Abstract
Adenosine Deaminases acting on RNA (ADARs) are RNA-editing enzymes responsible for the conversion of adenosine to inosine at specific locations in cellular RNAs. ADAR1 and ADAR2 are two members of the family that have been shown to be catalytically active. Earlier we reported a phenotypic screen for the study of human ADAR2 using budding yeast S. cerevisiae as the host system. While this screen has been successfully applied to the study of ADAR2, it failed with ADAR1. Here, we report a new reporter that uses a novel editing substrate and is suitable for the study of ADAR1. We screened plasmid libraries with randomized codons for two important residues in human ADAR1 (G1007 and E1008). The screening results combined with in vitro deamination assays led to the identification of mutants that are more active than the wild type protein. Furthermore, a screen of the ADAR1 E1008X library with a reporter construct bearing an A•G mismatch at the editing site suggests one role for the residue at position 1008 is to sense the identity of the base pairing partner for the editing site adenosine. This work has provided a starting point for future in vitro evolution studies of ADAR1 and led to new insight into ADAR’s editing site selectivity.
Introduction
Deamination of adenosine (A) in RNA is one of the most important post-transcriptional modifications occurring in human beings (1). This reaction generates an inosine (I) at the corresponding nucleotide position. Since inosine can mimic guanosine (G) during translation, this modification can lead to codon changes and the introduction of amino acids into a protein that were not originally encoded by the gene (recoding) (2, 3). Recoding is common in metazoa and appears to be important in generating the protein structural diversity necessary for normal life for these organisms (2, 4–6). Proteins that catalyze A to I RNA editing reactions are called Adenosine DeAminases acting on RNA (ADARs) (7). There are three members in the ADAR family: ADAR1, ADAR2 and ADAR3. Each ADAR is composed of double stranded RNA-binding domains (dsRBDs) and a deaminase domain. Of the three members, only ADAR1 and ADAR2 have been shown to be catalytically active. In addition to its recoding function, ADAR1 has an essential function in mammals editing endogenous dsRNAs to prevent activation of the cytosolic dsRNA response (8). Defects in this function in human ADAR1 lead to Aicardi-Goutierres Syndrome (AGS), an inherited autoimmune disease (9).
ADARs specifically edit certain adenosines over others (10–12). This editing site selectivity arises in part from selective binding by the dsRBDs (13–16). However, published results also suggest that the deaminase domain plays an important role in controlling editing site selectivity (10, 15). The deaminase domain harbors a zinc-containing active site that requires the reactive adenosine to be flipped out of the double helical RNA substrate to gain access (17, 18). The ease with which adenosines in different structural contexts can undergo this flipping step is believed to contribute to editing site selectivity, yet how an ADAR catalytic domain might promote this step in the reaction is not well understood (19). In the absence of crystal structures of catalytic domain-RNA complexes, investigators have turned to screening approaches to probe the role of individual amino acids (20–22). For instance, our lab developed a phenotypic screen in yeast that couples editing in an RNA derived from the human GluR B subunit pre-mRNA to expression of α-galactosidase and processing of the colorimetric substrate X-α-Gal (20). Screening libraries of mutant ADARs for those that give the colored colony phenotype identifies active clones. In a recent study using this approach, Kuttan and Bass showed that the catalytic domain residue E488 of hADAR2 is involved in flipping the editing site base out of the RNA duplex (22). These authors identified an ADAR2 mutant (E488Q) with increased deaminase activity compared to the wild type protein and induced a greater increase in fluorescence for a 2-aminopurine-substituted RNA, a measure of base-flipping activity (22). While screens of this type have been effective in the study of ADAR2, they have failed for ADAR1. This is likely due to hADAR1’s low editing efficiency on the GluR B substrate used in reporter constructs. A new RNA substrate was needed that is more efficiently deaminated by ADAR1. We recently discovered an ADAR editing site in the mRNA for S. cerevisiae bromo domain factor 2 (BDF2) (23). This substrate RNA is rapidly deaminated by hADAR2 and does not require the dsRBDs for efficient reaction. Here, we show that the BDF2 mRNA site is also an excellent substrate for the hADAR1 catalytic domain (hADAR1-D), and a new reporter plasmid utilizing a BDF2-derived substrate was constructed and applied to phenotypic screening in yeast. We screened plasmid libraries with randomized codons for two important residues in hADAR1-D (G1007 and E1008) and analyzed the editing efficiency and selectivity of the selected mutants. We also screened a library of E1008X mutants with a reporter construct bearing an A•G mismatch at the editing site to probe the role of this amino acid in sensing the base-pairing partner of the editing site adenosine. This work has provided a starting point for future in vitro evolution studies of ADAR1 and led to new insight into ADAR’s editing site selectivity.
