Abstract
Remarkable progress in structural biology has equipped virologists with insight into structures of viral proteins and virions at increasingly high resolution. Structural information has been used extensively to address fundamental questions about virtually all aspects of how viruses replicate in cells, interact with the host, and in the design of antiviral compounds. However, many critical aspects of virology exist outside the snapshots captured by traditional methods used to generate high-resolution structures. Like all proteins, viral proteins are not static structures. The conformational flexibility and dynamics of proteins play a significant role in protein-protein interactions, and in the structure and biology of virus particles. This review will discuss the implications of the dynamics of viral proteins on the biology, antigenicity, and immunogenicity of flaviviruses.
Keywords: flavivirus, West Nile virus, dengue virus, viral breathing, structural dynamics, antibody-mediated neutralization
An introduction to protein dynamics
It is well recognized that proteins exhibit liquid-like internal motions and that many biological processes require substantial plasticity in protein structure (reviewed by (Karplus and Kuriyan, 2005). Protein motions span a large range in scale from picosecond-nanosecond motions localized to side chain and backbone dihedral rotations to longer timescale motions leading to internal rearrangements of whole domains or polypeptide chains. Numerous studies, both experimental and computational, have characterized small-amplitude, fast-timescale internal motions, and elucidated the physical aspects and importance of localized fluctuations to protein function. Internal dynamics underlie induced-fit ligand binding, access to buried regions, and configurational entropy contributions to protein energetics (Borbat et al., 2001; Dodson et al., 2008; Kneller, 2005; Marsh et al., 2012; Osawa et al., 2012). By contrast, relatively little is known about the larger amplitude motions associated with conformational fluctuations involving large segments of polypeptide chains that deviate substantially from the equilibrium/time-averaged structures defined by crystallography or NMR spectroscopy. Possibly the first evidence of large amplitude excursions from the average structure of a protein molecule comes from hydrogen/deuterium exchange and pulsed proteolysis experiments; the results indicate a transient accessibility of buried regions of the polypeptide chain, described as a “local unfolding” event (Bai et al., 1995; Park and Marqusee, 2004; Thomsen and Poulsen, 1993). Of particular interest for this review are the proteins that comprise virus particles, as both non-enveloped and enveloped viruses undergo processes during the virus lifecycle that clearly require protein dynamics and large amplitude internal motions. The change from an immature to mature form of the virus, the entry into the host cell through interaction with cellular membranes, and the uncoating and release of the viral genome are all processes that involve large changes in virus structure.
The conformational flexibility of viral proteins
Viruses have evolved numerous strategies to deliver genetic material from one host to another within viral capsids or virions. Viruses balance a requirement to protect the viral genome within a structure that is sufficiently stable to withstand potentially harsh conditions experienced during transmission among cells and hosts with a requirement to disassemble that same virus particle following the infection of cells (reviewed by (Mateu, 2013)). The viral proteins that orchestrate the entry of many enveloped viruses into cells have been studied extensively and vary considerably in structure (reviewed by (Harrison, 2008)). These proteins drive the fusion of viral and cellular membranes that allows deposition of viral cargo into target cells. Critical events in the entry process are triggered by interactions with cellular macromolecules or environmental changes, and may exploit the conformational flexibility of viral proteins.
Entry of HIV-1 into cells is coordinated by a small number of trimers of gp120 and gp41 proteins incorporated into the viral membrane. This process has been studied extensively (reviewed by (Wilen et al., 2012; Wyatt and Sodroski, 1998)), and is the target of two classes of antiviral molecules used in the clinic (reviewed by (Haqqani and Tilton, 2013)). Interactions between CD4 and HIV-1 catalyze changes in the structure of gp120 that result in the formation of the binding site for viral co-receptors and the displacement of variable loops that function to shield critical surfaces of the viral entry machinery from the humoral response (Kwong et al., 1998; Munro and Mothes, 2014; Wyatt et al., 1995). Interactions between CD4-triggered gp120 and chemokine receptors precede the large conformational changes in gp41 that drive the formation of the energetically stable six-helix bundle and lipid mixing (Pancera et al., 2014; Wyatt et al., 1995; Wyatt and Sodroski, 1998). Of significant interest, a recent study identified three states of unliganded envelope trimers using single molecule fluorescence energy transfer (Munro et al., 2014). This exciting study suggests the frequency with which gp120 populates each state is modulated by receptor interactions. Thus an ensemble of envelope trimer structures may contribute to the mechanics of viral entry and sensitivity to neutralizing antibodies. While incompletely understood, studies of monoclonal antibody (mAb) reactivity suggest trimers of influenza HA also exist in multiple states (Bachi et al., 1985).
Viral breathing: Insights and concepts from the common cold
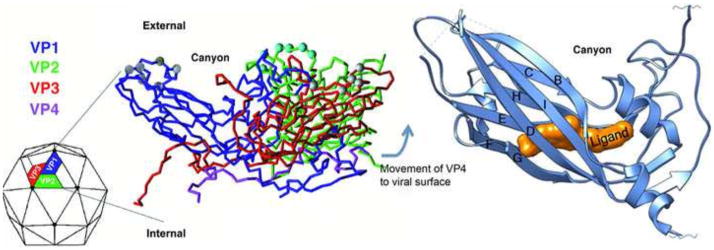
Despite the availability of elegant images of virions, they are not static structures. Virions undergo large amplitude, concerted fluctuations at equilibrium, a phenomenon referred to as viral “breathing” (Lewis et al., 1998). How breathing impacts viral structure and biology is understood in the greatest detail for non-enveloped viruses (reviewed by (Mateu, 2013; Witz and Brown, 2001)). Picornaviruses are a family of small non-enveloped icosahedral viruses that cause a variety of serious human illnesses, including poliomyelitis, meningitis, and symptoms secondary to respiratory infections. Most picornavirus capsids are composed of sixty copies each of four viral proteins (VP1-VP4) (Racaniello, 2007). The exterior surface is formed by VP1, VP2, and VP3 proteins arranged with pseudo T=3 symmetry (Figure 1) (Hogle et al., 1985; Rossmann et al., 1985). VP4 is internal in the virion as demonstrated by X-ray crystallography and cryo-electron microscopy. The five-fold symmetry axis of the virion of a sub-group of picornaviruses features a canyon that contains amino acids involved in binding the immunoglobulin-superfamily receptors used by many of these viruses for attachment to target cells (Kolatkar et al., 1999; Olson et al., 1993). Amino acids within the canyon are more conserved than residues that form its rim and the surface of the virion. Because antibody molecules may not efficiently access residues within the canyon, regions involved in receptor recognition may be largely unaffected by immune pressure (Rossmann and Palmenberg, 1988), although exceptions have been noted (Smith et al., 1996). A hydrophobic pocket in VP1 beneath the floor of the canyon is occupied by a “pocket factor” thought to promote particle stability (Hadfield et al., 1997). It is suggested that cellular receptors that bind the canyon compete with pocket factor, which is likely a lipid, for binding to the virion (Figure 1) (Kolatkar et al., 1999). Thus, the displacement of pocket factor reduces stability of the virion just prior to penetration of target cells.
