Abstract
The import of proteins into peroxisomes possesses many unusual features such as the ability to import folded proteins, and a surprising diversity of targeting signals with differing affinities that can be recognized by the same receptor. As understanding of the structure and function of many components of the protein import machinery has grown, an increasingly complex network of factors affecting each step of the import pathway has emerged. Structural studies have revealed the presence of additional interactions between cargo proteins and the PEX5 receptor that affect import potential, with a subtle network of cargo-induced conformational changes in PEX5 being involved in the import process. Biochemical studies have also indicated an interdependence of receptor–cargo import with release of unloaded receptor from the peroxisome. Here, we provide an update on recent literature concerning mechanisms of protein import into peroxisomes.
Keywords: mechanisms, models, peroxisome, PEX5, protein import cycle, targeting signal
Introduction
Peroxisomes are organelles found in all but the most primitive eukaryotic cells. They are surrounded by a single bilayer membrane which encloses a soluble matrix space. Peroxisomes contain no DNA and therefore all their constituent matrix proteins are imported post-translationally from the cytosol [1,2]. Peroxisome membrane proteins may be imported post-translationally, or in some cases after import into the endoplasmic reticulum followed by subsequent sorting pathways [3]. Peroxisomes are also able to divide and segregate to daughter cells upon cell division [4,5]. The list of peroxisome functions is long and ever growing. Peroxisomes have many different functions depending on cell type and environment, but fatty acid beta-oxidation and reactive oxygen species metabolism are common to most peroxisome types [6,7]. Peroxisomes can change their function by virtue of importing different proteins and enzymes [8]. The importance of peroxisomes for the proper development of multicellular organisms is underscored by the severe and frequently lethal phenotypes of both animal [9] and plant [10] peroxisome biogenesis mutants. Indeed, in humans the peroxisome biogenesis disorders have provided a strong motivation to study the underlying biochemistry, genetics and cell biology of peroxisomes [7].
The import of proteins into peroxisomes is quite different from the targeting and transport of proteins across other cellular membranes. This brief review provides an update on models and mechanisms of protein import into peroxisomes, focusing on the major matrix protein import pathway mediated by the import receptor PEX5.
Molecular recognition of the PTS1 by PEX5
The first peroxisome targeting signal comprising of a C-terminal tripeptide serine–lysine–leucine (SKL in the single letter amino acid code) was discovered in firefly luciferase and termed peroxisome targeting signal 1 (PTS1) [11]. It was soon realized that PTS1 was conserved between organisms [12] and the discovery of its receptor protein, subsequently termed PEX5, rapidly followed [13–16]. Comparison of the sequences of native peroxisomal proteins, along with mutational analysis coupled with targeting studies and assessment of PEX5 binding preferences, established that the PTS1 is in fact a family of sequences which generally conform to the pattern of [small side chain amino acid]–[basic amino acid]–[hydrophobic amino acid], but in some cases can be considerably more diverse [17] and hinted that residues immediately adjacent to the C-terminal tripeptide might have an auxiliary function [18–20].
PEX5 is a modular protein which shows conservation of its essential features between organisms. The C-terminal domain comprises of seven tetratricopeptide (TPR) repeats. These are repeats of 34 amino acids that form a pair of helices that are a common protein–protein interaction motif [21] and bind the PTS1 peptide. Determination of the X-ray structure of the C-terminal domain of PEX5 from human [22] and Trypanosoma brucei [23] revealed molecular details of the interaction between the PEX5 TPR domain and model PTS1 peptides. The PTS1 binds within a cavity formed by two sets of TPRs, 1–3 and 5–7, with TPR4 adopting a more extended conformation linking the two sets of three TPRs. This provides a funnel shaped binding site in which the PTS1 peptide sits. A series of highly conserved asparagine residues within the TPRs make hydrogen bonds to the peptide backbone of the PTS1 sequence. The side chain in position -2 (lysine) of the model peptide, containing the C-terminal tripeptide SKL, sits within a negatively charged pocket where it hydrogen bonds via a water molecule. Serine in position -3 also makes hydrogen bonds to the receptor indirectly via water, as does the terminal carboxyl group. The side chain of the terminal leucine is accommodated in a hydrophobic pocket. These features explain why PTS1 must be at the C-terminus of the protein and the strong preference for a hydrophobic C-terminal residue, along with the lack of specific side-chain interactions which may allow adaptability in the recognition of sequence variants.