Results and discussion
BDF2 is a superb substrate for human ADAR1 catalytic domain (hADAR1-D)
An adenosine found in the codon for R269 of S. cerevisiae BDF2 is rapidly deaminated by human ADAR2 (23). Since this substrate does not require ADAR2’s dsRBDs for rapid and selective reaction, we thought it could be a useful tool for further study of the properties of the ADAR catalytic domains. However, this RNA had not been tested for reaction with the ADAR1 deaminase domain. To this end, we carried out deamination reactions with a 232 nt BDF2-derived RNA (Figure 1A) and hADAR1-D. We observed nearly 90% editing of the BDF2 mRNA after 60 min at 75 nM hADAR1-D (Figure 1B). Therefore, like hADAR2-D, hADAR1-D is able to efficiently edit the BDF2 site. In order to determine the maximum rate of reaction of this RNA with hADAR1-D, we measured deamination rate constants (kobs) at various concentrations of hADAR1-D under single-turnover conditions. This allowed us to calculate a maximum rate constant (kmax = 1.3 ± 0.2 min−1), and an apparent affinity constant (Kd = 0.18 ± 0.04 μM) for this RNA (Figure 1C).
Figure 1.
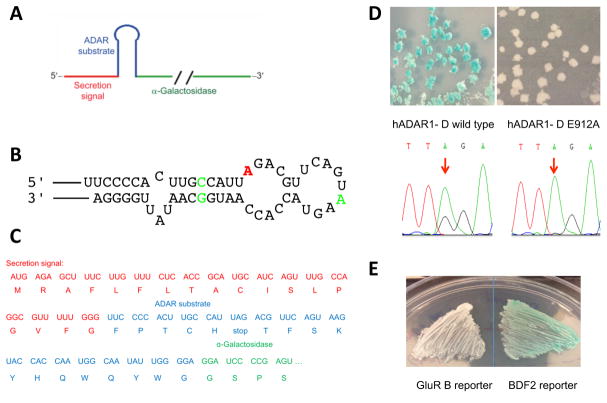
A) Predicted secondary structure surrounding the BDF2 editing site. B) Sequence traces for RT-PCR products of edited region in 232 nt BDF2-derived RNA incubated with 75 nM ADAR1-D for 0 or 60 minutes. C) Plot of kobs as a function of ADAR1-D concentration for reactions containing 10 nM 232 nt BDF2-derived RNA.
A new editing reporter using BDF2-derived RNA
We previously reported a screening assay to evaluate the A to I RNA editing activity of ADAR2 in yeast (20). The assay relies on a reporter plasmid that encodes a secretion signal, an ADAR substrate RNA and the reporter protein α-galactosidase (Figure 2A). In the substrate sequence, the editing site is within a UAG stop codon. The resulting transcript, when edited, will allow the translation of α-galactosidase. The reporter plasmid and an ADAR expression plasmid are sequentially transformed into yeast. If the substrate in the reporter plasmid is edited by ADAR, the α-galactosidase will be expressed, secreted and render yeast colonies green through reaction with 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal) present on the culture plate. To date, reporter plasmids containing GluR B sequence as the substrate have been used (20–22, 24, 25). Although GluR B-derived reporter plasmids work efficiently for ADAR2, they fail with ADAR1 (24, 25).
Figure 2.
BDF2 based α-galactosidase reporter for evaluating A to I RNA editing activity in S. cerevisiae. A) Schematic of reporter mRNA. B) BDF2-derived sequence used as substrate with edited A colored red. Green color indicates sequences changes to maintain open reading frame. C) Full sequence of reporter mRNA near editing site with the encoded protein sequence shown. D) Yeast colony color with BDF2 reporter expressed in the presence of wild type hADAR1-D or the inactive mutant E912A (above) and sequencing of RT-PCR products from reporter mRNA in these samples (bottom). E) Colored phenotype is only apparent using the new BDF2-based reporter and hADAR1-D.
We have demonstrated that the 232 nt truncation of BDF2 is efficiently edited by the ADAR1 catalytic domain (Figure 1). Additionally, the editing site in BDF2 substrate falls within a UAG nucleotide sequence. Taken together, this suggests that BDF2-derived RNA could be a good candidate for a screening assay with ADAR1. To test this idea, we generated a new reporter plasmid using BDF2-derived RNA as the substrate sequence. The 54 nt truncation of the BDF2 substrate sequence contains several modifications to form an open reading frame while maintaining its secondary structure required for efficient editing (Figures 2B, 2C). Because BDF2 is an excellent substrate for catalytic domains of both ADAR1 and ADAR2, this new reporter plasmid should allow for screens with both enzymes. Here we present the application of this new reporter plasmid to the study of hADAR1-D, since less is known about this enzyme. When hADAR1-D was expressed with the BDF2 reporter plasmid, yeast grown on X-α-Gal plates showed a green phenotype within 2–3 days, indicating editing and expression of α-galactosidase (Figure 2D). No color and no editing were observed when a known inactive mutant of hADAR1 (E912A) was expressed with the BDF2 reporter plasmid (Figure 2D). Additionally, we confirmed that this reporter plasmid can also be applied to full length ADAR1 (data not shown). We compared the effect of expression of hADAR1-D in the presence of either the BDF2 reporter plasmid or the GluR B reporter plasmid in parallel. As expected, yeast expressing the BDF2 reporter plasmid and hADAR1-D showed the green phenotype within 2–3 days while those with GluR B reporter plasmid remain white, even after a longer incubation time (Figure 2E). Clearly, the BDF2 reporter plasmid is superior to the GluR B reporter plasmid for study of ADAR1 in yeast. Previously, Garncarz et al. used a GluR B-derived substrate in a similar screen in yeast for enhancers of ADAR activity (24). However, the assay failed to detect any editing activity on GluR B by ADAR1 in yeast, both indirectly via yeast growth or directly via sequencing of the GluR B substrate RNA (24). It was not clear at the time whether the failure was due to the substrate RNA used or low ADAR1 activity in yeast. Our demonstration here that the BDF2-derived RNA can be used in a reporter for the study of ADAR1 in yeast suggests that substrate RNA was likely the issue in the earlier study (24). This observation emphasizes the importance of choice of RNA substrate in the design of ADAR activity reporters.
