Abstract
An ethylene-inducing xylanase (EIX) is a potent elicitor of plant defense responses in specific cultivars of tobacco (Nicotiana tabacum) and tomato (Lycopersicon esculentum). The LeEix locus in tomatoes was characterized by map-based cloning, which led to the identification of a novel gene cluster from which two members (LeEix1 and LeEix2) were isolated. Similar to the tomato Ve resistance genes in tomato plants, the deduced amino acid sequences encoded by LeEix1 and LeEix2 contain a Leu zipper, an extracellular Leu-rich repeat domain with glycosylation signals, a transmembrane domain, and a C-terminal domain with a mammalian endocytosis signal. Silencing expression of the LeEix genes prevented the binding of EIX to cells of an EIX-responsive plant and thus inhibited the hypersensitive response. Overexpression of either LeEix1 or LeEix2 genes in EIX-nonresponsive tobacco plants enabled the binding of EIX, although only LeEix2 could transmit the signal that induced the hypersensitive response. Overexpressing LeEix2 in mammalian COS-7 cells enables binding of EIX, indicating physical interaction between the EIX elicitor and LeEix2 gene product. Structural analysis of the LeEix proteins suggests that they belong to a class of cell-surface glycoproteins with a signal for receptor-mediated endocytosis. Mutating the endocytosis signal in LeEix2 (Tyr 993 to Ala) abolished its ability to induce the hypersensitive response, suggesting that endocytosis plays a key role in the signal transduction pathway.
INTRODUCTION
Plants are constantly under attack by such pathogens as bacteria, fungi, viruses, and nematodes, which can potentially cause significant crop losses. Plants have evolved numerous defense mechanisms to protect themselves from pathogens (Yang et al., 1997). These include the strengthening of mechanical barriers, oxidative burst, de novo production of antimicrobial compounds such as pathogenesis-related proteins and phytoalexins, and the induction of the hypersensitive response (HR) mechanism, where the tissue surrounding the infection site dies and confines pathogen growth (Dangl and Holub, 1997; Greenberg, 1997; Morel and Dangl, 1997; Somssich and Hahlbrock, 1998).
These defense mechanisms are triggered when a pathogen-derived signal (an Avr gene product) or other organic component (termed elicitor) is recognized by a plant disease resistance protein (R-protein). The elicitors belong to a diverse range of molecular types (Boller, 1995). Many do not appear to be determinants of race-specific cultivars and are considered non-race-specific elicitors (Ebel and Cosio, 1994; Boller, 1995). The ligand–receptor interaction has been used to explain the gene for gene specificity model. In this model, the R-protein acts as a receptor for the elicitor (Gabriel and Rolfe, 1990).
More than 40 R-genes have been isolated from different plant species, including both monocots and dicots, out of which the corresponding Avr factors have been isolated only for some. These R-genes are effective against bacterial, viral, nematode, and fungal pathogens (Jones et al., 1994; Bent, 1996; Baker et al., 1997; Martin et al., 2003). R-genes encode proteins with certain common motifs and have been divided into five classes (Dangl and Jones, 2001; Martin et al., 2003): (1) intracellular protein kinases (e.g., Pto); (2) receptor-like protein kinases with an extracellular Leu-rich repeat (LRR) domain (e.g., Xa21 and FLS2); (3) intracellular proteins with a region of LRRs, a nucleotide binding site, and a Leu zipper motif (e.g., Pi-ta, RPS2, and RPM1); (4) intracellular nucleotide binding site-LRR proteins with a region similar to the Toll and interleukin-1 receptor proteins (e.g., N, L6, and RPP5); and (5) LRR proteins that encode membrane-bound extracellular proteins (e.g., Cf-2 and Cf-9). Proteins with these motifs are known to play important roles in signal recognition and transduction in mammals. Despite these important insights into R-protein structures, much remains to be elucidated regarding the molecular mechanisms by which proteins encoded by R-genes recognize pathogen elicitors and transduce this information in the plant cell to initiate a cascade of defense response events.
The physical interaction between Avr gene products and their corresponding R-gene products has been demonstrated in a few cases (Scofield et al., 1996; Tang et al., 1996; Jia et al., 2000; Leister and Katagiri, 2000). Pto kinase interacts physically with AvrPto, and mutations in the AvrPto or Pto that disrupt the interaction were also found to abolish the induction of disease resistance (Scofield et al., 1996; Tang et al., 1996). Similarly, interaction between the cytoplasmic receptor Pi-ta and the corresponding AVR-Pita is required to induce resistance response (Jia et al., 2000). While these interactions are crucial for plant defense induction, the Avr9 elicitor exhibits high affinity binding to plasma membranes isolated from both resistant and susceptible tomato (Lycopersicon esculentum) cultivars (Kooman-Gersmann et al., 1996; Luderer et al., 2001).
The 22-kD fungal protein ethylene-inducing xylanase (EIX; Fuchs et al., 1989) induces ethylene biosynthesis, electrolyte leakage, pathogensis-related protein expression, and HR in specific plant species and/or varieties (Bailey et al., 1990, 1993; Ron et al., 2000; Elbaz et al., 2002). Analysis of EIX mutants lacking enzymatic activity (β-1-4-endoxylanase) but retaining elicitation activity showed that the xylanase activity is not required for the HR elicitation process because the protein per se functions as the elicitor (Enkerli et al., 1999; Furman-Matarasso et al., 1999; Rotblat et al., 2002). In this system, the EIX was shown to specifically bind to the plasma membrane of both the tomato and tobacco (Nicotiana tabacum) responding cultivars (Hanania and Avni, 1997).
The response to EIX in tobacco and tomato cultivars is controlled by a single dominant locus (Bailey et al., 1993; Ron et al., 2000). The EIX-responding locus (LeEix) was mapped to the short arm of chromosome 7 of the tomato cultivar, and a yeast artificial chromosome (YAC) clone carrying a 300-kb DNA segment, derived from the LeEix region (Ron et al., 2000), was isolated. Mapping the ends of this YAC clone showed that it spans the LeEix locus (Ron et al., 2000).