Structures of the PEX5 TPR domain in complex with whole proteins rather than just model peptides have given insights into contacts outside the PTS1. For both PEX5:sterol carrier protein 2 (SCP2), which has a consensus PTS1 AKL [24], and PEX5:alanine glyoxylate aminotransferase (AGT), which has the non-consensus PTS1 KKL [25], contacts between receptor and cargo protein extend beyond the PTS region; however, these differ between the two structures. In the SCP2:PEX5 complex this region of secondary contact to the receptor was distant from the PTS1 within the primary sequence [24] whereas in AGT the region immediately preceding the PTS1 makes extended contacts to the surface of PEX5 and mutations within this region result in a 2–5-fold reduction in affinity [25]. Mis-targeting of AGT to mitochondria due to mutations that increase mitochondrial targeting propensity and destabilize protein structure gives rise to the disorder primary hyperoxaluria type 1. The binding of variant AGT proteins and terminal octapeptides to the TPR domain of PEX5 was compared using isothermal titration calorimetry (ITC), molecular modelling and protein stability assays [26]. The authors concluded that a consensus PTS1 sequence increased the binding affinity principally by reducing the enthalpic penalty of binding, probably reflecting optimization of binding interactions through changes in conformation and buried surface area. The corresponding PTS1 peptides showed a similar order of binding affinities, but with approximately 10-fold weaker interaction, consistent with the idea that the C-terminus provides the main specificity but that other ancillary regions also contribute to the affinity. They also demonstrated that binding of peptides to PEX5 stabilized the receptor to thermal denaturation and proteolysis with the extent of protection correlating well with the measured binding affinity [26]. Direct evidence for structural reorganization of the receptor upon binding cargoes has been presented. For example the apo conformation of the PEX5 TPR domain is snail shaped and undergoes a conformational change to a ring shape upon binding SCP2 [24]. In the case of AGT binding to PEX5, mutation of the KKL targeting sequence to the consensus AKL removes a steric clash allowing the invariant PEX5 asparagine 534 to specifically bind to the -3 position of the PTS1. In turn this results in movement, especially of TPR6, leading to compaction of the PTS1 binding cavity [27]. By comparing the known PEX5:cargo protein structures it was proposed that high affinity binding correlates with a greater degree of compaction of the PTS1 binding pocket, and although less optimal PTS1 sequences are recognized, they are bound with lower affinity [27].
A site-directed photo-crosslinking approach was used to map interactions between Saccharomyces cerevisiae PEX5 and a peroxisomal oxalyl CoA synthetase (PCS60) which contains a consensus PTS1, SKL [28]. Crosslinks were formed predominantly between amino acids immediately preceding the PTS1 and TPRs 6, 7 and the 7C loop that is located C-terminal to TPR7. Placement of crosslinkers within the PTS1 tripeptide blocked interaction with PEX5, presumably through steric interference. Recombinant PCS60 binds PEX5 with a Kd of 0.19 μM as measured by ITC, but a variant in which the PTS1 was deleted was still able to bind with a measured Kd of 7.7 μM, even though this PTS1 deletion mutant could not import into peroxisomes in vivo. Using SPR, two binding events of wild-type PCS60 to PEX5 with different affinities could be detected. The high affinity binding event required the PTS1 sequence but the low affinity binding event was independent of the PTS1 and required the adjacent five residues. These results led to the proposal that interaction between PCS60 and PEX5 is a two-step process with an initial binding interaction followed by a ‘lock in’ [28].
Binding affinities of model peptides to the PEX5 TPR have been determined in several studies and cover several orders of magnitude [29–31]. It was proposed that proteins with low levels of expression may have evolved stronger PTS's to compensate for their lower abundance [29]; however, it is difficult if not impossible to obtain accurate measurements of the cytosolic abundance of peroxisome proteins prior to import. Our recent study compared in vivo targeting, in silico targeting predictions and quantitative in vitro targeting peptide binding to PEX5 [32]. Although all three methods agreed well in their rank order of prediction, import in vivo was possible with sequences that were below the detection limit (Kd >100 μM) for binding to PEX5 in vitro. It is hard to imagine that either PEX5 or cargo proteins could exist at these concentrations in vivo, leading us to propose that import may not be a simple pre-equilibrium model where steady state concentrations of cargo and receptor determine import [32]. If downstream steps leading to import are rapid and irreversible, the effective concentration of receptor–cargo complex will be kept low, therefore driving the equilibrium towards formation of the receptor–cargo complex (Figure 1). Thus a protein that associates with the receptor may be ‘captured’ and committed to the import pathway even if it might otherwise rapidly dissociate from the receptor. The in vivo data demonstrate that the weak sequences are imported much more slowly than the strong ones, which suggests that only a small number of such molecules succeed in entering the import pathway at any one time [32]. Of course other factors may also come into play with native proteins such as auxiliary binding sequences, interactions with other cellular components such as chaperones, or in the case of peroxisomal proteins that assemble as oligomers in the cytosol, multivalent interactions leading to cooperativity of binding.
Figure 1. Models of PEX5:cargo interaction regulating import.