Probing the role of ADAR1 residues G1007 and E1008 with library screens
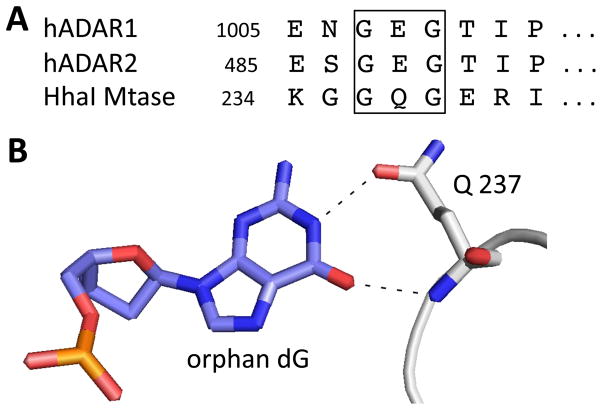
We chose to use the new ADAR1 activity reporter to probe the role of two amino acid residues in the protein that had previously been implicated in controlling ADAR activity; G1007 and E1008 (9, 22, 26). hADAR1 E1008 is found in a region of the protein where sequence is highly conserved in the ADAR family (Figure S2) and corresponds to E488 in hADAR2. hADAR2 E488 is believed to be involved in the base flipping step of the ADAR reaction (see above) (22). Furthermore, it has been suggested that this residue plays a role similar to Q237 of another base flipping enzyme, the cytosine-specific DNA methyltransferase M.HhaI, where the glutamine pushes the target cytosine out of the helix, penetrates deep into the resulting pocket and directly contacts the orphaned guanine base (22, 27, 28) (Figure 3). M.HhaI MTase Q237 is flanked by two glycine residues that are important for the deep penetration of the glutamine side chain into the DNA duplex (Figure 3A). hADAR1 E1008 is also flanked by two glycines, including G1007 (Figure 3A). The G1007R mutation in human ADAR1 is linked to both Aicardi-Goutierres Syndrome (AGS) and the skin disorder Dyschromatosis Symmetrica Hereditaria (DSH) (9, 26). Rice et al. showed this mutant is catalytically inactive and functions as a competitive inhibitor of wild type ADAR1 (9).
Figure 3.
A) Sequence context of E1008 of hADAR1, E488 of hADAR2 and Q237 of Hhal Mtase are similar B) Illustration of contacts made between Q237 of Hhal Mtase and the orphan dG after base flipping of that enzyme’s target cytosine.
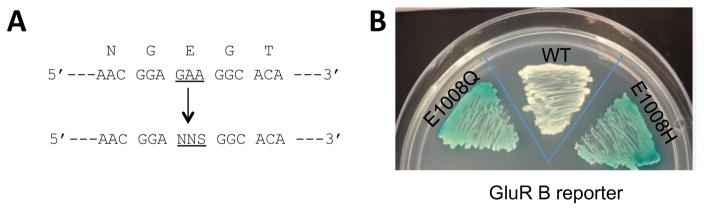
To define further the role of these residues, we generated mutant libraries E1008X and G1007X of hADAR1-D by site-saturation mutagenesis and screened the resulting libraries using the BDF2-based reporter plasmid described above (Figure 4A). For the purpose of identifying more active ADAR1 mutants, we also used the GluR B reporter plasmid with the E1008X library. As indicated, the GluR B reporter does not lead to the green color phenotype with wild type ADAR1; however, it might with highly active mutants. Among yeast expressing the BDF2 reporter plasmid and hADAR1-D library, green colonies with various color intensities developed rapidly. In contrast, yeast expressing the GluR B reporter plasmids and hADAR1-D library generated only a few green colonies among a majority of white colonies, and their color intensities were low, even when incubated for an extended period. This indicates that the BDF2 RNA is a superior substrate for the reporter compared to GluR B RNA by lowering the detection limit of the assay allowing active mutants that span a larger range of activities to be identified.
