX1 and CRKL with respect to cardiac development. Other features, such as CNS-related pathogenesis, may require the effects of multigenic reduced gene dosage within the 22q11.2 deletion interacting with permissive variants in modifier genes elsewhere in the genome.
Current research aims to unravel potential pathophysiological roles of allelic variation of genes within the 22q11.2 region of the non-deleted chromosome, modifier genes that reside outside of the deleted region, somatic mutations, epigenetic phenomena and individual characteristics or environmental factors226. Polygenic ‘background’ effects are already demonstrated in mouse models that show highly variable expression of key phenotypes when the same deletion or individual mutations of deletion region genes are engineered in different mouse strains92. The phenotypical variability, even within the same mouse strain, as in monozygotic twins with 22q11.2DS29, is also consistent with the role of stochastic effects, and potentially epigenetic and other mechanisms. It can be anticipated that ongoing genome-wide association, whole-genome sequencing and expression studies will identify genetic modifiers and mechanisms73,74,105,124,227,228.
Features that are easily visible in mouse embryos or human fetuses have been the main focus of research thus far. Advances in technologies, and interest, are gradually shifting focus to functional changes — that is, studies of neurobehaviour in humans and mouse models. 22q11.2DS is an interesting model to investigate possible genes, genetic mechanisms and central neurotransmitter systems associated with the 22q11.2 region that may contribute to the observed cognitive decline in childhood. Genetically engineered mouse models will continue to provide the foundation for mechanistic analysis to facilitate understanding of the pathogenesis of each of the associated syndromic features and accordingly provide promise for improving diagnosis and treatment79. The advantages of studying mouse models of 22q11.2DS, particularly for changes in cortical circuitry that underlie neurobehavioural phenotypes, have recently been summarized79. Large-scale prospective studies of key interacting factors will be needed to understand the complex interplay between cognitive, mood, psychotic, neuromotor and other neurological symptoms that contribute to the variable neuropsychiatric phenotypes associated with 22q11.2DS.
An outstanding question is why 22q11.2DS is the most frequent genomic disorder. Owing to the complexity of LCRs229, the rearrangement breakpoints and sequences driving non-allelic homologous recombination remain uncharted. Even the latest human genome assembly (GRCh38) contains gaps in the LCRs of the 22q11.2 region59,60. A more definitive map may help to pinpoint underlying variability in the population. Understanding this variability may not only be important for evaluating the reasons for the high prevalence of 22q11.2DS but may also stratify individuals at risk for de novo 22q11.2 deletions. In addition, variability of LCRs might explain part of the phenotypic variability. Copy number variation within the LCR subunits, the precise position of the LCR-mediated rearrangements and the broader structural variation among patients may directly affect the expression of the 90 genes and/or pseudogenes located within the LCRs and/or indirectly affect expression of flanking genes.
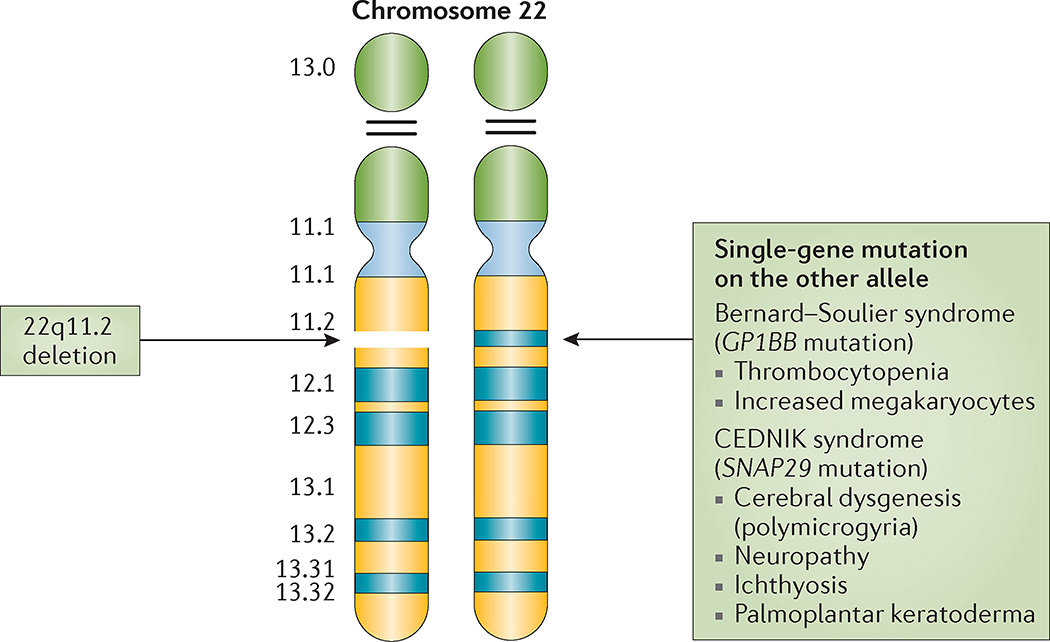
Importantly, if mutations that lead to other autosomal recessive disorders are present in the non-deleted allele, they can be unmasked owing to the deletion of the other 22q11.2 allele (for example, Bernard–Soulier and CEDNIK syndromes)68,230 (FIG. 8).
Figure 8. Associated autosomal recessive conditions on 22q11.2.
A deletion on 22q11.2 in combination with a mutation in a single gene on the other allele can unmask an autosomal recessive condition, for example, Bernard–Souilier syndrome (platelet glycoprotein Ib β-polypeptide (GP1BB)) and cerebral dysgenesis, neuropathy, ichthyosis and palmoplantar keratoderma (CEDNIK) syndrome (synaptosomal-associated protein 29 kDa (SNAP29)).
22q11.2DS as a model system
22q11.2DS has been shown to be a good model system over the past 25 years for human microdeletions and their related genomic disorders or syndromes. This work has helped to demonstrate how microdeletions could explain many syndromes with known, but extremely rare, microscopically visible deletions, similar to Williams–Beuren syndrome (7q11.23 deletion), WAGR syndrome (Wilms’ tumour, aniridia, genitourinary malformation and retardation; 11p13 deletion) and Wolf–Hirschhorn syndrome (4p16.3 deletion). In addition, 22q11.2DS has demonstrated how chromosomally engineered and subsequently transgenic mice could be used to identify a causative gene (or genes) in copy number variation disorders.
The combination of human genetics with developmental analysis of mouse models also demonstrated how phenotypic overlap at the clinical level could be recapitulated in specific anatomical defects at the embryonic level (for example, Tbx1-mutant and Chd7-mutant mice, and 22q11.2DS and CHARGE syndrome in humans, respectively). More-detailed analysis of the role of Tbx1 in mice has been fundamental in expanding our knowledge of the role of the second heart field lineage in congenital heart malformations and has informed other areas, such as thymic development (including the induction of thymic cells from stem cells).
22q11.2DS is also a very strong genetic cause of psychiatric abnormalities, in particular schizophrenia, and thus serves as a good model of this complex condition to study the trajectory from genetic risk to psychiatric expression231. Patients with 22q11.2DS with psychiatric disorders may have no formal intellectual disability. The association with a defined genetic cause, as for complex anatomic anomalies such as cardiac defects, has facilitated human and animal studies of neurodevelopment and neurofunctional changes to schizophrenia and other psychotic disorders.
Thus, 22q11.2DS is a model for understanding common congenital anomalies, rare and common psychiatric and other medical conditions and developmental differences, and may provide insight into translational strategies across the lifespan, not only for patients with 22q11.2DS but also for those with these individual conditions in the general population.
Management
Animal models provide promise in the development of pharmaceutical agents to treat 22q11.2DS-related phenotypes. Simple model organisms (for example, Caenorhabditis elegans, Drosophila and zebrafish) are particularly suited to high-throughput screening of chemical libraries for potential drug development89. More data are needed on treatment response and caveats in 22q11.2DS for all associated conditions and treatment modalities189,201,202. Importantly, current cohorts of patients with 22q11.2DS have become sufficiently large to begin examining the effect of current and novel treatment strategies in adequately powered clinical trials.
