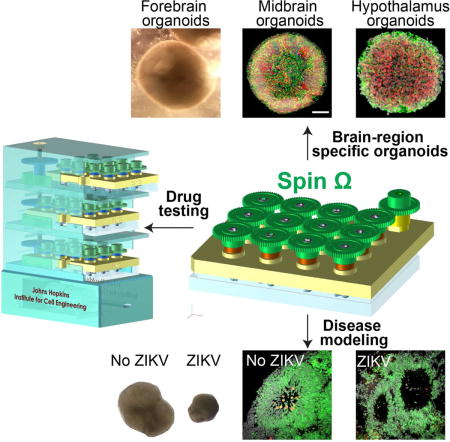
SUMMARY
Cerebral organoids, three-dimensional cultures that model organogenesis, provide a new platform to investigate human brain development. High cost, variability and tissue heterogeneity limit their broad applications. Here we developed a miniaturized spinning bioreactor (SpinΩ) to generate forebrain-specific organoids from human iPSCs. These organoids recapitulate key features of human cortical development, including progenitor zone organization, neurogenesis, gene expression, and notably, a distinct human-specific outer radial glia cell layer. We also developed protocols for midbrain and hypothalamic organoids. Finally, we employed the forebrain organoid platform to model Zika virus (ZIKV) exposure. Quantitative analyses revealed preferential, productive infection of neural progenitors with either African or Asian ZIKV strains. ZIKV infection leads to increased cell death and reduced proliferation, resulting in decreased neuronal cell layer volume resembling microcephaly. Together, our brain region-specific organoids and SpinΩ provide an accessible and versatile platform for modeling human brain development and disease, and for compound testing including potential ZIKV antiviral drugs.
eTOC
Zika virus preferentially infects neural progenitors in early stage cortical organoids, generated using cost-effective miniaturized spinning bioreactors, resulting in suppressed proliferation, increased cell death, and macroscopic features resembling microcephaly.
INTRODUCTION
Human induced pluripotent stem cells (iPSCs) can generate virtually any cell type in the body to model human development and disease, screen for therapeutic drugs, and develop cell replacement therapies. Traditional monolayer cultures allow for external control of targeted differentiation of human iPSCs to produce more uniform cell populations; however, these cultures lack 3D cell assembly properties that define endogenous biological systems. Structures resembling whole developing organs, named organoids, have recently been generated via 3D cultures and include intestinal, kidney, retinal, and cerebral organoids (Lancaster and Knoblich, 2014; Yin et al., 2016). Organoid technology evolved from embryoid body cultures, which are 3D aggregates of stem cells that self-organize to develop disparate tissues in vitro, similar to teratoma formation in vivo. Organoids provide a unique opportunity to model human organogenesis, which is not accessible to experimentation. An immediate application of organoid technology would be to address the current global public health emergency concerning a suspected link between Zika virus (ZIKV) and microcephaly, a neurodevelopmental disorder, by modeling human brain development.
One recent advance in cerebral organoid technology was the adoption of a spinning bioreactor to facilitate nutrient and oxygen absorption, which enables formation of longer neuroepithelium-like zones and supports growth of large, complex organoids that more closely resemble the developing human brain than had been achieved by previous approaches (Lancaster et al., 2013). Derived from an early NASA-designed rotating wall vessel bioreactor to simulate microgravity, this technology potentially offers two additional benefits: low fluid shear stress to promote cell-cell interactions and induction of differentiation (Goodwin et al., 1993); and randomized gravitational vectors that affect intracellular signal transduction and gene expression (Jessup et al., 1993). This and other human cerebral organoid technologies (Kadoshima et al., 2013; Lancaster et al., 2013; Mariani et al., 2015; Muguruma et al., 2015; Pasca et al., 2015) have generated much excitement for using organoids to model human brain development and disorders.
Despite the promise of these pioneering organoid technologies, there are several major challenges. First, available spinning bioreactors require a large volume of medium and incubator space (Figure S1Aa). With frequent media changes over several months of culturing, the system is cost-prohibitive for most laboratories and precludes scalability, use of growth factors, or chemical screening. It also presents a roadblock for testing different conditions to optimize protocols. Second, the current cerebral organoid methodology (“intrinsic protocol”) is based on cell self-assembly without external control, and thus each organoid is typically comprised of diverse cell types found in forebrain, hindbrain, and retina (Lancaster et al., 2013). Large sample to sample variability associated with current methods complicates quantitative analyses and limits applicability. Third, key features of human brain development have yet to be robustly recapitulated in cerebral organoids. For example, unlike rodents, the embryonic human cerebral cortex contains an abundant population of specialized outer radial glia cells (oRGCs) in the outer subventricular zone (oSVZ), the cellular population considered pivotal to the evolutionary increase in human cortex size and complexity (Lui et al., 2011). Current cerebral organoids contain only sparse progenitors that have morphological characteristics of oRGCs and none have exhibited a well-developed oSVZ layer. Taken together, there is a critical need to develop an organoid platform with reduced cost, higher throughput, increased reproducibility, and that better resembles critical aspects of human cortical development.
To address these challenges, we engineered a miniaturized spinning bioreactor using 3D design and printing technology, and developed a protocol to generate forebrain-specific organoids from human iPSCs, which recapitulate human embryonic cortical development in a reproducible and quantifiable manner. We also developed protocols for midbrain and hypothalamic organoids. For proof-of-principle applications of our platform, we performed chemical compound testing and modeled ZIKV exposure. Our versatile, simple to use, cost-effective, and reproducible brain region-specific organoid platform provides accessible and affordable technology to a broad scientific community for modeling human organogenesis, human disorders, and compound testing.
RESULTS
A Miniaturized Spinning Bioreactor to Optimize Organoid Cultures
To reduce cost of generating organoids under different conditions, we attempted to miniaturize the large spinning flask. Nonlinear fluid dynamics precluded simply scaling down the system. Instead, we engineered a multi-well spinning device to fit a standard 12-well tissue culture plate. Above the cover, spinning shafts are attached to a set of 13 interconnecting gears, driven by a single electric motor (Figure 1A). We used Computer-Aided Design software to design and 3D print each component. We assembled prototypes to optimize designs that sustain organoids of varying sizes in suspension under moderate spinning speed and prevent aggregation at the center of each well. After multiple rounds of systematic optimization of individual components, including number, shape, size and angle of leafs, and diameter, length and shape of shafts, we arrived at SpinΩ, a miniaturized spinning bioreactor unit that requires as little as 2 ml of media per well, a 50-fold reduction in media consumption and drastically reduced incubator space (Figure S1Ab). We further designed a modular stackable version with insertable cassettes driven by one common motor (Figure S1Ac). The miniaturized spinning bioreactor permits comparisons of a large number of conditions in parallel for protocol optimization.
Figure 1. SpinΩ Bioreactor-based Forebrain Organoid Culture System.
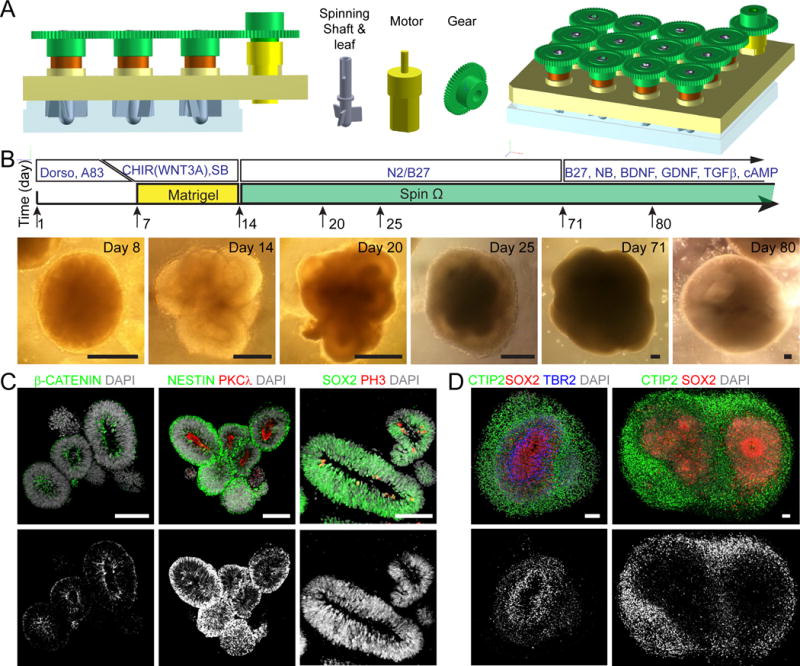
(A) Computer-Aided-Design drawings of 12-well version SpinΩ bioreactor and individual parts.
(B) Schematic diagram of forebrain organoid protocol and sample phase images at different stages. Scale bars: 200 μm.
(C–D) Immunostaining of forebrain organoids at days 14, 63 and 84. Scale bars: 100 μm.