Figure 4.
A) Site-saturation mutagenesis was carried out at hADAR1-D E1008 codon, and the resulting library was screened for the generation of differentially colored yeast colonies. The randomized codon is designed as NNS where N is any natural nucleotide and S is either guanosine or cytosine. B) The phenotypes of yeast co-expressing (1) hADAR1-D E1008Q and GluR B reporter plasmid; (2) hADAR1-D wild type (WT) and GluR B reporter plasmid; and (3) hADAR1-D E1008H and GluR B reporter plasmid.
In screening the E1008X library using the BDF2 reporter plasmid, 20 green colonies and 11 white colonies were identified and analyzed. As shown in Table 1, residues that lead to active ADAR1 are mostly polar and large (e.g. Glu, Gln, Arg, His, Lys). In contrast, negative hits have either small polar (Ser) or nonpolar (Phe, Trp, Gly, Ile, Val, Pro) residues. This suggests that the large size and the polarity of the residue at position 1008 of ADAR1 are important for proper function of the enzyme. This is consistent with the proposed helix penetration and orphan base contact role for this residue (22, 27, 28). In order to distinguish the relative activities of the positive hits, the color intensities of yeast colonies were categorized into intense green, medium green and light green based on visual inspection. In the intense green category, glutamine and arginine were selected four times and two times, respectively. This is in contrast to the wild type sequence that was absent in the most intensely colored colonies. This suggests that E1008Q and E1008R may be more active than the wild type protein in deaminating the BDF2 substrate.
Table 1.
Results of screening of hADAR1-D E1008X using BDF2 reporter and GluR B reporter.
| Reporter and library combination | Colony color | Codon | Number of colonies | Frequencies of amino acids |
|---|---|---|---|---|
| BDF2 reporter + hADAR1-D E1008X library | Intense green | CAG(Gln) | 4 | Glu(WT):7 Arg: 5 Gin: 4 Met: 1 His: 1 Lys: 1 Asn: 1 |
| AGG(Arg) | 2 | |||
| ATG(Met) | 1 | |||
| Medium green | GAA(Glu) | 4 | ||
| AGG(Arg) | 1 | |||
| CGC(Arg) | 1 | |||
| CAC(His) | 1 | |||
| Light green | GAA(Glu) | 3 | ||
| AAC(Asn) | 1 | |||
| AAG(Lys) | 1 | |||
| CGC(Arg) | 1 | |||
| white | GGG(Gly) | 3 | Gly:3 lie: 2 Pro: 1 Val: 1 Phe: 1 Trp: 1 Ser: 1 |
|
| ATC(lle) | 2 | |||
| CCG(Pro) | 1 | |||
| TTC(Phe) | 1 | |||
| GTC(Val) | 1 | |||
| TGG(Trp) | 1 | |||
| TCG(Ser) | 1 | |||
| GluR B reporter + hADAR1-D E1008X library | green | CAG(Gln) | 4 | Gin: 4 His: 5 |
| CAC(His) | 5 |
In the screening at E1008 using the GluR B reporter plasmid, nine green colonies were identified. Interestingly, only two mutants, E1008Q and E1008H were selected (Table 1). To eliminate the possibility that these are false positives, we reproduced the green phenotype exhibited by the two mutants that showed to be in clear contrast to the wild type protein (Figure 4B). The exclusive selection for E1008Q and E1008H using GluR B reporter plasmid suggests that these two mutants are more active than the wild type protein in deaminating the GluR B substrate.
From screening the G1007X library using the BDF2 reporter plasmid, 19 green colonies and 6 white colonies were identified. As shown in Table 2, the positive hits tend to be small and non-polar residues (e.g. Ala, Gly, Val), while the negative hits are relatively large residues (e.g Arg, Lys, Trp, Tyr). This suggests that the small size of the residue at site 1007 plays an important role in determining the activity of the protein, consistent with the notion that this residue is on the protein loop that promotes base flipping and adjacent to the residue that penetrates the RNA helix (i.e. E1008) (Figure 3) (17, 22). It should be noted that the G1007R mutant, implicated in both Aicardi-Goutierres Syndrome (AGS) and Dyschromatosis Symmetrica Hereditaria (DSH), is identified as inactive in our screening assay.
Table 2.