Prevention, diagnosis and screening
Lack of recognition of the condition and/or lack of familiarity with genetic testing methods, together with the wide variability of expression, delays diagnosis both prenatally and postnatally. Early diagnosis offers multiple advantages and could improve preparedness and outcomes while potentially reducing medical, emotional and fiscal (and therefore societal) costs.
Although helpful in the initial recognition of the syndrome, relying on congenital features as ‘characteristic’ of 22q11.2DS would now be considered inadequate to capture the variable expression of the 22q11.2 deletion across the lifespan. Old names for the syndrome perpetuate the myth that a patient ‘must’ have one or more of these features, promoting the often lengthy diagnostic odyssey endured by many patients, families and clinicians. Until there is completely unbiased ascertainment — that is, identification of the 22q11.2 deletion by screening all live births — with lifetime follow-up of emerging features, all prevalence figures for associated features must be assumed to be only gross estimates, probably biased to more-severe expression22,126.
Non-invasive prenatal screening for fetal aneuploidy detection is an increasingly offered service, with the ability to detect smaller fetal (and sometimes maternal) segmental aneuploidies, such as 22q11.2 deletions197,232. Further technological advances are likely to improve accuracy. Prenatal detection of 22q11.2DS will enable future parents to make informed choices, prepare for obstetrical and neonatal management, and provide the opportunity to improve survival and outcome157,179. The unbiased detection of 22q11.2DS will establish true incidence, will enable longitudinal medical course surveillance from the prenatal period and will allow for earlier intervention.
Several technologies have been tested for sensitivity and specificity in the setting of newborn screening233–235. All rely on DNA detection and are therefore both robust yet sensitive to settings where DNA may be limited or degraded (such as when there is low white blood cell count or improper sample storage). Strategies range from genome-wide deletion, duplication and rearrangement approaches to specific detection of the 22q11.2 deletion. All have shown technical feasibility. The larger hurdle will be to establish the political will to implement one or more strategies in a pilot approach in a real-life setting. The usual test for application of a new screening test is demonstration that early detection improves outcomes. Efforts are underway to establish this connection and a powerful grass roots movement has supported the newborn screening initiative.
Acknowledgments
The authors acknowledge grants from the National Institute of Mental Health (consortium grants U01MH101723, U01MH101720 and U01MH101719-01 to D.M.M.-M., N.P., A.S., J.A.S.V., B.S.E., J.R.V., B.E.M. and A.S.B., and grant U01MH087636 to D.M.M.-M.); NIH grant P01-HD070454 to D.M.M.-M., B.S.E. and B.E.M.; the Immune Deficiency Foundation, Baxalta and Janssen (to K.S.); Brain and Behavior Research Foundation (formerly NARSAD) 2010 Young Investigator Award (to J.A.S.V.); and the Canadian Institutes of Health Research (CIHR; MOP 97800 and MOP 111238), the Canada Research Chair in Schizophrenia Genetics and Genomic Disorders, and the Dalglish Chair in 22q11.2 Deletion Syndrome (to A.S.B.). The authors thank L. DiCairano, L. Lunny, A. Melchiorre, K. Schlechtweg, M. Torsan and G. Wong for assistance with manuscript formatting.
Footnotes
Competing interests
All other authors declare no competing interests.
D.M.M.-M. has presented lectures on 22q11.2 deletion syndrome for Natera.
Author contributions
Introduction (D.M.M.-M., K.E.S. and A.S.B.); Epidemiology (D.M.M.-M., K.E.S., B.M. and A.S.B.); Mechanisms/pathophysiology (D.M.M.-M., K.E.S., B.M., J.A.S.V., J.R.V., B.E.M., P.J.S. and A.S.B.); Diagnosis, screening and prevention (D.M.M.-M., K.E.S., B.M., N.P., A.S., J.A.S.V., E.H.Z., B.S.E. and A.S.B.); Management (D.M.M.-M., K.E.S., B.M., N.P., A.S., J.A.S.V., E.H.Z. and A.S.B.); Quality of life (D.M.M.-M., K.E.S., B.M., A.S., J.A.S.V. and A.S.B.); Outlook (D.M.M.-M., K.E.S., B.E.M, P.J.S., J.R.V. and A.S.B.); Overview of the Primer (D.M.M.-M.).
References
- 1.DiGeorge A. Discussion on a new concept of the cellular immunology. J. Pediatr. 1965;67:907–908. [Google Scholar]
- 2.Takao A, Ando M, Cho K, Kinouchi A, Murakami Y. In: Etiology and Morphogenesis of Congenital Heart Disease. Van Praagh R, Takao A, editors. Futura Pub. Co.; 1980. pp. 253–269. [Google Scholar]
- 3.Digilio MC, Marino B, Formigari R, Giannotti A. Maternal diabetes causing DiGeorge anomaly and renal agenesis. Am. J. Med. Genet. 1995;55:513–514. doi: 10.1002/ajmg.1320550427. [DOI] [PubMed] [Google Scholar]
- 4.Sulik KK, Johnston MC, Daft PA, Russell WE, Dehart DB. Fetal alcohol syndrome and DiGeorge anomaly: critical ethanol exposure periods for craniofacial malformations as illustrated in an animal model. Am. J. Med. Genet. Suppl. 1986;2:97–112. doi: 10.1002/ajmg.1320250614. [DOI] [PubMed] [Google Scholar]
- 5.Coberly S, Lammer E, Alashari M. Retinoic acid embryopathy: case report and review of literature. Pediatr. Pathol. Lab. Med. 1996;16:823–836. [PubMed] [Google Scholar]
- 6.Sanlaville D, et al. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J. Med. Genet. 2006;43:211–217. doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jyonouchi S, McDonald-McGinn DM, Bale S, Zackai EH, Sullivan KE. CHARGE (coloboma, heart defect, atresia choanae, retarded growth and development, genital hypoplasia, ear anomalies/deafness) syndrome and chromosome 22q11.2 deletion syndrome: a comparison of immunologic and nonimmunologic phenotypic features. Pediatrics. 2009;123:e871–e877. doi: 10.1542/peds.2008-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yagi H, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. Mutations were identified in TBX1 in two unrelated patients who do not have a 22q11.2 deletion but have some of the medical findings. This finding implicates TBX1 as a causative gene for 22q11.2DS.
- 9.Zweier C, Sticht H, Aydin-Yaylagul I, Campbell CE, Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am. J. Hum. Genet. 2007;80:510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daw SC, et al. A common region of 10p deleted in DiGeorge and velocardiofacial syndromes. Nat. Genet. 1996;13:458–460. doi: 10.1038/ng0896-458. This paper demonstrates that pathogenetic copy number variations elsewhere in the genome can cause similar phenotypes as in 22q11.2DS.
- 11.Grossfeld PD, et al. The 11q terminal deletion disorder: a prospective study of 110 cases. Am. J. Med. Genet. A. 2004;129A:51–61. doi: 10.1002/ajmg.a.30090. [DOI] [PubMed] [Google Scholar]
- 12.de la Chapelle A, Herva R, Koivisto M, Aula P. A deletion in chromosome 22 can cause DiGeorge syndrome. Hum. Genet. 1981;57:253–256. doi: 10.1007/BF00278938. [DOI] [PubMed] [Google Scholar]
- 13. Kelley RI, et al. The association of the DiGeorge anomalad with partial monosomy of chromosome 22. J. Pediatr. 1982;101:197–200. doi: 10.1016/s0022-3476(82)80116-9. This paper and reference 12 were seminal in elucidating the association of 22q11.2DS with the clinical features of DiGeorge syndrome.
- 14. Scambler PJ, et al. Microdeletions within 22q11 associated with sporadic and familial DiGeorge syndrome. Genomics. 1991;10:201–206. doi: 10.1016/0888-7543(91)90501-5. The development of FISH probes, as described in this seminal paper and in reference 15, changed our understanding of both the prevalence and the breadth of clinical variability for 22q11.2DS.
