Abstract
Heterotrimeric G proteins composed of α, β, and γ subunits link ligand perception by G protein–coupled receptors (GPCRs) with downstream effectors, providing a ubiquitous signaling mechanism in eukaryotes. The Arabidopsis thaliana genome encodes single prototypical Gα (GPA1) and Gβ (AGB1) subunits, and two probable Gγ subunits (AGG1 and AGG2). One Arabidopsis gene, GCR1, encodes a protein with significant sequence similarity to nonplant GPCRs and a predicted 7-transmembrane domain structure characteristic of GPCRs. However, whether GCR1 actually interacts with GPA1 was unknown. We demonstrate by in vitro pull-down assays, by yeast split-ubiquitin assays, and by coimmunoprecipitation from plant tissue that GCR1 and GPA1 are indeed physically coupled. GCR1–GPA1 interaction depends on intracellular domains of GCR1. gcr1 T-DNA insertional mutants exhibit hypersensitivity to abscisic acid (ABA) in assays of root growth, gene regulation, and stomatal response. gcr1 guard cells are also hypersensitive to the lipid metabolite, sphingosine-1-phosphate (S1P), which is a transducer of the ABA signal upstream of GPA1. Because gpa1 mutants exhibit insensitivity in aspects of guard cell ABA and S1P responses, whereas gcr1 mutants exhibit hypersensitivity, GCR1 may act as a negative regulator of GPA1-mediated ABA responses in guard cells.
INTRODUCTION
Ligand signaling via G protein–coupled receptors (GPCRs) is a widespread mechanism of signal perception in eukaryotic organisms ranging from slime molds to humans (Graul and Sadee, 2001; Pierce et al., 2002; Pin et al., 2003). GPCRs traditionally have been divided into six families (Kolakowski, 1994; Horn et al., 1998), although classification schemes continue to evolve (Foord, 2002; Graul and Sadee, 2001). GPCRs are typified by a conserved 7-transmembrane domain (7TM) structure composed of an extracellular N terminus, seven hydrophobic stretches of ∼20 amino acids linked by alternating intracellular and extracellular loops (il1-3 and ol1-3), and a cytoplasmic C-terminal tail (Strader et al., 1994; Pin et al., 2003). In the classic paradigm of G protein signaling (Gudermann et al., 1997; Morris and Malbon, 1999), the GPCR associates with heterotrimeric G proteins composed of α, β, and γ subunits. Ligand binding to the GPCR results in conformational changes that stimulate exchange of GTP for GDP at a guanine-nucleotide binding site on the Gα subunit. GTP binding disrupts Gα interaction with Gβ, thereby freeing both the α subunit and the βγ pair (which acts as a nondissociable dimer) to interact with a variety of downstream effectors. Intrinsic GTPase activity of the α subunit, which can be accelerated by RGS (regulator of G protein signaling) proteins (Ross and Wilkie, 2000; Chen et al., 2003), returns the G protein to the inactive trimeric state.
In plants, heterotrimeric G protein signaling has been implicated in transduction of several different phytohormone signals as well as in the control of cell division (reviewed in Ma, 1994; Assmann, 2002; Jones, 2002). In Arabidopsis thaliana, T-DNA insertional null mutants of the sole canonical Gα subunit GPA1 exhibit insensitivity to abscisic acid (ABA) inhibition of stomatal opening and altered ABA responsiveness of guard-cell inward K+ channels and slow anion channels (Wang et al., 2001). Previous studies have shown that ABA and drought stimulate production of the lipid metabolite sphingosine-1-phosphate (S1P) in plants and implicate S1P as a secondary messenger for guard cell ABA responses (Ng et al., 2001; Coursol et al., 2003; Worrall et al., 2003). gpa1 guard cells also show insensitivity to S1P regulation of stomatal apertures and ion channels (Coursol et al., 2003). However, in seed germination assays, gpa1 mutants exhibit hypersensitivity to ABA as well as hypersensitivity to sugars, hyposensitivity to gibberellic acid, and insensitivity to brassinolide (Ullah et al., 2002; Lapik and Kaufman, 2003). A cupin domain protein AtPirin1 has been identified recently on the basis of its interaction with GPA1 and is proposed to function immediately downstream of GPA1 in regulation of seed germination and early seedling development (Lapik and Kaufman, 2003).
gpa1 knockout plants also show reduced cell division in developing hypocotyls and leaves (Ullah et al., 2001), whereas mutants of the sole prototypical Arabidopsis Gβ subunit gene AGB1 show increased production of lateral root primordia, altered leaf and flower shape, and shorter siliques (Lease et al., 2001; Ullah et al., 2003). Plants or cultured cells overexpressing GPA1 show ectopic cell division and acceleration of the cell cycle, respectively (Ullah et al., 2001). Studies on mutants of the rice (Oryza sativa) Gα subunit RGA1 also implicate heterotrimeric G protein signaling in gibberellin responses and pathogen resistance (Ueguchi-Tanaka et al., 2000; Suharsono et al., 2002).
In previous efforts to identify plant GPCRs (Josefsson and Rask, 1997; Plakidou-Dymock et al., 1998; Kanyuka et al., 2001), BLAST searches of the database of ESTs using representative members of the six GPCR families (Kolakowski, 1994) as queries were executed, and a homologous Arabidopsis EST was identified. The associated complete cDNA sequence, named GCR1, is most similar to the GPCR Family E cAMP receptors (CARs) of Dictyostelium. Analyses revealed that GCR1 has 20 to 23% identity over its 7TM domain with CAR proteins as well as significant similarity in defined regions with calcitonin of the Family B glucagons/secretin-like GPCRs and serotonin of the Family A rhodopsin/adrenergic-like GPCRs (Josefsson and Rask, 1997; Plakidou-Dymock et al., 1998). GCR1 has a predicted 7TM structure and shows conservation of key amino acid residues (Josefsson and Rask, 1997; Plakidou-Dymock et al., 1998). Plants overexpressing GCR1 have been reported to show accelerated flowering, when plants are grown to maturity on plates, and reduced seed dormancy (Colucci et al., 2002). GCR1 overexpressing tobacco (Nicotiana tabacum) BY2 cells exhibit increased thymidine incorporation into DNA (Colucci et al., 2002; Apone et al., 2003). Colucci and coworkers (Apone et al., 2003) believe that phosphatidylinositol-specific phospholipase C may serve as an effector in this response.
The observations that overexpression of GCR1 or GPA1 affect the cell cycle (Ullah et al., 2001; Colucci et al., 2002; Apone et al., 2003) and that GCR1 overexpression and GPA1 elimination have opposite effects on the ABA sensitivity of seed germination are consistent with GCR1 and GPA1 functioning in the same pathway. However, GCR1 coupling with GPA1 has not been evaluated. If GCR1 functions as a genuine GPCR in Arabidopsis, we would hypothesize that GCR1 would physically interact with GPA1 in a manner dependent on the intracellular domains of GCR1, as has been shown for mammalian GPCRs (Wess, 1998). Further, we would predict that gcr1 knockout mutants would exhibit at least a subset of the phenotypes seen in gpa1 knockout lines. This research was designed to address these two hypotheses.
RESULTS
GCR1 Sequence Similarity
Earlier sequence-based homology searches identified GCR1 as the only protein in Arabidopsis that shows significant similarity to Dictyostelium CARs. However, in light of the complete sequencing of the Arabidopsis genome and the accumulation of a large amount of new sequence data for metazoan GPCRs, we reevaluated GCR1 sequence homologs. Using GCR1 protein as the query, BLAST (www.ncbi.nlm.nih.gov/BLAST) and FASTA (www.ebi.ac.uk.fasta33) analyses show that in addition to CAR receptors of Dictyostelium, GCR1 also shows significant sequence similarity to members of the Frizzled/Smoothened family of receptors and also to Methuselah-like proteins of Drosophila. Table 1 shows representatives of each family identified by this analysis with an e-value cutoff of 2e−2 or less.
Table 1.
Overall Similarity Prediction of GCR1
| G Protein–Coupled Receptors | Identity | Similarity |
|---|---|---|
| Slime mold (Dictyostelium discoideum) CAR1 | 25% | 41% |
| Slime mold (D. discoideum) CAR2 | 24% | 45% |
| Slime mold (D. discoideum) CAR3 | 25% | 41% |
| Slime mold (D. discoideum) CAR4 | 23% | 44% |
| Filamentous fungus (Polysphondylium pallidum) CAR | 24% | 44% |
| Slime mold (D. discoideum) frizzled protein | 22% | 42% |
| Drosophila melanogaster Methuselah-like 3 protein | 22% | 42% |
| D. melanogaster Methuselah-like 10 protein | 22% | 38% |
GCR1 and GPA1 Interact
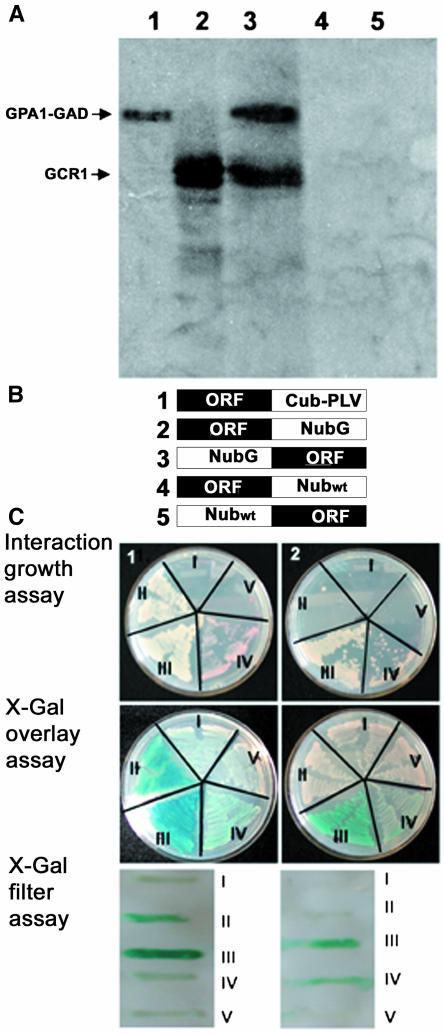
To test whether GCR1 is actually G protein coupled, we first used an in vitro system (Figure 1A). In vitro binding assays were performed with GPA1 fused with the GAL4 activation domain of yeast (Saccharomyces cerevisiae) (GPA1-GAD) and GCR1 obtained by coupled in vitro transcription/translation in a cell free system (lanes 1 and 2). Pull-down assays were performed with anti-GAD antibodies, and the antigen-antibody complex was precipitated with protein A–agarose beads. Anti-GAD antibodies could precipitate GCR1 only in the presence of GPA1 (lane 3) but not by itself (lane 4). Similar interaction assays with a GAD-tagged putative protein kinase (lane 5) failed to precipitate GCR1, confirming the specificity of the binding reaction.
Figure 1.
GCR1 Interacts with GPA1.