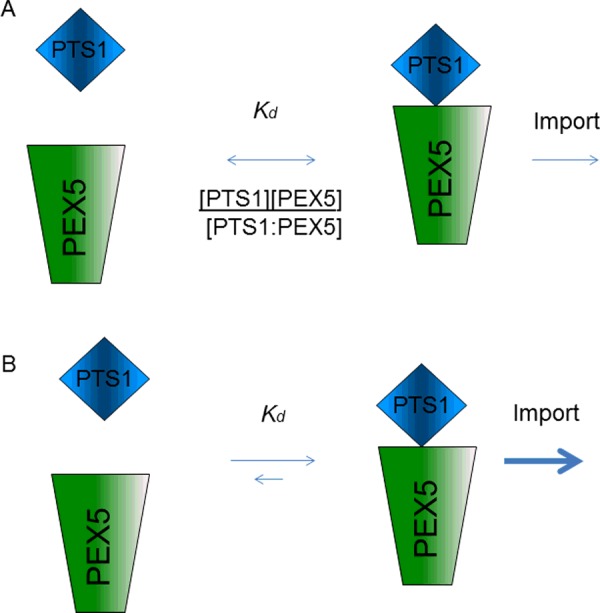
(A) In an equilibrium binding model the respective concentrations of PTS1 cargo protein [PTS1] and PEX5 receptor [PEX5] will determine the concentration of the receptor–cargo complex [PEX5:PTS1]. If PEX5:PTS1 must form for PTS1 cargo to be imported, any PTS1 cargo with Kd above the concentration of the receptor will not be imported. This model is not consistent with observed import of proteins in vivo which have Kd > 100 μM in vitro [32]. (B) If the PEX5:PTS1 complex is rapidly imported after formation, [PEX5:PTS1] in the cytosol will be kept low thus continually driving the equilibrium towards further formation of complex.
The role of the N terminus of PEX5 in protein import
The N-terminus of PEX5 is believed to be a natively unstructured domain [33] and contains a number of functionally significant motifs. In humans and plants PEX5 contains the binding site for PEX7, which is the import receptor for a second class of matrix proteins that carry the N-terminal PTS2 signal [34,35]. PEX5 also contains several lysines and a conserved cysteine residue that are targets for ubiquitination. This modification regulates receptor turnover and recycling [36].
PEX5 from all organisms has multiple repeats of a sequence WX3F/Y which bind to PEX14, the docking factor for PEX5 at the peroxisome membrane [37–39]. Human PEX5 also contains an LVXE/F motif which binds PEX14 but dissociates over 30 times more rapidly, and is essential for import [40]. The role of the multiple PEX14 binding motifs is still unclear, but may have an important role in translocation. For example models in which a single PEX14 binds and slides along the PEX5 N-terminus, or in which multiple molecules of PEX14 are recruited to form the import pore have been proposed [40,41].
Whether the N-terminal domain of PEX5 has a role in cargo binding is unclear. Some reports have indicated contacts between ‘atypical’ cargo proteins such as catalase and S. cerevisiae acyl CoA oxidase with PEX5 that lie outside of the TPR domain [42–44]. It has also been proposed that cargo binding causes a conformational change in PEX5 that releases the N-terminus for interaction with PEX14 [45]. The human PEX5 N526K mutant mimics the cargo bound state and therefore is a substrate for import [46]. It was further reported that the N- and C-terminal domains of PEX5 interact with one another in the absence of cargo [47]; however small angle X-ray scattering indicates that cargo-free PEX5 has an extended N terminal region that is free to bind PEX14 in a 1:6 ratio [48], and pull-down assays with recombinant proteins also show cargo-free interaction between PEX5 and PEX14 [49]. A lack of interaction of the N-terminal domain of PEX5 and the C-terminal TPR domain is also demonstrated by co-immunoprecipitation (Figure 2 and Supporting Information). Thus, whereas cargo binding clearly causes conformational changes within the PEX5 receptor, an increasing body of evidence disputes a model where cargo binding results in exposure of the N-terminus for interaction with PEX14.
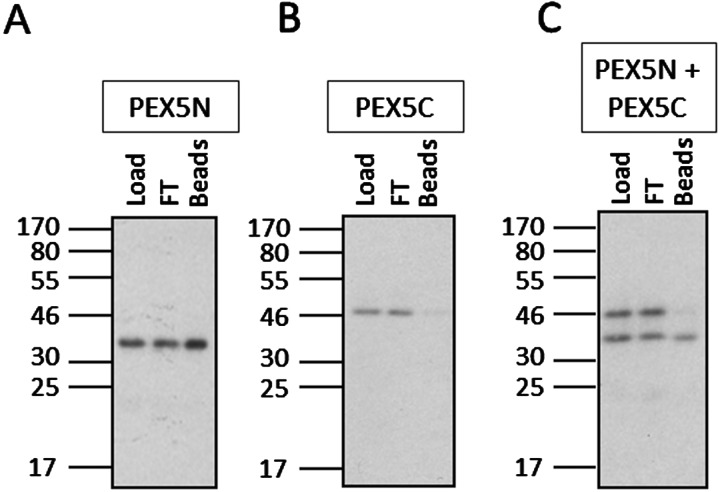
Figure 2. The N- and C-terminal domains of Arabidopsis thaliana PEX5 do not interact with one another in the absence of bound cargo.
Recombinant truncated PEX5 constructs, PEX5(1–304) (PEX5N) and PEX5(340–728) (PEX5C) both containing hexahistine tags were purified from Escherichia coli (see Supporting Information) and immunoprecipitated as follows. (A) PEX5N, α-PEX5N [69] positive control showing that PEX5N is immunoprecipitated. (B) PEX5C, α-PEX5N negative control showing that PEX5C is not immunoprecipitated. (C) PEX5N, PEX5C, α-PEX5N showing that PEX5N is immunoprecipitated but PEX5C is not co-immunoprecipitated, therefore the two domains of the protein do not interact. Proteins were incubated with antibody for 1 h before bound proteins were isolated from solution via protein A coupled beads. Load, sample of protein mixture; FT, sample of unbound proteins; Beads, immune isolated proteins.