This article describes the identification of a novel gene family corresponding to the LeEix locus of tomato. Two members of this family (LeEix1 and LeEix2) were isolated and show homology to the Ve and Cf resistance genes in tomato. Using silencing and complementation experiments, we show that both these genes are capable of binding the EIX elicitor independently. However, only LeEix2 can transmit the HR induction signal. Furthermore, induction of HR is dependent on the endocytosis signal.
RESULTS
Identification of a Cluster of Cf Homologs at the LeEix Locus
The LeEix tomato locus was previously mapped to the short arm of chromosome 7, between restriction fragment length polymorphism (RFLP) markers TG61 and TG131 (Ron et al., 2000). Two YACs, YAC 317E4 and YAC 294G5, containing tomato genomic DNA were isolated from this region. We showed that YAC 317E4 encompasses the LeEix locus, whereas the end of YAC 294G5 cosegregates with the LeEix locus (Ron et al., 2000). Positional cloning of the LeEix locus was performed as schematically shown in Figure 1. Using the end of YAC 294G5 (294R), we screened a tomato binary BAC2 (BiBAC2) library (Hamilton, 1997) and identified a single BAC (8L5) containing the 294R marker (Ron et al., 2000). Mapping the BAC end clones on the recombinant population showed the left end of BAC 8L5 (L5L) cosegregated with the LeEix, whereas the right end of the clone cosegregated with the RFLP marker TG131 (Figure 1). We searched the DNA databank using the sequence of the two clones (L5L and 294R). The two clones showed homology to many plant resistance genes and particularly to a family of extracellular transmembrane LRRs (e.g., Cf-9, Cf-2, Cf-5, Ve, and HcrVf). The L5L clone showed homology to the 3′ end of these genes, whereas 294R showed homology to their 5′ end (data not shown). To obtain a full-length cDNA clone of the candidate LeEix gene(s), we screened two different cDNA libraries generated from leaves of L. esculentum (106 clones from each library) using both probes. Seven full-length cDNA clones, hybridized to L5L and 294R, were identified. Partial sequencing of these clones and restriction analysis with several enzymes showed them to be identical (designated LeEix1).
Figure 1.
Schematic, Genetic, and Physical Representation of the LeEix Region.
(A) Genetic linkage map of chromosome 7, adapted from Eshed and Zamir (1995). The introgression regions of the introgression lines (ILs) are shown in black. Chromosome walking was initiated with the single-copy RFLP marker TG-61.
(B) YAC contig spanning the LeEix region (Ron et al., 2000); YACs are shown in gray.
(C) BiBAC clone isolated by hybridization with the right end clone of YAC 294G5 (294R). The BiBAC left end clone (L5L) cosegregates with the LeEix locus.
(D) Cosmid clones derived from YAC 317E4. Only cosmids hybridized to both 294R and L5L are shown.
(E) Position of LeEix1, LeEix2, and LeEix3 genes.
LeEix1 comprises 3247 nucleotides, has an open reading frame (ORF) of 1031 amino acids, and a 151-bp-long 3′ untranslated region (UTR). The LeEix1 cDNA deduced amino acid sequence shares 48% similarity and 31% identity with the Cf-2 resistance protein (Dixon et al., 1996) and 45% similarity and 30% identity with the tomato Ve1 resistance protein (Kawchuk et al., 2001).
The isolated cDNA clone shares 87% identity with L5L and 294R clones, suggesting that they are different genes. When the LeEix1 gene was mapped, it cosegregated with the LeEix locus (Figure 2). Several polymorphic fragments were detected when hybridizing the full-length LeEix1 cDNA with genomic DNA from both EIX-responding and EIX-nonresponding plants (Figure 2). This suggests that the LeEix1 clone may contain exons spanning a large region or a family of related genes. We tested whether the genomic and LeEix1 cDNA share a similar structure. Amplification of genomic DNA with LeEix1-specific primers amplified a DNA fragment with the same size as the cDNA, suggesting that the LeEix1 gene does not have introns (data not shown). The detection of several polymorphic bands in the DNA gel blot analysis therefore suggests that it represents a family of related genes, which cosegregate with the LeEix locus (Figures 2 and 3). In our attempts to identify additional members of the LeEix cluster, a cosmid library generated from YAC 317E4 DNA (Ron et al., 2000) was screened using the 294R and L5L markers as probes. Two of the hybridizing cosmids (Figure 1) were partially sequenced, and a new ORF was identified. Using specific primers, the new gene (LeEix2) was amplified from the cosmid and sequenced.
Figure 2.
Genetic Map of the LeEix Locus.
(A) The full-length cDNA of LeEix1 was hybridized to DNA from the near-isogenic lines L. esculentum cv M82, cv IL 7-5, F1 hybrid, and the two recombinant plants P5 and P75, which had been digested with HindIII. The responsiveness of the above lines to EIX is indicated above each lane. The triangle represents the band hybridizing with the 3′ UTR of LeEix1; the circle represents the band hybridizing with the 3′ UTR of LeEix2.
(B) A schematic representation of the LeEix gene family on the genetic map of chromosome 7. Black represents L. pennellii DNA and white L. esculentum DNA.
Figure 3.
Physical Map of the LeEix Region.
L. esculentum cv M82 genomic DNA (5 μg/lane) was digested with several restriction enzymes as indicated. Fragments were separated on a 1.0% agarose gel, transferred to a nylon membrane, and hybridized with a mixture of LeEix1, LeEix2, and the partial LeEix3 clones. The arrow represents the band hybridizing with the 3′ UTR of LeEix1 and LeEix2.
LeEix2 comprises 3213 nucleotides, has an ORF of 1021 amino acids, and a 144-bp-long 3′ UTR. The amino acid sequences of LeEix1 and LeEix2 are 81.4% identical and 85.1% similar. DNA gel blot analysis with the 3′ UTR of the two genes mapped them to different regions of the LeEix locus (Figure 2). Sequence analysis revealed that marker 294R is identical to LeEix2; however, the L5L marker showed 87% similarity to LeEix1 and 84% similarity to LeEix2, suggesting that it belongs to a third gene of the LeEix family (LeEix3).