(A) GCR1 interacts with GPA1 under in vitro conditions. GPA1 and a putative protein kinase (At5g66890) were prepared with the GAL4 activation domain (GAD) as fusion proteins. GPA1-GAD fusion proteins and GCR1 were synthesized by in vitro transcription/translation in the presence of 35S-Met (lanes 1 and 2). Kinase-GAD fusion protein was in vitro transcribed/translated in the presence of cold Met. GCR1 can be pulled down with GAD antibodies when incubated with GPA1 (lane 3) but not when incubated with the anti-GAD antibodies alone (lane 4) or the putative protein kinase (lane 5).
(B) Constructs used for split ubiquitin assays. Protein fusion with the C-terminal part of ubiquitin followed by PLV (Cub-PLV) was made at the C terminus of the bait protein (1). Protein fusions with the N-terminal part of ubiquitin (Nub) were made either at the C terminus of the prey protein with NubG or Nubwt (2 and 4, respectively) or at the N terminus of the prey protein with NubG or Nubwt (3 and 5, respectively).
(C) GCR1 and GPA1 interact in yeast-based split ubiquitin system. Interaction between GCR1 and GPA1 was determined by growth assay on media lacking His and Ade but containing 200 μM Met, by β-galactosidase activity using X-Gal overlay assay, and by X-Gal filter assay. (1) Interaction assays with GPA1-Cub as bait construct. The constructs were used in the following combinations: (I) GPA1-Cub + GCR1-NubG, (II) GPA1-Cub + NubG-GCR1, (III) GPA1-Cub + GCR1-Nubwt, (IV) GPA1-Cub + Nubwt-GCR1, and (V) GPA1-Cub + GPA1-NubG. GCR1-NubG and NubG-GCR1 were test constructs (I and II). GCR1-Nubwt constructs serve as positive controls (III and IV). The GPA1-NubG construct serves as a negative control (V). GPA1-Cub interacts with GCR1 only when the NubG fusion is at the N terminus of GCR1 (II) and not when the C terminus of GCR1 is fused with NubG (I). (2) Interaction assays with GCR1-Cub as bait construct. The constructs were used in the following combinations: (I) GCR1-Cub + GPA1-NubG, (II) GCR1-Cub + NubG-GPA1, (III) GCR1-Cub + GPA1-Nubwt, (IV) GCR1-Cub + Nubwt-GPA1, and (V) GCR1-Cub + GCR1-NubG. GPA1-NubG and NubG-GPA1 were test constructs (I and II). GPA1-Nubwt constructs serve as positive controls (III and IV). The GCR1-NubG construct serves as a negative control (V). GCR1 fused with Cub at its C terminus fails to interact with GPA1. Growth and β-galactosidase activity tests are positive only for the positive controls (III and IV) that have wild-type ubiquitin fused with GPA1.
We confirmed that GCR1 binds directly to GPA1 in vivo using a modified split ubiquitin system developed to assess interactions of membrane proteins (Ludewig et al., 2003). In this system, interactions between membrane-bound fusion proteins can be monitored by the release of an artificial transcription factor consisting of protein A, LexA, and VP16 (PLV). In vivo interaction between the two fusion proteins reconstitutes the N-terminal part of ubiquitin (Nub) with the C-terminal part of ubiquitin fused with PLV (CubPLV), leading to the cleavage and release of PLV by ubiquitin-specific proteases and, thus, to PLV-activated expression of lacZ, HIS3, and ADE reporter genes integrated in the yeast genome.
GCR1 and GPA1 open reading frames were fused to either the C-terminal part of ubiquitin (GCR1-Cub and GPA1-Cub) followed by PLV or to the N-terminal part of ubiquitin (Nubwt or NubG, I13G) (Stagljar et al., 1998) by in vivo recombination cloning. Nub fusions were made at both the C terminus and the N terminus of the gene. Thus, for each open reading frame (ORF), we made five fusion constructs: ORF-Cub, ORF-NubG, NubG-ORF, ORF-Nubwt, and Nubwt-ORF, where ORF is GCR1 or GPA1 (Figure 1B). NubG constructs served as test proteins. Nubwt constructs served as positive controls because these proteins will release active PLV even in the absence of interaction between the target and the bait proteins. Interactions were tested by mating of yeast strains AP4 (harboring Cub fusion proteins) and AP5 (harboring Nub fusion proteins), growth of diploid yeast cells on media lacking His and Ade, and also by LacZ expression and β-galactosidase activity in presence of 5-bromo-4-chloro-β-d-galactosidase (X-Gal). GPA1-Cub and GCR1-Cub were each used as baits in two separate assays. As shown in Figure 1C, GPA1-Cub protein interacts with NubG-GCR1 but not with GCR1-NubG (Figure 1C, plate 1, I and II). GCR1-Cub, however, does not show interaction with GPA1-Nub constructs (Figure 1C, plate 2, I and II), illustrating that a free GCR1 C terminus is required for interaction.
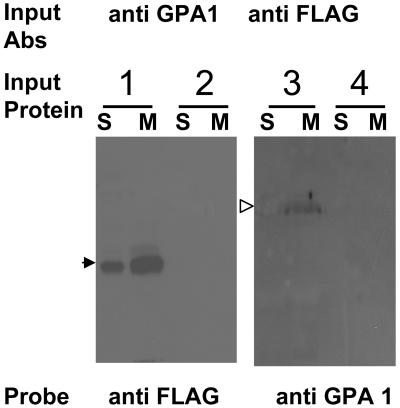
We further confirmed the interaction between GCR1 and GPA1 in planta. gcr1-3 mutant plants were transformed with a full-length GCR1 cDNA fused with a FLAG epitope tag under the control of a glucocorticoid-inducible promoter. Protein expression was induced by treating plants with dexmethasone (Dex) for 24 h. Noninduced and gcr1-3 knockout plants served as controls. As shown in Figure 2, GCR1 and GPA1 can be coimmunoprecipitated. By protein gel blotting using anti-FLAG antibodies as probe, we could detect GCR1 in the immunocomplex precipitated with anti-GPA1 antibodies, from GCR1:FLAG plants induced with Dex (lane 1, S and M), but not in the proteins precipitated from the noninduced plants (lane 2, S and M). Similarly, GPA1 could be detected by immunoblotting with anti-GPA1 antibodies, in the immunocomplex precipitated with anti-FLAG antibodies from GCR1:FLAG plants (lane 3, S and M), but not from the gcr1-3 plants (lane 4, S and M).
Figure 2.
GCR1 Interacts with GPA1 in Planta.
Soluble (S) and membrane (M) protein fractions isolated from gcr1-3 and GCR1:FLAG (Dex-induced [lane 3] and noninduced [lane 4]) plants were used for in vivo coimmunoprecipitation. Proteins immunoprecipitated with anti-GPA1 antibodies were probed with anti-FLAG antibodies by immunoblotting and vice versa. Anti-GPA1 antibodies could pull down GCR1 (closed arrowhead, lanes 1S and 1M) and anti-FLAG antibodies could pull down GPA1 (open arrowhead, lanes 3S and 3M). No signal was detected in gcr1-3 knockout plants (lanes 2S and 2M) or noninduced GCR1:FLAG plants (lanes 4S and 4M).
Interaction of Truncated GCR1 Proteins with GPA1
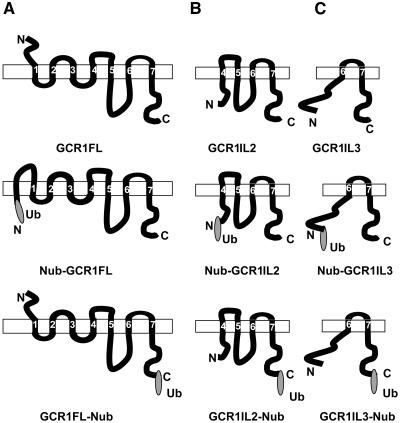
Results obtained with our initial split ubiquitin tests indicate that a free C terminus is required for GCR1 interaction with GPA1. GCR1 with ubiquitin-fused at its C terminus fails to interact with GPA1 (Figure 1C, plate 2, I and II), whereas an N-terminal ubiquitin fused GCR1 does interact (Figure 1C, plate 1, II). This result, however, raised a question about the topology of GCR1. GCR1 is predicted to have a classic 7TM structure with its N terminus outside and C terminus inside the plasma membrane. Ubiquitin reconstitution, however, takes place in the cytoplasm. Thus, based on the classic structure prediction, ubiquitin fused at the N terminus of GCR1 would not be available for reconstitution. To resolve this issue, we used several Web-based topology prediction programs (DAS, SMART, and TMHMM) to deduce the orientation of GCR1 full-length and truncated proteins in their native or ubiquitin-fused forms. Figure 3 shows schematic representations of model diagrams generated by the TMHMM program (Krogh et al., 2001). Identical results (data not shown) were obtained with DAS and SMART programs (Tusnady and Simon, 2001). As shown in Figure 3A, the full-length GCR1 has a prototypical 7TM structure with an N out, C in orientation. Nub-GCR1FL fusion protein (full-length GCR1 with ubiquitin fusion at the N terminus), however, is predicted to have eight transmembrane domains, with both the N and C termini inside. This thus justifies the interaction results we see in the split ubiquitin system. By contrast, GCR1FL-Nub fusion protein (full-length GCR1 with ubiquitin fusion at the C terminus) maintains the N out, C in orientation of native GCR1. Because the Nub-GCR1FL but not the GCR1FL-Nub fusion protein shows interaction with GPA1, we deduce that a free C terminus of GCR1 is required for this interaction.
Figure 3.
Topology Prediction of GCR1 Fragments Fused with Ubiquitin at the N or C Terminus as Used for Split Ubiquitin Assays.
Schematic representation of topology of GCR1 full-length and truncated proteins with or without a ubiquitin (N-terminal part, Nub) fusion. Topology was predicted using transmembrane domain hidden Markov model (TMHMM version 2.0). Rectangles represent plasma membrane. Predicted transmembrane regions are numbered from 1 to 7. Ub represents N-terminal portion of ubiquitin.
(A) GCR1 full-length protein (GCR1FL) without or with Ub fusion at the N terminus (Nub-GCR1FL) or at the C terminus (GCR1FL-Nub).
(B) GCR1IL2 protein (amino acids 105 to 326) without or with Ub fusion at the N terminus (Nub-GCR1IL2) or at the C terminus (GCR1IL2-Nub).
(C) GCR1IL3 protein (amino acids 271 to 326) without or with Ub fusion at the N terminus (Nub-GCR1IL3) or at the C terminus (GCR1IL3-Nub).
GCR1IL2 protein (amino acids 105 to 326) starts at the junction of predicted transmembrane 3 and intracellular loop 2. This truncated protein, lacking the transmembrane 3 region (amino acids 84 to 104), fails to integrate to the membrane from its N terminus and thus has only one intracellular loop (il3). The N-terminal region, from amino acids 105 to 119, is predicted to remain free in the cytoplasm along with the C terminus, with or without ubiquitin fusion (Figure 3B).