- 15.Driscoll DA, et al. Prevalence of 22q11 microdeletions in DiGeorge and velocardiofacial syndromes: implications for genetic counselling and prenatal diagnosis. J. Med. Genet. 1993;30:813–817. doi: 10.1136/jmg.30.10.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burn J, et al. Conotruncal anomaly face syndrome is associated with a deletion within chromosome 22q11. J. Med. Genet. 1993;30:822–824. doi: 10.1136/jmg.30.10.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka R, et al. Confirmation that the conotruncal anomaly face syndrome is associated with a deletion within 22q11.2. Am. J. Med. Genet. 1994;53:285–289. doi: 10.1002/ajmg.1320530314. [DOI] [PubMed] [Google Scholar]
- 18.McDonald-McGinn DM, et al. Autosomal dominant ‘Opitz’ GBBB syndrome due to a 22q11.2 deletion. Am. J. Med. Genet. 1995;59:103–113. doi: 10.1002/ajmg.1320590122. [DOI] [PubMed] [Google Scholar]
- 19.Giannotti A, Digilio MC, Marino B, Mingarelli R, Dallapiccola B. Cayler cardiofacial syndrome and del 22q11: part of the CATCH22 phenotype. Am. J. Med. Genet. 1994;53:303–304. doi: 10.1002/ajmg.1320530320. [DOI] [PubMed] [Google Scholar]
- 20.McDonald-McGinn DM, et al. The 22q11.2 deletion: screening, diagnostic workup, and outcome of results, report on 181 patients. Genet. Test. 1997;1:99–108. doi: 10.1089/gte.1997.1.99. [DOI] [PubMed] [Google Scholar]
- 21.McDonald-McGinn DM, Zackai EH, Low D. What’s in a name? The 22q11.2 deletion. Am. J. Med. Genet. 1997;72:247–249. [PubMed] [Google Scholar]
- 22.Bassett AS, et al. Practical guidelines for managing patients with 22q11.2 deletion syndrome. J. Pediatr. 2011;159:332.e331–339.e331. doi: 10.1016/j.jpeds.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald-McGinn DM, Sullivan K. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Medicine. 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 24.Botto LD, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112:101–107. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- 25.Devriendt K, Fryns JP, Mortier G, van Thienen MN, Keymolen K. The annual incidence of DiGeorge/velocardiofacial syndrome. J. Med. Genet. 1998;35:789–790. doi: 10.1136/jmg.35.9.789-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodship J, Cross I, LiLing J, Wren C. A population study of chromosome 22q11 deletions in infancy. Arch. Dis. Child. 1998;79:348–351. doi: 10.1136/adc.79.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oskarsdottir S, Vujic M, Fasth A. Incidence and prevalence of the 22q11 deletion syndrome: a population-based study in Western Sweden. Arch. Dis. Child. 2004;89:148–151. doi: 10.1136/adc.2003.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ténas Du Montcel S, Mendizabai H, Ayme S, Levy A, Philip N. Prevalence of 22q11 microdeletion. J. Med. Genet. 1996;33:719. doi: 10.1136/jmg.33.8.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald-McGinn DM, et al. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: cast a wide FISHing net! Genet. Med. 2001;3:23–29. doi: 10.1097/00125817-200101000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Costain G, Chow EW, Silversides CK, Bassett AS. Sex differences in reproductive fitness contribute to preferential maternal transmission of 22q11.2 deletions. J. Med. Genet. 2011;48:819–824. doi: 10.1136/jmedgenet-2011-100440. [DOI] [PubMed] [Google Scholar]
- 31.Repetto GM, et al. Case fatality rate and associated factors in patients with 22q11 microdeletion syndrome: a retrospective cohort study. BMJ Open. 2014;4:e005041. doi: 10.1136/bmjopen-2014-005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald-McGinn DM, et al. The perplexing prevalence of familial nested 22q11.2 deletions. ASGH. 2014 [online], http://www.ashg.org/2014meeting/abstracts/fulltext/f140120440.htm. [Google Scholar]
- 33.Wapner RJ, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012;367:2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grati FR, et al. Prevalence of recurrent pathogenic microdeletions and microduplications in over 9500 pregnancies. Prenat. Diagn. 2015;35:801–809. doi: 10.1002/pd.4613. [DOI] [PubMed] [Google Scholar]
- 35.Tomita-Mitchell A, et al. Multiplexed quantitative real-time PCR to detect 22q11.2 deletion in patients with congenital heart disease. Physiol. Genomics. 2010;42A:52–60. doi: 10.1152/physiolgenomics.00073.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chien YH, et al. Incidence of severe combined immunodeficiency through newborn screening in a Chinese population. J. Formos. Med. Assoc. 2015;114:12–16. doi: 10.1016/j.jfma.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Kaminsky EB, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwinger E, Devriendt K, Rauch A, Philip N. Clinical utility gene card for: DiGeorge syndrome, velocardiofacial syndrome, Shprintzen syndrome, chromosome 22q11.2 deletion syndrome (22q11.2 TBX1) Eur. J. Hum. Genet. 2010 doi: 10.1038/ejhg.2010.5. http://dx.doi.org/10.1038/ejhg.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald-McGinn DM, et al. The Philadelphia story: the 22q11.2 deletion: report on 250 patients. Genet. Couns. 1999;10:11–24. This paper and references 20 and 29 were the first to outline the broad scope and breadth of features associated with 22q11.2DS, both in a large cohort of patients as well as in affected family members.
- 40.Delio M, et al. Enhanced maternal origin of the 22q11.2 deletion in velocardiofacial and DiGeorge syndromes. Am. J. Hum. Genet. 2013;92:439–447. doi: 10.1016/j.ajhg.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald-McGinn DM, et al. The 22q11.2 deletion in African-American patients: an underdiagnosed population? Am. J. Med. Genet. A. 2005;134:242–246. doi: 10.1002/ajmg.a.30069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu AP, et al. Under-recognition of 22q11.2 deletion in adult Chinese patients with conotruncal anomalies: implications in transitional care. Eur. J. Med. Genet. 2014;57:306–311. doi: 10.1016/j.ejmg.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Goldmuntz E, et al. Microdeletions of chromosomal region 22q11 in patients with congenital conotruncal cardiac defects. J. Med. Genet. 1993;30:807–812. doi: 10.1136/jmg.30.10.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peyvandi S, et al. 22q11.2 deletions in patients with conotruncal defects: data from 1,610 consecutive cases. Pediatr. Cardiol. 2013;34:1687–1694. doi: 10.1007/s00246-013-0694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zori RT, et al. Prevalence of 22q11 region deletions in patients with velopharyngeal insufficiency. Am. J. Med. Genet. 1998;77:8–11. [PubMed] [Google Scholar]
- 46.Boorman JG, Varma S, Mackie Ogilvie C. Velopharyngeal incompetence and chromosome 22q11 deletion. Lancet. 2001;357:774. doi: 10.1016/S0140-6736(00)04183-0. [DOI] [PubMed] [Google Scholar]
- 47.Ruiter EM, Bongers EM, Smeets D, Kuijpers-Jagtman AM, Hamel BC. No justification of routine screening for 22q11 deletions in patients with overt cleft palate. Clin. Genet. 2003;64:216–219. doi: 10.1034/j.1399-0004.2003.00134.x. [DOI] [PubMed] [Google Scholar]
- 48.Rauch A, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am. J. Med. Genet. 2006;140:2063–2074. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- 49.Bassett AS, et al. Clinically detectable copy number variations in a Canadian catchment population of schizophrenia. J. Psychiatr. Res. 2010;44:1005–1009. doi: 10.1016/j.jpsychires.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horowitz A, Shifman S, Rivlin N, Pisante A, Darvasi A. A survey of the 22q11 microdeletion in a large cohort of schizophrenia patients. Schizophr. Res. 2005;73:263–267. doi: 10.1016/j.schres.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 51. Bassett AS, et al. Premature death in adults with 22q11.2 deletion syndrome. J. Med. Genet. 2009;46:324–330. doi: 10.1136/jmg.2008.063800. This paper was the first to systematically study mortality in adults with 22q11.2DS, identifying shortened longevity as an issue.
- 52. Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999;64:1076–1086. doi: 10.1086/302343. The molecular mechanism responsible for chromosome rearrangements leading to the 22q11.2 deletion was identified. The de novo deletion is caused by non-allelic recombination events between flanking LCRs during meiosis.