To reduce tissue heterogeneity, we pre-patterned embryoid bodies to the fate of a specific brain region. We first treated human iPSCs with dual SMAD inhibitors (dorsomorphin and A-83) for 7 days and then embedded embryoid bodies in Matrigel for another 7 days, followed by Matrigel removal and spinning in SpinΩ (Figure 1B). Compared to the “intrinsic protocol”, we could reliably generate organoids from multiple iPSC lines with reduced heterogeneity in organoid shape and size (Figure S1B–C). However, there was significant cell death within organoids as shown by activated Caspase-3 (CAS3) immunostaining (Figure S1D–E and Table S1). We then tested combinations of different signaling molecules for various durations. We found that treatment with three factors, GSK-3β inhibitor CHIR99021, recombinant WNT3A protein, and SMAD inhibitor SB-431542, during the Matrigel stage drastically reduced the number of CAS3+ cells at day 14 (Figure S1D–E). Later we determined that the WNT3A contribution was minimal, likely because WNT3A and CHIR99021 activate the same downstream signaling pathway (Figure S1D–F). At day 14, well-defined polarized neuroepithelium-like structures resembled neural tubes, with a nearly pure population of NESTIN+SOX2+ NPCs, and expression of adherent junction markers (β-CATENIN and PKCλ) and proliferation marker phospho-Histone H3 (PH3) near the ventricular surface (Figure 1C and S1F). Notably, individual neuroepithelium-like structures were consistently much larger than those generated without treatment of these factors (Figure S1D). Upon spinning in SpinΩ, organoids developed into multi-layer stratified structures, composed of SOX2+ NPCs, TBR2+ intermediate progenitor cells (IPCs) and CTIP2+ neurons (Figure 1D). With a small volume, it became affordable to supplement media with growth factors at later stages (Figure 1B).
In comparison, we maintained organoids in stationary cultures after day 14. At day 42, there was substantial cell death in the interior (Figure S1G). Ventricular structures were largely absent; instead, extensive neurogenesis without defined organization was observed (Figure S1H). We also cultured forebrain organoids using orbital shakers under a similar rotation speed as spinning in SpinΩ. At day 42, organoids showed substantial cell death in the neuronal layer despite retaining defined ventricular structures (Figure S1I). These results suggest that spinning cultures enhance cell viability and promote maintenance of the stem cell niche, at least for forebrain organoids generated using our protocol. The miniaturized spinning bioreactor platform opens doors for cost-effective generation of organoids and provides accessible and affordable organoid technology to a broader scientific community.
Organoids with a Forebrain Identity and Increased Homogeneity
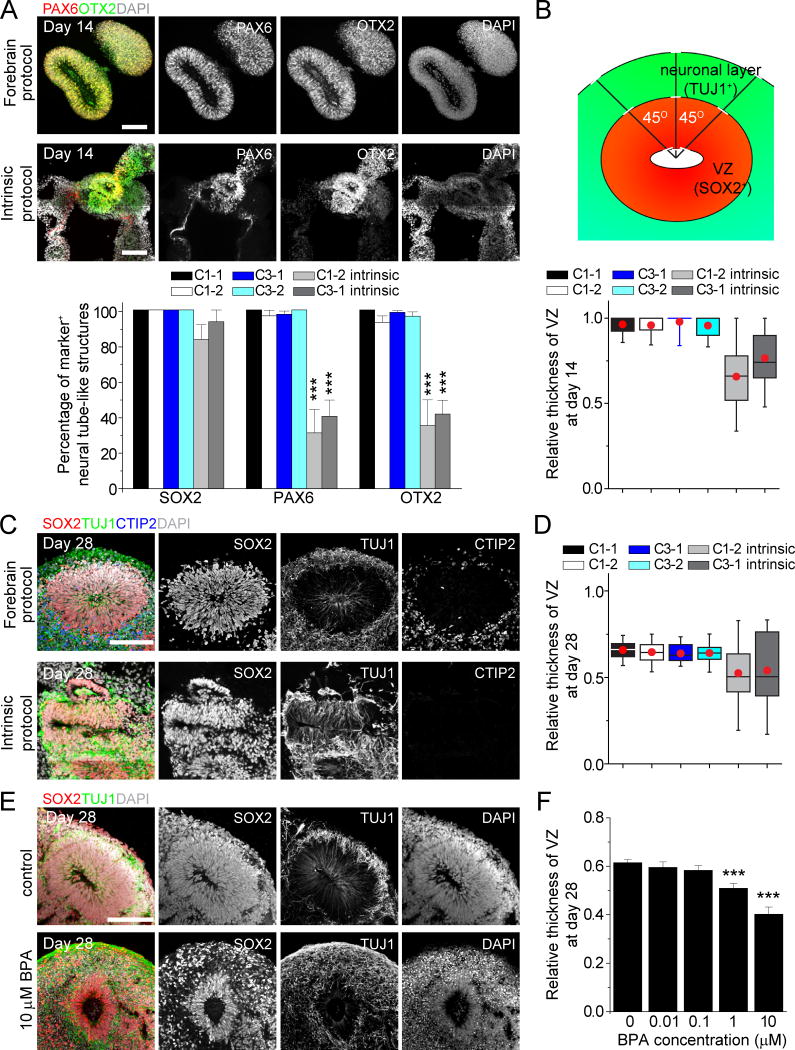
We next performed detailed characterizations of early stage forebrain organoids (Figure 1B). At day 14, immunohistological analysis showed almost exclusive expression of forebrain-specific progenitor markers, including PAX6, OTX2 and FOXG1, with minimal expression of markers for other brain regions tested (Figure 2A and S2A). We obtained similar results with multiple iPSC lines and with different clones (Figure 2A and S2A–B). Consistent with previous findings, cerebral organoids generated in large spinning flasks using the “intrinsic protocol” exhibited diverse brain region identities with less than 50% of rosettes expressing PAX6 or OTX2 (Figure 2A and S2A–B).
Figure 2. Homogeneity of Early Stage Forebrain Organoids and Effects of BPA.
(A) Sample images and quantification among multiple iPSC cell lines and clones for immunostaining of organoids at day 14. Scale bars: 100 μm. Values represent mean ± SEM (42–128 and 11- 30 total neural tube structures from at least 10 organoids each for the forebrain protocol and “intrinsic protocol”, respectively; ***P < 0.005, Student’s t-test).
(B) Schematic drawing for SOX2+ ventricular zone (VZ) and TUJ1+ neuronal layer measurement in cortical structures (top panel) and box plot for relative VZ thickness in day 14 organoids. For each cortical structure, three measurements were taken at 45 degree angles to obtain the mean value. Relative VZ thickness is the ratio of VZ thickness to total thickness from ventricular surface to pial surface. The red dot indicates mean; upper and lower error bars in each box plot represent the top whisker (maximum value) and bottom whisker (minimum value), respectively (n = 30 and 15 cortical structures from at least 10 organoids each for the forebrain and “intrinsic protocol”, respectively).
(C–D) Sample images of immunostaining of organoids at day 28 (C; Scale bars: 100 μm) and box plot for relative VZ thickness (D). Similar to (B) (n = 20 cortical structures from at least 10 organoids each).
(E–F) Effect of BPA treatment of forebrain organoids from day 14 to 28. Shown are sample images of immunostaining of control and BPA-treated forebrain organoids (E) and quantification of relative VZ thickness (F) at day 28. Scale bar: 100 μm. Values represent mean ± SEM. (n = 21 cortical structures from at least 10 organoids; ***P < 0.0005, Student’s t-test).
Also see Figure S2.
We further assessed the temporal consistency of neuronal differentiation by quantifying the relative thickness of SOX2+ ventricular zone-like (VZ) layer and TUJ1+ neuronal layer between apical and basal surfaces at specific time points (Figure 2B). At day 14, organoids generated using the “intrinsic protocol” exhibited varying degrees of neurogenesis with mixed cell types, whereas very few TUJ1+ neurons were detected in forebrain organoids (Figure S2A). As a result, our protocol produced organoids with nearly all cells organized in the VZ layer at this stage (Figure 2B). By day 28, we observed a consistent ratio between SOX2+ progenitor layer and TUJ1+/CTIP2+ neuronal layer in forebrain organoids, compared to the large variability using the “intrinsic protocol” (Figure 2C–D).
The apparent homogeneity of forebrain organoids, small volume per condition and multi-well format of SpinΩ comprise a platform that is amenable to chemical compound testing. As a proof-of-principle, we tested the effect of Bisphenol A (BPA), which is commonly found in household plastic products and known to affect rodent neural development (Kundakovic et al., 2013). Treatment of forebrain organoids from day 14 to 28 with BPA led to a dose-dependent decrease in the relative VZ thickness at day 28 (Figure 2E–F). With acute treatment of higher BPA concentrations for 24 hours and then pulse-labeling proliferating cells with EdU (Figure S2C), quantitative analysis showed decreased density of EdU+ or PH3+ NPCs (Figure S2D–E), indicating that reduced NPC proliferation contributes to decreased relative VZ thickness.
