Abstract
Although mechanisms of acquired resistance of EGFR mutant non-small cell lung cancers to EGFR inhibitors have been identified, little is known about how resistant clones evolve during drug therapy. Here, we observe that acquired resistance caused by the T790M gatekeeper mutation can occur either by selection of pre-existing T790M clones or via genetic evolution of initially T790M-negative drug tolerant cells. The path to resistance impacts the biology of the resistant clone, as those that evolved from drug tolerant cells had a diminished apoptotic response to third generation EGFR inhibitors that target T790M EGFR; treatment with navitoclax, an inhibitor of BCL-XL and BCL-2 restored sensitivity. We corroborated these findings using cultures derived directly from EGFR inhibitor-resistant patient tumors. These findings provide evidence that clinically relevant drug resistant cancer cells can both pre-exist and evolve from drug tolerant cells, and point to therapeutic opportunities to prevent or overcome resistance in the clinic.
Introduction
Despite the success of targeted cancer therapies, the duration of clinical response is limited by the inevitable development of acquired drug resistance, as in the case of EGFR mutant non-small cell lung cancers (NSCLC) treated with EGFR inhibitor therapy1–3. Although molecular mechanisms of acquired resistance to EGFR inhibitors have been identified4–6, little is known about how resistant clones evolve during drug therapy. In some cases, clones with clinically validated genetic resistance mechanisms may exist prior to drug exposure and may be selected by treatment7–10. Alternatively, it has been hypothesized that drug tolerant (or “persister”) cells without bona fide resistance mechanisms may survive initial drug treatment by epigenetic adaptations11–13, and undergo further evolution over time to acquire validated genetic resistance mechanisms (Supplementary Fig. 1). Although this would have immediate implications for new therapeutic strategies to prevent resistance, there has not been any direct evidence that drug tolerant cells can undergo such evolution.
To better understand the evolution of acquired resistance, we studied the development of resistance caused by the T790M gatekeeper mutation in EGFR, which occurs in 50–60% of EGFR mutant NSCLC patients with acquired resistance to EGFR inhibitor therapy4. By monitoring the development of large numbers of resistant clones in parallel, we were able to identify temporal patterns that reflected emergence of pre-existing resistant T790M clones as well as de novo acquisition of the T790M mutation within initially T790M-negative drug tolerant cells. Moreover, those that evolved from drug tolerant cells bear epigenetic hallmarks of the drug tolerant state and have a diminished apoptotic response to third generation EGFR inhibitors that target T790M EGFR. These findings provide evidence that drug resistant cancer cells bearing the identical clinically relevant genetic resistance mechanism can both pre-exist and evolve from drug tolerant cells, and suggest that cancer cells that survive initial therapy may serve as an important reservoir from which acquired resistance can emerge in the clinic.