GCR1IL3 truncated protein (amino acids 184 to 326) starts at the junction of predicted transmembrane 5 and il3. This truncated protein does not have the transmembrane region 5 (amino acids 161 to 183) and also fails to integrate to the membrane from its N terminus and has no intracellular loop. The N-terminal region, from amino acid 104 to 218, thus remains free in the cytoplasm along with the C terminus, with or without ubiquitin fusion (Figure 3C). GCR1Ct truncated protein (amino acids 271 to 326) is the cytoplasmic tail of GCR1, from the end of the 7th transmembrane region to the C-terminal end of the protein. This protein contains no transmembrane domains and is predicted to be cytoplasmic.
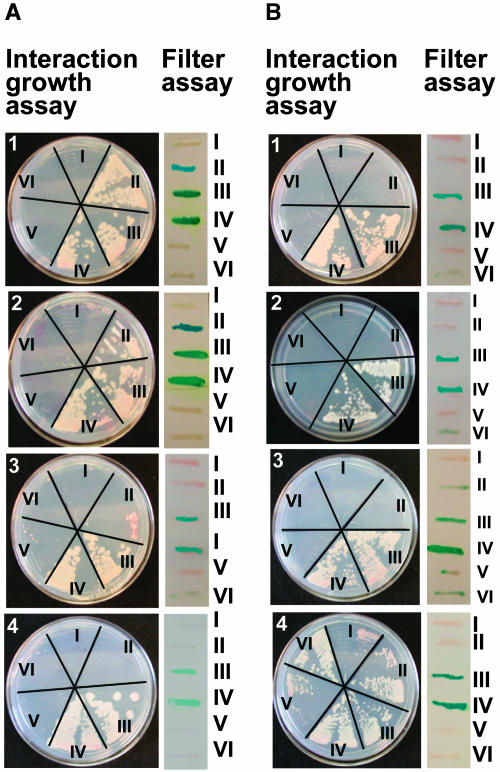
To ascertain the regions of GCR1 required for interaction with GPA1, we used these truncated constructs in split ubiquitin assays. Interaction growth assays on media lacking His and Ade and β-galactosidase activity by X-Gal filter assays were performed using GPA1-Cub protein as bait and GCR1FL (Figure 4A, plate 1), GCR1IL2 (Figure 4A, plate 2), GCR1IL3 (Figure 4A, plate 3), and GCR1Ct (Figure 4A, plate 4) as prey proteins. Assays were also performed with GCR1FL-Cub (Figure 4B, plate 1), GCR1IL2-Cub (Figure 4B, plate 2), GCR1IL3-Cub (Figure 4B, plate 3), and GCR1Ct-Cub (Figure 4B, plate 4) as bait proteins and GPA1 as prey protein. Results in Figure 4A, plates 1 to 4, show that GCR1FL interacts with GPA1 only when GCR1 has a free C terminus (as shown also in Figure 3B). GCR1IL2 also interacts with GPA1 in an identical fashion, showing that the first 105 amino acids are not required for interaction of GCR1 and GPA1. GCR1IL3 fails to interact with GPA1, showing either requirement of amino acids 105 to 119 and/or an intact il3 for the interaction. GCR1Ct Nub fusions also fail to interact with GPA1. None of the GCR1-Cub fusion proteins showed specific interaction with GPA1, confirming the requirement of a free C terminus, though nonspecific interactions were observed with GCR1Ct-Cub protein (Figure 4B, plate 4), possibly because of its very small size. GPA1 does not interact with itself (Figure 4, V, all plates) nor with an unrelated protein, the inward K+ channel protein KAT1 (Figure 4, VI, all plates), supporting the specific nature of the GPA1–GCR1 interactions observed.
Figure 4.
Interaction of Different Fragments of GCR1 with GPA1.
(A) Interaction using GPA1-Cub as bait construct with different fragments of GCR1 as prey constructs. (1) GPA1-Cub: (I) +GCR1FL-NubG, (II) +NubG-GCR1FL, (III) +GCR1FL-Nubwt, (IV) +Nubwt-GCR1FL, (V) +GPA1-NubG, and (VI) +NubG-KAT1. (2) GPA1-Cub: (I) +GCR1IL2-NubG, (II) +NubG-GCR1IL2, (III) +GCR1IL2-Nubwt, (IV) +Nubwt-GCR1IL2, (V) +GPA1-NubG, and (VI) +NubG-KAT1. (3) GPA1-Cub: (I) +GCR1IL3-NubG, (II) +NubG-GCR1IL3, (III) +GCR1IL3-Nubwt, (IV) +Nubwt-GCR1IL3, (V) +GPA1-NubG, and (VI) +NubG-KAT1. (4) GPA1-Cub: (I) +GCR1Ct-NubG, (II) +NubG-GCR1Ct, (III) +GCR1Ct-Nubwt, (IV) +Nubwt-GCR1Ct, (V) +GPA1-NubG, and (VI) +NubG-KAT1. Interaction was determined by growth of diploid yeast colonies on media lacking His and Ade but containing 200 μM Met and by β-galactosidase activity using X-Gal filter assay. I and II were test interactions, III and IV were positive controls, and V and VI were negative controls in all cases.
(B) Interaction using different fragments of GCR1-Cub as bait constructs and GPA1 as prey construct. Interactions were tested in the following combinations. (1) GCR1FL-Cub: (I) +GPA1-NubG, (II) +NubG-GPA1, (III) +GPA1-Nubwt, (IV) +Nubwt-GPA1, (V) +GCR1FL-NubG, and (VI) +NubG-KAT1. (2) GCR1IL2-Cub: (I) +GPA1-NubG, (II) +NubG-GPA1, (III) +GPA1-Nubwt, (IV) +Nubwt-GPA1, (V) +GCR1FL-NubG, and (VI) +NubG-KAT1. (3) GCR1IL3-Cub: (I) +GPA1-NubG, (II) +NubG-GPA1, (III) +GPA1Nubwt, (IV) +Nubwt-GPA1, (V) +GCR1FL-NubG, and (VI) +NubG-KAT1. (4) GCR1Ct-Cub: (I) +GPA1-NubG, (II) NubG-GPA1, (III) +GPA1-Nubwt, (IV) +Nubwt-GPA1, (V) +GCR1FL-NubG, and (VI) +NubG-KAT1. Interaction was determined by growth of diploid yeast colonies on media lacking His and Ade but containing 200 μM Met and by β-galactosidase activity using X-Gal filter assay. I and II were test interactions, III and IV were positive controls, and V and VI were negative controls in all cases.
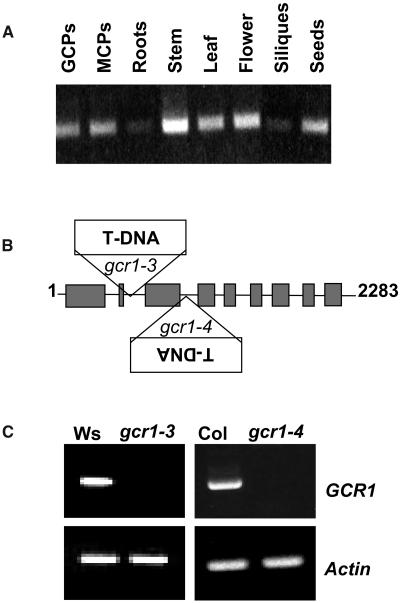
Identification of gcr1 T-DNA Insertional Mutants
To investigate the physiological role of GCR1, we first examined the expression pattern of GCR1. Expression could be detected in all cell/tissue types assessed by RT-PCR, including stomatal guard cells (Figure 5A). We then took a reverse genetic approach and isolated knockout mutants in this gene. Two different mutant lines, gcr1-3 and gcr1-4, that carry a T-DNA insertion in the GCR1 gene were obtained. The gcr1-3 mutant, harboring a T-DNA insertion in intron 2 (at 506 bp) of the GCR1 gene (Figure 5B), was found in a PCR screen of a collection of 72,960 T-DNA–inserted Arabidopsis lines (Arabidopsis Knockout Facility, University of Wisconsin, BASTA population) using a GCR1-specific primer and a T-DNA left border–specific primer (Krysan et al., 1999). The genetic background for this mutant line is ecotype Wassilewskija (Ws). The gcr1-4 mutant, harboring a T-DNA insertion in intron 3 (at 733 bp) of the GCR1 gene (Figure 5B), was obtained from the SAIL collection of Torrey Mesa Research Institute (TMRI; Syngenta). The genetic background for this mutant line is ecotype Columbia (Col). Both insertions were confirmed by sequencing of the genomic PCR products. To confirm that both gcr1-3 and gcr1-4 are transcript-null alleles, RT-PCR analysis was performed with RNA isolated from wild-type and mutant plants. Indeed, using primers that flank the insertion site, gcr1-3 and gcr1-4 plants did not yield an RT-PCR product under standard growth conditions that revealed the presence of GCR1 transcript in wild-type plants (Figure 5C).
Figure 5.
GCR1 Expression and Isolation of gcr1 Insertion Mutants.
(A) RT-PCR profiling of GCR1 expression. GCPs, guard cell protoplasts; MCPs, mesophyll cell protoplasts.
(B) T-DNA insertion sites in gcr1-3 (Ws ecotype; Wisconsin collection) and gcr1-4 (Col ecotype; Syngenta). Boxes and lines represent exons and introns, respectively (figure not drawn to the scale).
(C) RT-PCR analysis of GCR1 expression in wild-type Ws and Col and gcr1 homozygous mutant lines using primers flanking the T-DNA insertion sites. Actin primers served as control.
GCR1 Is Primarily Associated with the Membrane Fraction
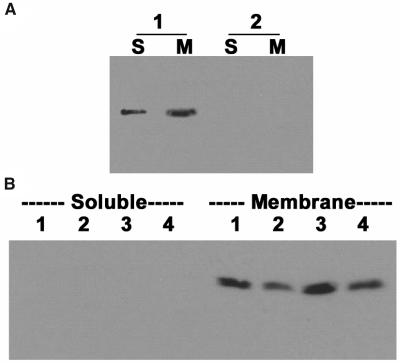
Because GCR1 is predicted to be an integral membrane protein with a 7TM structure, we determined the GCR1 protein expression level in soluble and membrane fractions of Arabidopsis leaves. Genetically complemented gcr1-3 plants expressing GCR1 as a FLAG-tagged protein under a glucocorticoid-inducible promoter were used as a source of protein after a 24-h induction with Dex. As shown in Figure 6A, most but not all GCR1 protein was detected in the membrane fractions isolated from the GCR1:FLAG plants (lanes 1, S and M). As expected, no protein could be detected with anti-FLAG antibodies in the noncomplemented gcr1-3 plants (lanes 2, S and M).
Figure 6.
GCR1 and GPA1 Both Associate with the Membrane Fraction.
(A) GCR1 is mostly associated with the membrane fraction. Total protein was isolated from GCR1:FLAG plants after 24 h of Dex induction (1) or from gcr1-3 mutant plants (2). Proteins were fractionated into soluble (S) or membrane (M) fractions. Equal amounts of protein were separated on SDS-PAGE and subjected to immunoblotting using anti-FLAG antibodies.