- 53.Shaikh TH, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 54.Saitta SC, et al. Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum. Mol. Genet. 2004;13:417–428. doi: 10.1093/hmg/ddh041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey JA, et al. Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am. J. Hum. Genet. 2002;70:83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Babcock M, et al. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome. Res. 2003;13:2519–2532. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrow B, et al. Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. Am. J. Hum. Genet. 1995;56:1391–1403. [PMC free article] [PubMed] [Google Scholar]
- 58. Rump P, et al. Central 22q11.2 deletions. Am. J. Med. Genet. A. 2014;164A:2707–2723. doi: 10.1002/ajmg.a.36711. Importantly, this paper reports cases of nested deletions of LCR22B–LCR22D, LCR22C–LCR22D and beyond, demonstrating that genes within the LCR22B–LCR22D regions result in features typically associated with the full LCR22A–LCR22D deletion.
- 59.Steinberg KM, et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome. Res. 2014;24:2066–2076. doi: 10.1101/gr.180893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaisson MJ, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellegood J, et al. Neuroanatomical phenotypes in a mouse model of the 22q11.2 microdeletion. Mol. Psychiatry. 2014;19:99–107. doi: 10.1038/mp.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukai J, et al. Molecular substrates of altered axonal growth and brain connectivity in a mouse model of schizophrenia. Neuron. 2015;86:680–695. doi: 10.1016/j.neuron.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Earls LR, Zakharenko SS. A synaptic function approach to investigating complex psychiatric diseases. Neuroscientist. 2013;20:257–271. doi: 10.1177/1073858413498307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karpinski BA, et al. Dysphagia and disrupted cranial nerve development in a mouse model of DiGeorge (22q11) deletion syndrome. Dis. Model. Mech. 2014;7:245–257. doi: 10.1242/dmm.012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meechan DW, Maynard TM, Tucker ES, Lamantia AS. Three phases of DiGeorge/22q11 deletion syndrome pathogenesis during brain development: patterning, proliferation, and mitochondrial functions of 22q11 genes. Int. J. Dev. Neurosci. 2011;29:283–294. doi: 10.1016/j.ijdevneu.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum. Mol. Genet. 2008;17:150–157. doi: 10.1093/hmg/ddm291. In this article, mouse models were used to demonstrate that Tbx1 function in individual tissues during embryonic development is sensitive to altered gene dosage.
- 67. Meechan DW, Maynard TM, Gopalakrishna D, Wu Y, LaMantia AS. When half is not enough: gene expression and dosage in the 22q11 deletion syndrome. Gene Expr. 2007;13:299–310. doi: 10.3727/000000006781510697. This review paper discusses the importance of gene dosage and 22q11.2DS.
- 68.McDonald-McGinn DM, et al. Hemizygous mutations in SNAP29 unmask autosomal recessive conditions and contribute to atypical findings in patients with 22q11.2DS. J. Med. Genet. 2013;50:80–90. doi: 10.1136/jmedgenet-2012-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amati F, et al. Dynamic changes in gene expression profiles of 22q11 and related orthologous genes during mouse development. Gene. 2007;391:91–102. doi: 10.1016/j.gene.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 70. Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev. Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. This article shows that both Tbx1 and Crkl genetically interact in mouse models during cardiac, thymus and parathyroid gland development.
- 71.Earls LR, et al. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 2012;32:14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brzustowicz LM, Bassett AS. miRNA-mediated risk for schizophrenia in 22q11.2 deletion syndrome. Front. Genet. 2012;3:291. doi: 10.3389/fgene.2012.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao D, et al. MicroRNA profiling of neurons generated using induced pluripotent stem cells derived from patients with schizophrenia and schizoaffective disorder, and 22q11.2 del. PLoS ONE. 2015;10:e0132387. doi: 10.1371/journal.pone.0132387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum. Mol. Genet. 2008;17:4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swillen A, et al. The behavioural phenotype in velo-cardio-facial syndrome (VCFS): from infancy to adolescence. Genet. Couns. 1999;10:79–88. [PubMed] [Google Scholar]
- 76.Arnold JS, et al. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development. 2006;133:977–987. doi: 10.1242/dev.02264. [DOI] [PubMed] [Google Scholar]
- 77.Pane LS, et al. Tbx1 is a negative modulator of Mef2c. Hum. Mol. Genet. 2012;21:2485–2496. doi: 10.1093/hmg/dds063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diogo R, et al. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature. 2015;520:466–473. doi: 10.1038/nature14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meechan DW, et al. Modeling a model: mouse genetics: 22q11.2 deletion syndrome, and disorders of cortical circuit development. Prog. Neurobiol. 2015;130:1–28. doi: 10.1016/j.pneurobio.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sivagnanasundaram S, et al. Differential gene expression in the hippocampus of the Df1/+ mice: a model for 22q11.2 deletion syndrome and schizophrenia. Brain. Res. 2007;1139:48–59. doi: 10.1016/j.brainres.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 81.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain. Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zou D, et al. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev. Biol. 2006;293:499–512. doi: 10.1016/j.ydbio.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kelly RG, Buckingham ME, Moorman AF. Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 2014;4:a015750. doi: 10.1101/cshperspect.a015750. The heart fields are crucial to form the aortic arch and conotruncal region of the heart, which are affected in 22q11.2DS. This paper explains the importance of the second heart field.
- 85.Milgrom-Hoffman M, Michailovici I, Ferrara N, Zelzer E, Tzahor E. Endothelial cells regulate neural crest and second heart field morphogenesis. Biol. Open. 2014;3:679–688. doi: 10.1242/bio.20148078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keyte AL, Alonzo-Johnsen M, Hutson MR. Evolutionary and developmental origins of the cardiac neural crest: building a divided outflow tract. Birth Defects. Res. C Embryo Today. 2014;102:309–323. doi: 10.1002/bdrc.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neeb Z, Lajiness JD, Bolanis E, Conway SJ. Cardiac outflow tract anomalies. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:499–530. doi: 10.1002/wdev.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lewin MB, et al. A genetic etiology for interruption of the aortic arch type B. Am. J. Cardiol. 1997;80:493–497. doi: 10.1016/s0002-9149(97)00401-3. [DOI] [PubMed] [Google Scholar]
- 89.Guna A, Butcher NJ, Bassett AS. Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J. Neurodev. Disord. 2015;7:18. doi: 10.1186/s11689-015-9113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene Tbx1. Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. By using mouse model approaches Tbx1 was found to be required for craniofacial, thymus and parathyroid gland as well as cardiac development. This is a seminal paper in the field.
- 91.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 92.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Z, et al. Tbx1 expression in pharyngeal epithelia is necessary for pharyngeal arch artery development. Development. 2005;132:5307–5315. doi: 10.1242/dev.02086. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Z, Huynh T, Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006;133:3587–3595. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papangeli I, Scambler P. The 22q11 deletion: DiGeorge and velocardiofacial syndromes and the role of TBX1. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:393–403. doi: 10.1002/wdev.75. [DOI] [PubMed] [Google Scholar]
- 96. Calmont A, et al. Tbx1 controls cardiac neural crest cell migration during arch artery development by regulating Gbx2 expression in the pharyngeal ectoderm. Development. 2009;136:3173–3183. doi: 10.1242/dev.028902. In mice, the gastrulation brain homeobox 2 (Gbx2) gene was found to be crucial in the pharyngeal ectoderm to signal to adjacent neural crest cells, which was required to form the aortic arch and branching vessels.
- 97.Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum. Mol. Genet. 2002;11:915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- 98. Caprio C, Baldini A. p53 suppression partially rescues the mutant phenotype in mouse models of DiGeorge syndrome. Proc. Natl Acad. Sci. USA. 2014;111:13385–13390. doi: 10.1073/pnas.1401923111. Genetic rescue is the gold standard for future therapeutics for 22q11.2DS. This is the first paper demonstrating that genetic rescue can take place by reducing the levels of p53.
- 99.Cioffi S, et al. Tbx1 regulates brain vascularization. Hum. Mol. Genet. 2014;23:78–89. doi: 10.1093/hmg/ddt400. [DOI] [PubMed] [Google Scholar]
- 100.Paylor R, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc. Natl Acad. Sci. USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 102.Chapnik E, Sasson V, Blelloch R, Hornstein E. Dgcr8 controls neural crest cells survival in cardiovascular development. Dev. Biol. 2012;362:50–56. doi: 10.1016/j.ydbio.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 103.Petri R, Malmevik J, Fasching L, Akerblom M, Jakobsson J. miRNAs in brain development. Exp. Cell. Res. 2014;321:84–89. doi: 10.1016/j.yexcr.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 104.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merico D, et al. MicroRNA dysregulation, gene networks, and risk for schizophrenia in 22q11.2 deletion syndrome. Front. Neurol. 2014;5:238. doi: 10.3389/fneur.2014.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sellier C, et al. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome. PLoS ONE. 2014;9:e103884. doi: 10.1371/journal.pone.0103884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet. 2001;27:293–298. doi: 10.1038/85855. This paper shows that, in addition to Tbx1, inactivation of Crkl on 22q11.2 can result in physical malformations observed in 22q11.2DS.