Multiple Progenitor Zones Recapitulating Human Embryonic Cortical Development
To characterize developmental dynamics, we systematically performed immunohistochemical analyses of day 28, 56 and 84 organoids. We observed well-defined VZ-like structures with packed SOX2+ NPCs near the lumen at all three time points (Figure 3A–C). At day 28, a layer containing a mixture of TBR2+ IPCs and CTIP2+ neurons formed above the VZ, reminiscent of the preplate (PP) in human cortical development (Figure 3A). By day 56, distinct SVZ-like structures containing a mixture of SOX2+ NPCs, TBR2+ IPCs and immature neurons formed above VZ, whereas cortical plate-like (CP) structures containing pure CTIP2+ neurons formed above VZ and SVZ (Figure 3B).
Figure 3. Organization and Marker Expression of Different Progenitor Zones and Cortical Neuron Subtypes.
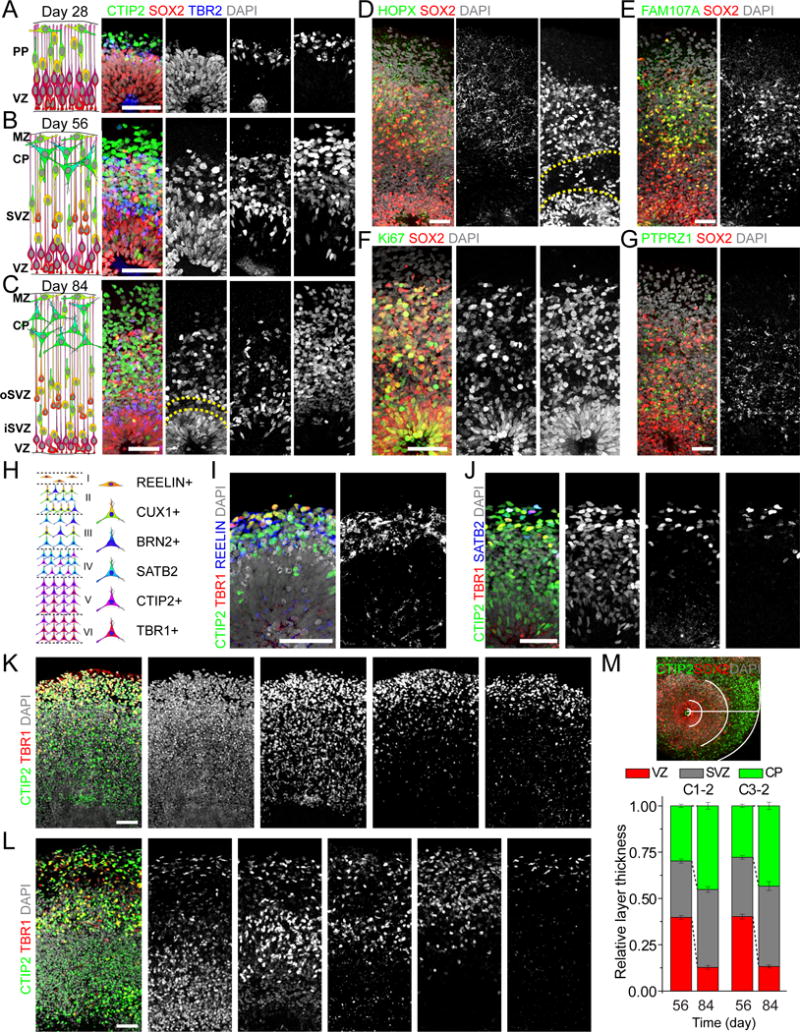
(A–C) Schematic representations and sample immunostaining images of forebrain organoids at days 28 (A), 56 (B) and 84 (C). Scale bars: 50 μm. Dash lines highlight a gap between oSVZ and iSVZ (C). PP: preplate; VZ: ventricular zone; MZ: marginal zone; CP: cortical plate; SVZ: subventricular zone; oSVZ: outer subventricular zone; iSVZ: inner subventricular zone.
(D–G) Sample images of immunostaining of oRGC markers HOPX (D), FAM107A (E) and PTPRZ1 (G), and Ki67 (F) in day 84 forebrain organoids. Scale bars: 50 μm.
(H) Schematic representation of marker expression for cortical neurons in the mature mammalian neocortex.
(I) Sample images of immunostaining for preplate Cajal-Retzius cell marker REELIN and deep layer neuron markers CTIP2 and TBR1 in day 28 forebrain organoids. Scale bar: 50 μm.
(J–K) Sample images of immunostaining for CTIP2, TBR1 and superficial layer neuron marker (SATB2) in forebrain organoids at days 56 (J) and 70 (K). Images shown in (K) are from consecutive sections for the same cortical structure. Scale bars: 50 μm.
(L) Sample images of immunostaining for CTIP2, TBR1 and superficial layer neuron markers (SATB2, BRN2 and CUX1) in forebrain organoids at day 84. Images shown are from consecutive sections for the same cortical structure. Scale bar: 50 μm.
(M) Sample image showing layer specification in forebrain organoids and quantification of the relative thickness of VZ, SVZ and CP at days 56 and 84 for two iPSC lines. For each cortical structure, three measurements were taken at 45 degree angles to obtain the mean. Values represent mean ± SEM (n ≥ 6 cortical structures from 6 organoids).
Also see Figure S3.
One hallmark of embryonic human cerebral cortex is the prominence of specialized oRGCs in the oSVZ layer (Lui et al., 2011). Similar to the developing human cortex, a thin gap appeared to separate the expanded SVZ in day 84 organoids into an inner SVZ-like (iSVZ) region that contained densely packed TBR2+ IPCs and an oSVZ-like region (Figure 3C). Recent studies have identified markers preferentially expressed by oRGCs in the developing human cortex, including HOPX, FAM107A and PTPRZ1 (Pollen et al., 2015; Thomsen et al., 2015). Using antibodies that we validated with gestational week 22 (GW22) human tissue (Figure S3A), we found a large number of SOX2+HOPX+ oRGCs in day 84 organoids (Figure 3D). Previous cerebral organoid protocols generated only sparse NPCs with apparent oRGC characteristics, which did not organize into a progenitor layer outside VZ. In contrast, our forebrain organoids exhibited a distinct SOX2+HOPX+ oSVZ-like layer separated from the SOX2+HOPX− VZ layer (Figure 3D). We sometimes observed HOPX+ radially-oriented basal processes from these oRGCs with pial contact but lacking an apical process (Figure S3B), a hallmark of human oRGCs (Hansen et al., 2010) (Figure S3A). Two other oRGC markers, FAM107A and PTPRZ1, were also specifically expressed in oSVZ (Figure 3E, G). Many oSVZ SOX2+ progenitors were Ki67+, indicating active cell division in this region (Figure 3F).
The presence of a prominent oRGC-like population in day 84 forebrain organoids offers an opportunity to track the time-course of oRGC marker expression during organoid development. A recent study showed that oSVZ-exclusive expression of HOPX, FAM107A and PTPRZ1 does not occur in the developing human cortex until gestational weeks 15–20 (Pollen et al., 2015). Interestingly, very limited HOPX expression was detected in day 28 organoids, while at day 56 its expression was prominent in both VZ and SVZ, but not exclusive to SVZ (Figure S3C).
Together, these results demonstrate that forebrain organoids exhibit multi-layer progenitor zone organization that recapitulates human cortical development, including a prominent oSVZ layer with oRGC-exclusive expression of defined molecular markers. Our system provides a platform to investigate the origin, properties and mechanisms that define and regulate human oRGCs.
Generation of Diverse Neuronal Subtypes of All Six Cortical Layers
Next, we performed detailed expression analyses of markers for different neuronal subtypes (Figure 3H). At day 28, we observed neurons expressing deep layer cortical neuron markers CTIP2 and TBR1, as well as neurons expressing the Cajal-Retzius cell marker REELIN (Figure 3I). At days 56 and 70, the SVZ contained neurons expressing low amount of CTIP2, a feature of migrating immature neurons found in this region (Lui et al., 2011) (Figure 3J–K). The CP-like structure hosted a dense population of neurons expressing CTIP2 and TBR1, as well as a sparser population of neurons expressing upper layer cortical neuron marker SATB2, which were localized close to the pial surface (Figure 3J–K). There was also a cell-sparse layer visualized by REELIN and DCX expression at the pial surface, resembling the marginal zone (MZ) that typically becomes layer I in vivo (Figure S3D). At day 84, late-born SATB2+ neurons formed a layer partially separated from the early-born CTIP2+ layer, suggesting specification of upper and deep cortical layers (Figure 3L). Furthermore, neurons expressing layer II/III markers CUX1 and BRN2 started to appear near the pial surface (Figure 3L). Quantification revealed CP and SVZ layer expansion and VZ layer reduction from day 56 to 84 (Figure 3M), resembling the developing human cortex.
Together, these results reveal the developmental time course of marker expression for neurons of all six cortical layers in forebrain organoids. Quantitative analysis of different organoids and human iPSC lines shows little variability in the relative thickness of different layers (Figure 3M), again indicating the robustness and reproducibility of our organoid system.