Results of screening of hADAR1-D G1007X using BDF2 reporter.
| Reporter and library combination | Colony color | codon | Number of colonies | Frequencies of amino acids |
|---|---|---|---|---|
| BDF2 reporter + hADAR1-D G1007X library | green | GGA(Gly) | 6 | Gly: 10 Ser: 6 Ala: 2 Val: 1 |
| GGG(Gly) | 3 | |||
| GGC(Gly) | 1 | |||
| TCG(Ser) | 4 | |||
| AGC(Ser) | 2 | |||
| GCG(Ala) | 2 | |||
| GTC(Val) | 1 | |||
| white | CGC(Arg) | 1 | Arg: 1 Lys: 1 Tyr: 1 Trp: 1 Leu: 1 Thr: 1 |
|
| AAG(Lys) | 1 | |||
| TAC(Tyr) | 1 | |||
| TGG(Trp) | 1 | |||
| TTG(Leu) | 1 | |||
| ACC(Thr) | 1 |
In vitro deamination activity of selected mutants
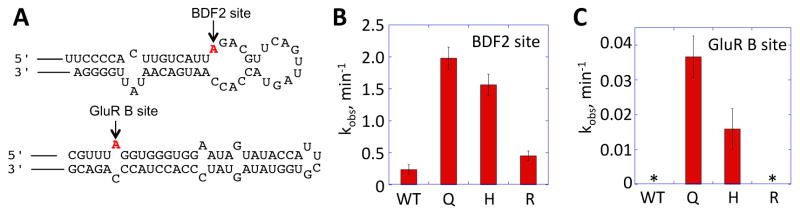
The screening results at site E1008 of hADAR1-D led us to predict that mutants E1008Q, E1008R and E1008H may be more active than the wild type protein. To further test this, we characterized these mutants together with the wild type protein using an in vitro deamination assay. Both the BDF2 and GluR B substrate RNAs were used in the deamination assay and the sequences surrounding the editing sites are shown in Figure 5A with editing sites highlighted. The mutants and wild type hADAR1-D were analyzed at a final concentration of 35 nM. We found that protein aggregation complicated the analysis at higher concentrations. The rate constant for product formation (kobs) was measured for each mutant using both the BDF2 and GluR B substrate RNAs (Figure 5B). These in vitro deamination reactions are more efficient with the BDF2 substrate RNA than with the GluR B substrate RNA, as expected (e.g. E1008Q edits the BDF2 site with a kobs = 2.0 ± 0.2 min−1 and the GluR B site with a kobs = 0.036 ± 0.006 min−1 under these conditions). Also, the results indicate that mutants E1008Q and E1008H have activities higher than the wild type protein and mutant E1008R has a similar activity to wild type.
Figure 5.
In vitro deamination of the BDF2 and GluR B substrate RNAs by hADAR1-D WT, hADAR1-D E1008Q, E1008H, and E1008R mutants. A) RNA sequences containing editing sites derived from BDF2 substrate (top) and GluRB substrate (bottom) used in the deamination assay. B) Comparison of observed rate constants (kobs) for deamination of the BDF2 substrate RNA by 35 nM hADAR1-D wild type (WT), E1008Q (Q), E1008H (H), and E1008R (R). Error bars indicate SD; n≥3. Two off target adenosines are also edited to low levels in this RNA under these conditions (see Supporting Information, Figure S3). C) Comparison of observed rate constants (kobs) for deamination of the GluR B substrate RNA by 35 nM hADAR1-D wild type (WT), E1008Q (Q), E1008H (H), and E1008R (R). Error bars indicate SD; n≥3. * indicates no product observed over 1 h reaction time.
The order of reactivity for the E1008X mutants is similar for the two RNA substrates (Q ~ H > R ~ E) in vitro. This is consistent with the screening results using the GluR B reporter where only the E1008Q and E1008H were found among the active clones. Screening with the BDF2 reporter suggested the E1008R mutant could have higher activity than the wild type enzyme whereas the E1008H mutant might have lower or similar activity since E1008R was found twice among the most intensely colored colonies and E1008H and the wild type sequence were not found in these colonies. These observations suggest caution when rank ordering the activity of mutants based only on the color intensity of colonies as varying ADAR expression levels, in addition to different editing activities, could contribute to variability in color intensity.
A previously reported screen of an ADAR2 E488X library showed that a variety of amino acid substitutions for glutamic acid support editing of an adenosine in the context of a favorable triplet, 5’-UAG-3’ (22). However, only glutamine and asparagine were able to edit adenosine in the context of an unfavorable triplet, 5’-GAC-3’(22). Similarly, our screening and the in vitro deamination assays identified the E1008Q mutant of ADAR1 as a more active protein than wild type with an adenosine in a 5’-UAG-3’ sequence. We also identified asparagine as a positive hit in our screening, but further in vitro characterization is needed to determine its activity relative to the wild type protein. A histidine mutation at 1008 of hADAR1 (E1008H) was shown here to increase the activity of the protein. The E488H mutant was not identified as a hit in the previous screen for active hADAR2 mutants (22). However, considering that glutamine and histidine are highly similar in terms of their physiochemical properties (29), we suspected ADAR2 E488H might also be more active than the wild type ADAR2 protein on these substrates. To test this hypothesis, we generated this mutation in the hADAR2-D protein and compared its activity to that of the wild type ADAR2 catalytic domain. Indeed, the hADAR2-D E488H mutant edited the BDF2 RNA with a rate constant three-fold higher than wild type hADAR2-D did under these conditions (Figure S4).