To estimate the number of genes in the LeEix locus, total genomic DNA from L. esculentum was digested with BamHI, DraI, EcoRI, EcoRV, HindIII, and XhoI, separated on an agarose gel, blotted onto a nylon filter, and probed with a mixture of LeEix1, LeEix2, and the partial clone LeEix3 (Figure 3). The combined size of all of the hybridizing bands ranged between 15 and 18 kb. The 3′ UTR of LeEix1 and LeEix2 hybridized to the same 4.8-kb EcoRI DNA fragment (Figure 3). Restriction map analysis of LeEix1 and LeEix2 combined with the hybridization data suggest that the distance between LeEix1 and LeEix2 is 4 kb. The combined size of LeEix1 and LeEix2 is 6 kb, and they are separated by a 4-kb region. We estimate that the size of LeEix3 is 3 kb. Therefore, we suggest that there are three genes in the LeEix locus.
Silencing the LeEix Gene Family Abolishes the Response to EIX Treatment
Our mapping experiments suggest that one or more members of the LeEix1 gene family control the plant's response to EIX treatment. To verify this hypothesis, a gene suppression approach using RNA interference (RNAi) was chosen. A segment of the LRR region of LeEix1 (684 bp from residue 1871 to residue 2554) was cloned in the pKANNIBAL vector in both sense and antisense orientation, flanking the Pdk intron (Wesley et al., 2001). This construct was subcloned into the binary vector pART27 (Gleave, 1992) and used for transforming N. tabacum cv Samsun (EIX-responding plants). Several independent transgenic plants harboring the RNAi construct were generated, and antibiotic resistance and DNA gel blot analysis confirmed transformation (Figure 4A). In responsive tobacco cultivars, EIX induces cell death within 48 h of treatment (Bailey et al., 1990) and complete tissue desiccation within 96 h. We examined the induction of cell death in 11 independent transgenic plants. Cell death by EIX was clearly suppressed compared with control plants (Figure 4B). Furthermore, fluorescin isothiocyanate labeled EIX (FITC-EIX) interacts only with the wild-type cells but not with the cells derived from the silenced transgenic plant (Figure 5A). RNA gel blot analysis indicated that the level of LeEix RNA in the transgenic cell suspension harboring the RNAi construct was reduced to levels below that found in wild-type plants (Figure 5B).
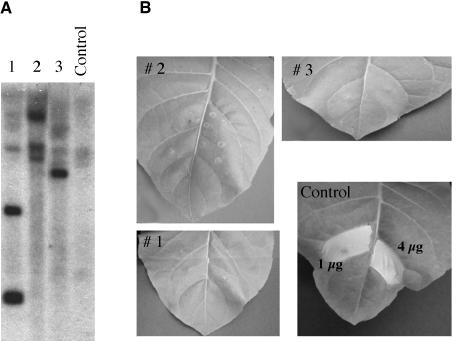
Figure 4.
LeEix Gene Silencing Suppresses EIX-Induced Cell Death.
(A) Genomic DNA (10 μg/lane) from three different transgenic tobacco plants (lanes 1 to 3) and a control (N. tabacum cv Samsun) was digested using XhoI, separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with the 684-bp fragment of LeEix1 used to create the silencing construct.
(B) Young, fully expanded leaves from three independent transgenic plants harboring the pKANNIBAL-LeEix1 or from control plants (N. tabacum cv Samsun) were injected with EIX (1 or 4 μg/mL) as described in Methods. Development of cell death was monitored 96 h after EIX treatment.
Figure 5.
LeEix Gene Silencing Abolishes FITC-EIX Binding to Tobacco Cells.
(A) Cell suspensions derived from the control (N. tabacum cv Samsun) or silenced plant (pKANNIBAL-LeEix1) were incubated with FITC-labeled EIX (0.5 μg/mL) for 30 min. Cells were washed three times and examined with a confocal laser-scanning microscope. DIC, Nomarsky differential interference contrast image; FITC-EIX labeling appears in white. Bars = 50 μm.
(B) Total RNA (15 μg/lane) was isolated from control (N. tabacum cv Samsun) and silenced cell suspensions, separated on a 1% agarose gel, transferred to a nylon filter, and hybridized to the full-length LeEix1 clone. Methylene blue staining of the 28S is shown.
These two experiments thus indicate that at least one of the LeEix genes controls the response to the elicitor.
Genetic Complementation
Transient expression assays (Bendahmane et al., 2000; Van der Hoorn et al., 2000) were conducted to identify which member(s) of the LeEix gene family control(s) the plant's response to the EIX elicitor. LeEix1 and LeLeEix2 cDNAs were cloned independently in the sense orientation into the binary vector pBINPLUS between the 35S-Ω promoter containing the translation enhancer signal and the Nos terminator. The cDNA encoding the EIX elicitor (tvEIX; Furman-Matarasso et al., 1999) was cloned in a similar vector. The constructs were electroporated into Agrobacterium tumefaciens GV3101 and the bacteria used for transient expression assays.
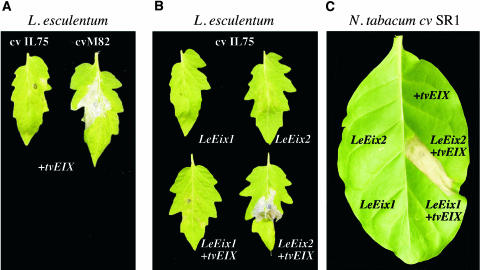
To test the specificity of the transient expression system, Agrobacterium GV3101 harboring Pro35S:tvEIX was injected into L. esculentum cv M82 (EIX-responding plants) and cv IL7-5 (EIX-nonresponding plants) (Figure 6A) or N. tabacum cv SR1 (EIX-nonresponding plants; Figure 6C). Leaves of the M82 tomato cultivar exhibited HR within 48 h of infiltration, whereas those of the EIX-nonresponding cultivar (IL7-5 and SR1) did not (Figures 6A and 6C). Two Agrobacterium GV3101 strains containing either a Pro35S:LeEix1 construct or a Pro35S:LeEix2 construct were each mixed separately with an Agrobacterium GV3101 strain containing Pro35S:tvEIX and then infiltrated into leaves of L. esculentum cv IL7-5. Tomato leaves infiltrated with a mixture of Pro35S:LeEix1 and Pro35S:tvEIX exhibited no response, whereas leaves infiltrated with a mixture of Pro35S:LeEix2 and Pro35S:tvEIX developed HR within 48 h (Figure 6B). Similarly, we examined the genetic complementation in N. tabacum cv SR1 (EIX-nonresponding cultivar). N. tabacum cv SR1 leaves infiltrated with a mixture of Pro35S:LeEix1 and Pro35S:tvEIX exhibited no response, whereas leaves infiltrated with a mixture of Pro35S:LeEix2 and Pro35S:tvEIX developed HR within 48 h (Figure 6C).