Molecular Signatures of Developing Forebrain Organoids
To further compare forebrain organoids to in vivo human brain development, we performed RNA-seq analyses of global transcriptomes from day 26, 40, 54 and 100 organoids. We compared organoid transcriptional profiles to datasets of 21 different human fetal organs during the first and second trimester (Roost et al., 2015). Pearson’s correlation analysis showed that organoids from all 4 time points strongly correlated with fetal brains and spinal cord, with less or no correlation with other fetal somatic tissues (Figure 4A and S4A). Further comparison with transcriptomes from human dorsolateral prefrontal cortex samples across 6 life stages, ranging from fetal development to aged human tissue (Jaffe et al., 2015) showed the highest correlation with fetal brain tissues, with the best correlation for day 100 organoids (Figure S4B). Collectively, these results suggest that organoid development is reminiscent of fetal human brain development at the molecular level.
Figure 4. Correlation of Global Transcriptomes between Forebrain Organoids and Fetal Human Brain Development.
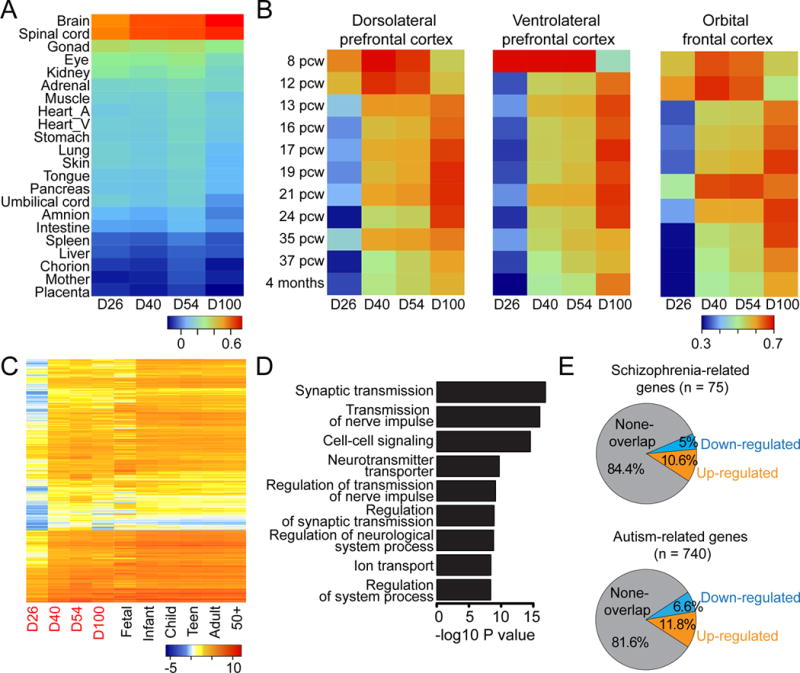
(A) Heatmap of Pearson’s correlation analysis of RNA-seq datasets from day 26, 40, 54 and 100 organoids and published datasets from 21 different human fetal organs (Roost et al., 2015). Shown are averaged values for biological replicates.
(B) Heatmaps of Pearson’s correlation analysis of RNA-seq datasets among forebrain organoids at different stages and published transcriptome datasets of 3 different cortical subregions at 11 developmental stages from Allen Brain Atlas. See Figure S5C for comparison of all 16 different human brain regions.
(C) Heatmap of gene expression dynamics from up-regulated genes between day 26 and later stages of organoid development.
(D) Gene Ontology analysis of upregulated genes. Nine top terms (in terms of P-values) are shown.
(E) Overlap of differentially expressed genes during organoid development with known schizophrenia related risk genes (from http://bioinfo.mc.vanderbilt.edu/SZGR/) and autism related risk genes (from https://gene.sfari.org/autdb/HG_Home.do). Overlapping genes are statistically significant (P < 0.001, chi-square test).
To pinpoint developmental stages and brain subregion identities of forebrain organoids, we performed large-scale comparisons with transcriptome datasets of 16 different human brain regions at 11 developmental stages (Figure S4C). These analyses revealed a temporal correlation between organoid and fetal human brain development, particularly for prefrontal cortex development (Figure 4B). For example, day 26–54 organoid profiles were closer to several subregions of prefrontal cortex at 8–9 PCW (post-conception week), whereas day 100 organoids were more closely related to 17–24 PCW, or even 35 PCW for some subregions (Figure 4B and S4C).
We also identified differentially expressed genes during organoid development (Table S2). These genes also displayed similar trends over the course of in vivo brain development (Figure 4C and S4D). Gene Ontology analysis revealed enrichment of many neuronal function pathways among upregulated genes (Figure 4D) and enrichment of cell cycle-related pathways among down-regulated genes (Figure S4E). Interestingly, differentially expressed genes during organoid development and risk genes for schizophrenia or autistic spectrum disorders showed significant overlap (P < 0.001, chi-square test; Figure 4E). Therefore, the organoid system can be used to study functional impact of dynamic expression of these disease risk genes in human brain development.
Together, our systematic and comprehensive transcriptome comparisons provide additional validation that forebrain organoids resemble normal human embryonic cortical development.
Functionally Connected Cortical Neurons and GABAergic Neuronal Subtypes
To assess physiological properties of cells in organoids, we performed electrophysiological whole-cell recording in slices acutely sectioned from organoids. Recorded neurons were capable of firing trains of TTX-sensitive action potentials (Figure 5A and S5A). Neurons showed rectifying membrane properties, Na+ and K+ currents in response to voltage ramps (Figure S5B). Cells with linear membrane properties were also observed, indicating presence of astrocytes (Figure 5B). We observed developmental changes of intrinsic properties in recorded neurons across different stages (Figure S5C–I).
Figure 5. Functional Characterization of Forebrain Organoids.
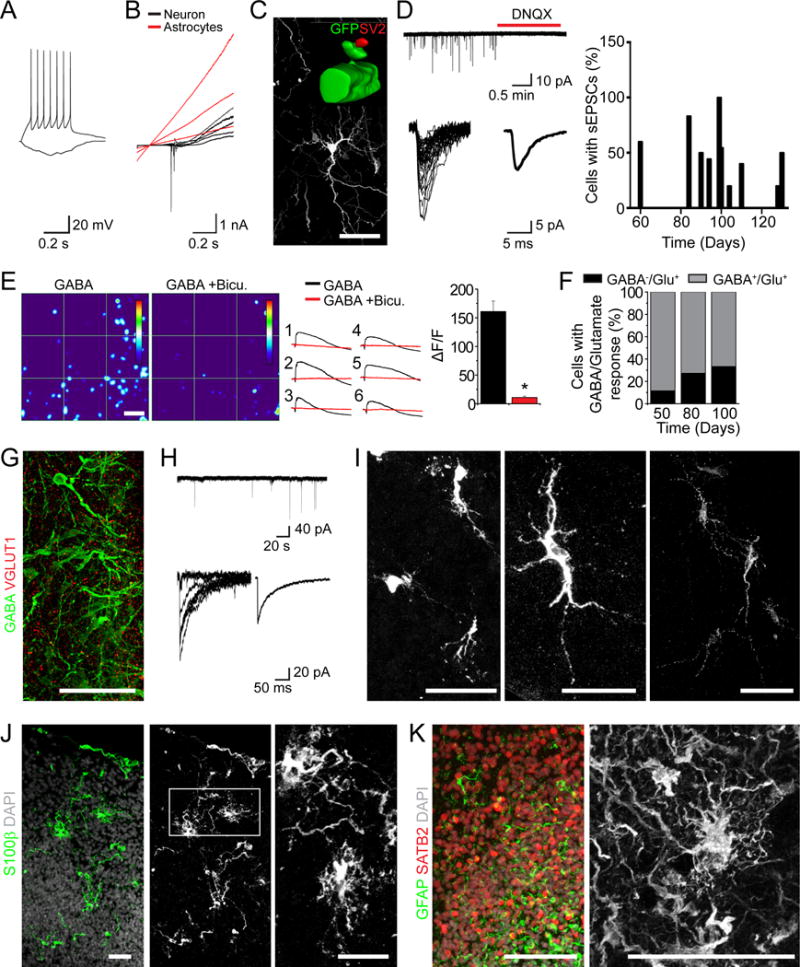
(A–D) Electrophysiological and morphological analyses of cells in forebrain organoids. Shown in (A) are sample current-clamp traces of a neuron firing a train of action potentials in response to 10 pA current injection. A hyperpolarizing step of - 5 pA is also shown. Shown in (B) are sample voltage-clamp traces showing currents in response to a ramp protocol (− 90 mV to 110 mV). Shown in (C) is a sample image of a neuron in a day 85 forebrain organoid labeled by GFP upon electroporation. The insert shows surface rendering of a dendritic spine structure on a GFP+ neuron with the pre-synaptic terminal labeled by SV2 staining in red. Scale bars: 50 μm. Shown in (D, left) are sample recording traces of sEPSCs and pharmacological blockade by DNQX. Identified sEPSC events are overlaid and the average sEPSC trace is shown. Also shown in (D, right) is the summary of the percentage of cells that exhibited detectable sEPSC events in organoids of different ages.