Results
Differential response of PC9 T790M cells to EGFR inhibition
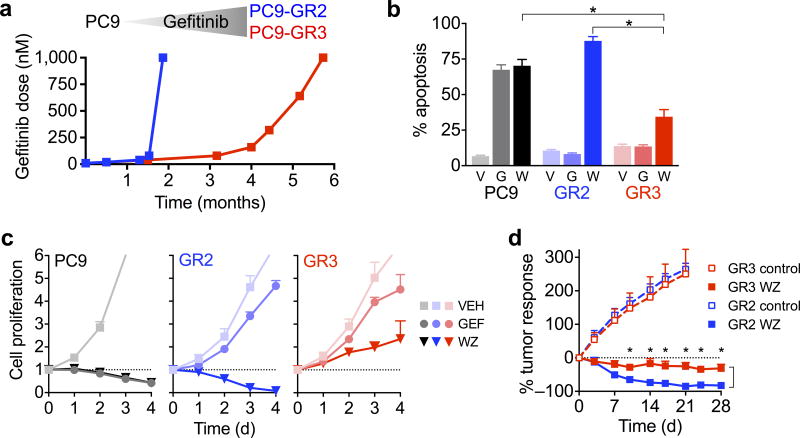
We previously cultured EGFR mutant NSCLC PC9 cells in escalating concentrations of the EGFR inhibitor, gefitinib, until resistant clones emerged14. In two resistant cell lines that acquired T790M, there was a marked difference in the time required to develop resistance, with the PC9-GR2 and PC9-GR3 lines developing in 6 and 24 weeks, respectively (Fig. 1a). Treatment with the third generation irreversible EGFR inhibitor WZ400215 suppressed EGFR phosphorylation and downstream MEK and PI3K signaling and induced cell cycle arrest in both resistant cell lines (Supplementary Fig. 2a–c). However, WZ4002 induced robust mitochondrial depolarization and subsequent apoptosis only in the PC9-GR2 cells (Supplementary Fig. 2d and Fig. 1b). Analysis of the expression of BCL-2 family genes, which regulate the mitochondrial apoptotic response induced by MEK/ERK and PI3K/AKT signaling pathways16, revealed that compared to parental and PC9-GR2 cells, PC9-GR3 cells had diminished upregulation of BIM (Supplementary Fig. 2e,f), a key mediator of apoptosis in EGFR mutant NSCLC17–20. Similarly, induction of BIM protein levels after drug treatment was significantly lower in PC9-GR3 cells compared with PC9-GR2 and parental cells (Supplementary Fig. 2a,g). Consistent with the differential levels of apoptosis following treatment with WZ4002, treatment induced a cytotoxic response in PC9-GR2 but not GR3 cells in vitro (Fig. 1c and Supplementary Fig. 2h). In vivo, WZ4002 suppressed the growth of sub-cutaneous PC9-GR3 xenograft tumors, but did not induce the degree of tumor regression observed for PC9-GR2 tumors (Fig. 1d), despite comparable inhibition of EGFR and ERK phosphorylation (Supplementary Fig. 3a).
Figure 1. Variable sensitivity of resistant T790M PC9 cell lines to EGFR inhibition.
(a) PC9 parental cells were cultured in escalating concentrations of gefitinib as tolerated until fully resistant, which was defined as lack of inhibitory effect of drug on cell proliferation.
(b) PC9-GR3, PC9-GR2 and parental PC9 cells were cultured with 1 μM gefitinib (G), WZ4002 (W) or vehicle (V) for 72 hours and apoptosis was determined by annexin staining (mean and s.e.m. of four independent experiments*P < 0.05, two-tailed t-test.).
(c) PC9-GR3, PC9-GR2 and parental PC9 cells were treated with 1 μM gefitinib (GEF), WZ4002 (WZ) or vehicle (VEH) and cell proliferation was determined by CellTiter-Glo assay at indicated time points (mean and s.e.m. of 4 independent experiments). The dotted line indicates relative cell number at time of drug addition.
(d) Mice bearing PC9-GR2 or PC9-GR3 subcutaneous xenograft tumors were treated with 50 mg/kg/day WZ4002. (PC9-GR2 - control (N=8), WZ (N=8); PC9-GR3 - control (N=8), WZ (N=8)). Tumors were measured with electronic calipers and % tumor response was calculated as the percentage change in tumor volume (V = 0.52 × L × W2) relative to the start of drug treatment (mean and s.e.m.). P < 0.01 (*) comparing WZ treatment arms at indicated time points by multiple t-tests with Sidak-Bonferroni multiple comparison.