Figure 6.
In Vivo Functional Analysis of LeEix Genes in Tomato and Tobacco.
(A) L. esculentum cv M82 and cv IL7-5 were injected with a suspension of Agrobacterium strain GV3101 (OD600 = 0.1) carrying a binary vector with tvEix driven by the 35S promoter of Cauliflower mosaic virus.
(B) L. esculentum cv IL7-5 was infiltrated with Agrobacterium GV3101 (OD600 = 0.1) strains containing either Pro35S:LeEix1 construct, Pro35S:LeEix2 construct, or a mixture of either Pro35S:LeEix1 construct or Pro35S:LeEix2 stains with Agrobacterium GV3101 strain containing Pro35S:tvEIX.
(C) N. tabacum cv SR1 were injected with a suspension of Agrobacterium GV3101 (OD600 = 0.1) strains containing either Pro35S:tvEIX construct, Pro35S:LeEix1 construct, or Pro35S:LeEix2 construct or a mixture of either Pro35S:LeEix1 construct or Pro35S:LeEix2 stains with Agrobacterium GV3101 strain containing Pro35S:tvEIX.
Development of HR was monitored 72 h after injection.
To further define the interaction among LeEix1, LeEix2, and the EIX elicitor, we used the above Agrobacterium strains carrying Pro35S:LeEix1 and Pro35S:LeEix2 to transform N. tabacum cv BY2 cell lines (an EIX-nonresponding cultivar). The resulting transgenic cells were tested for their ability to bind the EIX elicitor. Transgenic cells harboring Pro35S:LeEix1 and those harboring Pro35S:LeEix2 exhibited binding to the EIX elicitor, whereas the control BY2 cell suspension did not (Figure 7). These experiments indicate that both LeEix1 and LeEix2 can restore the binding of the EIX elicitor, although only LeEix2 can transmit the signal necessary to induce the HR response.
Figure 7.
LeEix Gene Expression Restores FITC-EIX Binding to Transgenic Cells.
Cell suspensions of control (N. tabacum cv BY2) or BY2 transgenic cells overexpressing either Pro35S:LeEix1 or Pro35S:LeEix2 were incubated with FITC-labeled EIX (0.5 μg/mL) for 30 min. Cells were washed three times and examined using a confocal laser-scanning microscope. FITC-EIX labeling appears in green; DIC, Nomarsky differential interference contrast image; Fluorescent+DIC, overlay of FITC-EIX and differential interference contrast images. Bars = 20 μm.
Binding between EIX and Mammalian COS-7 Cells Overexpressing LeEix2
Our data suggests physical interaction between the EIX elicitor and the LeEix proteins. LeEix2 was overexpressed in COS-7 cells to allow binding studies between the EIX elicitor and LeEix2 protein in the absence of other plant proteins. COS-7 cells were transfected with LeEix2 fused to a mammalian Igκ signal peptide. The resulting transgenic COS-7 cells were tested for their ability to bind the FITC-EIX elicitor. Transgenic cells transfected with LeEix2 exhibited binding to the EIX elicitor, whereas no such binding was detected when the cells were transfected with empty vector (Figure 8). These experiments indicate that binding between EIX elicitor and LeEix2 protein is not dependent on other plant proteins, suggesting direct interaction between the two proteins.
Figure 8.
Binding of FITC-EIX to COS-7 Cells Overexpressing LeEix2.
COS-7 cells were either transfected with pSeqTaq-LeEix2 (left panel) or mock transfected (right panel). Two days after transfection, FITC-labeled EIX (250 ng/mL) was added to COS-7 cells for 4 h. Cells were washed three times with PBS, fixed with 4% paraformaldehyde, and visualized by confocal laser microscopy. FITC-EIX labeling appears in white. Bars = 10 μm.
Deduced Primary Structure of LeEix1 and LeEix2
On average, the protein sequences of the LeEix1 and LeEix2 genes revealed a 31% identity with the tomato Cf family disease resistance proteins and shared the same domains as those predicted for the Cf9, Cf4, Cf2, and Cf5 resistance genes (Jones et al., 1994; Dixon et al., 1996, 1998) and the tomato Ve genes (Kawchuk et al., 2001). The predicted domains for LeEix1 and LeEix2 are shown in Figure 9. Domain A consists of the signal peptide and its cleavage site at the N terminus of the protein, whereas domain B consists of the predicted NH2 terminus of the mature protein and a Leu zipper motif. The LRR domain C contains 31 imperfect repeats of the consensus sequence LxxLxxLxxLxLSxNxLGxIP (Jones et al., 1994), often associated with protein–protein interaction and ligand binding. The presence of the amino acid Gly within the consensus sequence is consistent with the LRR region being extracytoplasmic, a location that facilitates the recognition of an extracellular ligand (Jones et al., 1994; Song et al., 1995). Within the predicted LRR region, 18 sequences, matching the N-glycosylation consensus sequence NX(S/T) were observed in LeEix1 and 17 in LeEix2. As often observed with membrane-spanning proteins, a hydrophobic sequence with a predicted α-helix secondary structure, predicted to be the transmembrane domain of the protein (domain E), was flanked by a negatively charged extracytoplasmic domain D and a positively charged cytoplasmic domain F. These domains are predicted to play a role in the orientation and anchoring of the protein to the cell membrane. The cytoplasmic domains possess a Tyr YXXø signal sequence, where ø is an amino acid with a hydrophobic side chain, which stimulates receptor-mediated endocytosis and degradation of mammalian cell-surface receptors (Letourneur and Klausner, 1992; Bonifacino and Traub, 2003). Thus, as with the Cf and Ve resistance genes, the LeEix1 and LeEix2 genes are likely to encode the extracytoplasmic glycoproteins anchored to the cell membrane, with the majority of the extracytoplasmic domain consisting of LRR motifs.