(E) Calcium imaging analysis of cellular response to GABA application (10 μM). Day 100 organoids were loaded with Fluo-4. Shown in left panels are sample heat maps of GABA-induced fluorescence changes (ΔF/F) within the same region in the absence or presence of Bicuculline (Bicu. 50 μM). The color scale at the right indicates a ΔF/F range of 0 to 250%. Scale bar: 50 μm. Shown in the middle panel are calcium response curves for individual cells indicated in the heatmap. Shown in the right panel is the summary of ΔF/F in response to GABA in absence or presence of bicuculline. Values represent mean ± SEM (n = 43 neurons from 3 organoids).
(F) Developmental shift of the percentage of cells in forebrain organoids that exhibit calcium rise in response to GABA (10 μM) and glutamate (20 μM). Value represent mean (n = 26, 77 and 69 neurons from 3 organoids at days 50, 80 and 100, respectively).
(G–I) GABAergic neurons in forebrain organoids. Shown are sample images of immunostaining for GABA and VGLUT1 (G) and GABAergic neuron subtypes (I). Scale bars: 50 μm. Shown in (H) are sample recording traces of sIPSCs. Identified sIPSC events are overlaid and the average sIPSC trace is shown.
(J–L) Sample images of immunostaining for astrocyte markers S100β (J) and GFAP (K) in organoids over 100 days. Scale bars: 50 μm.
Also see Figure S5.
To visualize morphology of individual neurons, we electroporated organoids to sparsely label cells with GFP. At day 85, GFP+ neurons exhibited complex neuronal morphology with spine-like structures in close association with presynaptic SV2+ puncta (Figure 5C). About 50% of cells recorded showed spontaneous excitatory postsynaptic current (sEPSC) that were sensitive to the glutamate receptor antagonist DNQX (Figure 5D). Both intrinsic properties and synaptic connectivity were similar between two iPSC clones (Figure S5J–K).
One hallmark of neuronal maturation is the switch from a depolarizing response to GABA to hyperpolarizing, due to developmentally regulated changes in intracellular Cl− concentration, mediated by NKCC1 down-regulation and KCC2 up-regulation (Ben-Ari and Spitzer, 2004). We found that NKCC1 was expressed at both days 56 and 84, whereas KCC2 was strongly expressed in the CP at day 84 but minimally at day 56 (Figure S5L). We further performed a functional assay to monitor Ca2+ rise in response to GABA-induced depolarization (Figure 5E). Quantification showed an increase over time in the percentage of neurons without GABA-induced Ca2+ rise among all neurons that responded to glutamate (Figure 5F). Therefore, forebrain organoids exhibit functional features of neuronal maturation found in vivo.
We also found GABA+VGLUT1− neurons in forebrain organoids after day 84 (Figure 5G). Electrophysiological recordings in the presence of DNQX to block all glutamatergic synaptic transmission also showed spontaneous postsynaptic currents with slower kinetics (Figure 5H). Immunohistological analysis further revealed the presence of at least three major subtypes of GABAergic neurons expressing parvalbumin, nNOS or somatostatin (Figure 5I). Consistent with electrophysiological recording results (Figure 5B), we observed S100β+ and GFAP+ astrocytes in close association with surrounding neurons (Figure 5J–K). Together, these findings demonstrate that forebrain organoids contain a diverse collection of neuronal and other cell types found in developing human brains.
Generation of Midbrain and Hypothalamic Organoids
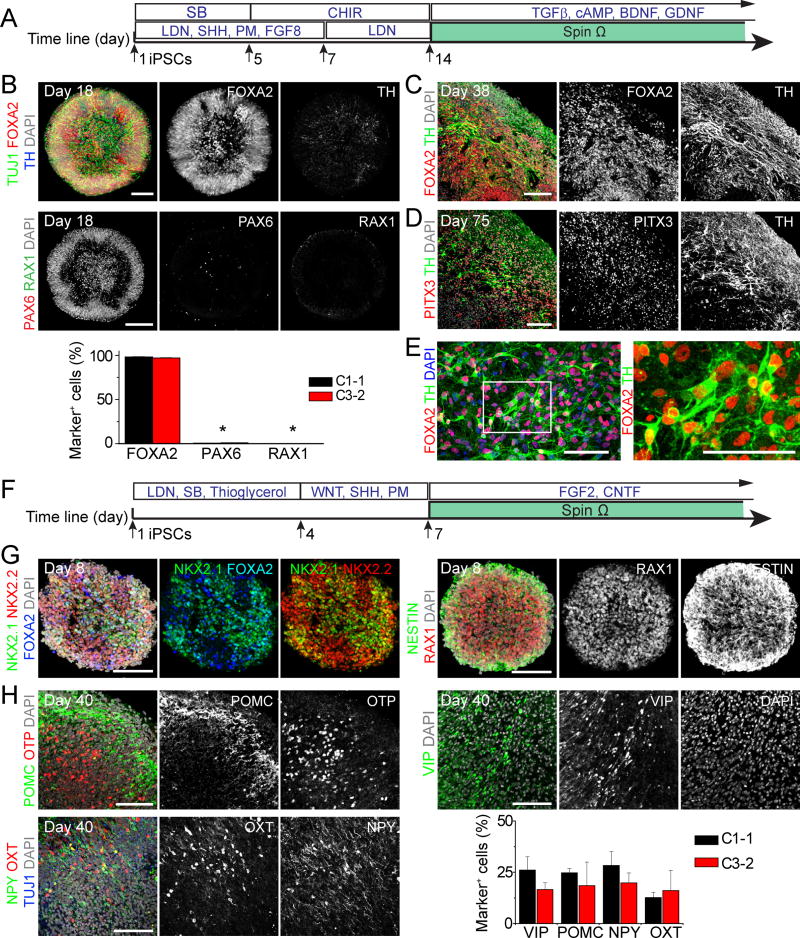
We next explored approaches to generate organoids with other brain region identities. Building upon a 2D differentiation protocol for generating midbrain dopaminergic (DA) neurons (Kriks et al., 2011), we applied Sonic Hedgehog (SHH) agonists (recombinant SHH and Purmorphamine), FGF-8, SMAD inhibitors (SB431542 and LDN193189), and GSK3β inhibitor (CHIR99021) to induce floor plate differentiation of human iPSCs, which were transferred to SpinΩ at day 14 (Figure 6A). At day 18, midbrain organoids showed organized neuroepithelium-like structures expressing NESTIN and floor-plate precursor marker FOXA2, but not DA neuron marker TH, whereas very few cells expressed forebrain marker PAX6 or hypothalamus progenitor marker RAX1 (Figure 6B and S6A). At day 38, we observed numerous TH+ DA neurons (Figure 6C). At day 56, the majority of TH+ neurons expressed FOXA2+ and dopamine transporter (DAT) (Figure S6B). In addition, midbrain organoids contained TH+ cells that expressed midbrain DA neuron markers NURR1 and PITX3 (Figure S6B). At day 75, PITX3 was robustly expressed by TH+ cells, suggesting specification of A9 DA neurons (Chung et al., 2005) (Figure 6D). To quantify TH and FOXA2 expression, we dissociated midbrain organoids at day 65. Upon culturing in monolayer for 5 days, we found that 95 ± 1% of cells were FOXA2+ and 55 ± 4% were TH+ DA neurons (n = 6; Figure 6E).
Figure 6. Generation of Midbrain and Hypothalamic Organoids.
(A–E) Midbrain organoids from human iPSCs. Shown in (A) is a schematic diagram of the midbrain organoid protocol. Shown in (B) are sample images of day 18 organoids (Scale bars: 100 μm) and quantifications. Values represent mean ± SEM (n = 4 organoids each; *P < 0.05, Student’s t-test). Also shown are sample images of immunostaining of midbrain organoids at day 38 (C) and day 75 (D), and monolayer cultures 5 days after dissociation and plating of day 65 midbrain organoids (E). Scale bars: 50 μm.
(F–H) Hypothalamic organoids. Shown in (F) is a schematic diagram of the hypothalamic organoid protocol. Shown are sample images of day 8 (G) and day 40 (H) organoids. Scale bars: 100 μm. Also shown in (H) is a summary of quantification for peptidergic neuronal markers expression in day 40 hypothalamic organoids from 2 iPSC lines. Values represent mean ± SEM (n = 3 organoids each).
Also see Figure S6.
We also explored methods to generate hypothalamic organoids from human iPSCs. We first treated human iPSCs with dual SMAD inhibitors (SB431542 and LDN193189) to pre-pattern them to the neuroectodermal fate (Figure 6F). After 3 days, embryoid bodies were treated with WNT3A, SHH and Purmorphamine to induce the hypothalamic lineage. At day 8, the majority of cells in organoids expressed NKX2.1, NKX2.2, RAX1, SOX2, NESTIN and FOXA2, markers that are consistently expressed during early hypothalamus development (Blackshaw et al., 2010) (Figure 6G). At day 40, peptidergic neuronal markers, including POMC, VIP, OXT and NPY, were detected in organoids generated from different iPSC lines (Figure 6H). At day 40, but not day 8, a subset of cell populations expressed OTP, a homeobox protein essential for specification of hypothalamic neuronal lineages (Wang and Lufkin, 2000) (Figure 6H and S6C). Together, these findings demonstrate the versatility of SpinΩ to support growth of organoids of different types.