Mechanisms of translocation
Peroxisomes are unusual among protein translocation systems in that they have the capacity to translocate folded, even oligomeric proteins [50,51]; however, recent evidence supports the notion that in many cases monomers may be the preferred clients of the import machinery [52] and indeed PEX5 may actively inhibit oligomerization of some proteins [43]. Nevertheless there are some clearly documented examples of proteins that lack a PTS ‘hitch hiking’ into peroxisomes via interaction with a protein partner that does contain a PTS [53].
The observations that peroxisomes transport folded proteins implies that the translocation machinery can accommodate a wide variety of sizes and shapes. Cargo-loaded PEX5 itself enters the peroxisomal membrane in an ATP independent manner with the C-terminal TPR domain protected from externally added protease [54]. As discussed in the previous section, the N-terminus of PEX5 binds multiple molecules of PEX14 and a purified complex containing only PEX5 and PEX14 can be reconstituted to form a gated ion conducting channel, which is opened by presentation of a PEX5:cargo complex [55]. How cargo is unloaded from PEX5 remains unresolved. The atypical cargo protein catalase is released from PEX5 by binding of PEX14 to the most C-terminal WX3F/Y repeat [43], but whether PEX14 can act as a general PTS1 cargo unloader is not clear. PEX14 binding to PEX5 could not displace a PTS1 model peptide [49], but as additional contacts between PEX5 and some cargo proteins have been reported this experimental set up may not be wholly representative of PEX5 loaded with a native protein. What is clear is that cargo unloading takes place before receptor ubiquitination [54], described in the next section. The retention within the translocation machinery of a PEX5 construct fused to a bulky C-terminal tag has also been invoked as evidence that cargo unloading is a prerequisite for receptor recycling [56].
Export coupled import
Protein import into peroxisomes is a cyclical process. Cargo binds the receptor in the cytosol, docks at the peroxisome membrane, is unloaded and the receptor is recycled for further rounds of import [57]. Receptor recycling requires the addition of ubiquitin to a conserved cysteine residue near the N-terminus of PEX5 by a peroxisome localized E3 ligase complex [58] followed by export of the ubiquitinated receptor by the AAA ATPase complex [59]. These are the two ATP-dependent steps of the overall import process. Many years ago experiments with peroxisome in vitro import systems pointed to an ATP dependence of protein import into peroxisomes [60,61], which subsequently appeared inconsistent with results demonstrating that import is driven solely by thermodynamically favourable binding events [54,62]. Better understanding of the receptor export cycle has led to an appreciation that the import and export processes must be coupled, and that the ATP-dependent export process is necessary to remove PEX5 from the translocation pore [63] to facilitate further rounds of import. Mutation of the cysteine required for ubiquitination of S. cerevisiae PEX5 inhibits receptor export and blocks subsequent import which is consistent with this proposal [64]. A recently developed methodology in our laboratory that allows interrogation of protein–protein interactions during in vitro import of proteins into peroxisomes has also provided evidence consistent with export-coupled import [65]. In this method a bait protein is covalently labelled with a biotinylated, photo-activatable and thiol-cleavable crosslinker (sulfo-SBED) (Figure 3A). Upon incubation of a sulfo-SBED derivatized PEX5 construct with peroxisomes under conditions compatible with protein import, subsequent UV irradiation forms crosslinks to adjacent proteins. Reduction of the disulfide bond within the crosslinker transfers the biotin label to the prey protein(s) allowing for detection. Using this method we were able to demonstrate specific label transfer from an N-terminally truncated PEX5 construct (which would not be capable of recycling) to a heterogeneous complex of proteins in the peroxisome membrane. This labelling was blocked by inclusion of competing PEX14 or by the omission of ATP (Figure 3B). The peroxisomes are isolated under conditions that would be expected to promote association of endogenous PEX5 with the import pore, namely low temperature and without ATP [66], thus the import pore is probably fully occupied with endogenous PEX5 and prevents further insertion of labelled PEX5. In the presence of ATP endogenous PEX5 can be released from the pore, allowing insertion of labelled PEX5 and label transfer to the peroxisomal translocation machinery [65] (Figure 3C). The concept of coupling import and export has also been examined using a modelling approach, which supports the notion that export of one PEX5 molecule requires its replacement by a second incoming molecule [67].
Figure 3. Label transfer approach to detect interactions with the import machinery.