Figure 9.
Primary Structures of LeEix Proteins.
LeEix1 (A) and LeEix2 (B) proteins deduced from the cDNA sequence. The polypeptides were divided into domains A to F as described in the text. A line is shown above the putative N-terminal Leu zipper in domain B and below the endocytosis signal in domain F. Hydrophobic amino acids (light green) of the putative signal peptide domain A and membrane-associated domain E are highlighted. In domain C, the conserved L of the LRRs (red) is often replaced by I, F, or V and occasionally by M (blue), whereas the conserved I (red) is replaced by L, F, or V (blue). Conserved N are highlighted in light blue, conserved G in purple, and conserved P in green. Neutral and acidic amino acids of domain D are highlighted in gray and neutral and basic amino acids of domain F in yellow.
Mutating the Endocytosis Signal Abolishes HR
In mammalian, receptor-mediated endocytosis acts as the mechanism for transducing the signal of an external stimuli (Goldstein et al., 1985; Ceresa and Schmid, 2000; Bonifacino and Traub, 2003). In soybean (Glycine max) cell culture, the Verticillium elicitor was shown to enter the cell by an endocytosis process (Horn et al., 1989). Interestingly, the tomato Ve2 resistance protein contains the conserved endocytosis signal (YXXø) within the short cytoplasmic domain (Kawchuk et al., 2001).
Hanania et al. (1999) showed that after binding the plant membrane, EIX is transported into the cytoplasm. The LeEix proteins contain the YXXø motif (Tyr at position 993 in LeEix2) within the short cytoplasmic domain similar to the Ve2 protein. The endocytosis motif was mutagenized to test the hypothesis that endocytosis plays a role in HR induction. Site-directed mutagenesis was used to modify YFTF, the epitope present in the LeEix2 protein, to AFTF. The mutated LeEix2-Y993A ORF was cloned into the binary vector pBINPLUS between the 35S-Ω promoter containing the translation enhancer signal and the Nos terminator. The construct was electroporated into Agrobacterium GV3101 and the bacteria used for transient expression assays. Two Agrobacterium GV3101 strains containing either a Pro35S:LeEix2 construct or a Pro35S:LeEix2-Y993A construct were mixed separately with an Agrobacterium GV3101 strain containing Pro35S:tvEIX and then infiltrated into leaves of N. benthamiana (EIX-nonresponding plant). Leaves infiltrated with a mixture of Pro35S:LeEix2-Y993A and Pro35S:tvEIX exhibited no response, whereas leaves infiltrated with a mixture of Pro35S:LeEix2 and Pro35S:tvEIX developed HR within 48 h (Figure 10A). RT-PCR analysis indicated that the two genes are expressed in leaf sections infiltrated with Pro35S:LeEix2-Y993A and Pro35S:LeEix2 compared with control leaf section infiltrated with medium (Figure 10B). These results indicate that endocytosis plays a key role in mediating the signal generated by EIX that leads to HR induction.
Figure 10.
Mutation in the Endocytosis Signal Abolish HR Induction.
(A) N. benthamiana (EIX-nonresponding plant) was injected with a suspension of Agrobacterium GV3101 (OD600 = 0.1) strains containing either Pro35S:tvEIX construct or induction medium (control) or a mixture of either Pro35S:LeEix2 construct or Pro35S:LeEix2Y993A stains with Agrobacterium GV3101 strain containing Pro35S:tvEIX. Development of HR was monitored 72 h after injection.
(B) Total RNA was isolated from leaves infiltrated with Pro35S:LeEix2-Y993A, Pro35S:LeEix2, or medium and was used to generate first-strand cDNA. The cDNA was used in RT-PCR reactions using specific primers to LeEix2 or N. tabacum actin genes. RT-PCR products were separated on an agarose gel and stained with ethiduim bromide.
DISCUSSION
Two LeEix genes from tomato, whose products act as receptors for the fungal elicitor EIX were positionally cloned, and the following was observed: LeEix gene family members cosegregate with the plant's response to the EIX elicitor; suppression of the LeEix genes expression abrogated the response to EIX; the LeEix2 complemented the EIX elicitor response in vivo; and both LeEix1 and LeEix2 proteins restored the binding of EIX. Partial sequencing of the LeEix locus and DNA gel blot analysis suggests the presence of three different genes in this locus.
LeEix genes show homology to genes encoding plant proteins with LRRs, including disease resistance genes that encode cytoplasmic proteins and membrane bound proteins (Dangl and Jones, 2001; Martin et al., 2003). Like the Ve resistance genes (Kawchuk et al., 2001), LeEix genes contain an endocytosis signal and a Leu zipper. Leu zippers are present in the cytoplasmic class of Arabidopsis thaliana resistance genes RPS2 and RPM1 for Pseudomonas syringae and can facilitate the dimerization of proteins through the formation of coiled-coil structures (Lupas et al., 1991). However, unlike the Ve resistance genes, the LeEix genes do not contain the PEST sequence that characterizes some rapidly degraded proteins (Rechsteiner and Rogers, 1996).
Physical interaction between the R-protein and its corresponding elicitor has been demonstrated only for a few cases (Scofield et al., 1996; Tang et al., 1996; Jia et al., 2000; Leister and Katagiri, 2000; Martin et al., 2003). The EIX elicitor was found to specifically bind to the membranes of EIX-responsive plants (Hanania and Avni, 1997).