(B) gcr1 mutant plants have lower levels of GPA1 protein. Total proteins were isolated from Ws, Col, gcr1-3, and gcr1-4 leaves and fractionated into soluble and membrane fractions. Equal protein amounts (20 μg/lane) were separated on SDS-PAGE and probed with anti-GPA1 antibodies. GPA1 could be detected only in the membrane fraction. Both of the mutant lines consistently show lower levels of GPA1 protein (gcr1-3, lane 2; gcr1-4, lane 4) compared with the wild-type plants (Ws, lane 1; Col, lane 3).
Because GCR1 and GPA1 proteins interact in planta, we also evaluated expression of GPA1 protein in the gcr1 mutant backgrounds. As shown in Figure 6B, GPA1 is a membrane-associated protein. Both gcr1-3 (lane 2) and gcr1-4 (lane 4) show lower levels of GPA1 protein compared with the respective wild-type ecotypes (lanes 1 and 3).
gcr1 Is Involved in ABA Response in Vegetative Tissues
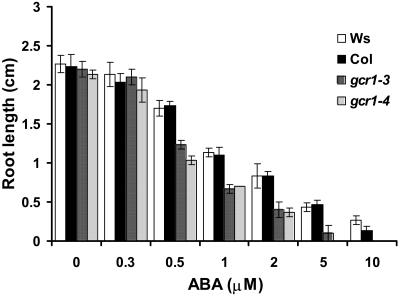
Because one of the signals transduced by GPA1 is ABA (Wang et al., 2001; Coursol et al., 2003), we examined responses of gcr1-3 and gcr1-4 mutants to this hormone. One of the classic responses regulated by ABA is inhibition of root growth (Finkelstein and Somerville, 1990). Wild-type seedlings germinated on control media show a marked inhibition of root growth when transferred to media containing 0.5 to 10 μM ABA compared with seedlings transferred to control (no ABA) media (Kang et al., 2002). Wild-type and gcr1 plants are identical when grown under normal growth conditions, but in the presence of ABA, root growth of gcr1 seedlings is markedly more sensitive to inhibition by ABA compared with wild-type plants (Figure 7). gcr1 root growth approached complete arrest when ABA concentration was >5 μM, whereas wild-type plants continued to show some growth.
Figure 7.
gcr1 Is Hypersensitive to ABA-Induced Inhibition of Root Length.
Inhibition of root length was measured after 5 d of growth. Each value is the mean ± 1 se of at least 60 plants. Data were compared using the Student's t test at the 95% significance level.
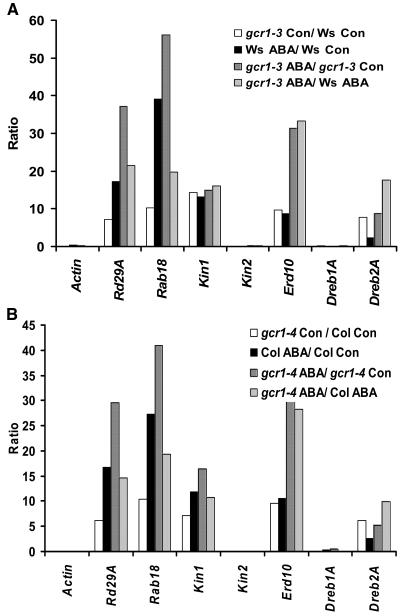
We also looked at the expression profiles of various stress- and ABA-regulated genes (Seki et al., 2002) in wild-type and gcr1 plants under control and ABA treated conditions. By RT-PCR, both gcr1-3 and gcr1-4 showed higher expression levels of a set of ABA and stress-regulated genes as compared with their wild-type counterparts (data not shown). We confirmed and extended these observations by real-time quantitative PCR. Ws and gcr1-3 and Col and gcr1-4 plants were grown under identical conditions (wild type and gcr1 side by side on the same media plates) on 0.5× MS media, 1% sucrose (control), or 0.5× MS media, 1% sucrose plates supplemented with 0.3 μM ABA (ABA). No visible phenotypic differences were seen between wild-type and mutant plants under these low ABA concentrations. Total RNA was isolated from 10-d-old seedlings, reverse transcribed, and used for real-time PCR in the presence of SYBR-Green intercalating dye. Amplification of the ACTIN2/8 gene under identical conditions served as an internal control. Results obtained are plotted as ratios of fold change in expression level of particular genes in gcr1-3 and Ws plants (Figure 8A) and gcr1-4 and Col plants (Figure 8B). The ratios of gcr1 controls to wild-type (WT) controls show higher basal levels of expression of most of these stress and ABA-regulated genes in gcr1 plants, without any exogenous ABA treatment. Ratios of WT-ABA to WT-control and gcr1-ABA to gcr1-control illustrate that these genes are expressed at higher levels after ABA treatment in both wild-type and gcr1 plants (with the exception of kin2 and DREB1A). Ratios of gcr1-ABA to WT-ABA show that these genes are expressed at a higher level in gcr1 plants compared with wild-type plants after ABA treatment. Comparison between gcr1-control/WT control and gcr1-ABA/WT-ABA also shows that the expression level in gcr1 lines is hypersensitive to ABA and is not solely because of initial differences in the expression levels of these genes. Similar trends were obtained with both gcr1-3 and gcr1-4, although quantitative differences, possibly because of differential ABA sensitivity of the different genotypic backgrounds (Ws versus Col) were also observed. Wild-type and gcr1 mutant plants show no difference in endogenous ABA levels, as detected by direct competitive ELISA measurements using the AGDIA immunodetection kit (data not shown).
Figure 8.
gcr1 Mutant Plants Have Higher Levels of Expression of Some Known ABA/Stress-Induced Genes.
Data represent means of ratios of three independent experiments. Ratio of (WT control) to (gcr1 control) shows higher levels of these genes (except kin2 and Dreb1A) in the mutant under normal conditions (no exogenous ABA). Ratio of (WT control) to (WT ABA) and (gcr1 control) to (gcr1 ABA) confirms these genes are expressed at higher levels by ABA treatment under these growth conditions. Ratio of (gcr1 ABA) to (WT ABA) shows higher expression levels of these genes in gcr1 by ABA treatment compared with wild-type plants. Amplification of actin gene under identical conditions served as control.
gcr1 Plants Have Improved Drought Tolerance and Lower Rates of Water Loss
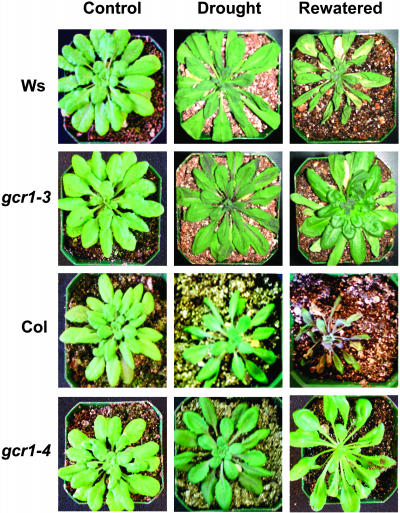
Because gcr1 plants exhibited relatively higher levels of ABA/stress-induced transcripts, we examined the effect of drought stress on these plants. Wild-type and mutant gcr1 plants show no difference in their growth and maturity under normal growth conditions; however, significant differences were observed when plants were drought stressed by withholding water for 12 d. As shown in Figure 9, wild-type plants dried faster and withered when withdrawn from water for 12 d. After rewatering, gcr1 plants showed highly improved recovery compared with wild-type plants. Only one of the 20 wild-type plants survived to maturity when rewatered, whereas all 20 gcr1 plants survived the treatment and completed their life cycle.
Figure 9.
gcr1 Plants Are More Resistant to Drought Stress.
Intact wild-type and gcr1 plants (control) were drought stressed by withholding water for 12 d (drought). Two days after rewatering (rewatered), gcr1 plants show better recovery than wild-type plants.
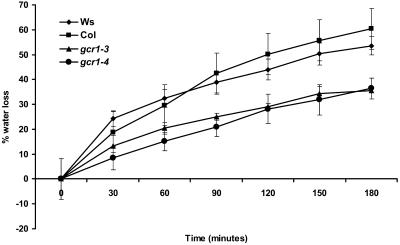
The enhanced drought tolerance of the gcr1 plants could result, at least in part, from the gcr1 plants having lower transpiration rates, as has been shown for other ABA hypersensitive mutants (Leung et al., 1997). When measured by the fresh weight loss of detached rosette leaves, the water loss rates of gcr1-3 and gcr1-4 mutant leaves were indeed ∼50% of those of the wild-type leaves (Figure 10).
Figure 10.
gcr1 Mutants Have Lower Rates of Water Loss.
Rate of water loss from detached leaves was measured as percentage of initial fresh weight after the indicated time periods. Values are mean ± se of three individual plants per genotype. The entire experiment was replicated four times. Data were compared using the Student's t test at the 95% significance level.
gcr1 Plants Are Hypersensitive to ABA and S1P Regulation of Stomatal Responses
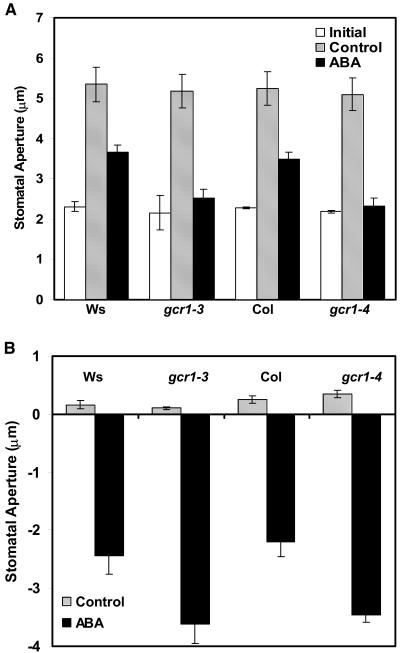
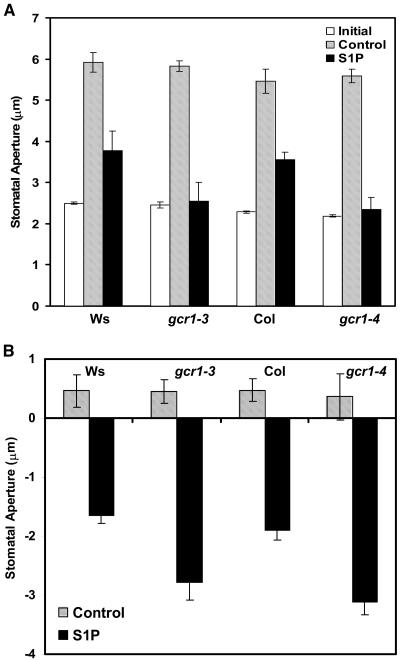
Based on the gcr1 drought stress and water loss phenotypes and also because GCR1 is expressed in guard cells and GPA1 is known to play a role in guard cell ABA responses (Wang et al., 2001; Coursol et al., 2003), we analyzed the responsiveness of gcr1 stomata to ABA in isolated epidermal peels. ABA at concentrations 20 to 50 μM inhibits opening of preclosed stomata and promotes closure of opened stomata in wild-type Arabidopsis plants (Pei et al., 1997). Both gcr1-3 and gcr1-4 guard cells were hypersensitive to both inhibition of opening of preclosed stomata (Figure 11A) and promotion of closure of preopened stomata (Figure 11B) by ABA. gcr1 plants respond to S1P in an identical fashion as they do to ABA: gcr1 guard cells are hypersensitive to S1P-induced inhibition of stomatal opening (Figure 12A) and S1P-induced promotion of stomatal closure (Figure 12B) compared with their respective wild-type ecotypes.