Figure 1. Structure of the picornavirus capsid and location of the hydrophobic pocket.
The left panel shows a color-coded C-alpha backbone model of the basic repeating unit of a picornavirus virion. The four capsid proteins, VP1, 2, 3, and 4 are color coded representing each of the proteins with VP4 being found on the interior of the particle apparently protected from immune surveillance. 60 copies of this repeating unit are organized as an icosahedron (shown as the particle on the left). At each vertex, five copies of VP1 come together and create a depression or “canyon” where many receptors have been shown to bind. The right panel shows a ribbon diagram of a picornavirus VP1 capsid protein showing the location of a small molecule inhibitor binding into the hydrophobic pocket normally occupied by pocket factor. The binding site is underneath the canyon floor and binding of inhibitor limits the flexibility and dynamics of the particle.
Structural changes of the picornavirus capsid drive penetration into cells. Numerous cellular receptors for picornaviruses have been identified. Many of these viruses bind receptors that interact with the canyon. In these instances, interactions with receptor result in the formation of an expanded, altered particle, or A-particle, characterized by exposure of the VP1 amino-terminus and a loss of the VP4 protein (Crowell and Philipson, 1971; Fricks and Hogle, 1990). In this configuration, the N-terminus of VP1, which has a sequence corresponding to an amphipathic helix, is in position to interact with cellular membranes. Subsequent injection of the viral genome across the cellular membrane is triggered by an unknown signal and is hypothesized to be facilitated by a channel formed by rearranged VP1 and VP4 (Ren et al., 2013; Strauss et al., 2013). Formation of the A-particle can occur spontaneously in the absence of receptor, a process that can be enhanced via incubation at higher temperature (Belnap et al., 2000; Curry et al., 1996). Conversion to the A-particle in solution is an irreversible first order process at 37°C that may be accelerated ~17-fold in the presence of receptor (Carson, 2014). Despite containing an intact positive-stranded RNA genome, the A-particle is poorly infectious because it no longer binds efficiently to receptor.
The breathing of picornaviruses has been inferred from several types of experiments. Limited proteolysis studies of purified poliovirus and human rhinovirus 14 identified exposure of the otherwise internal VP4 protein and the amino terminus of VP1 (Fricks and Hogle, 1990; Lewis et al., 1998). These protein regions are located deep inside the capsid and, because the protease cannot penetrate the protein boundary, it was reasoned that large-scale concerted fluctuations must occur in the native state of the viral capsid to transiently expose them to the outside of the capsid. Analogous results were observed for flock house virus, a non-enveloped particle with icosahedral symmetry that is classified within the nodavirus family (Bothner et al., 1998). Neutralizing antibodies are capable of trapping reversibly displayed structures (Li et al., 1994; Roivainen et al., 1993); epitopes on these reversibly exposed states are targets for antibody recognition following infection (Jimenez-Clavero et al., 2000; Sauter et al., 2008). For picornaviruses, viral breathing describes reversible transitions between the “closed” state of the mature infectious virion, and an “open” structure in which the amino-terminus of VP1 and VP4 are exposed. The open state of the virus particle may be uniquely capable of binding receptor with high affinity (McDermott et al., 2000). In this context, the role of receptor may be to shift the equilibrium towards the more open structure of the A-particle, as discussed by Organtini, Carson, and colleagues (Carson, 2014; Organtini et al., 2014).
Inhibiting viral breathing is an antiviral strategy
The significance of capsid dynamics during the infectious cycle of rhinoviruses is supported by the existence of antiviral compounds that inhibit viral breathing. Several small molecule “WIN compounds” (named for the Sterling Wintrop Research Institute) have been developed that block infection by picornaviruses (Heinz et al., 1989; Otto et al., 1985; Pevear et al., 1999; Turner et al., 1993), and one has advanced to clinical trials with results yet to be reported (NIH clinical trials identifier: NCT00394914). These compounds bind the same hydrophobic pocket beneath the canyon floor as the pocket factor, inducing a local conformational change (Figure 1) (Pevear et al., 1989) and the entropic stabilization of the capsid (Phelps and Post, 1995; Tsang et al., 2000). Viruses incubated with WIN compounds do not transiently display the otherwise internal N-terminus of VP1 and VP4 characteristic of capsids that breathe at equilibrium, indicating that WIN binding dampens the large-amplitude capsid fluctuations (Lewis et al., 1998; Reisdorph et al., 2003). Stabilization of picornavirus capsids has the potential to inhibit infection via several mechanisms. WIN compounds interfere with interactions of rhinoviruses that use ICAM-1 receptors that bind within the canyon (Pevear et al., 1989). However, as not all picornaviruses bind the same receptors nor do so via similar viral structures, blocking attachment is not a universal mode of antiviral action (Hewat et al., 2000). More generally, the stabilization conferred by WIN compound binding prevents the loss of VP4 that occurs during the viral uncoating process described above (Gruenberger et al., 1991; Heinz et al., 1989; McSharry et al., 1979). Thus the dampened motions of the native viral capsid in the presence of WIN compounds are consistent with these motions contributing to cell entry and uncoating. Beyond their potential utility as therapeutics, compounds that block viral breathing of picornaviruses have proven to be powerful tools for investigating the dynamics of viral proteins and the ensemble of structures sampled by viruses at different stages of the viral entry pathway (Roy and Post, 2012).
Flaviviruses
Flaviviruses are a large group of RNA viruses transmitted principally by arthropod vectors. Viruses within this group are responsible for considerable morbidity worldwide. For example, it is estimated that roughly 390 million dengue virus (DENV) infections occur each year, and one third of the population is at risk of infection (Bhatt et al., 2013). Flaviviruses are spherical, enveloped virions composed of three structural proteins (reviewed by (Mukhopadhyay et al., 2005)). The capsid protein is a small, alpha-helical molecule found within virions that interacts with lipids and the positive-stranded viral RNA genome (Ma et al., 2004). The capsid-RNA complex within the virus particle appears unstructured (Kuhn et al., 2002). The premembrane (prM) protein functions as a chaperone for the envelope (E) protein. prM binds E shortly after synthesis, facilitates protein folding, and is incorporated into the virus particle in complex with the E protein (Konishi and Mason, 1993; Lorenz et al., 2002). The E protein is an elongated class II viral membrane fusion protein that orchestrates viral assembly, budding, and entry into target cells. It is also the principal target of neutralizing antibodies (reviewed by (Diamond et al., 2008; Dowd and Pierson, 2011)).