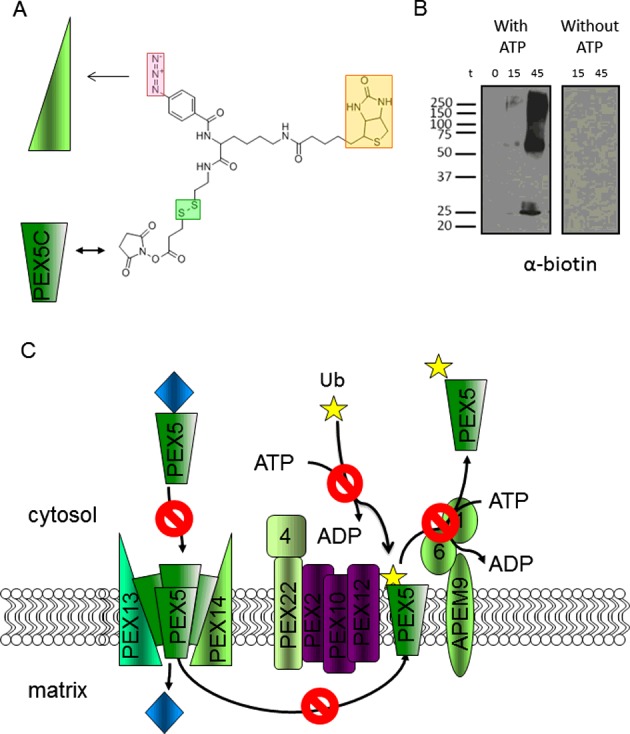
(A) Sulfo-SBED is a trifunctional reagent with an amine reactive sulfo-NHS group which was used for coupling to PEX5C. Sulfo-SBED contains an aryl azide (pink box) which upon UV irradiation covalently links to a nearby ‘prey’ protein (green triangle), a biotin moiety for detection (yellow) and a cleavable disulfide (green) to allow release of the bait and transfer of the biotin label to the prey. (B) Western blot of carbonate extracted peroxisome membranes probed with streptavidin-HRP after an import reaction carried out in the presence or absence of ATP followed by UV irradiation. The time import was allowed to proceed for prior to UV irradiation is indicated above lanes in minutes. (C) Schematic diagram of the import machinery illustrating how, in the absence of ATP, ubiquitination of PEX5 could not take place. This would prevent PEX5 export and hence lead to its accumulation in the peroxisome membrane, thus preventing cargo-loaded PEX5 from inserting, even though the insertion step itself is not ATP-dependent. Blue diamond=PTS1 cargo. PEX14, PEX13; components of the docking apparatus on the peroxisome membrane. PEX22/PEX4; ubiquitin E2 ubiquitin ligase. PEX2/PEX10/PEX12; E3 ligase complex. Yellow star; ubiquitin. APEM9/PEX1/PEX6 receptor export complex (adapted from [65]: Bhogal, M.S., Lanyon-Hogg, T., Johnston, K.A., Warriner, S.L. and Baker, A. (2016) Covalent label transfer between peroxisomal importomer components reveals export-driven import interactions. J. Biol. Chem. 291, 2460–2468).
Conclusions and future directions
Recent years have seen considerable progress in understanding of the mechanism of peroxisomal protein import. In particular, the application of biochemical approaches has been fundamental in identifying and testing components and breaking down the import process into discrete steps. As understanding of individual steps within the peroxisomal import cycle has grown, it has become clear that a range of factors influence many of these processes. For example, ancillary, non-PTS1 interactions with PEX5 influence PTS1 cargo import, PEX14 displays the ability to provide both docking and unloading functions, and import steps have been linked to concomitant export steps in the translocation cycle. The interplay of these factors adds additional layers of complexity to the interpretation of experimental results, and requires increasingly intricate experimental design to unravel such effects. The study of the peroxisomal import machinery in a holistic manner therefore presents an unprecedented challenge; however, the continuing development of new analytical tools to build on current understanding offers increased promise in this endeavour. As progress in understanding the structures of component proteins at an atomic level provides new insights into molecular recognition, the logical development is into the structural study of larger complexes. New label transfer methods to detect interaction with the translocation machinery also afford the opportunity to quantify inhibition of such interactions by various factors, through Western blotting followed by densitometric analysis to generate IC50 curves. Application of computational approaches also has the potential to bring new insights into mechanism, but they require quantitative experimental data for model building and validation. It must also be remembered that all these biochemical processes take place in a complex cellular environment where activities are regulated in response to intrinsic and extrinsic signals, so approaches such as optical control over protein import are potentially interesting [68]. New methods and technologies will also drive further innovation for example developments in super resolution imaging that may one day allow us to ‘see’ the import process.
Acknowledgments
We thank Dr Makoto Hayashi for the kind gift of the α-PEX5N antibody, Divya Ramkumar for cloning of the PEX5N construct, and Laura Cross and Dr Rupesh Paudyal for critical reading of the manuscript.
Abbreviations
- AGT
alanine glyoxylate aminotransferase
- ITC
isothermal titration calorimetry
- PTS1
peroxisome targeting signal 1
- SCP2
sterol carrier protein 2
- TPR
tetratricopeptide repeat
Footnotes
Membrane Proteins From A to Z: Held at University of Leeds, U.K., 16–17 December 2015
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/D000904/1]; and the Leverhulme Trust [grant number RPG-2012-516].