Modeling ZIKV Exposure during Cortical Neurogenesis
Our organoid system provides a quantitative platform to model human diseases. The World Health Organization recently declared ZIKV a Public Health Emergency of International Concern, due in part to the uncertainty surrounding increased reports of microcephaly and other neurological disorders coinciding with clusters of ZIKV outbreaks (Heymann et al., 2016). Recent studies of human NPCs in 2D and neurosphere cultures showed efficient infection by ZIKV, leading to increased cell death and attenuated growth (Tang et al., 2016)(Garcêz et al. 2016, PeerJ, preprint). Without organizational features unique to 3D brains, such as cortical layers, these initial studies in 2D cultures do not directly address the potential link between ZIKV and microcephaly. It also remains unknown whether ZIKV exhibits specific tropism for different neural cell types in more complex 3D tissue.
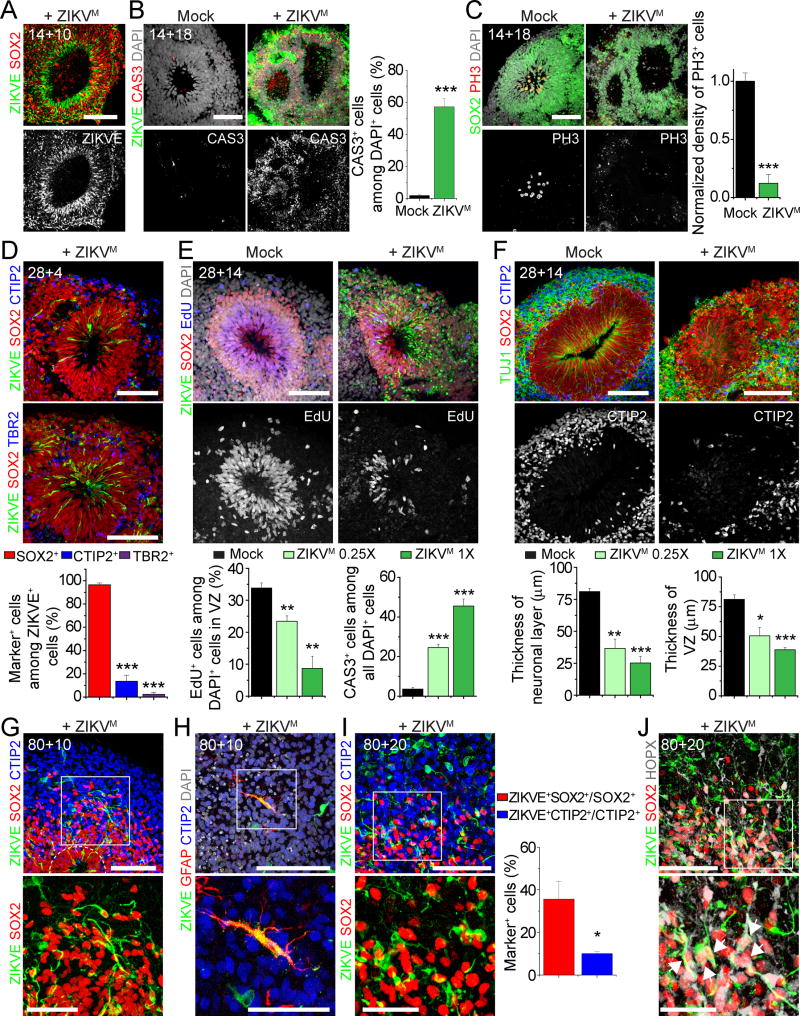
We performed a series of experiments to model transient ZIKV exposure at different stages of human cortical development by incubating forebrain organoids with ZIKV in medium for 24 hours in SpinΩ. We initially used a prototype ZIKV strain of African lineage (MR766, termed ZIKVM hereafter) (Haddow et al., 2012). ZIKVM readily infected SOX2+ NPCs in day 14 forebrain organoids (Figure 7A and S7A). After 18 days, ZIKVM infection resulted in overall decreased organoid size (Figure S7B–C). Quantitative analyses showed dramatically reduced VZ thickness and size (Figure S7D–E), likely due to significant cell death and suppression of NPC proliferation (Figure 7B–C). We also observed a significant increase in lumen size within ventricular structures (Figure S7F), reminiscent of dilated ventricles in a recently reported clinical case of a fetal brain infected with ZIKV (Driggers et al., 2016).
Figure 7. Modeling Impact of ZIKV Exposure using Forebrain Organoids.
(A) Sample immunostaining images of forebrain organoids exposed to ZIKVM (1X) or mock-treated at day 14 for 24 hours and analyzed 10 days later (14+10). Scale bar: 100 μm.
(B–C) Sample immunostaining images of forebrain organoids exposed to ZIKVM (1X) or mock-treated at day 14 for 24 hours and analyzed 18 days later (14+18) for CAS3 (B), or PH3 (C). Scale bars: 100 μm. Also shown are quantifications for the percentage of CAS3+ cells among the total number of nuclei stained by DAPI (B) and density of PH3+ cells within VZ (C). Values represent mean ± SEM (n = 5 organoids; *** P < 0.0005, Student’s t-test)
(D) Sample immunostaining image of a forebrain organoid exposed to ZIKVM (1X) at day 28 for 24 hours and analyzed 4 days later (28+4) (Scale bars: 100 μm) and quantifications. Values represent mean ± SEM (n = 5 cortical structures from 3 organoids; *** P < 0.0005, Student’s t-test).
(E–F) Forebrain organoids exposed to ZIKVM (1X or 0.25X) or mock-treated at day 28 for 24 hours and analyzed 14 days later (28+14). Shown are sample immunostaining images and quantification of cell proliferation and cell death in ZIKV-infected regions (E) and thickness of SOX2+ VZ layer and TUJ1+ neuronal layer (F). Scale bars: 100 μm. Values represent mean ± SEM (n = 5 cortical structures from 3 organoids; *P < 0.05; **P < 0.005, ***P < 0.0005, Student’s t-test).
(G–J) Sample immunostaining images (top; Scale bars: 100 μm) and magnified views (bottom; Scale bars: 50 μm) of forebrain organoid exposed to ZIKVM (1X) at day 80 for 24 hours and analyzed 10 days (80+10; G–H) or 20 days later (80+20; I–J). Arrows in (J) point to ZIKV+HOPX+SOX2+ oRGC-like cells in the oSVZ region. Also shown in (I) are quantifications for the percentage of ZIKV+ cells among the total number of SOX2+ or CTIP2+ cells in the whole cortical structure. Values represent mean ± SEM (n = 7 cortical structures from 5 organoids, *P < 0.05, Student’s t-test).
Also see Figure S7.
Next, we exposed day 28 organoids, which contained both progenitor and neuronal layers to two different doses of ZIKVM (1X and 0.25X; Figure S7G). Most ZIKVM infected cells were SOX2+ NPCs and very few were TBR2+ IPCs or CTIP2+ immature neurons when quantified 4 days later (Figure 7D), suggesting specific tropism of ZIKVM towards NPCs in the 3D tissue. By day 14, we observed a significantly increased number of ZIKVM infected cells (Figure S7H), consistent with productive infection by ZIKV. In addition to overall size reduction (Figure S7I), we observed a ZIKV dose-dependent decrease of EdU+ proliferating cells and increased CAS3+ cells (Figure 7E and S7J). Interestingly, many CAS3+ cells were ZIKV−, indicating a bystander effect (Figure S7J). As a result, ZIKV infection of early stage organoids, corresponding to the first trimester of human fetal development, led to a significant reduction in both VZ and neuronal layer thickness (Figure 7F), resembling microcephaly.
We also assessed the effect of ZIKVM on day 80 forebrain organoids (Figure S7K). After 10 days, we again observed preferential localization of ZIKVM in SOX2+ NPCs in VZ and oSVZ, but it was also detected in CTIP2+ neurons and occasionally in GFAP+ astrocytes (Figure 7G–H). The infection appeared less robust compared to that of earlier stages of organoids, possibly due to limited ZIKV penetration to the interior of organoids where NPCs reside. After 20 days, we observed an increased number of ZIKV+ cells (Figure 7I). Quantification showed a higher percentage of SOX2+ NPCs with ZIKVM than that for CTIP2+ neurons (Figure 7I). The presence of ZIKV+SOX2+HOPX+ cells indicates infection of oRGCs by ZIKV (Figure 7J).
Two recent studies have shown few differences between properties of different ZIKV strains in different models (Bayer et al., 2016; Lazear et al., 2016). We explored a ZIKV strain of Asian lineage that exhibits over 99% amino acid sequence similarity to strains currently circulating in Brazil (FSS13025, termed ZIKVC hereafter) (Haddow et al., 2012). Quantitative analysis showed similar enrichment of ZIKVC in SOX2+ NPCs, compared to CTIP2+ immature neurons or TBR2+ IPCs in early stage organoids (Figure S7L–M).