The COS-7 cell expression system has previously been shown to be suitable for expression of various functional receptor proteins (Mathews and Vale, 1991; Kieffer et al., 1992). Furthermore, Luderer et al. (2001) demonstrated that COS-7 cells, transfected with the Cf-9 gene, presented the protein on the plasma membrane. We showed that expression of LeEix2 in the COS-7 cells enables the binding of EIX in the absence of other plant proteins and hence indicates direct binding between the EIX and the LeEix2 protein. Thus, LeEix2 acts as a functional receptor for the EIX elicitor.
Recent studies suggest that R-proteins are part of multiprotein complexes (Bogdanove, 2002; Ellis et al., 2002). The inability of the LeEix1 protein to induce HR may be because of poor interaction with other membrane-associated proteins, resulting in a malfunctioning complex. Alternatively, differences in the cytoplasmic domain between LeEix1 and LeEix2 may account for the inability of LeEix1 to transduce the signal, which triggers HR induction.
All eukaryotic cells exhibit receptor-mediated endocytosis as a mechanism of communication for external stimuli and as regulation of signal transduction (Goldstein et al., 1985; Bonifacino and Traub, 2003). In plant cells, indirect evidence of receptor-mediated endocytosis and the presence of clathrin-coated pits has been obtained (Battey et al., 1999). The YXXø motif has been found in mammalian endocytic receptors (Bonifacino and Traub, 2003). The presence of YFKF and YFTF sequences in the short cytoplasmic domains of LeEix1 and LeEix2 suggests that plant and mammalian cell-surface receptors share similar endocytosis signals (Mellman, 1996; Bonifacino and Traub, 2003).
Compartmentalization may play a crucial role in the initiation of R-gene dependent signaling. Following the demonstration that mutating the endocytosis signal inhibits HR induction and that the EIX elicitor interacts with the cytoplasmic protein T-SUMO (Hanania et al., 1999), we propose that the binding of the EIX elicitor to the LeEix2 protein causes ligand-induced conformational change spreading from the extracytoplasmic to the cytoplasmic domain. These conformational changes may induce signals similar to those suggested for the Cf-9 resistance protein (Romeis et al., 1999) and the Clavata receptor (Trotochaud et al., 1999). Alternatively, the binding of EIX to LeEix2 protein may induce receptor-mediated endocytosis, thus allowing the receptor and/or EIX to interact with the cytoplasmic proteins and hence generate a signal to induce the defense response.
The identification of the EIX receptor provides the biochemical foundation for future experiments aimed at understanding the regulatory pathway leading to induction of plant defense responses.
METHODS
Plant and Cell Culture Material and Growth Conditions
Nicotiana tabacum cv Samsun and cv SR1 and N. benthamiana and Lycopersicon esculentum cv M82 and cv IL7-5 plants (Eshed and Zamir, 1995) were grown from seeds under greenhouse conditions. N. tabacum cv Samsun and cv Bright Yellow 2 (BY2) cells were maintained by weekly dilution in fresh MS medium (Sigma, St. Louis, MO) supplemented with 100 μg/L of 2,4-D and 30 g/L of sucrose for N. tabacum or 200 mg/L of KH2PO4, 1 mg/L of thiamine HCl, 100 mg/L of myoinositol, 0.2 mg/L of 2,4 D, and 30 g/L of sucrose for BY2. The medium was adjusted to pH 5.7. The cells were maintained with shaking at 110 rpm at 25°C.
DNA and RNA Analysis
Standard methods were used for DNA and RNA manipulation (Bernatzky and Tanksley, 1986; Ausubel et al., 1987; Sambrook et al., 1989). The cosmid library was generated from the YAC 317E4 (Ron et al., 2000). YAC DNA was isolated from agarose gel, partially digested with Sau3AI, and inserted into the binary cosmid vector pCLD04541 (Jones et al., 1992). DNA packaging was performed using commercial extracts (Gigapack; Strategene, La Jolla, CA) as per the manufacturer's instructions. The L. esculentum mixed elicitors (T1297) and Pseudomonas syringae resistance (T1080) cDNA libraries screening was performed as described previously (Sambrook et al., 1989).
For RT-PCR analysis, total RNA was extracted from leaves 28 h after Agrobacterium tumefaciens infiltration. Two micrograms of RNA were converted to cDNA using M-MLV reverse transcriptase (Promega, Madison, WI). One microliter of each RT reaction was used as template in a 50-μL PCR reaction containing LeEix2-specific primers or Actin-specific primers as control.
Sequence Analysis
DNA sequencing was performed using the dideoxy chain termination method (Sanger, 1981). Sequence analysis was performed using the GCG sequence analysis software package (version 10.0; Accelrys, San Diego, CA). The BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/) was used to search the DNA and protein databases for similarity. Motifs were identified using the SMART program (http://smart.embl-heidelberg.de/) and the PROSITE program (http://www.expasy.ch/prosite).
Construction of Silencing and Complementation Plasmids
The pKANNIBAL vector (Wesley et al., 2001) was used for silencing. A fragment of 684 bp from the LRR region of the LeEix1 gene was amplified by PCR using the following primers: 5′-CTCGAGGATCCATCTCTTCAATTTGTCG-3′ and 5′-GGTACCATCGATTCTCATATCAGCTATTTCTTTAG-3′.
The sense and antisense orientations were cloned into a pKANNIBAL vector (Wesley et al., 2001). The pKANNIBAL containing the sense and antisense fragments was digested with NotI, and the fragment was subcloned into the binary vector pART27 (Gleave, 1992). The resulting construct was introduced by electroporation into Agrobacterium GV3101.
For the complementation assays, the cDNAs of LeEix1 and LeEix2 and the cDNA encoding the EIX elicitor (Furman-Matarasso et al., 1999) were cloned separately into pBINPLUS (van Engelen et al., 1995) between the 35S-Ω promoter containing the translation enhancer signal and the Nos terminator (Hanania et al., 1999). The resulting constructs were electroporated into Agrobacterium GV3101.
For mutagenesis, we applied the mega-primer method (Sarkar and Sommer, 1990). Site-directed mutagenesis was generated by PCR, using LeEix2 as template and the primer 5′-GCCGCTTTCACATTCTTAACAG-3′ to change Tyr 993 into Ala. The resulting clone was sequenced to verify the mutation. LeEix2-Y993A was then cloned into pBINPLUS as described above. The resulting construct was electroporated into Agrobacterium GV3101.