Figure 11.
gcr1 Mutants Show Hypersensitivity to ABA-Induced Inhibition of Stomatal Opening and Promotion of Stomatal Closure.
(A) Inhibition of opening of preclosed stomata in response to ABA. Measurements were taken after 2 h of incubation in light after addition of ABA.
(B) Promotion of closure of open stomata in response to ABA. Measurements were taken after 3 h of incubation in light after ABA treatment. Values represent means ± 1 se from three independent experiments; n = 40 apertures per experiment. Data were compared using the Student's t test at the 95% significance level.
Figure 12.
gcr1 Mutants Show Hypersensitivity to S1P-Induced Inhibition of Stomatal Opening and Promotion of Stomatal Closure.
(A) Inhibition of opening of preclosed stomata in response to S1P. Measurements were taken after 2 h of incubation in light after addition of S1P.
(B) Promotion of closure of open stomata in response to S1P. Measurements were taken after 3 h of incubation in light after S1P treatment. Values represent means ± 1 se from three independent experiments; n = 40 apertures per trial. Data were compared using the Student's t test at the 95% significance level.
DISCUSSION
Arabidopsis GPCRs
The GPCR superfamily contains >1000 members (Bockaert and Pin, 1999; Hall et al., 1999). There is little sequence conservation across the six identified GPCR families (Kolakowski, 1994; Horn et al., 1998, 2003), and sequence conservation within a family is typically limited to ∼25% sequence identity within the transmembrane domains (Pierce et al., 2002). Such diversity is presumably important for the ability of the GPCR superfamily as a whole to perceive a great diversity of signals, ranging from light, odorants, and tastants to numerous hormones and even Ca2+ ions (Gether, 2000; Pin et al., 2003). In Arabidopsis, however, previous homology searches of the EST databases identified only one potential GPCR, GCR1 (Josefsson and Rask, 1997; Plakidou-Dymock et al., 1998). Our reanalysis based on current database information still identifies GCR1 as a strong candidate for a plant GPCR. Table 1 shows that in addition to previously identified similarity of GCR1 to CAR-type receptors, GCR1 also harbors significant similarity to members of a new family of GPCRs, the Frizzled/Smoothened family, as well as to Methuselah-like proteins of the secretin GPCR family. Interestingly, mutation of Methuselah leads to stress resistance in Drosophila (Lin et al., 1998).
Previous analyses identified only GCR1 as a putative Arabidopsis GPCR. Similarly, only one prototypical Gα subunit (GPA1; Ma et al., 1990), one prototypical Gβ subunit (AGB1; Weiss et al., 1994), two probable Gγ subunits (AGG1 and AGG2; Mason and Botella, 2000, 2001), and one RGS protein (Chen et al., 2003) have been identified in Arabidopsis. Although the coupling of multiple GPCRs with a single Gα is an established phenomenon in animal systems (Albert and Robillard, 2002), if this is the explanation for the multiple signals transduced via the G protein heterotrimer in Arabidopsis, then the commonality unifying these GPCRs is unlikely to be sequence similarity. Therefore, future studies on proteins upstream of the heterotrimer in plants may focus on other predicted 7TM proteins, regardless of whether sequence similarity to nonplant GPCRs is present. In fact, the 7TM topology of one such family, the MLOs, has been experimentally verified (Devoto et al., 1999; Cohen et al., 2003), although at least one MLO family member may not be G protein coupled according to recent experimental analyses (Kim et al., 2002).
GCR1 and GPA1 Interact
Theoretical models devised from various fold recognition servers predict that the three Arabidopsis subunits form a valid heterotrimer, similar to heterotrimers formed by mammalian G protein subunits (Ullah et al., 2003), and interaction between Gβ and Gγ subunits has been verified experimentally (Mason and Botella, 2000, 2001), predicting a functional heterotrimeric G protein signaling complex in plants. Our data now show that the putative GPCR, GCR1, satisfies one of the most essential criteria for identification of a protein as a GPCR: interaction with a Gα subunit. GCR1 and GPA1 interact in vitro (Figure 1A) and in vivo both in the yeast-based split ubiquitin system (Figures 1B and 4) and in planta (Figure 2).
Using the classic yeast two-hybrid method, we did not observe interaction between GCR1 and GPA1 (data not shown), as was also reported previously by Humphrey and Botella (2001). This is possibly because of the inefficiency of the standard yeast two-hybrid method to detect interaction between membrane proteins, although some membrane protein interactions have been detected using this method, including interaction with G protein subunits (e.g., Mason and Botella, 2000, 2001; Lapik and Kaufman, 2003), which are not intrinsic membrane proteins but are usually membrane-associated via lipid modifications (Casey, 1994; Morris and Malbon, 1999). Because the majority of the GCR1 protein is indeed localized to the membrane in Arabidopsis (Figure 6A) and because our analysis shows that GPA1 is a bona fide membrane-associated protein (Figure 6B), we instead used a split ubiquitin based system, optimized to test interactions between membrane proteins (Ludewig et al., 2003), to assess potential interaction between GCR1 and GPA1. Positive results were obtained in the split ubiquitin system and were further validated by demonstration of in planta interaction of GCR1 and GPA1 (Figure 2). FLAG epitope–tagged GCR1 could be detected in the immunocomplex precipitated with anti-GPA1 antibodies and vice versa, confirming that GCR1 is indeed coupled to the G protein α subunit of Arabidopsis. The success of the coimmunoprecipitation experiments may indicate the presence in the tissue sample of an active ligand associated with GCR1; alternatively, it could reflect the phenomenon of precoupling, in which the GPCR and the heterotrimer form a stable complex even in the absence of a ligand (Wess, 1998). It is also interesting to note that we obtained negative results in tests of interaction between GPA1 and the KAT1 inward K+ channel (Figure 4). Pharmacological studies have implicated a membrane-delimited pathway of G protein regulation of the guard-cell inward K+ current (Wu and Assmann, 1994). These results suggest that this regulation may not be executed by direct interaction of GPA1 with the KAT1 channel, with the caveat that the present split ubiquitin data most likely reflect interactions of the inactive, GDP-bound form of GPA1.
GCR1 and GPA1 Interaction Depends on Intracellular Domains of GCR1
We further analyzed the interaction between GCR1 and GPA1 with respect to the GCR1 domains required for this interaction. Previous studies in animal systems with chimeric and mutagenized GPCRs have shown the il2 and in particular the il3 to be the major determinants of the coupling specificity between GPCRs and G proteins (Strader et al., 1994; Wess, 1998). Our analysis of GCR1–GPA1 interaction using the split ubiquitin system shows that, as in mammalian systems, the presence of an intact il3 structure in GCR1 and/or some key amino acids from the beginning of il2 to the beginning of il3 are essential for its interaction with GPA1: among the deletion constructs GCR1IL2, GCR1IL3, and GCR1Ct (Figure 4), interaction was not seen with GCR1IL3 or GCR1Ct, which lack the il3 (Figure 3). The second requirement for GCR1–GPA1 interaction that can be deduced from our experiments is the presence of a free C terminus in GCR1 (Figures 1B and 4B). This result stands in contrast with data from mammalian systems, in which the C-terminal cytoplasmic portion (with the exception of the membrane proximal 8 to 16 amino acids), is not required for receptor/G protein interactions (Wess, 1998), notwithstanding the vital importance of the C-terminal tail for interactions with other types of regulatory proteins (Wess, 1998; Hall et al., 1999; Bockaert et al., 2003). Future studies in which individual residues within the C-terminal tail and intracellular loops of GCR1 are mutagenized will help to pinpoint the exact amino acid sequences required for GCR1–GPA1 interaction.
In animal systems, GPCRs have been frequently shown to dimerize, and this dimerization modulates activity (Dean et al., 2001; Brady and Limbird, 2002; Pierce et al., 2002). Our data with GCR1-Cub and GCR1-Nub fusion proteins do not show GCR1 dimerization (Figures 1B, V, and 4B, V). However, caution must be exercised in interpreting the significance of this result for signaling in planta because the GCR1-Cub fusion might not interact with GCR1-Nub simply because of the presence of ubiquitin at the C terminus of GCR1-Nub.
GCR1 Is a Ubiquitous Membrane Protein
By RT-PCR, we could detect GCR1 transcript in all the tissue types tested (cf. Colucci et al., 2002) and also in guard cell protoplasts and mesophyll cell protoplasts (Figure 5A). However, GCR1 transcript is present at a very low level and was difficult to detect by RNA gel blot hybridization (Colucci et al., 2002; our unpublished data). Protein gel blot analysis of GCR1 protein in GCR1:FLAG plants after Dex induction revealed the presence of GCR1 predominantly in the membrane fraction (Figure 6A). These results are comparable to the expression pattern of GCR1:GFP fusion protein shown previously by Humphrey and Botella (2001). They reported that GCR1-GFP fusions localize to discrete regions in the outermost portion of Arabidopsis leaf epidermal cells, interpreted to be either plasma membrane domains or tonoplast abutting the plasma membrane. From recent studies on metazoan GPCRs, it is now known that these receptors do not obligately reside in the cell membrane and that they can be internalized in response to various stimuli (Drmota et al., 1998; McLean and Milligan, 2000; Daly and McGrath, 2003). We also confirm membrane localization for GPA1, as would be expected given that GCR1 and GPA1 interact in planta. Arabidopsis GPA1 and tobacco Gβ subunits have been localized in the plasma membrane in previous biochemical studies (Weiss et al., 1997; Peskan and Oelmuller, 2000). Interestingly, we observe lower apparent expression of GPA1 in the gcr1 background (Figure 6), consistent with the hypothesis that these two proteins are functionally linked.