Together, our forebrain organoid system allowed quantitative investigation of consequences of ZIKV exposure and our results suggest that ZIKV, upon access to the fetal brain, targets NPCs and causes microcephalic-like deficits in cortical development.
DISCUSSION
We have developed a cost effective, simple to use system for 3D organoid cultures by designing a miniaturized multi-well spinning bioreactor, SpinΩ, which can be used with standard cell culture plates. The low cost of the platform allowed us to optimize protocols to generate forebrain organoids with minimized heterogeneity and variability that enables quantitative analyses and better recapitulation of the developing human cortex. Specifically, these forebrain organoids exhibit a well-developed oSVZ-like region containing NPCs that share molecular and morphological features of human oRGCs, organized neuronal subtypes found in all six cortical layers, and GABAergic neuronal subtypes. We further demonstrated SpinΩ’s versatility by developing protocols to generate organoids recapitulating characteristics of other brain regions. Finally, we applied our forebrain organoid platform for chemical compound testing and modeling ZIKV infection.
SpinΩ, a Miniaturized Spinning Bioreactor for Cost-effective Organoid Culturing
Several pioneering studies showed that cerebral organoid systems offer improved growth conditions for 3D tissue, leading to a more representative model of the developing human brain (Danjo et al., 2011; Kadoshima et al., 2013; Lancaster et al., 2013; Mariani et al., 2015; Pasca et al., 2015). In particular, the use of a spinning flask provides a 3D low-shear stress suspension culture with enhanced diffusion of oxygen and nutrients that supports formation of larger, continuous cortical structures (Lancaster et al., 2013). Under our culture conditions, direct comparison with stationary and orbital shaker cultures confirmed the beneficial effect of spinning for forebrain organoids. However, maintaining organoids in standard spinning flasks makes it cost-prohibitive to supplement the media with small molecules and growth factors to promote growth and differentiation of organoids. Our miniaturized spinning bioreactor SpinΩ addresses this limitation by dramatically reducing the required media volume, allowing for systematic and efficient testing of culture conditions in parallel. Moreover, SpinΩ’s small footprint and compact shape reduces the incubator space required, a feature that is further highlighted by the stackable version (Figure S1A). Many of the design parameters of SpinΩ, including number and size of wells, rotation speed, shaft angle and shape, can be customized based on specific needs. Together, the SpinΩ system provides better accessibility and higher efficiency for developing 3D tissue cultures for applications related to the brain and other organs.
Features of Forebrain Organoids and Areas for Improvements
Compared to several pioneering cerebral organoid systems, our forebrain organoids show high reproducibility, which is critical to realize its promise as a standardized model for human cortical development. Two rounds of patterning factors effectively induce forebrain differentiation and significantly reduce both tissue and temporal development heterogeneity. Our proof-of-principle study with BPA, although with concentrations likely higher than normal human exposure, demonstrates that many parameters in these organoids can be reliably quantified; therefore this platform can be broadly used for drug testing, compound screening and disease modeling.
Forebrain organoids better recapitulate developing human cortex along multiple dimensions as compared to previously reported methods. First, it produces a well-defined oSVZ-like region with a prominent oRGC-like NPC layer, which are distinct features of developing human cortex that are absent in rodents and previous organoid models. Time-course of SVZ and oSVZ layer formation and progression also models dynamic changes during human cortical development. Moreover, oRGCs in forebrain organoids express all three recently identified human oRGC markers. Second, forebrain organoids robustly generate organized cortical neurons expressing markers found in all six layers of human cortex, including a layer of CUX1+ neurons destined for layer II. The peak in production of late-born neurons expressing the upper layer neuron marker SATB2 occurred after day 56, coinciding with oSVZ specification and expansion. Because the peak of oSVZ proliferation coincides in time with formation of upper cortical layers, which are particularly cell dense in human cortex, it has been suggested that the abundant oRGC population in human oSVZ is responsible for this evolutionary distinction (Lui et al., 2011). Therefore, the presence of well-developed oSVZ may be responsible for robust generation of upper layer neurons in forebrain organoids. Our electrophysiology and calcium imaging analyses revealed functional neuronal properties, active synaptic transmission, and recapitulation of neuronal maturation characteristics similar to those observed in vivo. We show the presence of GABAergic neuronal subtypes in organoids. The apparent absence of NKX2.1+ ventral progenitors during early differentiation suggest a possible dorsal origin of GABAergic neurons, a distinct feature of primates and humans (Petanjek et al., 2009; Yu and Zecevic, 2011). Lastly, large-scale comparisons of global transcriptome analyses confirm that forebrain organoid development closely correlates with human cortical development at the molecular level. Forebrain organoids with a well-developed oSVZ will significantly expand our ability to study distinct characteristics of human cortical development that cannot be represented in rodent models. Compared to studies of postmortem human tissues, forebrain organoids offer a model to investigate embryonic human cortical development as a continuous dynamic process in live cells, and allow pharmacological and genetic manipulations to investigate underlying mechanisms.
It is likely that continued optimization can further improve the forebrain organoid system. First, depletion of nutrients and oxygen in the interior of organoids is one factor limiting our ability to model human brain development beyond the second trimester. Due to dramatic CP expansion, progenitor zones in forebrain organoids become gradually depleted after day 100. One potential solution is to engineer vascularized 3D tissue by endothelial cell co-cultures, or implementing microfluidic perfusion networks. An alternative approach would be to explore culture conditions that can accelerate forebrain organoid development to produce features of late-stage cortical development with smaller overall organoid size. Second, forebrain organoids do not contain well-defined regions representing the intermediate zone (IZ) and subplate, which play important roles in neuronal migration during cortical development. Intriguingly, a previously reported cortical neuroepithelial system showed formation of a cell-sparse IZ-like region despite lacking oSVZ (Kadoshima et al., 2013). Third, although we have identified cortical neurons expressing markers found in all six human cortical layers, they display only rudimentary separation. Additional chemical and physical cues may be required to better regulate neuronal migration and positioning.
Modeling ZIKV Exposure during Different Stages of Cortical Neurogenesis
As an application of our organoid platform for disease modeling, we modeled the impact of ZIKV exposure at different stages of pregnancy. Recent clinical studies have established that ZIKV can pass through placenta to gain access to the developing fetal brain (Calvet et al., 2016; Driggers et al., 2016; Mlakar et al., 2016). We show that among different cell types in 3D tissue, ZIKV exhibits specific tropism towards NPCs, including oRGCs, although ZIKV could be detected in immature neurons, IPCs, and astrocytes. Time-course analysis further shows that ZIKV infection in NPCs is productive, resulting in more infected cells over time. Therefore, even a very low dose and transient ZIKV exposure in utero may have a prolonged and increasingly severe effect over time. Consistent with clinical findings that first trimester infections are the most dangerous (Cauchemez et al., 2016; Faria et al., 2016), exposure of early stage forebrain organoids to ZIKV for only one day leads to detrimental effects, mimicking many features of microcephaly, including decreased neuronal layer thickness and overall size as well as enlarged lumen/ventricles. Mechanistically, we show increased cell death and suppressed proliferation of infected NPCs. The same ZIKV treatment of day 80 organoids, which are more complex and resemble the second trimester, also leads to preferential infection of SOX2+ NPCs, including HOPX+ oRGCs. Together, our results provide compelling evidence that, upon access to the fetal brain, productive and preferential infection of NPCs by ZIKV leads to characteristic features resembling microcephaly. Forebrain organoids therefore provide a quantitative experimental platform for future studies to investigate the impact of ZIKVs, identify cellular and molecular mechanisms, and screen for therapeutic interventions, issues that are critical to resolving the current global health emergency related to ZIKV.
Additional Future Applications
Brain organoids also provide a renewable source of human neurons and other cell types, such as DA neurons for transplantation in models of Parkinson’s disease. Organoid growth is coupled with dramatic expansion in cell numbers. For example, embryoid bodies of around 300 μm in diameter could expand to organoids that are up to 3 mm in diameter, achieving a 1000-fold expansion in cell mass. Just as the cerebral organoid methodology was inspired by self-organizing tissue organoids developed for other organs, SpinΩ has the potential to be broadly applied to other types of 3D tissue cultures beyond the nervous system, where SpinΩ’s advantages in reduced cost, increased throughput, enhanced cell survival and improved factor absorption would prove beneficial. The modular stackable version of SpinΩ allows for consistent culture conditions for multiple plates simultaneously and potential large-scale 3D tissue cultures and drug screening.
EXPERIMENTAL PROCEDURES
Bioreactor Design and 3D Printing
We used SolidWorks for design and drawings of all components for 3D printing. Modular individual bioreactors were made to fit into a stackable bioreactor with some modifications. The blueprints for 3D printing of the 12-wells version and the stackable version of the bioreactor are provided in Supplementary File 1. Each folder contains the CAD files for design of each component as well as a PDF reference file to view the full assembly of the pieces.