Plant and Cell Suspension Transformation
N. tabacum cv Samsun was transformed as previously described (Horsch et al., 1985). N. tabacum cv BY2 cell suspension were transformed as described by Savaldi-Goldstein et al. (2003).
For transient expression assays, the LeEix1 and LeEix2 constructs that were cloned in pBINPLUS (van Engelen et al., 1995) were introduced by electroporation into Agrobacterium strain GV3101. Agrobacterium was grown in LB medium overnight, diluted into an induction medium (50 mM Mes, pH 5.6, 0.5% (w/v) glucose, 1.7 mM NaH2PO4, 20 mM NH4Cl, 1.2 mM MgSO4, 2 mM KCl, 17 μM FeSO4, 70 μM CaCl2, and 200 μM acetosyringone) and grown for an additional 6 h until OD600 reached 0.4 to 0.5. The Agrobacterium culture was diluted to OD600 = 0.1, and the suspensions expressing Pro35S:tvEIX, Pro35S:LeEix1, Pro35S:LeEix2, or a mixture thereof were injected with a needleless syringe into the leaves of 7- to 8-week-old L. esculentum plants. Leaves were observed for HR induction 48 to 96 h after injection.
Generation of Cell Cultures from Transgenic Plants
The leaves or stems of the transgenic plants were washed in 70% ethanol for 30 s, followed by surface sterilization using 3% bleach and 0.1% Tween-20 for 20 min, and rinsed three times with sterile water. Leaf disks or stem disks were spread on MS plates supplemented with 2 mg/L of 2,4-D and 0.2 mg/L of 6-benzylaminopurine riboside or 3 mg/L of a-naphtalene acetic acid and 1 mg/L 6-benzylaminopurine riboside. The plates were incubated at 25°C until the formation of calli. The calli were transferred to a fresh liquid MS medium supplemented with 1:5 of the original hormones and maintained with shaking at 110 rpm at 25°C. Cell cultures were further maintained as described for the N. tabacum cv Samsun.
EIX Treatments
Xylanase (Fluka, Milwaukee, WI) was purified as previously described (Dean and Anderson, 1991). In the plants, EIX (1 to 4 μg/mL) was injected into leaves using needleless syringes as previously described (Hanania et al., 1999). The development of cell death was monitored 24 to 96 h after treatment.
Binding of FITC-Labeled EIX to Plant Cells
EIX was labeled with FITC as previously described (Hanania and Avni, 1997). Cells were incubated for 30 min with 0.1% BSA followed by the addition of EIX-FITC (500 ng/mL) for 30 min. The cells were subsequently washed three times in washing buffer (10 mM phosphate buffer, pH 7, and 0.1% BSA) and visualized by confocal laser microscopy. Confocal imaging was performed using a Zeiss LSM510 confocal laser-scanning microscope (Jena, Germany). Excitation was performed using an argon laser set to 488 nm, and emission was detected with a 525 nm ± 15 nm bandpath filter. Image analysis was performed using Zeiss CLSM-5 and Adobe Photoshop 7.0 (Mountain View, CA).
COS-7 Cell Culture Transfection and EIX Binding
COS-7 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum (Paz et al., 2001). For transfections, cells were plated on 20 × 20-mm cover slides in a six-well plate. Cells were transfected with 0.5 μg of DNA (pSeqTag vector containing LeEix2; Invitrogen, Carlsbad, CA) by dextran (Amersham Biosciences, Uppsala, Sweden) as previously described (Seed and Aruffo, 1987). Two days after transfection, 0.1% BSA was added to the medium 30 min before adding FITC-labeled EIX (250 ng/mL). Cells were further incubated for 4 h at 37°C in the presence of the FITC-EIX followed by 3 washes with PBS. Cells were fixed with 4% paraformaldehyde and visualized by confocal laser microscopy.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY359965 (LeEix1) and AY359966 (LeEix2).
Acknowledgments
We thank Dvora Aviv for assistance with the tobacco cell suspension transformation and Barak Rotblat for transfecting COS-7 cells. We are grateful to Silvia Schuster for invaluable assistance and helpful suggestions and Uzi Cohen for plant maintenance. We also thank Greg Martin for supplying us with the cDNA libraries and Commonwealth Scientific and Industrial Research Organization Plant Industry for providing us with the pKANNIBAL and PART27 vectors. We are grateful to A. Mattoo and G. Sessa for helpful suggestions and discussions. This research was supported in part by the Israel Science Foundation administrated by the Israel Academy of Science and Humanities.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Adi Avni (lpavni@post.tau.ac.il).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.022475.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1987). Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Bailey, B.A., Dean, J.F.D., and Anderson, J.D. (1990). An ethylene biosynthesis-inducing endoxylanase elicits electrolyte leakage and necrosis in Nicotiana tabacum cv Xanthi leaves. Plant Physiol. 94, 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, B.A., Korcak, R.F., and Anderson, J.D. (1993). Sensitivity to an ethylene biosynthesis-inducing endoxylaanase in Nicotiana L. cv. Xanthi is controlled by a single dominant gene. Plant Physiol. 101, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interaction. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Battey, N.H., James, N.C., Greenland, A.J., and Brownlee, C. (1999). Exocytosis and endocytosis. Plant Cell 11, 643–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A., Querci, M., Kanyuka, K., and Baulcombe, D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (1996). Plant disease resistance: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatzky, R., and Tanksley, S.D. (1986). Methods for detection of single or low copy sequences in tomato on Southern blots. Plant Mol. Biol. Rep. 4, 37–41. [Google Scholar]
- Bogdanove, A.J. (2002). Protein-protein interactions in pathogen recognition by plants. Plant Mol. Biol. 50, 981–989. [DOI] [PubMed] [Google Scholar]
- Boller, T. (1995). Chemoperception of microbial signals in plant-cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 189–214. [Google Scholar]
- Bonifacino, J.S., and Traub, L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447. [DOI] [PubMed] [Google Scholar]
- Ceresa, B.P., and Schmid, S.L. (2000). Regulation of signal transduction by endocytosis. Curr. Opin. Cell Biol. 12, 204–210. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Holub, E. (1997). La dolce vita: A molecular feast in plant-pathogen interactions. Cell 91, 17–24. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dean, J.F.D., and Anderson, J.D. (1991). Ethylene biosynthesis-inducing endoxylanase. II. Purification and physical characterization of the enzyme produced by Trichoderma viride. Plant Physiol. 95, 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Hatzixanthis, K., Jones, D.A., Harrison, K., and Jones, J.D. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Jones, D.A., Keddie, J.S., Thomas, C.M., Harrison, K., and Jones, J.D. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. [DOI] [PubMed] [Google Scholar]