References
- 1.Lazarow P.B., Fujiki Y. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 2.Lanyon-Hogg T., Warriner S.L., Baker A. Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biol. Cell. 2010;102:245–263. doi: 10.1042/BC20090159. [DOI] [PubMed] [Google Scholar]
- 3.Kim P.K., Hettema E.H. Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 2015;427:1176–1190. doi: 10.1016/j.jmb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fagarasanu A., Fagarasanu M., Rachubinski R.A. Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 5.Hu J.P. Molecular basis of peroxisome division and proliferation in plants. Int. Rev. Cell Mol. Biol. 2010;279:79–99. doi: 10.1016/S1937-6448(10)79003-1. [DOI] [PubMed] [Google Scholar]
- 6.Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., Zolman B.K. Plant peroxisomes: biogenesis and function. Plant Cell. 2012;24:2279–2303. doi: 10.1105/tpc.112.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waterham H.R., Wanders R.J.A. Metabolic functions and biogenesis of peroxisomes in health and disease. Biochim. Biophys. Acta. 2012;1822:1325–1325. doi: 10.1016/j.bbadis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Goto-Yamada S., Mano S., Yamada K., Oikawa K., Hosokawa Y., Hara-Nishimura I., Nishimura M. Dynamics of the light-dependent transition of plant peroxisomes. Plant Cell Physiol. 2015;56:1264–1271. doi: 10.1093/pcp/pcv081. [DOI] [PubMed] [Google Scholar]
- 9.Waterham H.R., Ebberink M.S. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta. 2012;1822:1430–1441. doi: 10.1016/j.bbadis.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Baker A., Sparkes I.A., Brown L.-A., O'Leary-Steele C., Warriner S.L. Peroxisome biogenesis and positioning. Biochem. Soc. Trans. 2010;38:807–816. doi: 10.1042/BST0380807. [DOI] [PubMed] [Google Scholar]
- 11.Gould S.J., Keller G.A., Hosken N., Wilkinson J., Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J. Cell. Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould S.J., Keller G.A., Schneider M., Howell S.H., Garrard L.J., Goodman J.M., Distel B., Tabak H., Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terlecky S.R., Nuttley W.M., McCollum D., Sock E., Subramani S. The Pichia-Pastoris peroxisomal protein Pas8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fransen M., Terlecky S.R., Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kragler F., Lametschwandtner G., Christmann J., Hartig A., Harada J.J. Identification and analysis of the plant peroxisomal targeting signal 1 receptor NtPEX5. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13336–13341. doi: 10.1073/pnas.95.22.13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brocard C., Kragler F., Simon M.M., Schuster T., Hartig A. The tetratricopeptide repeat-domain of the Pas10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-Skl. Biochem. Biophys. Res. Commun. 1994;204:1016–1022. doi: 10.1006/bbrc.1994.2564. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhary G., Kataya A.R.A., Lingner T., Reumann S. Non-canonical peroxisome targeting signals: identification of novel PTS1 tripeptides and characterization of enhancer elements by computational permutation analysis. BMC Plant Biol. 2012;12:142. doi: 10.1186/1471-2229-12-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lametschwandtner G., Brocard C., Fransen M., Van Veldhoven P., Berger J., Hartig A. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J. Biol. Chem. 1998;273:33635–33643. doi: 10.1074/jbc.273.50.33635. [DOI] [PubMed] [Google Scholar]
- 19.Brocard C., Hartig A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta. 2006;1763:1565. doi: 10.1016/j.bbamcr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Lingner T., Kataya A.R., Antonicelli G.E., Benichou A., Nilssen K., Chen X.Y., Siemsen T., Morgenstern B., Meinicke P., Reumann S. Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell. 2011;23:1556–1572. doi: 10.1105/tpc.111.084095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Andrea L.D., Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Gatto G.J., Geisbrecht B.V., Gould S.J., Berg J.M. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 23.Sampathkumar P., Roach C., Michels P.A.M., Hol W.G.J. Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5. J. Mol. Biol. 2008;381:867–880. doi: 10.1016/j.jmb.2008.05.089. [DOI] [PubMed] [Google Scholar]
- 24.Stanley W.A., Filipp F.V., Kursula P., Schueller N., Erdmann R., Schliebs W., Sattler M., Wilmanns M. Recognition of a functional peroxisome type 1 target by the dynamic import receptor Pex5p. Mol. Cell. 2006;24:653–663. doi: 10.1016/j.molcel.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fodor K., Wolf J., Erdmann R., Schliebs W., Wilmanns M. Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1. PLoS Biol. 2012;10:e1001309. doi: 10.1371/journal.pbio.1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesa-Torres N., Tomic N., Albert A., Salido E., Pey A.L. Molecular recognition of PTS-1 cargo proteins by Pex5p: implications for protein mistargeting in primary hyperoxaluria. Biomolecules. 2015;5:121–141. doi: 10.3390/biom5010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fodor K., Wolf J., Reglinski K., Passon D.M., Lou Y., Schliebs W., Erdmann R., Wilmanns M. Ligand-induced compaction of the PEX5 receptor-binding cavity impacts protein import efficiency into peroxisomes. Traffic. 2015;16:85–98. doi: 10.1111/tra.12238. [DOI] [PubMed] [Google Scholar]