Culture of Brain-region Specific Organoids, Immunohistology and Quantification
All studies were performed with approved protocols of Johns Hopkins University School of Medicine. Human iPSC lines were previously characterized (Wen et al., 2014). See detailed protocols to generate forebrain (Figure 1B), midbrain (Figure 7A) and hypothalamic organoids (Figure 7F) in Supplementary Information.
Whole organoids were processed for immunocytochemistry as previously described (Yoon et al., 2014). See list of antibodies and their information in Table S1. For cell fate quantifications of day 14 organoids, neural tube structures were counted as positive for forebrain markers when more than 80% of all nuclei were positive for respective markers. Markers for different brain regions were quantified by measuring the area stained positive for markers and normalized to DAPI in ImageJ software. VZ was defined by SOX2 immunoreactivity and neural-tube morphology and the outer layer was defined by the area outside the VZ to the nearest pial surface. The relative VZ thickness was defined as the ratio of VZ thickness to VZ plus outer layer thickness. Layer thickness measurements at days 56 and 84 in forebrain organoids were performed similarly with the addition of SVZ. SVZ was defined by the region within mixed population of SOX2+ and CTIP2+ nuclei outside VZ. CP was defined by the region from the boundary of SVZ to the pial surface with exclusive CTIP2+ nuclei.
RNA-Seq and Bioinformatics Analyses
Forebrain organoids at days 26, 40, 54 and 100 were processed for RNA-seq and bioinformatics analyses as previously described (Tang et al., 2016). Sequence read counts for 22 different human fetal organs were obtained from GSE66302. Human dorsolateral prefrontal cortex RNA-seq datasets from six different life stages were obtained from (http://www.nature.com/neuro/journal/v18/n1/extref/nn.3898-S9.zip). RNA-seq gene expression for 11 time points of fetal development and 16 different brain regions were obtained from Allen Brain Atlas (http://www.brain-map.org/). Schizophrenia-related risk genes were obtained from (http://bioinfo.mc.vanderbilt.edu/SZGR/). Autism-related risk genes were obtained from (https://gene.sfari.org/autdb/HG_Home.do). R programming language was used to perform all data analysis and generate the figures.
Electrophysiology and Calcium Imaging
Organoid slices were prepared by embedding organoids in 4% low melting point agarose cooled to approximately 32°C. Slices (250 μm) were sectioned and were immediately ready for recording. Calcium imaging was performed similarly as previously described (Kim et al., 2012).
Modeling ZIKV Exposure
ZIKV was prepared and titered as previously described (Tang et al., 2016). Supernatant from ZIKV-infected mosquito C6/36 cells (ZIKVM) or Vero cells (ZIKVC) was diluted 1:10 (1X) or 1:40 (0.25X) and applied directly in SpinΩ for 24 hours and then replaced with fresh medium. Forebrain organoids infected at day 28 were pulsed with 10 μM EdU for 2 hours on day 42, and fixed for analysis 24 hours later. Quantitative analyses were conducted on randomly picked cortical structures. Cell death was quantified by counting CAS3+ nuclei over total nuclei stained by DAPI. Area of VZ and lumen, and thickness of VZ and neuronal layers were measured using ImageJ software. Overall size of organoids was measured under calibrated 4X bright field microscope.
Supplementary Material
Highlights.
A miniaturized spinning bioreactor for cost effective culturing of organoids
Generation of brain region-specific organoids from human iPSCs
ZIKV causes decrease of neuronal cell layer volume resembling microcephaly
Both African and Asian ZIKV promote neural progenitor death in organoids
Acknowledgments
We thank Hopkins WSE machine shop and Nathaniel Leon for help of 3D printing, and members of Ming and Song laboratories for discussion, L. Liu and Y. Cai for technical support, and Dr. Robert B. Tesh at NIAID and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) for the Cambodian strain ZIKVC. This work was supported by grants from NIH (NS048271, MH105128, and NS095348 to G.L.M., NS047344 and ES021957 to H.S., AI119530 and AI111250 to H.T, NS051630, NS079625, and MH102690 to P.J., MH104593 to B.J.M.), MSCRF (to H.S.), and SFARI (308988 to H.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The accession number for RNA-seq data reported in this paper is GEO: GSE 80073.
SUPPLEMENTAL INFORMATION
Supplemental information includes seven figures, two tables, experimental procedures, and references.
AUTHOR CONTRIBUTIONS
X.Q., H.N.N., H.S. and G-L.M. conceived of the research, designed the study, and wrote the manuscript. M.M.S., C. Hadiono and W.J. designed SpinΩ. X.Q., S.C.O., C. Hammack and H.T. performed ZIKV experiments. P.Y.S. provided ZIKVC. B.Y., L.L., Y.L., H.W. and P.J. performed RNA-seq analyses. G.H. and B.J.M. performed electrophysiology analysis. C.Y.H. and D.N. contributed human tissue samples. F.J., C.Z., J.T., K-j.Y., D.B., C.Z., E.K., M.C., Z.W., K.M.C. contributed to additional data collection and writing.
References
- Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ETJ, Cherry S, Sadovsky Y, Coyne CB. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host & Microbe. 2016 doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y, Spitzer NC. Nature and nurture in brain development. Trends Neurosci. 2004;27:361. doi: 10.1016/j.tins.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Scholpp S, Placzek M, Ingraham H, Simerly R, Shimogori T. Molecular pathways controlling development of thalamus and hypothalamus: from neural specification to circuit formation. J Neurosci. 2010;30:14925–14930. doi: 10.1523/JNEUROSCI.4499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet G, Aguiar RS, Melo AS, Sampaio SA, de Filippis I, Fabri A, Araujo ES, de Sequeira PC, de Mendonca MC, de Oliveira L, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016 doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- Cauchemez S, Besnard M, Bompard P, Dub T, Guillemette-Artur P, Eyrolle-Guignot D, Salje H, Van Kerkhove MD, Abadie V, Garel C, et al. Association between Zika virus and microcephaly in French Polynesia, 2013–15: a retrospective study. Lancet. 2016 doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Hedlund E, Hwang M, Kim DW, Shin BS, Hwang DY, Kang UJ, Isacson O, Kim KS. The homeodomain transcription factor Pitx3 facilitates differentiation of mouse embryonic stem cells into AHD2-expressing dopaminergic neurons. Molecular and cellular neurosciences. 2005;28:241–252. doi: 10.1016/j.mcn.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Danjo T, Eiraku M, Muguruma K, Watanabe K, Kawada M, Yanagawa Y, Rubenstein JL, Sasai Y. Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J Neurosci. 2011;31:1919–1933. doi: 10.1523/JNEUROSCI.5128-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jaaskelainen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016 doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- Faria NR, Azevedo RD, Kraemer MU, Souza R, Cunha MS, Hill SC, Theze J, Bonsall MB, Bowden TA, Rissanen I, et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science. 2016 doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Schroeder WF, Wolf DA, Moyer MP. Rotating-wall vessel coculture of small intestine as a prelude to tissue modeling: aspects of simulated microgravity. Proc Soc Exp Biol Med. 1993;202:181–192. doi: 10.3181/00379727-202-43525. [DOI] [PubMed] [Google Scholar]
- Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Heymann DL, Hodgson A, Sall AA, Freedman DO, Staples JE, Althabe F, Baruah K, Mahmud G, Kandun N, Vasconcelos PF, et al. Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016 doi: 10.1016/S0140-6736(16)00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, Gao Y, Jia Y, Maher BJ, Hyde TM, et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2015;18:154–161. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup JM, Goodwin TJ, Spaulding G. Prospects for use of microgravity-based bioreactors to study three-dimensional host-tumor interactions in human neoplasia. J Cell Biochem. 1993;51:290–300. doi: 10.1002/jcb.240510308. [DOI] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Liu CY, Zhang F, Duan X, Wen Z, Song J, Feighery E, Lu B, Rujescu D, St Clair D, et al. Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell. 2012;148:1051–1064. doi: 10.1016/j.cell.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Gudsnuk K, Franks B, Madrid J, Miller RL, Perera FP, Champagne FA. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc Natl Acad Sci U S A. 2013;110:9956–9961. doi: 10.1073/pnas.1214056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A mouse model of Zika virus pathogenesis. Cell Host & Microbe. 2016 doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016 doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Berger B, Esclapez M. Origins of cortical GABAergic neurons in the cynomolgus monkey. Cereb Cortex. 2009;19:249–262. doi: 10.1093/cercor/bhn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost MS, van Iperen L, Ariyurek Y, Buermans HP, Arindrarto W, Devalla HD, Passier R, Mummery CL, Carlotti F, de Koning EJ, et al. KeyGenes, a Tool to Probe Tissue Differentiation Using a Human Fetal Transcriptional Atlas. Stem Cell Reports. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, et al. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Methods. 2015 doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lufkin T. The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev Biol. 2000;227:432–449. doi: 10.1006/dbio.2000.9902. [DOI] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering Stem Cell Organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zecevic N. Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J Neurosci. 2011;31:2413–2420. doi: 10.1523/JNEUROSCI.5249-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.