- 108. Racedo SE, et al. Mouse and human CRKL is dosage sensitive for cardiac outflow tract formation. Am. J. Hum. Genet. 2015;96:235–244. doi: 10.1016/j.ajhg.2014.12.025. The combination of human and mouse genetics has shed new light on the function of CRKL in the formation of the cardiac outflow tract.
- 109.Zheng P, et al. Molecular mechanisms of functional natural killer deficiency in patients with partial DiGeorge syndrome. J. Allergy Clin. Immunol. 2015;135:1293–1302. doi: 10.1016/j.jaci.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bedeschi MF, et al. Unmasking of a recessive SCARF2 mutation by a 22q11.12 de novo deletion in a patient with Van den Ende–Gupta syndrome. Mol. Syndromol. 2010;1:239–245. doi: 10.1159/000328135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bassett AS, Caluseriu O, Weksberg R, Young DA, Chow EW. Catechol-O-methyl transferase and expression of schizophrenia in 73 adults with 22q11 deletion syndrome. Biol. Psychiatry. 2007;61:1135–1140. doi: 10.1016/j.biopsych.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 113.Gothelf D, et al. Risk factors and the evolution of psychosis in 22q11.2 deletion syndrome: a longitudinal 2-site study. J. Am. Acad. Child Adolesc. Psychiatry. 2013;52:1192.e3–1203.e3. doi: 10.1016/j.jaac.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 114.Philip N, Bassett AS. Cognitive, behavioural and psychiatric phenotype in 22q11.2 deletion syndrome. Behav. Genet. 2011;41:403–412. doi: 10.1007/s10519-011-9468-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goodman BK, Rutberg J, Lin WW, Pulver AE, Thomas GH. Hyperprolinaemia in patients with deletion (22)(q11.2) syndrome. J. Inherit. Metab. Dis. 2000;23:847–848. doi: 10.1023/a:1026773005303. [DOI] [PubMed] [Google Scholar]
- 116.Magnée MJ, Lamme VA, de Sain-van der Velden MG, Vorstman JA, Kemner C. Proline and COMT status affect visual connectivity in children with 22q11.2 deletion syndrome. PLoS ONE. 2011;6:e25882. doi: 10.1371/journal.pone.0025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paronett EM, Meechan DW, Karpinski BA, LaMantia A-S, Maynard TM. Ranbp1, deleted in DiGeorge/22q11.2 deletion syndrome, is a microcephaly gene that selectively disrupts layer 2/3 cortical projection neuron generation. Cereb. Cortex. 2014;25:3977–3993. doi: 10.1093/cercor/bhu285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raux G, et al. Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum. Mol. Genet. 2007;16:83–91. doi: 10.1093/hmg/ddl443. [DOI] [PubMed] [Google Scholar]
- 119.Vorstman JAS, et al. Proline affects brain function in 22q11DS children with the low activity COMT158 allele. Neuropsychopharmacology. 2008;34:739–746. doi: 10.1038/npp.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Newbern J, et al. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc. Natl Acad. Sci. USA. 2008;105:17115–17120. doi: 10.1073/pnas.0805239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dykes IM, et al. HIC2 is a novel dosage-dependent regulator of cardiac development located within the distal 22q11 deletion syndrome region. Circ. Res. 2014;115:23–31. doi: 10.1161/CIRCRESAHA.115.303300. [DOI] [PubMed] [Google Scholar]
- 122.Toritsuka M, et al. Deficits in microRNA-mediated Cxcr4/Cxcl12 signaling in neurodevelopmental deficits in a 22q11 deletion syndrome mouse model. Proc. Natl Acad. Sci. USA. 2013;110:17552–17557. doi: 10.1073/pnas.1312661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Swaby JA, et al. Complex congenital heart disease in unaffected relatives of adults with 22q11.2 deletion syndrome. Am. J. Cardiol. 2011;107:466–471. doi: 10.1016/j.amjcard.2010.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Merico D, et al. Whole-genome sequencing suggests schizophrenia risk mechanisms in humans with 22q11.2 deletion syndrome. G3 (Bethesda) 2015;5:2453–2461. doi: 10.1534/g3.115.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Swillen A, McDonald-McGinn D. Developmental trajectories in 22q11.2 deletion syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2015;169:172–181. doi: 10.1002/ajmg.c.31435. This paper provides a current state of the art perspective on developmental trajectories with appropriate interventions.
- 126. Fung WL, et al. Practical guidelines for managing adults with 22q11.2 deletion syndrome. Genet. Med. 2015;17:599–609. doi: 10.1038/gim.2014.175. This paper (for adults) and reference 22 (for children) provide a comprehensive overview of all domains to be covered in the multidisciplinary management of patients with 22q11.2DS.
- 127.Vergaelen E, et al. 3 generation pedigree with paternal transmission of the 22q11.2 deletion syndrome: intrafamilial phenotypic variability. Eur. J. Med. Genet. 2015;58:244–248. doi: 10.1016/j.ejmg.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 128.McElhinney DB, McDonald-McGinn D, Zackai EH, Goldmuntz E. Cardiovascular anomalies in patients diagnosed with a chromosome 22q11 deletion beyond 6 months of age. Pediatrics. 2001;108:E104. doi: 10.1542/peds.108.6.e104. [DOI] [PubMed] [Google Scholar]
- 129.John AS, McDonald-McGinn DM, Zackai EH, Goldmuntz E. Aortic root dilation in patients with 22q11.2 deletion syndrome. Am. J. Med. Genet. A. 2009;149A:939–942. doi: 10.1002/ajmg.a.32770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Piliero LM, Sanford AN, McDonald-McGinn DM, Zackai EH, Sullivan KE. T-cell homeostasis in humans with thymic hypoplasia due to chromosome 22q11.2 deletion syndrome. Blood. 2004;103:1020–11025. doi: 10.1182/blood-2003-08-2824. The consequences of thymic hypoplasia were elucidated in this paper. It highlights the dynamic nature of immunodeficiency over time in this syndrome.