The E protein is comprised of three domains connected by short flexible linkers (Figure 2A) (Kanai et al., 2006; Luca et al., 2012; Modis et al., 2003). This elongated molecule is anchored to the viral membrane by a helical stem and two antiparallel transmembrane domains. E protein domain III (E-DIII) is an immunoglobulin-fold that, in some laboratory adapted strains, contains sites of interactions with negatively charged heparin sulfate molecules on target cells (Chen et al., 1997). E-DIII contains epitopes recognized by potently neutralizing antibodies (Beasley and Barrett, 2002; Crill and Roehrig, 2001; Gromowski and Barrett, 2007; Oliphant et al., 2005; Sanchez et al., 2005; Shrestha et al., 2010), although the contribution of antibodies of these specificities in vivo is questionable (Wahala et al., 2012; Wahala et al., 2009; Williams et al., 2012). Domain II (E-DII) is a long finger-like domain containing residues involved in the formation of the antiparallel dimers present on mature viruses. A highly conserved fusion loop (DII-FL) is located at the distal end of this domain; the majority of antibodies that bind the DII-FL are cross-reactive and vary considerably in neutralization potency (Nelson et al., 2008; Oliphant et al., 2006; Stiasny et al., 2006). E-DI is a beta barrel structure positioned between DIII and DII. Depending on the viral species and strain, the E protein may contain asparagine-linked sugars on DI and DII. One essential role of the E protein is to catalyze fusion between viral and cellular membranes in the acidic environment of the endosome (reviewed by (Pierson and Kielian, 2013)). Under these low pH conditions the E protein homodimers rearrange into fusion-competent E protein homotrimers. This rearrangement is accomplished by considerable movement among the three E protein domains made possible by the hinges that connect them. This flexibility is reflected by differences in the intra-domain angles captured by the numerous E protein structures now available (Bressanelli et al., 2004; Luca et al., 2012; Modis et al., 2003, 2004; Zhang et al., 2004). Of interest, recent studies suggest that some antibodies recognize epitopes not captured by soluble E proteins, indicative of quaternary epitopes expressed only in the context of the intact virion (Beltramello et al., 2010; de Alwis et al., 2012; Kaufmann et al., 2010).
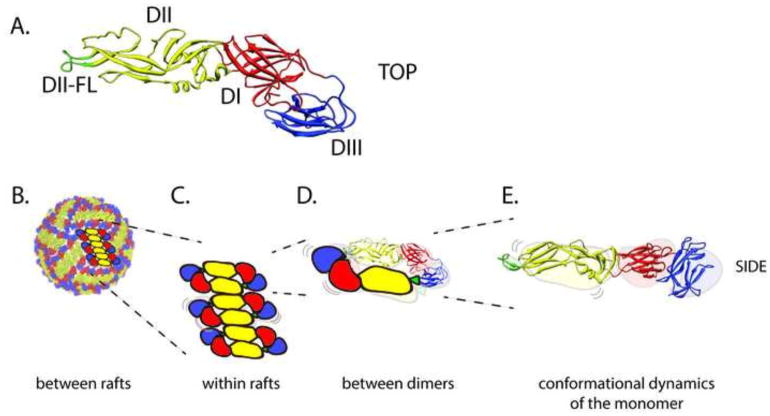
Figure 2. The structure, arrangement, and dynamic motion of E proteins on the mature flavivirus virion.
(A) Top view of the flavivirus E protein monomer shown as a ribbon diagram, with domains I, II, and III (DI-DIII) colored in red, yellow, and blue, respectively. The conserved fusion loop at the distal tip of domain II (DII-FL) is colored in green. Not shown are the helical stem and transmembrane anchor located at the carboxy terminus of DIII. (B) The surface of the mature virion contains 90 sets of head to tail E protein dimers arranged with pseudo T=3 icosahedral symmetry. Domains are colored as in panel A. The flavivirus virion must strike a balance between protection of the RNA genome during transmission and the ability to disassemble the virus structure during the entry process. The strength of the contacts between E proteins that hold together the virion structure may differ among viral strains due to sequence variation, with the potential to increase or decrease the extent of conformational flexibility. The effects of this may be relevant in terms of amino acid contacts (B) between E protein rafts, (C) between the E homodimers that comprise the rafts, (D) at the E dimer interface, and (E) at the level of an individual E protein.
Flaviviruses assemble at membranes of the endoplasmic reticulum as non-infectious immature viruses. prM and E proteins are present on immature virions as an icosahedral array of spikes composed of three prM-E heterodimers (Zhang et al., 2007). In this arrangement, prM is positioned close to the DII-FL and prevents adventitious fusion of the virus during egress (Guirakhoo et al., 1991; Yu et al., 2009). While immature viruses are incapable of fusion (Elshuber et al., 2003), the presence of prM does not lock E proteins of the virus into a single configuration. Exposure of spikey immature virions to an acidic environment during egress results in a reorganization of prM and E that orients E proteins roughly flat against the surface of the virion as homodimers, and exposes a cleavage site on prM recognized by furin-like proteases (Yu et al., 2008). Cleavage of prM by furin is required for the formation of infectious virions, but can be incomplete (Cherrier et al., 2009; Zhang et al., 2007). The result of this virion maturation process is a membrane anchored M peptide, and a soluble “pr” molecule that remains associated with the virus particle until it is released into the neutral pH of the extracellular space. The mature virus is a relatively smooth virion on which E proteins are arranged in rafts of three antiparallel dimers (Kuhn et al., 2002; Mukhopadhyay et al., 2003).
Structural heterogeneity arising from incomplete prM cleavage
Flaviviruses are secreted from cells as a structurally heterogeneous population of virus particles that retain uncleaved prM protein in varying amounts (reviewed by (Pierson and Diamond, 2012)). Partially mature viruses are defined herein as prM+ virions with less than 180 copies of uncleaved prM. Because the amount of uncleaved prM retained on flaviviruses differs by virus type, is influenced by viral sequence diversity, and is impacted by the cell line in which virions are produced, the structures of partially mature virions are expected to vary (Junjhon et al., 2010). Cryo-electron tomographic analysis of partially mature viruses identified a mosaic structure of discrete regions of mature-like and immature-like structure (Plevka et al., 2011). The prM content of virions has been shown to markedly impact sensitivity to neutralization by many classes of antibodies (Oliphant et al., 2006; Pierson et al., 2007), and some polyclonal mixtures of antibody elicited by candidate flavivirus vaccines (Nelson et al., 2008). Because the process of virion maturation is irreversible, changes in the structure or antigenic surface of the virion after release must be attributed to other processes.
Structural hints of a dynamic virion
While biological processes cannot be inferred solely from static structures, interest in the dynamics of flaviviruses arose from the cryo-electron reconstruction of DENV serotype 2 (DENV2) with Fab fragments of a cross-reactive DENV antibody. 1A1D-2 is a murine mAb capable of potently neutralizing DENV serotypes 1, 2, and 3, but not 4 (Lok et al., 2008). This mAb binds an epitope that includes the β-strand of DIII, which is not predicted to be accessible for binding on the mature virus particle. Interestingly, efficient binding of 1A1D-2 Fab fragments to the virion was achieved at 37°C, but not 4°C. Reconstruction studies of the virus-Fab complex revealed that an unexpected degree of structural changes in DENV was required for binding. These findings suggest 1A1D-2 trapped a temperature-dependent conformation of the virus on which the otherwise cryptic DIII β-strand epitope was accessible.