Early resistant clones derive from pre-existing T790M cells
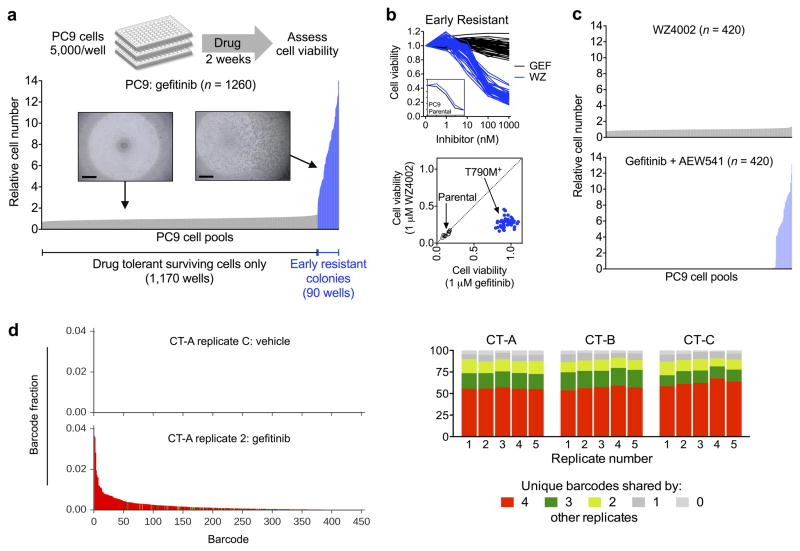
The variable time to resistance led us to question whether the GR2 and GR3 cells may have developed T790M via different mechanisms (i.e. pre-existing versus drug tolerant evolution (Supplementary Fig. 1)). To explore this possibility, we cultured over 1,200 small pools (5,000 cells each) of parental PC9 cells in the presence of gefitinib and monitored for emergence of resistant clones. After two weeks of drug exposure, 5–10% of the wells contained a rapidly growing resistant colony (Fig. 2a), whereas the other wells contained a small number of surviving drug tolerant cells similar to those described previously by Settleman and colleagues11. Allele specific quantitative PCR revealed that all “early” resistant colonies examined (n = 50) harbored the T790M mutation (Supplementary Fig. 4a), and all were sensitive to WZ4002 but not gefitinib (Fig. 2b). In contrast, treatment of PC9 pools with WZ4002 for two weeks yielded drug tolerant cells but completely suppressed emergence of the early T790M colonies (Fig. 2c). Conversely, treatment of PC9 pools with gefitinib + the IGF-1R inhibitor AEW541, which was previously shown to abrogate the survival of PC9 drug tolerant cells11, eliminated drug tolerant cells without affecting the emergence of early resistant T790M clones. These results demonstrate that the early resistant T790M clones emerged independently from drug tolerant cells.
Figure 2. Early T790M acquired resistance results from selection of pre-existing T790M clones.
(a) PC9 parental cell pools were treated with 300 nM gefitinib for two weeks and cell viability was determined by CellTiter-Glo assay. 7% of wells contained a rapidly proliferating resistant colony, while the remaining wells contained a small number of drug tolerant cells (inset, scale bar = 500 μm). Each bar represents one well; data shown are combined from two independent experiments and are normalized to the mean viability of drug tolerant wells.
(b) 50 early resistant clones were treated with gefitinib (GEF) or WZ4002 (WZ) for 72 hours and cell viability determined (upper panel). Inset shows dose response of PC9 parental cells. Lower panel compares cell viability of each early resistant T790M clone after treatment with 1 μM WZ4002 or gefitinib relative to vehicle control. Diagonal axis indicates equal sensitivity to both drugs (e.g., non-T790M), whereas sensitivity to WZ4002 but not gefitinib (lower right quadrant) indicates T790M. Parental cells sensitive to both drugs are shown for comparison.
(c) PC9 cell pools (5,000 cells) were treated with 1 μM WZ4002 or 300 nM gefitinib + 500 nM AEW541 for two weeks and cell viability determined.
(d) The barcode distribution (fraction of each unique barcode in the total barcode reads) of one representative replicate of ClonTracer barcoded PC9 cells (CT-A) treated with gefitinib compared with vehicle treated control. The x-axes of the histograms are identical; each bar represents one unique barcode. Colors denote the number of other replicates in which the barcodes were identified as in Figure 2e.
(e) Percentage of enriched barcodes shared by each replicate (1–5) for each independently barcoded experiment (CT-A, B, C).
We hypothesized that the early resistant T790M clones derived from rare pre-existing T790M cells that existed in our PC9 parental cell line. Direct detection of genetically distinct rare sub-populations using currently available standard next-generation sequencing is limited to allelic frequencies of approximately 0.1%21, whereas droplet digital PCR has a sensitivity of 0.01–0.005%22. In our PC9 parental cell line, we estimated that approximately 1 out of 25,000–50,000 cells harbor a T790M allele prior to treatment (see Online methods for calculation). Because PC9 cells carry 8–10 copies of EGFR (Supplementary Fig. 4b), and only one T790M allele is required to confer resistance (Supplementary Fig. 4c–d), direct detection of the pre-existing T790M clones in our parental PC9 population would necessitate a sensitivity at least an order of magnitude beyond what is routinely achievable.