- 131.Sullivan KE, et al. Lack of correlation between impaired T cell production, immunodeficiency, and other phenotypic features in chromosome 22q11.2 deletion syndromes. Clin. Immunol. Immunopathol. 1998;86:141–146. doi: 10.1006/clin.1997.4463. [DOI] [PubMed] [Google Scholar]
- 132.Sullivan KE, McDonald-McGinn D, Zackai EH. CD4+CD25+ T-cell production in healthy humans and in patients with thymic hypoplasia. Clin. Diagn. Lab. Immunol. 2002;9:1129–1131. doi: 10.1128/CDLI.9.5.1129-1131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sullivan KE, et al. Longitudinal analysis of lymphocyte function and numbers in the first year of life in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Clin. Diagn. Lab. Immunol. 1999;6:906–911. doi: 10.1128/cdli.6.6.906-911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jawad AF, et al. A prospective study of influenza vaccination and a comparison of immunologic parameters in children and adults with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) J. Clin. Immunol. 2011;31:927–935. doi: 10.1007/s10875-011-9569-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Perez EE, Bokszczanin A, McDonald-McGinn D, Zackai EH, Sullivan KE. Safety of live viral vaccines in patients with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Pediatrics. 2003;112:e325. doi: 10.1542/peds.112.4.e325. [DOI] [PubMed] [Google Scholar]
- 136.Smith CA, et al. Increased prevalence of immunoglobulin A deficiency in patients with the chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Clin. Diagn. Lab. Immunol. 1998;5:415–417. doi: 10.1128/cdli.5.3.415-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Staple L, Andrews T, McDonald-McGinn D, Zackai E, Sullivan KE. Allergies in patients with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) and patients with chronic granulomatous disease. Pediatr. Allergy Immunol. 2005;16:226–230. doi: 10.1111/j.1399-3038.2005.00259.x. [DOI] [PubMed] [Google Scholar]
- 138.Zemble R, et al. Secondary immunologic consequences in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Clin. Immunol. 2010;136:409–418. doi: 10.1016/j.clim.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sullivan KE, et al. Juvenile rheumatoid arthritis-like polyarthritis in chromosome 22q11.2 deletion syndrome (DiGeorge anomalad/velocardiofacial syndrome/conotruncal anomaly face syndrome) Arthritis Rheum. 1997;40:430–436. doi: 10.1002/art.1780400307. [DOI] [PubMed] [Google Scholar]
- 140.Lawrence S, McDonald-McGinn DM, Zackai E, Sullivan KE. Thrombocytopenia in patients with chromosome 22q11.2 deletion syndrome. J. Pediatr. 2003;143:277–278. doi: 10.1067/S0022-3476(03)00248-8. [DOI] [PubMed] [Google Scholar]
- 141.Kratz CP, et al. Evans syndrome in a patient with chromosome 22q11.2 deletion syndrome: a case report. Pediatr. Hematol. Oncol. 2003;20:167–172. doi: 10.1080/0880010390158685. [DOI] [PubMed] [Google Scholar]
- 142.Kawame H, et al. Graves’ disease in patients with 22q11.2 deletion. J. Pediatr. 2001;139:892–895. doi: 10.1067/mpd.2001.119448. [DOI] [PubMed] [Google Scholar]
- 143.Bale PM, Sotelo-Avila C. Maldescent of the thymus: 34 necropsy and 10 surgical cases, including 7 thymuses medial to the mandible. Pediatr. Pathol. 1993;13:181–190. doi: 10.3109/15513819309048205. [DOI] [PubMed] [Google Scholar]
- 144.Chinen J, Rosenblatt HM, Smith EO, Shearer WT, Noroski LM. Long-term assessment of T-cell populations in DiGeorge syndrome. J. Allergy Clin. Immunol. 2003;111:573–579. doi: 10.1067/mai.2003.165. [DOI] [PubMed] [Google Scholar]
- 145.Dyce O, et al. Otolaryngologic manifestations of the 22q11.2 deletion syndrome. Arch. Otolaryngol. Head Neck Surg. 2002;128:1408–1412. doi: 10.1001/archotol.128.12.1408. [DOI] [PubMed] [Google Scholar]
- 146.Hamilton S, Husein M, Dworschak-Stokan A. Velopharyngeal insufficiency clinic: the first 18 months. J. Otolaryngol. Head Neck Surg. 2008;37:586–590. [PubMed] [Google Scholar]
- 147.Solot CB, et al. Communication issues in 22q11.2 deletion syndrome: children at risk. Genet. Med. 2001;3:67–71. doi: 10.1097/00125817-200101000-00015. [DOI] [PubMed] [Google Scholar]
- 148.Ruotolo RA, et al. Velopharyngeal anatomy in 22q11.2 deletion syndrome: a three-dimensional cephalometric analysis. Cleft Palate Craniofac. J. 2006;43:446–456. doi: 10.1597/04-193R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Widdershoven JC, et al. A candidate gene approach to identify modifiers of the palatal phenotype in 22q11.2 deletion syndrome patients. Int. J. Pediatr. Otorhinolaryngol. 2013;77:123–127. doi: 10.1016/j.ijporl.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stransky C, et al. Perioperative risk factors in patients with 22q11.2 deletion syndrome requiring surgery for velopharyngeal dysfunction. Cleft Palate Craniofac. J. 2015;52:183–191. doi: 10.1597/13-206. [DOI] [PubMed] [Google Scholar]
- 151.Forbes BJ, et al. Ocular findings in the chromosome 22q11.2 deletion syndrome. J. AAPOS. 2007;11:179–182. doi: 10.1016/j.jaapos.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 152.Cheung EN, et al. Prevalence of hypocalcemia and its associated features in 22q11.2 deletion syndrome. Clin. Endocrinol. 2014;81:190–196. doi: 10.1111/cen.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bassett AS, et al. Clinical features of 78 adults with 22q11 deletion syndrome. Am. J. Med. Genet. A. 2005;138:307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weinzimer SA. Endocrine aspects of the 22q11.2 deletion syndrome. Genet. Med. 2001;3:19–22. doi: 10.1097/00125817-200101000-00005. [DOI] [PubMed] [Google Scholar]
- 155.Digilio MC, et al. Auxological evaluation in patients with DiGeorge/velocardiofacial syndrome (deletion 22q11.2 syndrome) Genet. Med. 2001;3:30–33. doi: 10.1097/00125817-200101000-00007. [DOI] [PubMed] [Google Scholar]
- 156.Habel A, McGinn M-J, 2nd, Zackai EH, Unanue N, McDonald-McGinn DM. Syndrome-specific growth charts for 22q11.2 deletion syndrome in Caucasian children. Am. J. Med. Genet. A. 2012;158A:2665–2671. doi: 10.1002/ajmg.a.35426. [DOI] [PubMed] [Google Scholar]
- 157.Van L, et al. Fetal growth and gestational factors as predictors of schizophrenia in 22q11.2 deletion syndrome. Genet. Med. 2015 doi: 10.1038/gim.2015.84. http://dx.doi.org/10.1038/gim.2015.84 [DOI] [PubMed] [Google Scholar]
- 158.Eicher PS, et al. Dysphagia in children with a 22q11.2 deletion: unusual pattern found on modified barium swallow. J. Pediatr. 2000;137:158–164. doi: 10.1067/mpd.2000.105356. [DOI] [PubMed] [Google Scholar]
- 159.Digilio MC, Marino B, Bagolan P, Giannotti A, Dallapiccola B. Microdeletion 22q11 and oesophageal atresia. J. Med. Genet. 1999;36:137–139. [PMC free article] [PubMed] [Google Scholar]
- 160.Oskarsdottir S, Belfrage M, Sandstedt E, Viggedal G, Uvebrant P. Disabilities and cognition in children and adolescents with 22q11 deletion syndrome. Dev. Med. Child Neurol. 2005;47:177–184. doi: 10.1017/s0012162205000320. [DOI] [PubMed] [Google Scholar]
- 161.Wu H-Y, et al. Genitourinary malformations in chromosome 22q11.2 deletion. J. Urol. 2002;168:2564–2565. doi: 10.1016/S0022-5347(05)64215-2. [DOI] [PubMed] [Google Scholar]
- 162.Devriendt K, Swillen A, Fryns JP, Proesmans W, Gewillig M. Renal and urological tract malformations caused by a 22q11 deletion. J. Med. Genet. 1996;33:349. doi: 10.1136/jmg.33.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sundaram UT, et al. Primary amenorrhea and absent uterus in the 22q11.2 deletion syndrome. Am. J. Med. Genet. A. 2007;143A:2016–2018. doi: 10.1002/ajmg.a.31736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barnett C, Langer JC, Hinek A, Bradley TJ, Chitayat D. Looking past the lump: genetic aspects of inguinal hernia in children. J. Pediatr. Surg. 2009;44:1423–1431. doi: 10.1016/j.jpedsurg.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 165.Binenbaum G, et al. Sclerocornea associated with the chromosome 22q11.2 deletion syndrome. Am. J. Med. Genet. A. 2008;146A:904–909. doi: 10.1002/ajmg.a.32156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bingham PM, Lynch D, McDonald-McGinn D, Zackai E. Polymicrogyria in chromosome 22 delection syndrome. Neurology. 1998;51:1500–1502. doi: 10.1212/wnl.51.5.1500. [DOI] [PubMed] [Google Scholar]
- 167.Ming JE, et al. Skeletal anomalies and deformities in patients with deletions of 22q11. Am. J. Med. Genet. 1997;72:210–215. doi: 10.1002/(sici)1096-8628(19971017)72:2<210::aid-ajmg16>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 168.Ricchetti ET, et al. Radiographic study of the upper cervical spine in the 22q11.2 deletion syndrome. J. Bone Joint Surg. Am. 2004;86:1751–1760. doi: 10.2106/00004623-200408000-00020. [DOI] [PubMed] [Google Scholar]
- 169.McDonald-McGinn DM, et al. Malignancy in chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Am. J. Med. Genet. A. 2006;140:906–909. doi: 10.1002/ajmg.a.31199. [DOI] [PubMed] [Google Scholar]