Two recent follow-up studies of DENV structure at 37°C in the absence of antibody suggest the structure of the virus particle changes at physiological temperatures (Fibriansah et al., 2013; Zhang et al., 2013). Exposure of DENV2 to 36°C or 37°C results in the rapid (within minutes) formation of an extended “bumpy” structure. The structure of DENV2 in this configuration is a more open conformation created by the rotation of E protein dimers outwards, creating greater access to the viral lipid membrane. Incubation of DENV2 at physiological temperatures was also associated with a marked increase in the heterogeneity of the virus population. While interpreting these findings against the backdrop of the modest particle to infectious particle ratio of flaviviruses is challenging, it is clear that changes in temperature have the potential to alter the structure(s) of DENV in ways that could impact the biology of the virion. Because “bumpy” particles have not been observed for all DENV serotypes studied (Kostyuchenko et al., 2014), future studies are required to identify viral features responsible for temperature-dependent changes in structure.
The dynamics of flaviviruses
The dynamic properties of flaviviruses have been inferred principally from the analysis of antibody reactivity. Epitope mapping studies have been completed for large panels of human and murine antibodies (de Alwis et al., 2012; Oliphant et al., 2007; Shrestha et al., 2010; Sukupolvi-Petty et al., 2010; Throsby et al., 2006). The utility of static models of flavivirus structure for translating this information into a detailed understanding of the molecular basis of viral recognition has been limited. Epitopes recognized by many mAbs with neutralizing activity are located on surfaces of the E protein not expected to be displayed on the surface of the virus particle (cryptic epitopes), or are occluded by the close proximity of neighboring E proteins (Austin et al., 2012; Lok et al., 2008; Nybakken et al., 2005; Oliphant et al., 2006; Stiasny et al., 2006). For example, the neutralizing DENV mAb E111 binds DIII on its lateral face roughly along the plane of the viral membrane. It is difficult to imagine how an intact antibody could access this structure on a static virus particle (Austin et al., 2012). Epitopes may also be displayed in a non-equivalent fashion among the 180 E proteins that comprise the virion. mAb E16 is a West Nile virus (WNV)-specific potent neutralizing antibody capable of binding only two thirds of the E-DIII lateral ridge (DIII-LR) epitopes on the mature virion. DIII-LR epitopes located adjacent to the five-fold symmetry axis are not bound by E16 due to steric constraints (Kaufmann et al., 2006; Nybakken et al., 2005). Not all epitopes are equally accessible for antibody binding; epitope accessibility is a principal factor that defines the potency of neutralizing antibodies (reviewed by (Dowd and Pierson, 2011)).
The conformational flexibility of flaviviruses has the potential to impact sensitivity to antibody-mediated neutralization through changes in epitope accessibility (Figure 3). Thus, antibodies are a powerful tool to probe the structure(s) of infectious flavivirus virions. The DII-FL epitope of WNV is not predicted to be accessible for antibody recognition on the mature virion. The neutralization potency of the DII-FL-reactive mAb E53 is very sensitive to the maturation state of the virion: both functional and structural studies suggest this antibody cannot bind the mature virion because its epitope is non-accessible (Cherrier et al., 2009; Nelson et al., 2008). Incubation of E53 with preparations of mature WNV under conditions sufficient to achieve steady-state binding results in very little inhibition of infection. However, prolonged incubation of WNV in the presence of antibody resulted in marked increases in the neutralization activity of E53 (Dowd et al., 2011). Because the E53 epitope is not displayed on the mature virion, time- and temperature-dependent increases in neutralization are thought to reflect changes in presentation of an otherwise structurally inaccessible epitope on mature viruses. Increased neutralization sensitivity may reflect a change in the number of epitopes available for recognition and/or changes in structure that allow for higher affinity or potentially bivalent binding. Time- and temperature-dependent patterns of neutralization are a characteristic of all anti-flavivirus E protein antibodies tested to date (Austin et al., 2012; Dowd et al., 2011; Dowd et al., 2014; Sabo et al., 2012). The magnitude of the shift in neutralization potency among flavivirus antibodies correlates generally with the predicted accessibility of an epitope on the mature virus. For example, prolonged incubation of E16, which binds the relatively accessible DIII lateral ridge epitope results in just a few fold increase in neutralization, whereas other antibodies that bind cryptic determinants become more than 100-fold more potent. Importantly, the extent to which neutralization sensitivity can be increased by prolonged incubation is finite; once all epitopes are engaged by antibody, further incubation would not be expected to impact antibody potency (Dowd et al., 2011; Dowd et al., 2014).
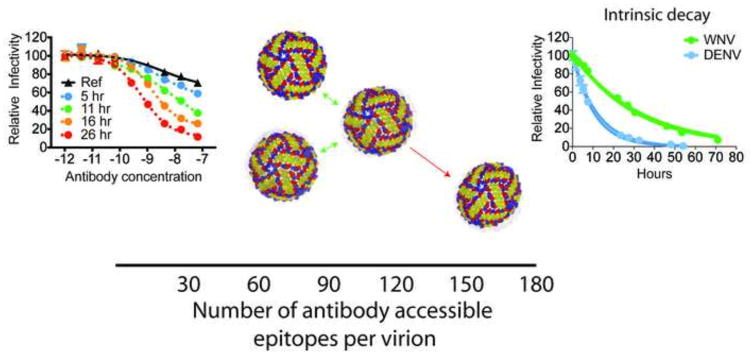
Figure 3. Flavivirus dynamics results in reversible and non-reversible conformational changes.
Flavivirus neutralization is governed by a stoichiometric threshold and requires binding by a critical number of antibody molecules per virion. Of the 180 potential E protein epitopes (x-axis, bottom of figure), estimates suggest that this threshold corresponds to docking on the virion by 30 antibodies (Pierson et al., 2007). In addition to antibody affinity, epitope accessibility is a critical factor that determines whether neutralization occurs. Because of the dense arrangement of E proteins on the surface of the mature virion, not all E protein epitopes are equally accessible for antibody binding. For some characterized antibodies, neutralization cannot be achieved even in the presence of saturating concentrations of antibody due to the lack of accessible epitopes (Nelson et al., 2008). Flavivirus dynamics can modulate the landscape available for antibody binding by transiently exposing otherwise inaccessible epitopes and increasing the absolute number available for binding. Time- and temperature-dependent increases in neutralization are observed when virus and antibody are incubated for increasing lengths of time (left panel; Ref= reference curve obtained after pre-incubation of virus with serial dilutions of antibody for 1 hour; additional antibody virus complexes were further incubated at 37°C for 5–26 hours before infecting target cells). This particular antibody binds an epitope shown to be inaccessible on the mature form of the virus. Time-dependent increases in neutralization potency reflect exposure of this otherwise ‘cryptic’ epitope through the process of virus breathing. In addition to reversible changes in virus structure (green arrows), we hypothesize that a subset of conformational transitions sample non-infectious structures that can no longer return to an infectious state (red arrow). These ‘dead-end’ structures are detected experimentally as a loss of virus infectivity over time in solution, referred to as intrinsic decay. Differences in the rate of intrinsic decay (shown for WNV and DENV, right panel) among virus strains indicate that sequence variation dictates the steady state virus structures and/or the ensembles available for virus breathing.