- Ebel, J., and Cosio, E.G. (1994). Elicitors of plant defense responses. Int. Rev. Cytol. 148, 1–36. [Google Scholar]
- Elbaz, M., Avni, A., and Weil, M. (2002). Constitutive caspase-like machinery executes programmed cell death in plant cells. Cell Death Differ. 9, 726–733. [DOI] [PubMed] [Google Scholar]
- Ellis, C., Turner, J.G., and Devoto, A. (2002). Protein complexes mediate signalling in plant responses to hormones, light, sucrose and pathogens. Plant Mol. Biol. 50, 971–980. [DOI] [PubMed] [Google Scholar]
- Enkerli, J., Felix, G., and Boller, T. (1999). The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol. 121, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, Y., Saxena, A., Gamble, H.R., and Anderson, J.D. (1989). Ethylene biosynthesis-inducing protein from cellulysin is an endoxylanase. Plant Physiol. 89, 138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman-Matarasso, N., Cohen, E., Du, Q., Chejanovsky, N., Hanania, U., and Avni, A. (1999). A point mutation in the ethylene-inducing xylanase elicitor inhibits the beta-1-4-endoxylanase activity but not the elicitation activity. Plant Physiol. 121, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, D.W., and Rolfe, B.G. (1990). Working models of specific recognition in plant-microbe interactions. Annu. Rev. Phytopathol. 28, 365–391. [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Goldstein, J.L., Brown, M.S., Anderson, R.G., Russell, D.W., and Schneider, W.J. (1985). Receptor-mediated endocytosis: Concepts emerging from the LDL receptor system. Annu. Rev. Cell Biol. 1, 1–39. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997). Programed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Hamilton, C.M. (1997). A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene 200, 107–116. [DOI] [PubMed] [Google Scholar]
- Hanania, U., and Avni, A. (1997). High affinity binding site for ethylene inducing xylanase elicitor on Nicotiana tabacum membranes. Plant J. 12, 113–120. [Google Scholar]
- Hanania, U., Furman-Matarasso, N., Ron, M., and Avni, A. (1999). Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 19, 533–541. [DOI] [PubMed] [Google Scholar]
- Horn, M.A., Heinstein, P.F., and Low, P.S. (1989). Receptor-mediated endocytosis in plant cells. Plant Cell 1, 1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch, R.B., Rogers, S.G., and Fraley, R.T. (1985). Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., Shlumukov, L., Carland, F., English, J., Scofield, S.R., Bishop, G.J., and Harrison, K. (1992). Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1, 285–297. [DOI] [PubMed] [Google Scholar]
- Kawchuk, L.M., Hachey, J., Lynch, D.R., Kulcsar, F., van Rooijen, G., Waterer, D.R., Robertson, A., Kokko, E., Byers, R., Howard, R.J., Fischer, R., and Prufer, D. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 98, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer, B.L., Befort, K., Gaveriaux-Ruff, C., and Hirth, C.G. (1992). The delta-opioid receptor: Isolation of a cDNA by expression cloning and pharmacological characterization. Proc. Natl. Acad. Sci. USA 89, 12048–12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann, M., Honee, G., Bonnema, G., and De Wit, P. (1996). A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, R.T., and Katagiri, F. (2000). A resistance gene product of the nucleotide binding site–leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 22, 345–354. [DOI] [PubMed] [Google Scholar]
- Letourneur, F., and Klausner, R.D. (1992). A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell 69, 1143–1157. [DOI] [PubMed] [Google Scholar]
- Luderer, R., et al. (2001). No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol. Plant-Microbe Interact. 14, 867–876. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Bogdanov, A.J., and Sessa, G. (2003). Understanding the function of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Mathews, L.S., and Vale, W.W. (1991). Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell 65, 973–982. [DOI] [PubMed] [Google Scholar]
- Mellman, I. (1996). Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575–625. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., and Dangl, J.L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Paz, A., Haklai, R., Elad-Sfadia, G., Ballan, E., and Kloog, Y. (2001). Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 20, 7486–7493. [DOI] [PubMed] [Google Scholar]
- Rechsteiner, M., and Rogers, S.W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267–271. [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., Zhang, S., Klessig, D.F., Hirt, H., and Jones, J.D. (1999). Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, M., Kantety, R., Martin, G.B., Avidan, N., Eshed, Y., Zamir, D., and Avni, A. (2000). High-resolution linkage analysis and physical characterization of the EIX-responding locus in tomato. Theor. Appl. Genet. 100, 184–189. [Google Scholar]
- Rotblat, B., Enshel-Seijffers, D., Gershoni, M.J., Schuster, S., and Avni, A. (2002). Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 32, 1–7. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanger, F. (1981). Determination of nucleotide sequence in DNA. Science 214, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Sarkar, G., and Sommer, S.S. (1990). The megaprimer method of site-directed mutagenesis. Biotechniques 8, 404–407. [PubMed] [Google Scholar]
- Savaldi-Goldstein, S., Aviv, D., Davydov, O., and Fluhr, R. (2003). Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Seed, B., and Aruffo, A. (1987). Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc. Natl. Acad. Sci. USA 84, 3365–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, I.E., and Hahlbrock, K. (1998). Pathogen defence in plants: A paradigm of biological complexity. Trends Plant Sci. 3, 86–90. [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R.A., Laurent, F., Roth, R., and De Wit, P.J. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant-Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.-P., Pereira, A., and Stiekema, W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]