Isolation and Characterization of GCR1 Knockout Mutants
In work published previously, GCR1 antisense plants showed no obvious developmental phenotypes (Humphrey and Botella, 2001; Kanyuka et al., 2001); however, this might be because of incomplete abrogation of gene expression. Overexpression studies have revealed phenotypes (Colucci et al., 2002; Apone et al., 2003), although on their own these studies run the risk of neomorphic effects. Thus, we decided to assess the phenotypes of gcr1 T-DNA insertional mutants. We isolated gcr1-3 from the T-DNA insertion mutant population from the Arabidopsis knockout facility at the University of Wisconsin and obtained gcr1-4 from the SAIL collection of T-DNA mutants from TMRI (Syngenta). As shown in Figure 5B, the gcr1-3 insertion is in the 2nd intron of GCR1 at position 506 bp. The gcr1-4 insertion is in the 3rd intron at position 733 bp (based on the genomic clone U95142). DNA gel blot analyses of homozygous gcr1-3 and gcr1-4 mutants revealed that there is only one T-DNA insertion in each of these mutant lines (data not shown). Both gcr1-3 and gcr1-4 plants did not produce GCR1 transcript with primers flanking the T-DNA insertion site (Figure 5C). We also could not detect GCR1 transcript in the homozygous mutants using two primers, both of which were located downstream of the T-DNA insertion site (data not shown). Transcript-null status of gcr1-3 was also confirmed by RNA gel blotting and by DNA gel blotting of the RT-PCR product (data not shown).
gcr1 Plants Are ABA Hypersensitive and Drought Tolerant
Classic responses mediated by ABA include inhibition of root elongation, drought tolerance, and altered expression of a set of stress-related genes (Finkelstein and Somerville, 1990; Lopez-Molina et al., 2001; Finkelstein et al., 2002; Seki et al., 2002). gcr1-3 and gcr1-4 exhibit these phenotypes. gcr1 plants showed significant hypersensitivity to ABA inhibition of root growth at ABA concentrations above 0.3 μM (Figure 7). gcr1 mutants also have higher basal levels of expression of six out of seven genes (kin2 being the exception) previously shown to be induced by ABA/dehydration stress (Lang and Palva, 1992; Liu et al., 1998; Seki et al., 2002). This gene set generally also showed higher levels of induction by ABA in gcr1 plants compared with wild-type plants (Figure 8). By contrast, we did not observe elevated expression levels in gcr1 lines of Dreb1A, a transcription factor that is induced by cold stress but not by ABA (Liu et al., 1998). We further observed that gcr1 plants tolerate drought stress better and show improved recovery after rewatering compared with wild-type plants (Figure 9). The endogenous levels of ABA were not significantly different in wild-type versus gcr1 mutant plants, demonstrating that gcr1 plants are ABA hypersensitive, thus implicating GCR1 function in stress response.
Based on observation of a whole-plant drought-tolerance phenotype in gcr1 plants, we next assessed stomatal responses of gcr1 knockout plants (Figures 10 to 12). Detached leaves of gcr1 mutants lose water more slowly than wild-type plants. These results suggested that gcr1 stomata might be more sensitive to ABA because increased stomatal closure could account for the observed lower rates of water loss in the gcr1 lines relative to the wild type (Figure 10). In support of this hypothesis, both gcr1-3 and gcr1-4 mutants showed hypersensitivity to ABA and S1P inhibition of stomatal opening, and ABA and S1P promotion of stomatal closure compared with the corresponding wild-type Ws or Col plants (Figure 11).
Roles of GPA1 and GCR1 in ABA Signaling in Guard Cells
In Arabidopsis, null mutants of the sole canonical Gα subunit GPA1 exhibit insensitivity to ABA inhibition of stomatal opening but wild-type ABA promotion of stomatal closure. Underlying abrogation of ABA sensitivity in the opening response, gpa1 plants show loss of inward K+ channel inhibition by ABA (Wang et al., 2001). ABA activation of slow anion channels in guard cells appears to rely on two pathways: one dependent on elevation of cytosolic pH and the other on GPA1. gpa1 knockouts show normal ABA-induced stomatal closure despite loss of GPA1-mediated anion channel activation, suggesting that the pH and GPA1-dependent pathways of ABA action may function redundantly (Wang et al., 2001).
In guard cells, ABA activates sphingosine kinase, leading to production of the lipid metabolite S1P (Coursol et al., 2003). S1P appears to function upstream of Gα because gpa1 knockout plants are insensitive to S1P inhibition of stomatal opening and promotion of stomatal closure as well as to S1P inhibition of inward K+ channels and activation of slow anion channels. In support of positioning S1P upstream of GPA1, ABA stimulation of sphingosine kinase activity is retained in gpa1 mutants (Coursol et al., 2003). S1P plays important signaling roles in animal systems, where it acts both intracellularly to promote release of Ca2+ from intracellular stores and extracellularly by serving as a ligand for a set of GPCR receptors, previously called the EDG receptors but recently renamed the S1P receptors (Spiegel and Milstien, 2003).
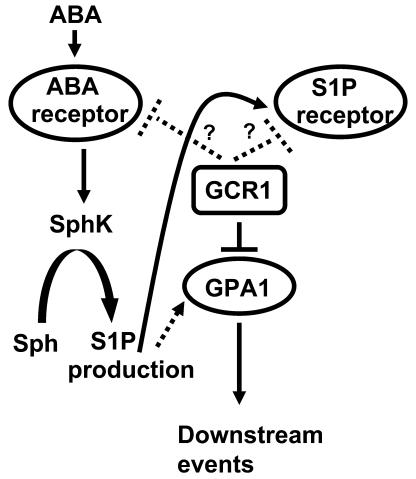
Based on our previous results from gpa1 knockouts (Wang et al., 2001; Coursol et al., 2003), on the fact that the GPCR-like protein GCR1 interacts with GPA1, and on the fact that S1P is perceived by GPCRs in mammalian cells, a straightforward hypothesis would be that GCR1 functions as either an S1P receptor or an ABA receptor in guard cells. If this were the case, a simple model would predict that guard cell phenotypes of gpa1 knockouts and gcr1 knockouts would be similar, if not identical. This model is not, however, supported. The ABA and S1P responses of gcr1 mutant guard cells (Figures 10 to 12) are opposite to those of gpa1 mutants: gcr1 guard cells exhibit hypersensitivity, whereas gpa1 guard cells exhibit insensitivity. To reconcile these observations, we hypothesize that in guard cells, GCR1 is a negative regulator of GPA1 (Figure 13). Molecular genetic studies have shown that ABA signaling involves a complex network of negative regulation. Loss of function mutations in protein phosphatases such as ABI1 and ABI2 (Merlot et al., 2001), enzymes such as farnesyl transferase (ERA1) (Cutler et al., 1996; Pei et al., 1998), and the RNA binding protein ABH1 (Hugouvieux et al., 2001) result in hypersensitivity to ABA, emphasizing the importance of negative regulation of ABA signaling pathways. The most parsimonious model predicts that negative regulation of GPA1 by GCR1 is accomplished by the direct binding of GCR1 with GPA1 that we observe. However, we cannot formally rule out the possibility that GCR1 negatively regulates ABA signaling via a mechanism independent of its binding to GPA1. One possible scenario is GCR1 regulation of proposed S1P or ABA receptors. Many mammalian GPCRs, including S1P receptors, have been shown to homodimerize or heterodimerize (Dean et al., 2001; Devi, 2001; Van Brocklyn et al., 2002). Thus, a possible interaction of GCR1 with Arabidopsis 7TM S1P or ABA receptors might alter ABA signal transduction in guard cells. Alternatively, GCR1 might interact with ABA or S1P receptors that are not GPCRs (Brady and Limbird, 2002). One example of such a mechanism found commonly in both invertebrates and vertebrates is the Hedgehog signaling pathway, in which the GPCR Smoothened is tonically inhibited by the 12-membrane-pass protein Patched as long as the Patched ligand, Hedgehog, is absent (Frank-Kamenetsky et al., 2002; Nybakken and Perrimon, 2002). Coupling of Smoothened to heterotrimeric G proteins has been established (DeCamp et al., 2000), but the endogenous ligand of Smoothened remains unknown. Interestingly, Smoothened falls in the same GPCR family as Frizzled (Foord et al., 2002), to which GCR1 has similarity (Josefsson, 1999; Table 1).
Figure 13.
Proposed Model for Interaction among GPA1, GCR1, and Possible ABA and S1P Receptors.
Solid lines indicate interactions confirmed by biochemical assays. Dotted lines indicate interaction implicated by genetic analyses. Question marks indicate hypothetical interactions consistent with current experimental data.
To summarize, based on the data presented herein, we conclude that GCR1 interacts with GPA1 in Arabidopsis and is a component of the ABA perception complex. Experimental identification of a ligand for GCR1 would unequivocally establish this protein as a genuine GPCR. Further work is required in this direction.
METHODS
Isolation of gcr1-3 and gcr1-4 Mutants
The Arabidopsis thaliana gcr1-3 insertion mutant (Ws background) was isolated by screening the collection of activation tagged T-DNA insertion lines (BASTA population) available at the Arabidopsis Knockout Facility (University of Wisconsin–Madison; www.biotech.wisc.edu/arabidopsis). Primers specific for the T-DNA left (5′-cattttataataacgctgcggacatctac-3′) and T-DNA right (5′-tgggaaaacctggcgttacccaacttaat-3′) borders were used in combination with GCR1 specific forward primer 5′-gaaatcgtcaattcaatctctagatcagt-3′ and reverse primer 5′-ttcgtgttcccaaagaatgtttcatatac-3′ to identify the insertion. The gcr1-4 T-DNA insertional mutant (Col background) was obtained from the SAIL collection of TMRI (Syngenta, Research Triangle Park, NC). Both the insertions were confirmed by sequencing. Full-length GCR1 cDNA fused with a C-terminal FLAG tag cloned in the glucocorticoid-inducible vector pTA7002 (Aoyama and Chua, 1997) was introduced into gcr1-3 plants via Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Transgenic plants were selected by hygromycin resistance and confirmed by PCR with gene-specific primers.
Plant Growth
Wild-type (Ws and Col) and homozygous gcr1 mutant seeds were first germinated on 0.5× MS media plates containing 1% sucrose. Robust plants were transferred to soil after 10 d of growth on plates. Plants were grown in controlled environment growth chambers under a 16-h-dark (20°C)/8-h-light (22°C) cycle. The light intensity was 150 μmol·m−2·s−1 for Ws and gcr1-3 plants and 120 μmol·m−2·s−1 for Col and gcr1-4 plants. For ABA inhibition of root growth, wild-type and mutant seeds were germinated in darkness on 0.5× MS plates containing 1% sucrose. After 2 d, germinated seeds were transferred to 0.5× MS, 1% sucrose plates containing different concentrations of ABA (AG Scientific, San Diego, CA). Root length was recorded after 5 d of growth.
In Vitro Transcription/Translation and Binding Assays
Full-length and truncated GCR1 cDNA fragments were cloned in pCite (2b) vector (Novagen, Madison, WI). GPA1 full-length cDNA and a putative protein kinase cDNA were fused in frame with the GAL4 activation domain of yeast (Saccharomyces cerevisiae), and the whole cassette was cloned in pCite (2b) vector. The constructs were used for in vitro transcription/translation reactions using the STP3 kit (Novagen) according to the manufacturer's instructions using translation grade 35S-Met (Amersham Biosciences, Piscataway, NJ) except for the kinase-GAD fusion protein. For Kinase-GAD, the transcription/translation was performed in the presence of cold Met. The binding reaction was performed by mixing 20 μL of anti-GAD Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) with 20 μL of GCR1 translation mix and 20 μL of GPA1-GAD translation mix in 500 μL of binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% BSA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× protease cocktail mix (Roche, Indianapolis, IN), and 0.01% Triton X-100). The reaction mix was agitated at 4°C overnight. The beads were washed five times with 3 mL of washing buffer (binding buffer without BSA, PMSF, and protease cocktail). Beads were spun down and mixed with SDS-PAGE sample buffer and boiled for 5 min and proteins separated by SDS-PAGE. After electrophoresis, the gel was dried and exposed for autoradiography.