As introduced above, the accessibility of many epitopes is modulated by the maturation state of the virus particle. The process of virion maturation reduces the potency of many neutralizing antibodies because epitopes become less accessible on the dense array of E proteins that comprise the mature virion. In contrast, prM+ partially mature viruses are generally more sensitive to neutralization (Dowd et al., 2014; Nelson et al., 2008). Recent studies demonstrate that both mature and prM+ virions are dynamic structures. Time-dependent changes in antibody potency were observed in studies of populations of WNV or DENV produced in a manner that allows the extent of prM cleavage to be controlled (Dowd et al., 2014). Of interest, even very long incubations of mature virus with antibody were not sufficient to render these virions as sensitive to neutralization as prM+ immature-like virus particles.
The rapid on-rate of antibodies provides a means to bind and trap transiently exposed regions otherwise buried in the equilibrium structure. Viral breathing in the presence of neutralizing antibodies has the potential to display an epitope for recognition, which in turn provides a mechanism to contribute to the stoichiometry required for virus neutralization (Pierson et al., 2007). Neutralization studies of picornaviruses suggest the VP4 protein is exposed via reversible processes on dynamic, breathing virus particles. Likewise, structural transitions that expose epitopes on infectious mature flaviviruses also appear to be reversible. Time-dependent changes in sensitivity to neutralization by antibodies that recognize poorly accessible epitopes required the presence of antibody during the prolonged incubation. Prolonged incubation in the absence of antibody resulted in neutralization titers similar to those of the conventional neutralization assays run in parallel (Dowd et al., 2014). The impact of “viral breathing” on the neutralization potency of antibodies does not require extended incubation in all circumstances. mAb E111 is a potent DENV type-specific mAb that binds an epitope on DIII not predicted to be exposed on mature or immature forms of the virion (Austin et al., 2012). The accessibility of this epitope reflects the dynamic properties of the virion, which have been reported to vary among DENV strains (Austin et al., 2012; Sukupolvi-Petty et al., 2013). Thus, relationships between neutralization potency and the predicted accessibility of epitopes on flaviviruses requires a deeper understanding of factors that govern the ensemble of structures that exist at steady state under physiologically-relevant conditions.
Alternatively, beyond revealing otherwise buried antibody epitopes, the conformational dynamics of the virus particle may play an important role in the formation of complex quaternary epitopes on the surface of the mature virus particle. Several recent studies have identified mAbs that appear to bind the virus particle, but not soluble E proteins. Antibody mapping and structural studies of WNV and DENV identify a class of epitopes composed of residues found on multiple E proteins (de Alwis et al., 2012; Dejnirattisai et al., 2015; Fibriansah et al., 2014; Fibriansah et al., 2015; Kaufmann et al., 2010; Rouvinski et al., 2015; Teoh et al., 2012). For example, a recent class of mAbs has been described that bind E dimer-dependent epitopes (EDE), resulting in potent neutralization of all four serotypes of DENV. A similar pattern of reactivity was recently described in serologic studies of tick-borne encephalitis virus infection and vaccination (Jarmer et al., 2014; Kiermayr et al., 2009). This EDE epitope includes multiple features of the dimer interface including the conserved DII-FL, and overlaps the portion of E bound by prM on immature or partially mature DENV virions (Rouvinski et al., 2015). Of significant interest, EDE mAbs are equally potent when uncleaved prM is retained on the virion (Dejnirattisai et al., 2015). These findings raise the possibility that E protein dimers and trimers exist as a dynamic equilibrium between these oligomeric states. The displacement of prM by EDE antibodies is thought to stabilize the surface accessible dimer-interface epitope, trapping the virus particle in a mature conformation.
Intrinsic decay
Like many viruses, the prolonged incubation of flaviviruses at physiological temperatures results in a loss of infectivity (Figure 3) (Ansarah-Sobrinho et al., 2008; Dowd et al., 2014). This process is referred to herein as intrinsic decay. Practical considerations have prompted studies to define conditions to enhance the half-life of infectious virions (Wiggan et al., 2011). However, insights into why flaviviruses become non-infectious in solution have not been reported. We hypothesize that the intrinsic decay of flaviviruses is a characteristic of a dynamic virus particle. From this perspective, it is possible that not all structural pathways sampled by the virus (or its E proteins) at equilibrium will be reversible, and these in turn may not be compatible with virus attachment or entry. The intrinsic decay rate for WNV and DENV differ significantly, as does the rate with which these viruses become more sensitive to neutralization when incubated with antibody for prolonged periods (Dowd et al., 2014). Furthermore, in recent studies, E protein variants found to alter the neutralization potency of mAbs at a distance (outside the epitope) also modulate intrinsic decay (Dowd and Pierson, unpublished data). The structural basis for intrinsic decay is not known. While it is attractive to imagine that flaviviruses adopt a non-infectious structure equivalent to the A-particle of picornaviruses, this may be difficult to support experimentally. Flaviviruses are structurally heterogeneous due to the presence of uncleaved prM protein, and may be infectious depending upon the efficiency of virion maturation (reviewed by (Pierson and Diamond, 2012)). Recent studies suggest they may rapidly become more structurally heterogeneous upon incubation at physiological temperatures (Fibriansah et al., 2013; Zhang et al., 2013). Identifying a non-infectious virion among a heterogeneous population by virtue of its structure is not possible.