- 28.Hagen S., Drepper F., Fischer S., Fodor K., Passon D., Platta H.W., Zenn M., Schliebs W., Girzalsky W., Wilmanns M., et al. Structural insights into cargo recognition by the yeast PTS1 receptor. J. Biol. Chem. 2015;290:26610–26626. doi: 10.1074/jbc.M115.657973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh D., Berg J.M. A proteome-wide perspective on peroxisome targeting signal 1 (PTS1)-Pex5p affinities. J. Am. Chem. Soc. 2010;132:3973–3979. doi: 10.1021/ja9109049. [DOI] [PubMed] [Google Scholar]
- 30.Gatto G.J., Maynard E.L., Guerrerio A.L., Geisbrecht B.V., Gould S.J., Berg J.M. Correlating structure and affinity for PEX5 : PTS1 complexes. Biochemistry. 2003;42:1660–1666. doi: 10.1021/bi027034z. [DOI] [PubMed] [Google Scholar]
- 31.Maynard E.L., Gatto G.J., Berg J.M. Pex5p binding affinities for canonical and noncanonical PTS1 peptides. Proteins. 2004;55:856–861. doi: 10.1002/prot.20112. [DOI] [PubMed] [Google Scholar]
- 32.Skoulding N.S., Chowdhary G., Deus M.J., Baker A., Reumann S., Warriner S.L. Experimental validation of plant peroxisomal targeting prediction algorithms by systematic comparison of in vivo import efficiency and in vitro PTS1 binding affinity. J. Mol. Biol. 2015;427:1085–1101. doi: 10.1016/j.jmb.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Carvalho A.F., Costa-Rodrigues J., Correia I., Pessoa J.C., Faria T.Q., Martins C.L., Fransen M., Sa-Miranda C., Azevedo J. E. The N-terminal half of the peroxisomal cycling receptor Pex5p is a natively unfolded domain. J. Mol. Biol. 2006;356:864–875. doi: 10.1016/j.jmb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Otera H., Harano T., Honsho M., Ghaedi K., Mukai S., Tanaka A., Kawai A., Shimizu N., Fujiki Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p center dot PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 2000;275:21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- 35.Woodward A.W., Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell. 2005;16:573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francisco T., Rodrigues T.A., Pinto M.P., Carvalho A.F., Azevedo J.E., Grou C.P. Ubiquitin in the peroxisomal protein import pathway. Biochimie. 2014;98C:29–35. doi: 10.1016/j.biochi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Schliebs W., Saidowsky J., Agianian B., Dodt G., Herberg F.W., Kunau W.H. Recombinant human peroxisomal targeting signal receptor PEX5–structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- 38.Neufeld C., Filipp F.V., Simon B., Neuhaus A., Schueller N., David C., Kooshapur H., Madl T., Erdmann R., Schliebs W., et al. Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19. EMBO J. 2009;28:745–754. doi: 10.1038/emboj.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su J.R., Takeda K., Tamura S., Fujiki Y., Miki K. Crystal structure of the conserved N-terminal domain of the peroxisomal matrix protein import receptor, Pex14p. Proc. Natl. Acad. Sci. U.S.A. 2009;106:417–421. doi: 10.1073/pnas.0808681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuhaus A., Kooshapur H., Wolf J., Meyer N.H., Madl T., Saidowsky J., Hambruch E., Lazam A., Jung M., Sattler M., et al. A novel Pex14 protein-interacting site of human Pex5 is critical for matrix protein import into peroxisomes. J. Biol. Chem. 2014;289:437–448. doi: 10.1074/jbc.M113.499707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emmanouilidis L., Gopalswamy M., Passon D.M., Wilmanns M., Sattler M. Structural biology of the import pathways of peroxisomal matrix proteins. Biochim. Biophys. Acta. 2016;1863:804–813. doi: 10.1016/j.bbamcr.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 42.Oshima Y., Kamigaki A., Nakamori C., Mano S., Hayashi M., Nishimura M., Esaka M. Plant catalase is imported into peroxisomes by pex5p but is distinct from typical PTS1 import. Plant Cell Physiol. 2008;49:671–677. doi: 10.1093/pcp/pcn038. [DOI] [PubMed] [Google Scholar]
- 43.Freitas M.O., Francisco T., Rodrigues T.A., Alencastre I.S., Pinto M.P., Grou C.P., Carvalho A.F., Fransen M., Sa-Miranda C., Azevedo J.E. PEX5 protein binds monomeric catalase blocking its tetrarnerization and releases it upon binding the N-terminal domain of PEX14. J. Biol. Chem. 2011;286:40509–40519. doi: 10.1074/jbc.M111.287201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein A.T.J., van den Berg M., Bottger G., Tabak H.F., Distel B. Saccharomyces cerevisiae Acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 2002;277:25011–25019. doi: 10.1074/jbc.M203254200. [DOI] [PubMed] [Google Scholar]
- 45.Gouveia A.M., Guimaraes C.P., Oliveira M.E., Sa-Miranda C., Azevedo J.E. Insertion of Pex5p into the peroxisomal membrane is cargo protein-dependent. J. Biol. Chem. 2003;278:4389–4392. doi: 10.1074/jbc.C200650200. [DOI] [PubMed] [Google Scholar]