Structure, Topology, and Similarity Prediction for GCR1
Transmembrane regions and topology of GCR1 were predicted using Simple Modular Architecture Retrieval Tool (SMART version 3.5) and transmembrane hidden Markov model (TMHMM version 2.0) programs. Overall similarity of GCR1 with other GPCR-like proteins was determined by PSI-BLAST.
Split Ubiquitin Assays
Vectors and interaction assays used for split ubiquitin analysis have been described elsewhere (Ludewig et al., 2003; Schulze et al., 2003). Briefly, GCR1FL (full-length cDNA), GCR1IL2 (sequence from amino acid 105 to the C terminus), GCR1IL3 (sequence from amino acid 184 to the C terminus), and GCR1Ct (sequence from amino acid 271 to the C terminus) were cloned in Cub, NubG, and Nubwt vectors by in vivo recombination cloning. GPA1 full-length sequence was also cloned in both Cub and Nub vectors. Interaction was determined by growth of diploid yeast colonies on media lacking His and Ade but containing 200 μM Met and also by β-galactosidase activity by X-Gal filter assay (Ludewig et al., 2003).
In Vivo Coimmunoprecipitation Assay
Total proteins were extracted from 4-week-old soil-grown gcr1-3 and GCR1:FLAG complemented gcr1-3 plants. For induction of GCR1-FLAG protein, plants were treated with 30 mM Dex for 24 h. Noninduced (control) plants were left untreated. Protein was extracted by grinding the leaves (0.4 g) on ice in 3 volumes of extraction buffer. Protein concentration in each lysate was adjusted to the same value, and equal volumes (1 mL) of lysates were transferred to new centrifuge tubes. Supernatants were mixed with anti-GPA1 or anti-FLAG antibodies and incubated at 4°C overnight with agitation. After incubation, 100 μL of protein A–Sepharose (Amersham Biosciences) was added to precipitate the antigen-antibody complex. The protein A–agarose beads were collected after 1 h of incubation at 4°C by centrifugation at 10,000g for 20 s. The beads were then washed five times with 5 mL of extraction buffer. The antigen-antibody complex was eluted by boiling in SDS sample buffer (Laemmli, 1970) and run on a 12% SDS-PAGE gel. Proteins immunoprecipitated with anti-GPA1 antibodies were probed with anti-FLAG antibodies and vice versa by immunoblotting.
Extraction of Arabidopsis Proteins and Protein Gel Blot Analysis
Total protein extracts were obtained from Arabidopsis plants by grinding whole seedlings or leaf tissue first in liquid nitrogen and then on ice in 3 volumes of extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 50 mM sucrose, 1 mM PMSF, 0.1% Triton X-100, and 1× plant protease inhibitor cocktail; Roche). Lysates (1 mL each) were cleared of debris by centrifugation at 12,000g for 20 min at 4°C. Cleared lysate was centrifuged at 100,000g at 4°C for 1 h to separate membrane (pellet) and soluble fractions. Protein concentrations in the extracts were measured by the Bradford (1976) assay (Bio-Rad, Hercules, CA). Twenty micrograms of proteins from each extract was separated on a 12% SDS-PAGE gel and then transferred onto a nitrocellulose membrane. The blots were probed with polyclonal antibodies against GPA1 (from A.M. Jones, University of North Carolina, Chapel Hill) or anti-FLAG Monoclonal M2 antibody (Sigma, St. Louis, MO). The antibodies were diluted in blocking buffer (1× Tris-buffered saline [TBS] containing 5% nonfat milk and 0.5% Tween 20) to 1:10,000. After washing in 1× TTBS (TBS containing 0.5% Tween 20), the blots were probed with appropriate secondary antibodies conjugated with horseradish peroxidase (1:10,000 dilution). The antibody-bound proteins were detected by a chemiluminescence reaction using the SuperSignal Kit (Pierce, Rockford, IL).
RT-PCR and Quantitative Real-Time RT-PCR
RNA was isolated from different plant tissue or cell types (for RT-PCR analysis) or from 10-d-old seedlings grown on 0.5× MS plates containing or lacking 0.3 μM ABA (for quantitative real-time PCR analysis). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) and treated with RNase free DNase I (Qiagen, Valencia, CA). Two micrograms of total RNA was reverse transcribed using the Superscript II RT kit (Invitrogen) according to the manufacturer's instructions. cDNA was diluted at a concentration of 1:100, aliquoted, and kept at 4°C throughout each experiment to avoid discrepancy in the data because of freeze-thaw cycles. To determine expression of GCR1 in different plant parts, GCR1 was amplified using primers GCR1-RTF 5′-acagcggaaatcgtcaattc-3′ and GCR1-RTR 5′-ccccaaatgtgagtggttgt-3′. To assess whether gcr1-3 and gcr1-4 are transcript null alleles, cDNA from wild-type and mutant seedlings was amplified using gene-specific primers flanking the insertion sites: GCR1-KORTF (5′-agcaaatcagatcgccttgt-3′) and GCR1-KORTR (5′-gaacgtatgacagtcacaac-3′). For quantitative real-time PCR, amplification was performed with oligonucleotides specific for various ABA- and stress-induced genes: RD29A forward (5′-atcacttggctccactgttgttc-3′) and RD29A reverse (5′-acaaaacacacataaacatccaaagt-3′); RAB18 forward (5′-cagcagcagtatgacgagta-3′) and RAB18 reverse (5′-cagttccaaagccttcagtc-3′); KIN1 forward (5′-accaacaagaatgccttcca-3′) and KIN1 reverse (5′-ccgcatccgatacactcttt-3′); KIN2 forward (5′-accaacaagaatgccttcca-3′) and KIN2 reverse (5′-actgccgcatccgatatact-3′); DREB1A forward (5′-gatcagcctgtctcaatttc-3′) and DREB1A reverse (5′-cttctgccatattagccaac-3′); DREB2A forward (5′-aaggtaaaggaggaccagag-3′) and DREB2A reverse (5′-acacaaccaggagtctcaac-3′); ERD10 forward (5′-tctctgaaccagagtcgttt-3′) and ERD10 reverse (5′-cttcttctcaccgtcttcac-3′). Amplification of ACTIN2/8 (forward primer 5′-ggtaacattgtgctcagtggtgg-3′ and reverse primer 5′-aacgaccttaatcttcatgctgc-3′) genes was used as an internal control (Charrier et al., 2002). The position of the oligonucleotides used for real-time PCR was chosen so that the size of all PCR products was between 200 and 250 bp. The suitability of the oligonucleotide sequences in term of efficiency of annealing was evaluated in advance using the Primer 3 program. Real-time quantitative PCR experiments were repeated thrice independently, and the data were averaged. For real-time quantitative-PCR, the cDNA was amplified in the presence of SYBR-GreenR I intercalating dye (Molecular Probes, Eugene, OR) at 0.125× final concentration using a DNA Engine Opticon 2 thermal cycler (MJ Research, Watertown, MA). The data obtained were analyzed with Opticon 2 software (MJ Research).
Drought Treatment
Four-week-old plants were drought treated by withholding water for 12 d. Plants were rewatered from the bottom on the 13th day to determine recovery after drought. Observations were recorded 2 d after rewatering.
Guard Cell Assays
Guard cell protoplasts and mesophyll cell protoplasts were isolated according to Pandey et al. (2002). Assays of water loss and stomatal responses were performed according to Pei et al. (1997), Wang et al. (2001), and Coursol et al. (2003). Briefly, for water loss experiments, detached leaves of Ws, Col, gcr1-3, and gcr1-4 were kept under controlled humidity and air flow, and rate of water loss was measured as percentage of initial fresh weight. For stomatal aperture measurements, leaves from wild-type Ws and Col and gcr1-3 and gcr1-4 plants were kept in darkness for 2 h and then transferred to light (450 μmol·m−2s−1) for 3 h in the presence of ABA (20 μM for Ws and gcr1-3 and 50 μM for Col and gcr1-4) or S1P (10 μM) to study inhibition of opening. For promotion of closure experiments, leaves were first kept in light for 2 h followed by addition of ABA (20 μM for Ws and gcr1-3; 50 μM for Col and gcr1-4) or S1P (10 μM). Apertures were recorded after 3 h of further incubation in light.
ABA Level Measurement
Wild-type and gcr1 mutant plants were grown on 0.5× MS media plates containing 1% sucrose. Seedlings (3 g fresh weight for each sample) were harvested after 10 d of growth and ground in liquid N2. Samples were prepared according to Ullah et al. (2002). ABA levels in the samples were determined by competitive ELISA using the AGDIA immunodetection kit (Elkhart, IN) according to manufacturer's instructions. The experiment was repeated twice.
Material Availability
The TMRI/Syngenta gcr1-4 line is available by request from Syngenta.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At1g48270 for GCR1, At2g26300 for GPA1, and At5g66890 for the putative protein kinase.
Acknowledgments
We thank Esther Siegfried for enlightening discussions on the analogy between our system and the Hedgehog signaling pathway. We thank Wolf Frommer and Petr Obrdlik for making their split ubiquitin system available and for advice concerning the assays, Jerry Workman and Philippe Prochasson for use of their real-time thermocycler and advice on real-time PCR experiments, and Alan Jones for anti-GPA1 antibodies. We also thank Cha-Young Kim for Flag-tagged pTA7002 vector and Liza Hall for excellent technical assistance. This work was supported by National Science Foundation Grant (MCB-0209694) to S.M.A.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Sona Pandey (sxp49@psu.edu) and Sarah M. Assmann (sma3@psu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020321.