Unanswered questions
The dynamics of non-enveloped viruses has been studied using numerous techniques and exploited for the development of antivirals. In contrast, very little is understood about the breathing of enveloped virions. When considered in the context of the flavivirus replication cycle, numerous questions remain, including:
What interactions control the dynamics of the virion? The structure of flaviviruses changes markedly during the maturation process of virions. As immature virus particles, E proteins exist as trimers in complex with prM. E proteins are present on mature viruses as anti-parallel dimers; the surface of the virus is composed of “rafts” of three dimers (Figure 2B) (Kuhn et al., 2002). Both immature and mature forms of the virion appear to sample multiple conformations at physiological temperatures (Dowd et al., 2014). The molecular interactions that govern the ensemble of structures sampled by each form of the virion are unknown. On mature virions, the conformational flexibility of E proteins may be a key feature of a dynamic virus particle (Figure 2B–E). The linkers connecting the three domains of each E protein monomer confer an ability to rotate that enable the conformational changes that drive fusion during viral entry (Figure 2E) (reviewed by (Pierson and Kielian, 2013)). Local motion of the E protein on the mature virus particle may be sufficient to expose otherwise cryptic neutralizing antibody epitopes and may be required for the virus to sample its environment in its search to bind and enter cells efficiently. However, this in turn may be modulated by the strength of interactions that maintain the E protein dimers that comprise the mature virion (Figure 2D). Of significant interest, Luca, Fremont, and colleagues hypothesized that the contribution of E protein dimers to the overall stability of the virion may vary among flaviviruses, consistent with differences in intrinsic decay (Luca et al., 2012). The interface that mediates E protein dimers is not of uniform size among flaviviruses. For example, structural features that contribute to tick-borne encephalitis virus (TBEV) dimers do not exist at the interface of Japanese encephalitis virus (JEV) E protein dimers. The conclusion that E protein dimers of viruses of the JEV serocomplex are of lower affinity is supported by the observation that their E proteins are monomers in solution, as compared to the dimers of DENV and tick-borne encephalitis viruses (Luca et al., 2012). This raises the yet unexplored possibility that stability of JEV serocomplex viruses may instead be governed by interactions among dimers on the mature virion. Finally, E protein contacts also orchestrate in the herringbone arrangement of E protein rafts on the surface of the mature virion, providing another potential avenue for modulating virion stability (Figure 2B–C).
How does sequence diversity among viruses impact dynamics? Recent findings suggest sequence variation among flaviviruses has the potential to shape the structural ensemble of the virus particle. The potency of the DENV1-reactive mAb E111 varies significantly among related strains (Shrestha et al., 2010). Structural and functional studies demonstrated that sequence variation within the E111 DIII epitope could not explain genotypic differences in the sensitivity of DENV1 strains Western Pacific and 16007 to neutralization (Austin et al., 2012). Because this epitope is predicted to be inaccessible on the mature virion, a role for dynamics was hypothesized and supported by the observation that time- and temperature-dependent patterns of neutralization sensitivity differed for these viruses. Exposure of the Western Pacific strain to elevated temperatures resulted in rapid changes in neutralization sensitivity of significant magnitude, whereas parallel studies with 16007 revealed only modest changes (Austin et al., 2012). The precise determinants of E111 sensitivity are currently being investigated. The identification and characterization of mutations in WNV or DENV that alter both intrinsic decay and kinetic patterns of neutralization will be instructive.
An impact of viral sequence diversity on the dynamics of virions has been studied previously. Adeno-associated virus (AAV) is a non-enveloped, single-stranded DNA virus with a T=1 icosahedral capsid. AAV exists as at least thirteen serotypes (reviewed by (Wu et al., 2006)). Recent biophysical studies of the dynamics of AAV capsids discovered strain-dependent differences in the behavior of AAV in solution (Rayaprolu et al., 2013). Using multiple approaches, Rayaprolu and colleagues identified large differences in the thermal stability of AAV in solution that did not correlate with the overall sequence divergence among the representative strains analyzed. While limited proteolysis studies suggested the capsids of all AAV are characterized by conformational flexibility, the patterns of cleavage sensitivity varied among the viruses studied.
Are dynamics something to be considered in the context of flavivirus vaccine design? Many epitopes recognized by neutralizing antibodies are poorly accessible on the mature virus particle, and are exposed for recognition only on breathing virus particles, or those that retain uncleaved prM protein. Whether structural heterogeneity is a beneficial property of immunogens or live-attenuated virus vaccines has not been evaluated. Would more dynamic immunogens elicit more potent cross-reactive antibody responses? Alternatively, might enhanced exposure of otherwise cryptic epitopes prime a response that targets epitopes available for antibody binding only at low occupancy? Antibodies with these characteristics have been hypothesized to contribute to enhanced disease following secondary DENV infections (Halstead, 2003; Kliks et al., 1988). Modulating the dynamic characteristics of live-attenuated vaccine candidates has the potential to enrich for more desirable antibody responses. Future studies that identify and characterize the underlying mechanisms behind flavivirus breathing and the specific amino acids or protein structures involved in dynamics will guide these exciting new prospects.
Highlights.
Virus particles may exist as an ensemble of conformations
Structural dynamics of the virion may be required for virus entry or uncoating
Particle instability is a consequence of “viral breathing”
The conformational dynamics of a virion modulates antibody epitope presentation
Acknowledgments
We thank members of our laboratories for their comments on this manuscript. This work was supported by the intramural program of the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Richard J. Kuhn, Email: kuhnr@purdue.edu.
Kimberly A. Dowd, Email: dowdka@mail.nih.gov.
Carol Beth Post, Email: cbp@purdue.edu.
Theodore C. Pierson, Email: piersontc@mail.nih.gov.
References
- Ansarah-Sobrinho C, Nelson S, Jost CA, Whitehead SS, Pierson TC. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology. 2008;381:67–74. doi: 10.1016/j.virol.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 2012;8:e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi T, Gerhard W, Yewdell JW. Monoclonal antibodies detect different forms of influenza virus hemagglutinin during viral penetration and biosynthesis. J Virol. 1985;55:307–313. doi: 10.1128/jvi.55.2.307-313.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley DW, Barrett AD. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J Virol. 2002;76:13097–13100. doi: 10.1128/JVI.76.24.13097-13100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Filman DJ, Trus BL, Cheng N, Booy FP, Conway JF, Curry S, Hiremath CN, Tsang SK, Steven AC, Hogle JM. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell host & microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbat PP, Costa-Filho AJ, Earle KA, Moscicki JK, Freed JH. Electron spin resonance in studies of membranes and proteins. Science. 2001;291:266–269. doi: 10.1126/science.291.5502.266. [DOI] [PubMed] [Google Scholar]
- Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. The Journal of biological chemistry. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. The EMBO journal. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson SD. Kinetic models for receptor-catalyzed conversion of coxsackievirus B3 to A-particles. J Virol. 2014;88:11568–11575. doi: 10.1128/JVI.01790-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nature medicine. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. The EMBO journal. 2009;28:3269–3276. doi: 10.1038/emboj.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell RL, Philipson L. Specific alterations of coxsackievirus B3 eluted from HeLa cells. J Virol. 1971;8:509–515. doi: 10.1128/jvi.8.4.509-515.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry S, Chow M, Hogle JM. The poliovirus 135S particle is infectious. J Virol. 1996;70:7125–7131. doi: 10.1128/jvi.70.10.7125-7131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinsky A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, Grimes JM, Tsai WY, Lai CY, Wang WK, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J, Screaton GR. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nature immunology. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Pierson TC, Fremont DH. The structural immunology of antibody protection against West Nile virus. Immunological reviews. 2008;225:212–225. doi: 10.1111/j.1600-065X.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson GG, Lane DP, Verma CS. Molecular simulations of protein dynamics: new windows on mechanisms in biology. EMBO reports. 2008;9:144–150. doi: 10.1038/sj.embor.7401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 2011;7:e1002111. doi: 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, Mukherjee S, Kuhn RJ, Pierson TC. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J Virol. 2014;88:11726–11737. doi: 10.1128/JVI.01140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, Pierson TC. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. The Journal of general virology. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J Virol. 2013;87:7585–7592. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, Crowe JE, Jr, Lok SM. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO molecular medicine. 2014;6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, Lok SM. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nature communications. 2015;6:6341. doi: 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricks CE, Hogle JM. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromowski GD, Barrett AD. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology. 2007;366:349–360. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- Gruenberger M, Pevear D, Diana GD, Kuechler E, Blaas D. Stabilization of human rhinovirus serotype 2 against pH-induced conformational change by antiviral compounds. The Journal of general virology. 1991;72( Pt 2):431–433. doi: 10.1099/0022-1317-72-2-431. [DOI] [PubMed] [Google Scholar]
- Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. The Journal of general virology. 1991;72( Pt 6):1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- Hadfield AT, Lee W, Zhao R, Oliveira MA, Minor I, Rueckert RR, Rossmann MG. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure. 1997;5:427–441. doi: 10.1016/s0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Advances in virus research. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Haqqani AA, Tilton JC. Entry inhibitors and their use in the treatment of HIV-1 infection. Antiviral research. 2013;98:158–170. doi: 10.1016/j.antiviral.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nature structural & molecular biology. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz BA, Rueckert RR, Shepard DA, Dutko FJ, McKinlay MA, Fancher M, Rossmann MG, Badger J, Smith TJ. Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J Virol. 1989;63:2476–2485. doi: 10.1128/jvi.63.6.2476-2485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewat EA, Neumann E, Conway JF, Moser R, Ronacher B, Marlovits TC, Blaas D. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. The EMBO journal. 2000;19:6317–6325. doi: 10.1093/emboj/19.23.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Jarmer J, Zlatkovic J, Tsouchnikas G, Vratskikh O, Strauss J, Aberle JH, Chmelik V, Kundi M, Stiasny K, Heinz FX. Variation of the specificity of the human antibody responses after tick-borne encephalitis virus infection and vaccination. J Virol. 2014;88:13845–13857. doi: 10.1128/JVI.02086-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Clavero MA, Douglas A, Lavery T, Garcia-Ranea JA, Ley V. Immune recognition of swine vesicular disease virus structural proteins: novel antigenic regions that are not exposed in the capsid. Virology. 2000;270:76–83. doi: 10.1006/viro.2000.0256. [DOI] [PubMed] [Google Scholar]
- Junjhon J, Edwards TJ, Utaipat U, Bowman VD, Holdaway HA, Zhang W, Keelapang P, Puttikhunt C, Perera R, Chipman PR, Kasinrerk W, Malasit P, Kuhn RJ, Sittisombut N. Influence of pr-M cleavage on the heterogeneity of extracellular dengue virus particles. J Virol. 2010;84:8353–8358. doi: 10.1128/JVI.00696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12400–12404. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18950–18955. doi: 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermayr S, Stiasny K, Heinz FX. Impact of quaternary organization on the antigenic structure of the tick-borne encephalitis virus envelope glycoprotein E. J Virol. 2009;83:8482–8491. doi: 10.1128/JVI.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliks SC, Nimmanitya S, Nisalak A, Burke DS. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. The American journal of tropical medicine and hygiene. 1988;38:411–419. doi: 10.4269/ajtmh.1988.38.411. [DOI] [PubMed] [Google Scholar]
- Kneller GR. Quasielastic neutron scattering and relaxation processes in proteins: analytical and simulation-based models. Physical chemistry chemical physics : PCCP. 2005;7:2641–2655. doi: 10.1039/b502040a. [DOI] [PubMed] [Google Scholar]
- Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. The EMBO journal. 1999;18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi E, Mason PW. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J Virol. 1993;67:1672–1675. doi: 10.1128/jvi.67.3.1672-1675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chew PL, Ng TS, Lok SM. Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J Virol. 2014;88:477–482. doi: 10.1128/JVI.02641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JK, Bothner B, Smith TJ, Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yafal AG, Lee YM, Hogle J, Chow M. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nature structural & molecular biology. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Lorenz IC, Allison SL, Heinz FX, Helenius A. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J Virol. 2002;76:5480–5491. doi: 10.1128/JVI.76.11.5480-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca VC, AbiMansour J, Nelson CA, Fremont DH. Crystal structure of the Japanese encephalitis virus envelope protein. J Virol. 2012;86:2337–2346. doi: 10.1128/JVI.06072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Teichmann SA, Forman-Kay JD. Probing the diverse landscape of protein flexibility and binding. Current opinion in structural biology. 2012;22:643–650. doi: 10.1016/j.sbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Mateu MG. Assembly, stability and dynamics of virus capsids. Archives of biochemistry and biophysics. 2013;531:65–79. doi: 10.1016/j.abb.2012.10.015. [DOI] [PubMed] [Google Scholar]
- McDermott BM, Jr, Rux AH, Eisenberg RJ, Cohen GH, Racaniello VR. Two distinct binding affinities of poliovirus for its cellular receptor. The Journal of biological chemistry. 2000;275:23089–23096. doi: 10.1074/jbc.M002146200. [DOI] [PubMed] [Google Scholar]