- 170.Butcher N, et al. Association between early-onset Parkinson disease and 22q11.2 deletion syndrome: identification of a novel genetic form of Parkinson disease and its clinical implications. JAMA Neurol. 2013;70:1359–1366. doi: 10.1001/jamaneurol.2013.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Swillen A, et al. Early motor development in young children with 22q.11 deletion syndrome and a conotruncal heart defect. Dev. Med. Child Neurol. 2005;47:797–802. doi: 10.1017/S0012162205001696. [DOI] [PubMed] [Google Scholar]
- 172.Solot CB, et al. Communication disorders in the 22Q11.2 microdeletion syndrome. J. Commun. Disord. 2000;33:187–203. doi: 10.1016/s0021-9924(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 173.Swillen A, et al. Intelligence and psychosocial adjustment in velocardiofacial syndrome: a study of 37 children and adolescents with VCFS. J. Med. Genet. 1997;34:453–458. doi: 10.1136/jmg.34.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.De Smedt B, et al. Intellectual abilities in a large sample of children with velo-cardio-facial syndrome: an update. J. Intellect. Disabil. Res. 2007;51:666–670. doi: 10.1111/j.1365-2788.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- 175.De Smedt B, Swillen A, Verschaffel L, Ghesquiere P. Mathematical learning disabilities in children with 22q11.2 deletion syndrome: a review. Dev. Disabil. Res. Rev. 2009;15:4–10. doi: 10.1002/ddrr.44. [DOI] [PubMed] [Google Scholar]
- 176.Wang PP, Woodin MF, Kreps-Falk R, Moss EM. Research on behavioral phenotypes: velocardiofacial syndrome (deletion 22q11.2) Dev. Med. Child Neurol. 2000;42:422–427. doi: 10.1017/s0012162200000785. [DOI] [PubMed] [Google Scholar]
- 177.Glaser B, et al. Language skills in children with velocardiofacial syndrome (deletion 22q11.2) J. Pediatr. 2002;140:753–758. doi: 10.1067/mpd.2002.124774. [DOI] [PubMed] [Google Scholar]
- 178.Evers LJ, et al. Psychopathology in adults with 22q11 deletion syndrome and moderate and severe intellectual disability. J. Intellect. Disabil. Res. 2014;58:915–925. doi: 10.1111/jir.12117. [DOI] [PubMed] [Google Scholar]
- 179.Cheung EN, et al. Neonatal hypocalcemia, neonatal seizures, and intellectual disability in 22q11.2 deletion syndrome. Genet. Med. 2014;16:40–44. doi: 10.1038/gim.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Duijff SN, et al. Cognitive development in children with 22q11.2 deletion syndrome. Br. J. Psychiatry. 2012;200:462–468. doi: 10.1192/bjp.bp.111.097139. This paper presents a longitudinal data study on cognitive development in a large cohort of children.
- 181. Vorstman JA, et al. Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry. 2015;72:377–385. doi: 10.1001/jamapsychiatry.2014.2671. Using data from a large collaborative effort (The International 22q11.2 Brain and Behavior Consortium), this paper shows the association of cognitive decline as an antecedent to the onset of psychosis. Drawing from this same consortium, reference 182, reports on the psychiatric disorders observed in this population.
- 182.Schneider M, et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 deletion syndrome. Am. J. Psychiatry. 2014;171:627–639. doi: 10.1176/appi.ajp.2013.13070864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fung WL, et al. Elevated prevalence of generalized anxiety disorder in adults with 22q11.2 deletion syndrome. Am. J. Psychiatry. 2010;167:998. doi: 10.1176/appi.ajp.2010.09101463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bassett AS, Chow EW. 22q11 deletion syndrome: a genetic subtype of schizophrenia. Biol. Psychiatry. 1999;46:882–891. doi: 10.1016/s0006-3223(99)00114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Amelsvoort TV, et al. Cognitive deficits associated with schizophrenia in velo-cardio-facial syndrome. Schizophr. Res. 2004;70:223–232. doi: 10.1016/j.schres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 186.Bassett AS, et al. The schizophrenia phenotype in 22q11 deletion syndrome. Am. J. Psychiatry. 2003;160:1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chow EW, Watson M, Young DA, Bassett AS. Neurocognitive profile in 22q11 deletion syndrome and schizophrenia. Schizophr. Res. 2006;87:270–278. doi: 10.1016/j.schres.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stoddard J, Niendam T, Hendren R, Carter C, Simon TJ. Attenuated positive symptoms of psychosis in adolescents with chromosome 22q11.2 deletion syndrome. Schizophr. Res. 2010;118:118–121. doi: 10.1016/j.schres.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Butcher NJ, et al. Response to clozapine in a clinically identifiable subtype of schizophrenia. Br. J. Psychiatry. 2015;206:484–491. doi: 10.1192/bjp.bp.114.151837. This paper is notable as it is the first report on the functional outcome of a large group (>100 subjects) of adults with 22q11.2 DS.
- 190.Baker K, Vorstman JAS. Is there a core neuropsychiatric phenotype in 22q11.2 deletion syndrome? Curr. Opin. Neurol. 2012;25:131–137. doi: 10.1097/WCO.0b013e328352dd58. [DOI] [PubMed] [Google Scholar]
- 191.Chan C, Costain G, Chow EWC, Bassett AS. Reproductive health issues for adults with a common genomic disorder. J. Genet. Couns. 2015;24:810–821. doi: 10.1007/s10897-014-9811-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Balci A, et al. Prospective validation and assessment of cardiovascular and offspring risk models for pregnant women with congenital heart disease. Heart. 2014;100:1373–1381. doi: 10.1136/heartjnl-2014-305597. [DOI] [PubMed] [Google Scholar]
- 193.Grewal J, Silversides CK, Colman JM. Pregnancy in women with heart disease: risk assessment and management of heart failure. Heart Fail. Clin. 2014;10:117–129. doi: 10.1016/j.hfc.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 194.Sorensen KM, et al. Detecting 22q11.2 deletions by use of multiplex ligation-dependent probe amplification on DNA from neonatal dried blood spot samples. J. Mol. Diagn. 2010;12:147–151. doi: 10.2353/jmoldx.2010.090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vorstman JA, et al. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum. Mut. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sandrin-Garcia P, et al. Recurrent 22q11.2 deletion in a sibship suggestive of parental germline mosaicism in velocardiofacial syndrome. Clin. Genet. 2002;61:380–383. doi: 10.1034/j.1399-0004.2002.610511.x. [DOI] [PubMed] [Google Scholar]
- 197.Gross SJ, et al. Clinical experience with single-nucleotide polymorphism-based noninvasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obstet. Gynecol. 2015 doi: 10.1002/uog.15754. http://dx.doi.org/10.1002/uog.15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bretelle F, et al. Prenatal and postnatal diagnosis of 22q11.2 deletion syndrome. Eur. J. Med. Genet. 2010;53:367–370. doi: 10.1016/j.ejmg.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 199.Carotti A, et al. Cardiac defects and results of cardiac surgery in 22q11.2 deletion syndrome. Dev. Disabil. Res. Rev. 2008;14:35–42. doi: 10.1002/ddrr.6. [DOI] [PubMed] [Google Scholar]
- 200.Michielon G, et al. Impact of DEL22q11, trisomy 21, and other genetic syndromes on surgical outcome of conotruncal heart defects. J. Thorac. Cardiovasc. Surg. 2009;138:565.e2–570.e2. doi: 10.1016/j.jtcvs.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 201.Mercer-Rosa L, Pinto N, Yang W, Tanel R, Goldmuntz E. 22q11.2 deletion syndrome is associated with perioperative outcome in tetralogy of Fallot. J. Thorac. Cardiovasc. Surg. 2013;146:868–873. doi: 10.1016/j.jtcvs.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.O’Byrne ML, et al. 22q11.2 deletion syndrome is associated with increased perioperative events and more complicated postoperative course in infants undergoing infant operative correction of truncus arteriosus communis or interrupted aortic arch. J. Thorac. Cardiovasc.Surg. 2014;148:1597–1605. doi: 10.1016/j.jtcvs.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Warnes CA, et al. ACC/AHA 2008 guidelines for the management of adults with congenital heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. College Cardiol. 2008;52:e143–e263. doi: 10.1016/j.jacc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 204.Lin AE, et al. Adults with genetic syndromes and cardiovascular abnormalities: clinical history and management. Genet. Med. 2008;10:469–494. doi: 10.1097/GIM.0b013e3181772111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Butcher N, et al. Functional outcomes of adults with 22q11.2 deletion syndrome. Genet. Med. 2012;14:836–843. doi: 10.1038/gim.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hofstetter AM, et al. Live vaccine use and safety in DiGeorge syndrome. Pediatrics. 2014;133:e946–e954. doi: 10.1542/peds.2013-0831. [DOI] [PubMed] [Google Scholar]
- 207. Moylett EH, Wasan AN, Noroski LM, Shearer WT. Live viral vaccines in patients with partial DiGeorge syndrome: clinical experience and cellular immunity. Clin. Immunol. 2004;112:106–112. doi: 10.1016/j.clim.2004.02.008. This paper and reference 135 were the first to define the safety of live viral vaccines in this syndrome.