Screening identifies an ADAR1 mutant active on an A•G mismatch
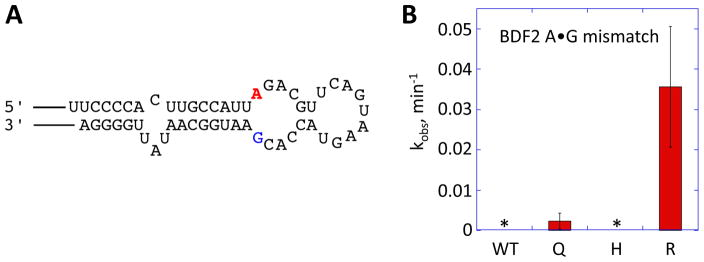
In deamination reactions processed by ADARs, the adenosine to be edited is flipped out of the RNA helix and placed into the active site buried deeply inside the protein structure (18). As stated above, E488 of hADAR2 (and E1008 of hADAR1 by analogy) has been implicated in the base-flipping step (22). If ADAR uses a similar mechanism as the M.Hhal Mtase, as has been suggested (22, 27, 28), the residue that promotes base flipping may also contact the orphaned base remaining in the helix. This suggests the preferred residue at this site could be dependent on the base-pairing partner of the edited adenosine in the substrate. To test this idea, we rescreened the E1008X library using a modified BDF2 reporter with an A•G mismatch within the BDF2 substrate (BDF2 (A•G) reporter) (Figure 6A). Wild type ADARs do not edit adenosines in A•G mismatches efficiently (10, 30) and the C to G mutation makes the BDF2 reporter no longer sensitive in the screening assay. Indeed, yeast expressing the E1008X library and the BDF2 (A•G) reporter took more than one week for green color to develop. Interestingly, all nine green colonies identified had arginine at position 1008 suggesting that arginine is favored over the other amino acids at this position when the base opposite the editing site is G.
Figure 6.
A) Predicted RNA secondary structure surrounding the BDF2 recoding site. The base opposite the editing site is mutated to a guanine in this RNA. B) Comparison of observed rate constants (kobs) for deamination of the BDF2 A•G mismatch substrate RNA by 35 nM hADAR1-D wild type (WT), E1008Q (Q), E1008H (H), and E1008R (R). Error bars indicate SD; n≥3. * indicates no product observed over 1 h reaction time.
We also performed in vitro deamination with the BDF2 RNA substrate containing an A•G mismatch and the proteins with varying 1008 side chains: the wild type hADAR1-D protein and its mutants (E1008Q, E1008H and E1008R). All four proteins tested deaminated this substrate less efficiently than the RNA with an A•C mismatch at the editing site. However, a new pattern of preference for the 1008 site chain is observed (R > Q > E ~ H) with E1008R the most active among the four proteins tested (Figure 6B). This agrees with the screening data where arginine was shown to be the preferred side chain when the base opposite the editing site is G. It is worth noting that with the GluR B substrate that also bears an A•C mismatch at the editing site, the relative activities of the four enzymes (Q ~ H > R ~ E) is the same as found with the BDF2 substrate (Figure 5B). This indicates the preference of 1008 side chain is dependent on the identity of the base opposite the editing site within editing substrates. Again, this is consistent with the proposed role for this residue in promoting base flipping, penetrating the RNA duplex and contacting the orphaned base. Defining how specific amino acids might be best equipped to do this for substrates with the reactive adenosine paired with different nucleotides will require additional studies. This will undoubtedly be dependent on a residue’s ability to both effectively fill the space left behind by the flipped out adenosine (e.g. stack into the helix) and engage the orphaned base in stabilizing interactions. It is tempting to suggest that the well-established ability of the arginine side chain to hydrogen bond to the Hoogsteen face of guanine may be a contributing factor for the reactivity of the E1008R mutant with the BDF2 (A•G) mismatch substrate (31).
Conclusion
In conclusion, we report an improved phenotypic screen in yeast that uses a novel editing substrate and can be applied to study of human ADAR1. Two residues found in the ADAR1 catalytic domain (G1007 and E1008) were chosen for study using the new screen. The results of screens of the G1007X and E1008X hADAR1-D libraries are consistent with proposed roles of these residues in the base-flipping step of the ADAR reaction. Interestingly, we also show that the identity of the residue at position 1008 that leads to highest ADAR1 activity is dependent on the base opposite the editing site. We suggest this correlation could arise from direct contact between the side chain of the position 1008 residue of ADAR1 and the base opposite the editing site. In addition, this work has provided a starting point for future in vitro evolution studies of ADAR1 and other ADAR-related proteins (32). Finally, simple modifications to the screen should allow for the discovery of proteins or small molecules that modulate ADAR1 activity.