To overcome these limitations, we used a recently reported a high-complexity DNA barcode library (ClonTracer) designed to track the evolution of pre-existing resistant clones too rare to be detected by NGS or ddPCR8. In this system, when a population of barcoded cells is made resistant to drug, enrichment of a shared set of barcodes between multiple replicates indicates selection of pre-existing resistant clones. We transduced 1–2 million PC9 cells with lentiviral Clontracer barcodes, expanded the cultures, and split them (CT-A) into multiple biological replicates of 20 million cells each to ensure 10-fold representation of barcode complexity (Supplementary Fig. 5a). Cells were treated with 300 nM gefitinib, and consistent with our prior experiments, rapidly proliferating early resistant clones were observed at 2–3 weeks. These clones were T790M positive (Supplementary Fig. 5b), and we observed a consistent number of unique barcodes enriched in each replicate (Fig. 2d and Supplementary Fig. 5c). Approximately 90% of enriched barcodes in each sample were shared by at least two other replicates and over 50% were shared by all five replicates (Fig. 2e and Supplementary Fig. 5d). Notably, the most highly shared barcodes were also the most highly enriched. We repeated the entire experiment with two additional, independently barcoded PC9 populations (CT-B and CT-C) that yielded the same results (Fig. 2e and Supplementary Fig. 5b–d). These results confirm that the early resistant T790M clones were derived from pre-existing T790M mutant cells that were selected during gefitinib treatment.
Late emerging T790M clones derive from drug tolerant cells
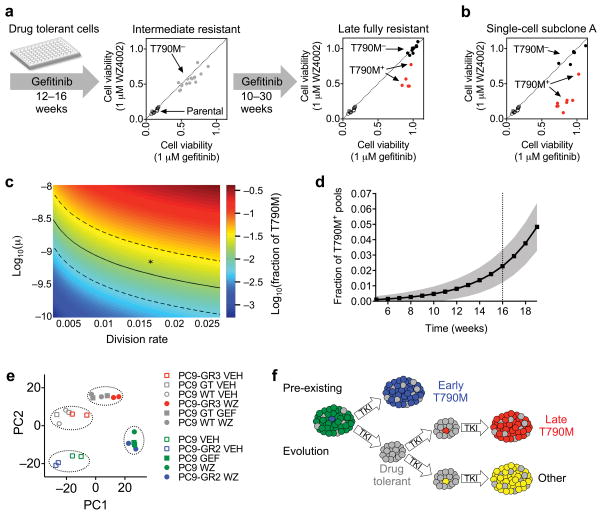
To determine whether T790M could also evolve during treatment from initially T790M-negative drug tolerant cells, we cultured 16 PC9 drug tolerant cell pools that showed no evidence of early resistant clones in the continuous presence of gefitinib until they had reached numbers sufficient for genotyping and drug sensitivity testing, which required 12–16 weeks. These pools—which we refer to as intermediate resistant—remained partially resistant to both gefitinib and WZ4002, and T790M allele-specific PCR was negative, indicating non-T790M mediated resistance (Fig. 3a and Supplementary Fig. 6a). These cells were further cultured in gefitinib until they became fully resistant, and five of the 16 resistant pools acquired T790M and demonstrated increased sensitivity to WZ4002 compared to gefitinib (Fig. 3a and Supplementary Fig. 6b). To gain a better understanding of the kinetics of evolution of T790M resistance in these cells, we examined additional intermediate time points during the development of the original PC9-GR3 resistant line and several of the late T790M clones. For the PC9-GR3 line, T790M was first detected at 22 weeks, at a fraction of 6% of the total cell population, and within one month had overtaken the population as the dominant clone (Supplementary Fig. 6c,d and Supplementary Fig. 4d). For three late T790M clones examined, a minority T790M clone was observed at 34, 43 and 47 weeks, respectively, and by one month later had overtaken the population (Supplementary Fig. 6d). Thus, these results suggest that the T790M resistance mutation can develop de novo in drug tolerant cells during the course of prolonged exposure to EGFR inhibitor.
Figure 3. Late emerging T790M acquired resistance results from evolution of drug tolerant cells.