- 46.Carvalho A.F., Grou C.P., Pinto M.P., Alencastre I.S., Costa-Rodrigues J., Fransen M., Sa-Miranda C., Azevedo J.E. Functional characterization of two missense mutations in Pex5p-C11S and N526K. Biochim. Biophys. Acta. 2007;1773:1141–1148. doi: 10.1016/j.bbamcr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Harano T., Nose S., Uezu R., Shimizu N., Fujiki Y. Hsp70 regulates the interaction between the peroxisome targeting signal type 1 (PTS1)-receptor Pex5p and PTS1. Biochem. J. 2001;357:157–165. doi: 10.1042/bj3570157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiozawa K., Konarev P.V., Neufeld C., Wilmanns M., Svergun D.I. Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering. J. Biol. Chem. 2009;284:25334–25342. doi: 10.1074/jbc.M109.002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanyon-Hogg T., Hooper J., Gunn S., Warriner S.L., Baker A. PEX14 binding to Arabidopsis PEX5 has differential effects on PTS1 and PTS2 cargo occupancy of the receptor. FEBS Lett. 2014;588:2223–2229. doi: 10.1016/j.febslet.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glover J.R., Andrews D.W., Rachubinski R.A. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNew J.A., Goodman H.M. An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freitas M.O., Francisco T., Rodrigues T.A., Lismont C., Domingues P., Pinto M.P., Grou C.P., Fransen M., Avezevedo J.E. Monomeric proteins are the preferential substrates of the peroxisomal protein import machinery. FEBS J. 2015;282:402–402. doi: 10.1098/rsob.140236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dias A.F., Francisco T., Rodrigues T.A., Grou C.P., Azevedo J.E. The first minutes in the life of a peroxisomal matrix protein. Biochim. Biophys. Acta. 2016;1863:814–820. doi: 10.1016/j.bbamcr.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 54.Francisco T., Rodrigues T.A., Freitas M.O., Grou C.P., Carvalho A.F., Sa-Miranda C., Pinto M.P., Azevedo J.E. A cargo-centered perspective on the PEX5 receptor-mediated peroxisomal protein import pathway. J. Biol. Chem. 2013;288:29151–29159. doi: 10.1074/jbc.M113.487140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meinecke M., Cizmowski C., Schliebs W., Kruger V., Beck S., Wagner R., Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 2010;12:273. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 56.Nordgren M., Francisco T., Lismont C., Hennebel L., Brees C., Wang B., Van Veldhoven P.P., Azevedo J.E., Fransen M. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy. 2015;11:1326–1340. doi: 10.1080/15548627.2015.1061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cross L.L., Ebeed H.T., Baker A. Peroxisome biogenesis, protein targeting mechanisms and PEX gene functions in plants. Biochim. Biophys. Acta. 2016;1863:850–862. doi: 10.1016/j.bbamcr.2015.09.027. [DOI] [PubMed] [Google Scholar]
- 58.Platta H.W., Brinkmeier R., Reidick C., Galiani S., Clausen M.P., Eggeling C. Regulation of peroxisomal matrix protein import by ubiquitination. Biochim. Biophys. Acta. 2016;1863:838–849. doi: 10.1016/j.bbamcr.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 59.Ciniawsky S., Grimm I., Saffian D., Girzalsky W., Erdmann R., Wendler P. Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat. Commun. 2015;6:7331–7331. doi: 10.1038/ncomms8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Behari R., Baker A. The carboxyl terminus of isocitrate lyase is not essential for import into glyoxysomes in an in vitro system. J. Biol. Chem. 1993;268:7315–7322. [PubMed] [Google Scholar]
- 61.Imanaka T., Small G.M., Lazarow P.B. Translocation of acyl-CoA oxidase into peroxisomes requires ATP hydrolysis but not a membrane-potential. J. Cell Biol. 1987;105:2915–2922. doi: 10.1083/jcb.105.6.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gouveia A.M., Guimaraes C.P., Oliveira M.E., Reguenga C., Sa-Miranda C., Azevedo J.E. Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J. Biol. Chem. 2003;278:226–232. doi: 10.1074/jbc.M209498200. [DOI] [PubMed] [Google Scholar]
- 63.Schliebs W., Girzalsky W., Erdmann R. Peroxisomal protein import and ERAD: variations on a common theme. Nat. Rev. Mol. Cell. Biol. 2010;11:885–890. doi: 10.1038/nrm3008. [DOI] [PubMed] [Google Scholar]
- 64.Schwartzkopff B., Platta H.W., Hasan S., Girzalsky W., Erdmann R. Cysteine-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation. Biosci. Rep. 2015;35:e00215. doi: 10.1042/BSR20150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhogal M.S., Lanyon-Hogg T., Johnston K.A., Warriner S.L., Baker A. Covalent label transfer between peroxisomal importomer components reveals export-driven import interactions. J. Biol. Chem. 2016;291:2460–2468. doi: 10.1074/jbc.M115.686501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dodt G., Gould S. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown A.I., Kim P.K., Rutenberg A.D. PEX5 and ubiquitin dynamics on mammalian peroxisome membranes. PLoS Comput. Biol. 2014;10:e1003426–e1003426. doi: 10.1371/journal.pcbi.1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiltoir J.I., Strickland D., Glotzer M., Tucker C.L. Optical control of peroxisomal trafficking. ACS Synth. Biol. 2015 doi: 10.1021/acssynbio.5b00144. doi: 10.1021/acssynbio.5b00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nito K., Hayashi M., Nishimura M. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:355–366. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]