Materials and methods
Analysis of reaction kinetics for hADAR1-D and a BDF2-derived RNA substrate
Editing of the BDF2 RNA derived substrate by ADAR1-D was evaluated by the following reaction. ADAR1-D (25, 50, 75, 150, 300 and 500 nM final concentrations) was mixed with 10 nM RNA in assay buffer containing 15 mM Tris–HCl, pH 7.8, 26 mM KCl, 5 mM NaCl, 40 mM potassium glutamate, 1.5 mM EDTA, 4% glycerol, 0.003% Nonidet P-40, 0.6 mM DTT, 160 U/ml RNasin, and 1.0 μg/ml yeast tRNAPhe in a final volume of 20 μL. Reaction solutions were pre-incubated at 30 °C for about 30 minutes before adding enzyme and then continued to incubate at 30 °C for varying times before quenched by the addition of 0.5% SDS at 95 °C. Deaminated RNA was purified by phenol-chloroform extraction and ethanol precipitation. The RNA solution was lyophilized to dryness and re-suspended in 200 μL nuclease free water. RT-PCR was used to generate cDNA from deaminated RNA and the resulting cDNA were subjected to Sanger sequencing. The sequencing data were quantified using Chromas Lite (Technelysium) and ImageJ and were fit to the equation [P]t = α[1 −e(−kobs*t)], where [P]t is the fraction of deamination product at time t, α is the fitted reaction end point, and kobs is the fitted rate constant using KaleidaGraph. Each experiment was carried out in triplicate. For enzyme saturation, kobs was measured as a function of [hADAR1-D]. The values of apparent Kd and kmax were obtained by fitting the data to the equation: kobs= kmax[hADAR1-D]/ (Kd+[hADAR1-D]).
Construction of the BDF2 reporter plasmid
The BDF2 reporter plasmid was generated based on the GluR B substrate/α-galactosidase reporter plasmid pR/GaGal (GluR B reporter plasmid) previously described (20). Plasmid preparation and full sequence of the new reporter is described in the Supporting Information.
Screening for functional hADAR1-D mutants in yeast
S. cerevisiae INVSC1 strain was first transformed with BDF2 plasmid with TRP1 selection followed by transformation of hADAR1-D mutant library with URA3 selection using a lithium acetate protocol. A description of the hADAR1-D mutant library generation is found in the Supporting Information. The double transformants were plated on CM –ura –trp + 2% glucose plates. After incubation at 30 °C for 2–3 days, the yeast colonies growing on the glucose plates were printed to agar plates containing complete medium –ura –trp +2% raffinose +3% galactose and X-α-Gal by replica plating. The galactose plates were then incubated at 30 °C for 2–4 days to allow the green color to develop. For the green or white colonies on the galactose plate to be further studied, their corresponding colonies on the glucose plate were identified. Those single colonies were then harvested from the glucose plate and transferred to CM –ura –trp + 2% glucose media for overnight culture at 30 °C with shaking. Yeast cells were harvested and lysed using glass beads (Life technology). The plasmid DNA was extracted from yeast lysate using QIAprep Miniprep kit (Qiagen). PCR was performed on the isolated plasmids to amplify the region encompassing the mutant site in hADAR1-D and the PCR product was purified by agarose gel. The identity of the hADAR1-D mutant in the green or white colony was then determined by sequencing the PCR product.
Deamination assay with hADAR1-D and hADAR1-D mutants
Details for the generation of hADAR1-D mutants, overexpression and purification of mutant proteins and a description of the generation of substrate RNAs by in vitro transcription are found in the Supporting Information. Deamination assays were performed under single turnover conditions with 15 mM Tris-HCl, pH=7.8, 26 mM KCl, 5 mM NaCl, 40 mM potassium glutamate, 4% glycerol, 1.5 mM EDTA, 0.003% Nonidet P-40, 0.6 mM DTT, 1.0 μg/ml yeast tRNAPhe, 160 U/mL RNAsin, 10 nM RNA and 35 nM enzyme. Each reaction solution was incubated at 30 °C for 45 minutes before adding enzyme and each reaction was allowed to proceed for 60 minutes at 30 °C prior to stopping with 10 μl 1% SDS and heating at 95 °C for 2 minutes. Deaminated RNA was purified by phenol-chloroform extraction and ethanol precipitation. RT-PCR was performed on the extracted RNA and the PCR products were purified using an agarose gel. The editing level was then determined by Sanger sequencing of the RT-PCR products. The sequencing data were quantified using Chromas Lite (Technelysium) and ImageJ and were fit to the equation [P]t = α[1 −e(−kobs*t)], where [P]t is the fraction of deamination product at time t, α is the fitted reaction end point, and kobs is the fitted rate constant using KaleidaGraph. Each experiment was carried out in triplicate.
Supplementary Material
Table 3.
Results of screening of E1008X using BDF2 (A•G) reporter
| Reporter and library combination | Colony color | Codon | Number of colonies | Frequencies of amino acids |
|---|---|---|---|---|
| BDF2 (A•G) reporter + hADARl-D E1008X | green | AGG(Arg) | 5 | Arg:9 |
| CGC(Arg) | 2 | |||
| CGG(Arg) | 2 |
Acknowledgments
P. Beal acknowledges the National Institutes of Health for financial support in the form of grant R01-GM061115. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIH. The authors acknowledge Y. Zheng for technical assistance.