- 208.Bjork AH, Oskarsdottir S, Andersson BA, Friman V. Antibody deficiency in adults with 22q11.2 deletion syndrome. Am. J. Med. Genet. A. 2012;158A:1934–1940. doi: 10.1002/ajmg.a.35484. [DOI] [PubMed] [Google Scholar]
- 209.Gennery AR, et al. Antibody deficiency and autoimmunity in 22q11.2 deletion syndrome. Arch. Dis. Child. 2002;86:422–425. doi: 10.1136/adc.86.6.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Maggadottir SM, Sullivan KE. The diverse clinical features of chromosome 22q11.2 deletion syndrome (DiGeorge syndrome) J. Allergy Clin. Immunol. Pract. 2013;1:589–594. doi: 10.1016/j.jaip.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 211.Basta MN, et al. A 35-year experience with syndromic cleft palate repair: operative outcomes and long-term speech function. Ann. Plast. Surg. 2014;73:S130–S135. doi: 10.1097/SAP.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 212.Kennedy WP, et al. 22q11.2 deletion syndrome and obstructive sleep apnea. Int. J. Pediatr. Otorhinolaryngol. 2014;78:1360–1364. doi: 10.1016/j.ijporl.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 213.Sobin C, Monk SH, Kiley-Brabeck K, Khuri J, Karayiorgou M. Neuromotor deficits in children with the 22q11 deletion syndrome. Mov. Disord. 2006;21:2082–2089. doi: 10.1002/mds.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Van Aken K, Caeyenberghs K, Smits-Engelsman B, Swillen A. The motor profile of primary school-age children with a 22q11.2 deletion syndrome (22q11.2DS) and an age- and IQ-matched control group. Child Neuropsychol. 2009;15:532–542. doi: 10.1080/09297040902740678. [DOI] [PubMed] [Google Scholar]
- 215.Vorstman JAS, et al. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 216.Dori N, Green T, Weizman A, Gothelf D. The effectiveness and safety of antipsychotic and antidepressant medications in individuals with 22q11.2 deletion syndrome. J. Child Adolesc. Psychopharmacol. 2015 doi: 10.1089/cap.2014.0075. http://dx.doi.org/10.1089/cap.2014.0075. [DOI] [PubMed] [Google Scholar]
- 217.Gothelf D, et al. Obsessive–compulsive disorder in patients with velocardiofacial (22q11 deletion) syndrome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;126B:99–105. doi: 10.1002/ajmg.b.20124. [DOI] [PubMed] [Google Scholar]
- 218.Gothelf D, et al. Methylphenidate treatment for attention-deficit/hyperactivity disorder in children and adolescents with velocardiofacial syndrome: an open-label study. J. Clin. Psychiatry. 2003;64:1163–1169. doi: 10.4088/jcp.v64n1004. [DOI] [PubMed] [Google Scholar]
- 219.Karas DJ, Costain G, Chow EW, Bassett AS. Perceived burden and neuropsychiatric morbidities in adults with 22q11.2 deletion syndrome. J. Intellect. Disabil. Res. 2014;58:198–210. doi: 10.1111/j.1365-2788.2012.01639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mercer-Rosa L, et al. 22q11.2 deletion status and disease burden in children and adolescents with tetralogy of Fallot. Circ. Cardiovasc. Genet. 2015;8:74–81. doi: 10.1161/CIRCGENETICS.114.000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Briegel W, Schneider M, Schwab KO. 22q11.2 deletion: handicap-related problems and coping strategies of primary caregivers. Z. Kinder Jugendpsychiatr. Psychother. 2009;37:535–540. doi: 10.1024/1422-4917.37.6.535. (in German) [DOI] [PubMed] [Google Scholar]
- 222.Looman WS, Thurmes AK, O’Conner-Von SK. Quality of life among children with velocardiofacial syndrome. Cleft Palate Craniofac. J. 2010;47:273–283. doi: 10.1597/09-009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mahle WT, et al. Deletion of chromosome 22q11.2 and outcome in patients with pulmonary atresia and ventricular septal defect. Ann. Thorac. Surg. 2003;76:567–571. doi: 10.1016/s0003-4975(03)00516-2. [DOI] [PubMed] [Google Scholar]
- 224.Woodin M, et al. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet. Med. 2001;3:34–39. doi: 10.1097/00125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 225.Jacobson C, et al. Core neuropsychological characteristics of children and adolescents with 22q11.2 deletion. J. Intellectual Disabil. Res. 2010;54:701–713. doi: 10.1111/j.1365-2788.2010.01298.x. [DOI] [PubMed] [Google Scholar]
- 226.Driscoll DA. Molecular and genetic aspects of DiGeorge/velocardiofacial syndrome. Methods. Mol. Med. 2006;126:43–55. doi: 10.1385/1-59745-088-X:43. [DOI] [PubMed] [Google Scholar]
- 227.Mlynarski EE, et al. Copy-number variation of the glucose transporter gene SLC2A3 and congenital heart defects in the 22q11.2 deletion syndrome. Am. J. Hum. Genet. 2015;96:753–764. doi: 10.1016/j.ajhg.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chung JH, et al. Whole-genome sequencing and integrative genomic analysis approach on two 22q11.2 deletion syndrome family trios for genotype to phenotype correlations. Hum. Mut. 2015;36:797–807. doi: 10.1002/humu.22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Guo T, et al. Genotype and cardiovascular phenotype correlations with TBXn 1,022 velo-cardio-facial/DiGeorge/22q11.2 deletion syndrome patients. Hum. Mut. 2011;32:1278–1289. doi: 10.1002/humu.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Budarf ML, et al. Identification of a patient with Bernard–Soulier syndrome and a deletion in the DiGeorge/velo-cardio-facial chromosomal region in 22q11.2. Hum. Mol. Genet. 1995;4:763–766. doi: 10.1093/hmg/4.4.763. This paper and reference 68 highlight the possibility of unmasking an autosomal recessive condition to explain atypical phenotypes and to identify important genes associated with 22q11.2DS beyond TBX1.
- 231.Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- 232.Wapner RJ, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am. J. Obstet. Gynecol. 2015;212:332.e1–332.e9. doi: 10.1016/j.ajog.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 233.Vialard F, et al. Prenatal BACs-on-Beads™: the prospective experience of five prenatal diagnosis laboratories. Prenat. Diagn. 2012;32:329–335. doi: 10.1002/pd.2934. [DOI] [PubMed] [Google Scholar]
- 234.Koontz D, Baecher K, Kobrynski L, Nikolova S, Gallagher M. A pyrosequencing-based assay for the rapid detection of the 22q11.2 deletion in DNA from buccal and dried blood spot samples. J. Mol. Diagn. 2014;16:533–540. doi: 10.1016/j.jmoldx.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Pretto D, Maar D, Yrigollen CM, Regan J, Tassone F. Screening newborn blood spots for 22q11.2 deletion syndrome using multiplex droplet digital PCR. Clin. Chem. 2015;61:182–190. doi: 10.1373/clinchem.2014.230086. [DOI] [PubMed] [Google Scholar]