- McSharry JJ, Caliguiri LA, Eggers HJ. Inhibition of uncoating of poliovirus by arildone, a new antiviral drug. Virology. 1979;97:307–315. doi: 10.1016/0042-6822(79)90342-8. [DOI] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nature reviews Microbiology. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, Blanchard SC, Mothes W. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Mothes W. The HIV-1 Env trimer in HD. Structure. 2014;22:935–936. doi: 10.1016/j.str.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S, Jost CA, Xu Q, Ess J, Martin JE, Oliphant T, Whitehead SS, Durbin AP, Graham BS, Diamond MS, Pierson TC. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 2008;4:e1000060. doi: 10.1371/journal.ppat.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nature medicine. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol. 2007;81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson NH, Kolatkar PR, Oliveira MA, Cheng RH, Greve JM, McClelland A, Baker TS, Rossmann MG. Structure of a human rhinovirus complexed with its receptor molecule. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol. 2014;88:5755–5765. doi: 10.1128/JVI.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Takeuchi K, Ueda T, Nishida N, Shimada I. Functional dynamics of proteins revealed by solution NMR. Current opinion in structural biology. 2012;22:660–669. doi: 10.1016/j.sbi.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Otto MJ, Fox MP, Fancher MJ, Kuhrt MF, Diana GD, McKinlay MA. In vitro activity of WIN 51711, a new broad-spectrum antipicornavirus drug. Antimicrobial agents and chemotherapy. 1985;27:883–886. doi: 10.1128/aac.27.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Marqusee S. Probing the high energy states in proteins by proteolysis. Journal of molecular biology. 2004;343:1467–1476. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- Pevear DC, Fancher MJ, Felock PJ, Rossmann MG, Miller MS, Diana G, Treasurywala AM, McKinlay MA, Dutko FJ. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J Virol. 1989;63:2002–2007. doi: 10.1128/jvi.63.5.2002-2007.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevear DC, Tull TM, Seipel ME, Groarke JM. Activity of pleconaril against enteroviruses. Antimicrobial agents and chemotherapy. 1999;43:2109–2115. doi: 10.1128/aac.43.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps DK, Post CB. A novel basis of capsid stabilization by antiviral compounds. Journal of molecular biology. 1995;254:544–551. doi: 10.1006/jmbi.1995.0637. [DOI] [PubMed] [Google Scholar]
- Pierson TC, Diamond MS. Degrees of maturity: the complex structure and biology of flaviviruses. Current opinion in virology. 2012;2:168–175. doi: 10.1016/j.coviro.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Kielian M. Flaviviruses: braking the entering. Current opinion in virology. 2013;3:3–12. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Xu Q, Nelson S, Oliphant T, Nybakken GE, Fremont DH, Diamond MS. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell host & microbe. 2007;1:135–145. doi: 10.1016/j.chom.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevka P, Battisti AJ, Junjhon J, Winkler DC, Holdaway HA, Keelapang P, Sittisombut N, Kuhn RJ, Steven AC, Rossmann MG. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO reports. 2011;12:602–606. doi: 10.1038/embor.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello VR. Picornaviridae: The Viruses and Their Replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields virology. 5. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- Rayaprolu V, Kruse S, Kant R, Venkatakrishnan B, Movahed N, Brooke D, Lins B, Bennett A, Potter T, McKenna R, Agbandje-McKenna M, Bothner B. Comparative analysis of adeno-associated virus capsid stability and dynamics. J Virol. 2013;87:13150–13160. doi: 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph N, Thomas JJ, Katpally U, Chase E, Harris K, Siuzdak G, Smith TJ. Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virology. 2003;314:34–44. doi: 10.1016/s0042-6822(03)00452-5. [DOI] [PubMed] [Google Scholar]
- Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. Picornavirus uncoating intermediate captured in atomic detail. Nature communications. 2013;4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roivainen M, Piirainen L, Rysa T, Narvanen A, Hovi T. An immunodominant N-terminal region of VP1 protein of poliovirion that is buried in crystal structure can be exposed in solution. Virology. 1993;195:762–765. doi: 10.1006/viro.1993.1427. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Palmenberg AC. Conservation of the putative receptor attachment site in picornaviruses. Virology. 1988;164:373–382. doi: 10.1016/0042-6822(88)90550-8. [DOI] [PubMed] [Google Scholar]
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Despres P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015 doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- Roy A, Post CB. Long-distance correlations of rhinovirus capsid dynamics contribute to uncoating and antiviral activity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5271–5276. doi: 10.1073/pnas.1119174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo MC, Luca VC, Ray SC, Bukh J, Fremont DH, Diamond MS. Hepatitis C virus epitope exposure and neutralization by antibodies is affected by time and temperature. Virology. 2012;422:174–184. doi: 10.1016/j.virol.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MD, Pierson TC, McAllister D, Hanna SL, Puffer BA, Valentine LE, Murtadha MM, Hoxie JA, Doms RW. Characterization of neutralizing antibodies to West Nile virus. Virology. 2005;336:70–82. doi: 10.1016/j.virol.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Sauter P, Chehadeh W, Lobert PE, Lazrek M, Goffard A, Soumillon M, Caloone D, Vantyghem MC, Weill J, Fajardy I, Alm G, Lucas B, Hober D. A part of the VP4 capsid protein exhibited by coxsackievirus B4 E2 is the target of antibodies contained in plasma from patients with type 1 diabetes. Journal of medical virology. 2008;80:866–878. doi: 10.1002/jmv.21171. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O'Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Chase ES, Schmidt TJ, Olson NH, Baker TS. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Kiermayr S, Holzmann H, Heinz FX. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J Virol. 2006;80:9557–9568. doi: 10.1128/JVI.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. Functional Analysis of Antibodies against Dengue Virus Type 4 Reveals Strain-Dependent Epitope Exposure That Impacts Neutralization and Protection. J Virol. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Science translational medicine. 2012;4:139ra183. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- Thomsen NK, Poulsen FM. Low energy of activation for amide hydrogen exchange reactions in proteins supports a local unfolding model. Journal of molecular biology. 1993;234:234–241. doi: 10.1006/jmbi.1993.1577. [DOI] [PubMed] [Google Scholar]
- Throsby M, Geuijen C, Goudsmit J, Bakker AQ, Korimbocus J, Kramer RA, Clijsters-van der Horst M, de Jong M, Jongeneelen M, Thijsse S, Smit R, Visser TJ, Bijl N, Marissen WE, Loeb M, Kelvin DJ, Preiser W, ter Meulen J, de Kruif J. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile Virus. J Virol. 2006;80:6982–6992. doi: 10.1128/JVI.00551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang SK, Danthi P, Chow M, Hogle JM. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. Journal of molecular biology. 2000;296:335–340. doi: 10.1006/jmbi.1999.3483. [DOI] [PubMed] [Google Scholar]
- Turner RB, Dutko FJ, Goldstein NH, Lockwood G, Hayden FG. Efficacy of oral WIN 54954 for prophylaxis of experimental rhinovirus infection. Antimicrobial agents and chemotherapy. 1993;37:297–300. doi: 10.1128/aac.37.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Huang C, Butrapet S, White LJ, de Silva AM. Recombinant dengue type 2 viruses with altered e protein domain III epitopes are efficiently neutralized by human immune sera. J Virol. 2012;86:4019–4023. doi: 10.1128/JVI.06871-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology. 2009;392:103–113. doi: 10.1016/j.virol.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggan O, Livengood JA, Silengo SJ, Kinney RM, Osorio JE, Huang CY, Stinchcomb DT. Novel formulations enhance the thermal stability of live-attenuated flavivirus vaccines. Vaccine. 2011;29:7456–7462. doi: 10.1016/j.vaccine.2011.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen CB, Tilton JC, Doms RW. Molecular mechanisms of HIV entry. Advances in experimental medicine and biology. 2012;726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- Williams KL, Wahala WM, Orozco S, de Silva AM, Harris E. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology. 2012;429:12–20. doi: 10.1016/j.virol.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witz J, Brown F. Structural dynamics, an intrinsic property of viral capsids. Archives of virology. 2001;146:2263–2274. doi: 10.1007/s007050170001. [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Yu IM, Holdaway HA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J Virol. 2009;83:12101–12107. doi: 10.1128/JVI.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. Dengue structure differs at the temperatures of its human and mosquito hosts. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Structure of immature West Nile virus. J Virol. 2007;81:6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]