Footnotes
Supporting Information Available: Sequences for all primers used, descriptions of reporter plasmid construction and full insert sequence, generation of hADAR1-D plasmid libraries, alignment of ADAR sequences around E1008 of hADAR1, off target editing in reporter RNA, kinetic data for hADAR2-D E488H, overexpression and purification of ADAR proteins along with in vitro transcription of RNA substrates. This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 3.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA Editing of AMPA Receptor Subunit GluR-B: A Base-Paired Intron-Exon Structure Determines Position and Efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 4.Hoopengardener B, Bhalla T, Staber C, Reenan R. Nervous System Targets of RNA Editing Identified by Comparative Genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 5.Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Spengle R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 7.Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 8.Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, Dorin J, Adams IR, Scadden AD, Ohman M, Keegan LP, O'Connell MA. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanielli M, De-Laet C, de Lonay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JPSM, Lourenco CM, Male AM, Marques W, Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe W, Vanderver A, Vassalo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O'Connell M, Lovell SC, Crow YJ. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genetics. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 12.Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens OM, Haudenschild BL, Beal PA. The binding selectivity of ADAR2's dsRBMs contributes to RNA-editing selectivity. Chem & Biol. 2004;11:1–20. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, Allain FH-T. The Solution Structure of the ADAR2 dsRBM-RNA Complex Reveals a Sequence-Specific Readout of the Minor Groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double stranded RNA. Nat Commun. 2011;2 doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FH-T. Structure and specific RNA-binding of ADAR2 double-stranded RNA-binding motifs. Structure. 2006;14:345–355. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Macbeth MR, Schubert HL, VanDemark AP, Lingam AT, Hill CP, Bass BL. Inositol Hexakisphosphate Is Bound in the ADAR2 Core and Required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens OM, Yi-Brunozzi H-Y, Beal PA. Analysis of the RNA-Editing Reaction of ADAR2 with Structural and Fluorescent Analogues of the GluR-B R/G Editing Site. Biochemistry. 2000;39:12243–12251. doi: 10.1021/bi0011577. [DOI] [PubMed] [Google Scholar]
- 19.Hart K, Nystrom B, Ohman M, Nilsson L. Molecular dynamics simulations and free energy calculations of base flipping in dsRNA. RNA. 2005;11:609–618. doi: 10.1261/rna.7147805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokharel S, Beal PA. High throughput screening for functional A to I RNA editing systems. ACS Chem Biol. 2006;1:761–765. doi: 10.1021/cb6003838. [DOI] [PubMed] [Google Scholar]
- 21.Pokharel S, Jayalath P, Maydanovych O, Goodman RA, Wang SC, Tantillo DJ, Beal PA. Matching Active Site Structure to Substrate Analog for an RNA Editing Reaction. J Am Chem Soc. 2009;131:11882–11891. doi: 10.1021/ja9034076. [DOI] [PubMed] [Google Scholar]
- 22.Kuttan A, Bass BL. Mechanistic Insights into edtiing-site specificity of ADARs. Proc Natl Acad Sci USA. 2012;109:E3295–3304. doi: 10.1073/pnas.1212548109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eifler T, Pokharel S, Beal PA. RNA-Seq Analysis Identifies A Novel Set of Editing Substrates for Human ADAR2 Present in Saccharomyces cerevisiae. Biochemistry. 2013;52:7857–7869. doi: 10.1021/bi4006539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gommans WM, McCane J, Nacarelli GS, Maas S. A mammalian reporter system for fast and quantitative detection of intracellular A to I RNA editing levels. Anal Biochem. 2010;399:230–236. doi: 10.1016/j.ab.2009.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tojo K, Sekijima Y, Suzuki T, Suzuki N, Tomita Y, Yoshida K, Hashimoto T, Ikeda S. Dystonia, mental deterioration, and dyschromatosis symmetrica hereditaria in a family with ADAR1 mutation. Mov Disord. 2006;21:1510–1513. doi: 10.1002/mds.21011. [DOI] [PubMed] [Google Scholar]
- 27.Daujotyte D, Serva S, Vilkaitis G, Merkiene E, Venclovas C, Klimasauskas S. HhaI DNA Methyltransferase Uses the Protruding Gln237 for Active Flipping of Its Target Cytosine. Structure. 2004;12:1047–1055. doi: 10.1016/j.str.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Klimasauskas S, Kumar P, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 29.Braun W, Venkatarajan MS. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical/chemical properties. J Mol Model. 2001;7:445–453. [Google Scholar]
- 30.Kallman AM, Sahlin M, Ohman M. ADAR2 A-->I editing: site selectivity and editing efficiency are separate events. Nucleic Acids Res. 2003;31:4874–4881. doi: 10.1093/nar/gkg681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Cho DC, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.