References
- Albert, P.R., and Robillard, L. (2002). G protein specificity: Traffic direction required. Cell. Signal. 14, 407–418. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Apone, F., Alyeshmerni, N., Wiens, K., Chalmers, D., Chrispeels, M.J., and Colucci, G. (2003). The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol. 133, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann, S.M. (2002). Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14 (suppl.), S355–S373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert, J., Marin, P., Dumuis, A., and Fagni, L. (2003). The ‘magic tail’ of G protein-coupled receptors: An anchorage for functional protein networks. FEBS Lett. 546, 65–72. [DOI] [PubMed] [Google Scholar]
- Bockaert, J., and Pin, J.P. (1999). Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 18, 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brady, A.E., and Limbird, L.E. (2002). G protein-coupled receptor interacting proteins: Emerging roles in localization and signal transduction. Cell. Signal. 14, 297–309. [DOI] [PubMed] [Google Scholar]
- Casey, P.J. (1994). Lipid modifications of G proteins. Curr. Opin. Cell Biol. 6, 219–225. [DOI] [PubMed] [Google Scholar]
- Charrier, B., Champion, A., Henry, Y., and Kreis, M. (2002). Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 130, 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.G., Willard, F.S., Huang, J., Liang, J., Chasse, S.A., Jones, A.M., and Siderovski, D.P. (2003). A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 30, 1728–1731. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, F.E., Emerson, B.C., Schulze-Lefert, P., and Panstruga, R. (2003). Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 56, 77–88. [DOI] [PubMed] [Google Scholar]
- Colucci, G., Apone, F., Alyeshmerni, N., Chalmers, D., and Chrispeels, M.J. (2002). GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc. Natl. Acad. Sci. USA 99, 4736–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol, S., Fan, L.M., Le Stunff, H., Spiegel, S., Gilroy, S., and Assmann, S.M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423, 651–654. [DOI] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Daly, C.J., and McGrath, J.C. (2003). Fluorescent ligands, antibodies, and proteins for the study of receptors. Pharmacol. Ther. 100, 101–118. [DOI] [PubMed] [Google Scholar]
- Dean, M.K., Higgs, C., Smith, R.E., Bywater, R.P., Snell, C.R., Scott, P.D., Upton, G.J., Howe, T.J., and Reynolds, C.A. (2001). Dimerization of G-protein-coupled receptors. J. Med. Chem. 44, 4595–4614. [DOI] [PubMed] [Google Scholar]
- DeCamp, D.L., Thompson, T.M., de Sauvage, F.J., and Lerner, M.R. (2000). Smoothened activates Gαi-mediated signaling in frog melanophores. J. Biol. Chem. 275, 26322–26327. [DOI] [PubMed] [Google Scholar]
- Devi, L.A. (2001). Heterodimerization of G-protein-coupled receptors: Pharmacology, signaling and trafficking. Trends Pharmacol. Sci. 22, 532–537. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Piffanelli, P., Nilsson, I., Wallin, E., Panstruga, R., von Heijne, G., and Schulze-Lefert, P. (1999). Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34993–35004. [DOI] [PubMed] [Google Scholar]
- Drmota, T., Gould, G.W., and Milligan, G. (1998). Real time visualization of agonis-mediated redistribution and internalization of a green fluorescent protein-tagged form of the thyrotropin-releasing hormone receptor. J. Biol. Chem. 273, 24000–24008. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Somerville, C.R. (1990). Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 105, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foord, S.M. (2002). Receptor classification: Post genome. Curr. Opin. Pharmacol. 2, 561–566. [DOI] [PubMed] [Google Scholar]
- Foord, S.M., Jupe, S., and Holbrook, J. (2002). Bioinformatics and type II G-protein-coupled receptors. Biochem. Soc. Trans. 30, 473–479. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky, M., Zhang, X.M., Bottega, S., Guicherit, O., Wichterle, H., Dudek, H., Bumcrot, D., Wang, F.Y., Jones, S., Shulok, J., Rubin, L.L., and Porter, J.A. (2002). Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J. Biol. 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether, U. (2000). Uncovering molecular mechanisms involved in activation of G protein–coupled receptors. Endocr. Rev. 21, 90–113. [DOI] [PubMed] [Google Scholar]
- Graul, R.C., and Sadee, W. (2001). Evolutionary relationships among G protein-coupled receptors using a clustered database approach. AAPS PharmSci. 3, E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudermann, T., Schoneberg, T., and Schultz, G. (1997). Functional and structural complexity of signal transduction via G-protein-coupled receptors. Annu. Rev. Neurosci. 20, 399–427. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., Premont, R.T., and Lefkowitz, R.J. (1999). Heptahelical receptor signaling: Beyond the G protein paradigm. J. Cell Biol. 145, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, F., Bettler, E., Oliveira, L., Campagne, F., Cohen, F.E., and Vriend, G. (2003). GPCRDB information system for G protein–coupled receptors. Nucleic Acids Res. 31, 294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, F., Weare, J., Beukers, M.W., Horsch, S., Bairoch, A., Chen, W., Edvardsen, O., Campagne, F., and Vriend, G. (1998). GPCRDB: An information system for G protein-coupled receptors. Nucleic Acids Res. 26, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. [DOI] [PubMed] [Google Scholar]
- Humphrey, T.V., and Botella, J.R. (2001). Re-evaluation of the cytokinin receptor role of the Arabidopsis gene GCR1. J. Plant Physiol. 158, 645–653. [Google Scholar]
- Jones, A.M. (2002). G-protein-coupled signaling in Arabidopsis. Curr. Opin. Plant Biol. 5, 402–407. [DOI] [PubMed] [Google Scholar]
- Josefsson, L.G. (1999). Evidence for kinship between diverse G-protein coupled receptors. Gene 239, 333–340. [DOI] [PubMed] [Google Scholar]
- Josefsson, L.G., and Rask, L. (1997). Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur. J. Biochem. 249, 415–420. [DOI] [PubMed] [Google Scholar]
- Kang, J.-Y., Choi, H., Im, M.-Y., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanyuka, K., Couch, D., and Hooley, R. (2001). Correction: A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr. Biol. 11, 535. [DOI] [PubMed] [Google Scholar]
- Kim, M.C., Panstruga, R., Elliott, C., Muller, J., Devoto, A., Yoon, H.W., Park, H.C., Cho, M.J., and Schulze-Lefert, P. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451. [DOI] [PubMed] [Google Scholar]
- Kolakowski, L.F., Jr. (1994). GCRDb: A G-protein-coupled receptor database. Recept. Channels 2, 1–7. [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lang, V., and Palva, E.T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Lapik, Y.R., and Kaufman, L.S. (2003). The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15, 1578–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, K.A., Wen, J., Li, J., Doke, J.T., Liscum, E., and Walker, J.C. (2001). A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower and fruit development. Plant Cell 13, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.J., Seroude, L., and Benzer, S. (1998). Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282, 943–946. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, U., Wilken, S., Wu, B., Jost, W., Obrdlik, P., El Bakkoury, M., Marini, A.M., Andre, B., Hamacher, T., Boles, E., Von Wiren, N., and Frommer, W.B. (2003). Homo- and hetero-oligomerization of AMT1 NH4+-uniporters. J. Biol. Chem. 278, 45603–45610. [DOI] [PubMed] [Google Scholar]
- Ma, H. (1994). GTP-binding proteins in plants: New members of an old family. Plant Mol. Biol. 26, 1611–1636. [DOI] [PubMed] [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1990). Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 87, 3821–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, M.G., and Botella, J.R. (2000). Completing the heterotrimer: Isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc. Natl. Acad. Sci. USA 97, 14784–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, M.G., and Botella, J.R. (2001). Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with G β. Biochim. Biophys. Acta 1520, 147–153. [DOI] [PubMed] [Google Scholar]
- McLean, A.J., and Milligan, G. (2000). Ligand regulation of green fluorescent protein-tagged forms of human β1-and β2-adrenoreceptors; comparison with the unmodified receptors. Br. J. Pharmacol. 130, 1825–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J. 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Morris, A.J., and Malbon, C.C. (1999). Physiological regulation of G protein-linked signaling. Physiol. Rev. 79, 1373–1430. [DOI] [PubMed] [Google Scholar]
- Ng, C.K., Carr, K., McAinsh, M.R., Powell, B., and Hetherington, A.M. (2001). Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410, 596–599. [DOI] [PubMed] [Google Scholar]
- Nybakken, K., and Perrimon, N. (2002). Hedgehog signal transduction: Recent findings. Curr. Opin. Genet. Dev. 12, 503–511. [DOI] [PubMed] [Google Scholar]
- Pandey, S., Wang, X.-Q., Coursol, S.A., and Assmann, S.M. (2002). Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol. 153, 517–526. [DOI] [PubMed] [Google Scholar]
- Pei, Z.-M., Ghassemian, M., Kwak, J.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyl transferase in ABA regulation of guard cell anion channel and plant water loss. Science 282, 287–290. [DOI] [PubMed] [Google Scholar]
- Pei, Z.M., Kuchitsu, K., Ward, J.M., Schwarz, M., and Schroeder, J.I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskan, T., and Oelmuller, R. (2000). Heterotrimeric G-protein beta-subunit is localized in the plasma membrane and nuclei of tobacco leaves. Plant Mol. Biol. 42, 915–922. [DOI] [PubMed] [Google Scholar]
- Pierce, K.L., Premont, R.T., and Lefkowitz, R.J. (2002). Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650. [DOI] [PubMed] [Google Scholar]
- Pin, J.P., Galvez, T., and Prezeau, L. (2003). Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 98, 325–354. [DOI] [PubMed] [Google Scholar]
- Plakidou-Dymock, S., Dymock, D., and Hooley, R. (1998). A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr. Biol. 8, 315–324. [DOI] [PubMed] [Google Scholar]
- Ross, E.M., and Wilkie, T.M. (2000). GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827. [DOI] [PubMed] [Google Scholar]
- Schulze, W.X., Reinders, A., Ward, J., Lalonde, S., and Frommer, W.B. (2003). Interactions between co-expressed Arabidopsis sucrose transporters in the split ubiquitin system. BMC Biochem. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 3, 279–292. [DOI] [PubMed] [Google Scholar]
- Spiegel, S., and Milstien, S. (2003). Sphingosine-1-phosphate: An enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407. [DOI] [PubMed] [Google Scholar]
- Stagljar, I., Korostensky, C., Johnsson, N., and te Heesen, S. (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95, 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, C.D., Fong, T.M., Tota, M.R., Underwood, D., and Dixon, R.A. (1994). Structure and function of G protein-coupled receptors. Annu. Rev. Biochem. 63, 101–132. [DOI] [PubMed] [Google Scholar]
- Suharsono, U., Fujisawa, Y., Kawasaki, T., Iwasaki, Y., Satoh, H., and Shimamoto, K. (2002). The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 99, 13307–13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady, G.E., and Simon, I. (2001). Topology of membrane proteins. J. Chem. Inf. Comput. Sci. 41, 364–368. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., and Matsuoka, M. (2000). Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97, 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Temple, B., Boyes, D.C., Alonso, J.M., Davis, K.R., Ecker, J.R., and Jones, A.M. (2003). The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15, 393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Wang, S., and Jones, A.M. (2002). Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 129, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Young, J.C., Im, K.H., Sussman, M.R., and Jones, A.M. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292, 2066–2069. [DOI] [PubMed] [Google Scholar]
- Van Brocklyn, J.R., Behbahani, B., and Lee, N.H. (2002). Homodimerization and heterodimerization of S1P/EDG sphingosine-1-phosphate receptors. Biochim. Biophys. Acta 1582, 89–93. [DOI] [PubMed] [Google Scholar]
- Wang, X.Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292, 2070–2072. [DOI] [PubMed] [Google Scholar]
- Weiss, C.A., Garnaat, C.W., Mukai, K., Hu, Y., and Ma, H. (1994). Isolation of cDNAs encoding guanine nucleotide-binding protein beta-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1). Proc. Natl. Acad. Sci. USA 91, 9554–9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, C.A., White, E., Huang, H., and Ma, H. (1997). The G protein α subunit (GPA1) is associated with the ER and the plasma membrane in meristematic cells of Arabidopsis and cauliflower. FEBS Lett. 407, 361–367. [DOI] [PubMed] [Google Scholar]
- Wess, J. (1998). Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 80, 231–264. [DOI] [PubMed] [Google Scholar]
- Worrall, D., Ng, C.K., and Hetherington, A.M. (2003). Sphingolipids, new players in plant signaling. Trends Plant Sci. 8, 317–320. [DOI] [PubMed] [Google Scholar]
- Wu, W.H., and Assmann, S.M. (1994). A membrane-delimited pathway of G-protein regulation of the guard-cell inward K+ channel. Proc. Natl. Acad. Sci. USA 91, 6310–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]