(a) PC9 cell pools comprised of only drug tolerant cells were cultured continuously in gefitinib and T790M status and sensitivity to gefitinib and WZ4002 were determined at the indicated time points (see Supplementary Fig 6a,b).
(b) Pools of PC9 single cell-derived subclone A were cultured in gefitinib until fully resistant and T790M status and sensitivity to gefitinib and WZ4002 were determined (see Supplementary Fig. 7b).
(c) The frequency of PC9 drug tolerant cell pools developing a T790M mutation during 16 weeks of drug treatment mathematically modeled as a function of varying mutational rates (μ) and cell division rates. Frequency values were calculated from the analytical solution of the mathematical model and are depicted as log10 of the fraction of wells predicted to develop T790M. Parameters that yield the experimentally observed frequency of 1.5% are indicated by the solid (mean) and dashed (95% confidence) lines. The asterisk denotes parameters used in Fig. 3d. The scale of the x-axis corresponds to a division rate of once every two weeks (low) to the rate of PC9 cells in the absence of drug (high).
(d) Predicted frequency of acquiring T790M as a function of time (black line is mean, gray represents standard deviation). Parameters for drug tolerant cells: initial population size N0 = 150, birth rate b = 0.0162, death rate d = 0.015, mutation rate μ = 7×10−10. Birth and death rates of T790M mutant cells are br = 0.04 and dr = 0.0015, respectively.
(e) Principal component analysis of transcriptional profiles of PC9 parental, drug tolerant (selected for 2 weeks in gefitinib (GT) or WZ4002 (WT), early T790M PC9-GR2 and late T790M PC9-GR3 cells determined by RNA-Seq. Prior to harvesting RNA, cells were treated with 1 μM gefitinib (GEF), WZ4002 (WZ) or vehicle (VEH) for 24 hours.
(f) Model for development of T790M acquired resistance from pre-existing T790M cells as well as evolution of initially T790M-negative drug tolerant cells.
Resistant cells that remained T790M-negative were subjected to a clinical assay that detects single nucleotide variants and insertions/deletions in 39 cancer-related genes23. Notably, these T790M-negative resistant cells had mutations in NRAS, KRAS, BRAF and RET (Supplementary Fig. 7a), albeit at low allele frequencies suggesting mixed clonal populations. We did not observe MET or HER2 amplification, and one resistant line had low-level KRAS amplification (Supplementary Fig. 7b) in addition to a KRAS mutation. Although some of these mutations have been previously observed in PC9 cells and in resistant EGFR mutant NSCLC tumors24,25, the significance of these particular mutations driving resistance in the clinic is unclear. Of note, the gefitinib resistant PC9-GR1 cell line developed in parallel with the PC9-GR2 and PC9-GR3 lines that we previously reported did not harbor a T790M mutation but was sensitive to combination EGFR+MEK inhibition14 developed at a rate nearly identical to the drug-tolerant derived T790M PC9-GR3 cell line (Supplementary Fig. 7c). Taken together, these results suggest that drug tolerant cells have the potential to develop both T790M and non-T790M mechanisms of resistance, which may contribute to the heterogeneity of acquired resistance observed in the clinic4,26.
To further demonstrate that T790M could evolve during treatment from initially T790M-negative drug tolerant cells, we established PC9 subclone lines derived from single cells to eliminate pre-existing T790M cells. Indeed, in ten independent single cell sub-clone lines, no early T790M colonies emerged after two weeks of gefitinib treatment (Supplementary Fig. 8a) in contrast to the parental PC9 cells which harbored pre-existing T790M cells (Fig. 2a). We then cultured 14 pools (5,000 cells each) from single cell subclone A (Supplementary Fig. 8a) in gefitinib up to 40 weeks until fully resistant cells emerged. Eight acquired T790M and six remained T790M-negative (Fig. 3b and Supplementary Fig. 8b). Similarly, three additional single cell subclones (B–D) were cultured in gefitinib for several months, and all developed late resistant T790M clones (data not shown). To provide statistical evidence that these late T790M clones did not derive from undetected pre-existing T790M cells, we spiked single RFP-labeled T790M PC9 cells into pools of unlabeled subclones A–D (5,000 cells) and measured the kinetics of emergence of resistant cells. Over a ten week time period, all resistant clones were derived from RFP-positive T790M cells, with the majority emerging within the first two weeks (Supplementary Fig. 8c). From this, we derived a cumulative density function (CDF) describing the probability of detecting the emergence of pre-existing resistant T790M cells as a function of time (Supplementary Fig. 8d,e). Since none of the single cell-derived subclones A–D gave rise to early T790M clones at 2 weeks (Supplementary Fig. 8a), the CDF determines that the probability that the late T790M clones that eventually emerged derived from undetected pre-existing T790M cells is 0.006 (see Supplementary Fig. 8 legend for calculation). Taken together, these results confirm that T790M acquired resistance can evolve from initially T790M-negative drug tolerant cells.
Considering the time scale and cell population sizes of our experiments, we were initially surprised that the T790M mutation could arise in the slow growing drug tolerant cells. Taking into account reasonable estimates of mutation and cell division rates and experimentally determined growth rates of drug tolerant cells, we mathematically modeled de novo acquisition of T790M during drug treatment (Supplementary Fig. 9a,b; see Online methods for details). Figure 3c displays the fraction of drug tolerant pools predicted to acquire T790M during 16 weeks of gefitinib treatment for a range of mutation and division rates. This fraction grows linearly with the initial population size, division and mutation rates, and exponentially with the growth rate and time (for fractions of less than 10%). Using a reasonable set of parameters, the mathematical model did predict emergence of T790M over a time period corresponding to several months (Fig. 3d). When we tested this model experimentally, we found that 1.5% of PC9 drug tolerant pools developed T790M over the equivalent time period, which is in reasonable agreement with the modeling results (Supplementary Fig. 9c).
To examine whether T790M can evolve from drug tolerant cells in other EGFR mutant NSCLC models, we examined MGH119 cells that were previously derived in our laboratory from a treatment naïve EGFR mutant NSCLC patient14. MGH119 cells were cultured in gefitinib until resistant, and two independent T790M lines (GR1, GR2) developed after 5–6 months. To determine if T790M cells pre-exist in this model, we treated MGH119 cell pools with gefitinib for three weeks, but no early resistant colonies were observed (Supplementary Fig. 10a). In contrast, when we simulated the presence of pre-existing T790M cells by introducing RFP-labeled MGH119-GR1 cells into MGH119 parental cell cultures, emerging gefitinib resistant RFP-positive colonies were detectable starting at 2–3 weeks (Supplementary Fig 10b). At 10 weeks, only RFP-positive resistant clones were observed. These results suggest that the MGH119 cell line does not harbor pre-existing T790M cells, and that the MGH119-GR1 and GR2 resistant cells arose from drug tolerant cells by acquiring T790M during drug treatment.
Late resistant T790M cells bear hallmarks of drug tolerant state
We next sought to determine if late resistant T790M cells exhibit molecular evidence of evolution from the drug tolerant state. Using RNA-Seq, we compared the transcriptional profiles of PC9 drug tolerant cells with PC9-GR3, PC9-GR2 and PC9 parental cells (Supplementary Fig. 11a). Principal component analysis (PCA) of mRNA expression at baseline and after EGFR inhibitor treatment revealed that the late T790M PC9-GR3 cells clustered with drug tolerant cells, and the early T790M PC9-GR2 cells clustered with parental PC9 cells (Fig. 3e). Whereas principal component 1 (PC1) primarily differentiated between baseline and drug treated samples, principal component 2 (PC2) segregated PC9-GR3 and drug tolerant cells from early PC9-GR2 and parental cells (Supplementary Fig. 11b). Gene set enrichment analysis revealed upregulation of genes related to epithelial-to-mesenchymal transition in drug tolerant and PC9-GR3 cells relative to parental and PC9-GR2; this pathway has been linked to resistance to EGFR inhibitor therapy in the clinic4. These results demonstrate that the late emerging PC9-GR3 cells share a transcriptional profile most similar to drug tolerant cells. This further supports the model that the late resistant T790M cells evolved from drug tolerant cells (Fig. 3f) and suggests that molecular features of the drug tolerant state that impact drug sensitivity may be maintained even after acquisition of the T790M mutation.
Late resistant T790M cells have reduced apoptosis upon EGFR inhibition
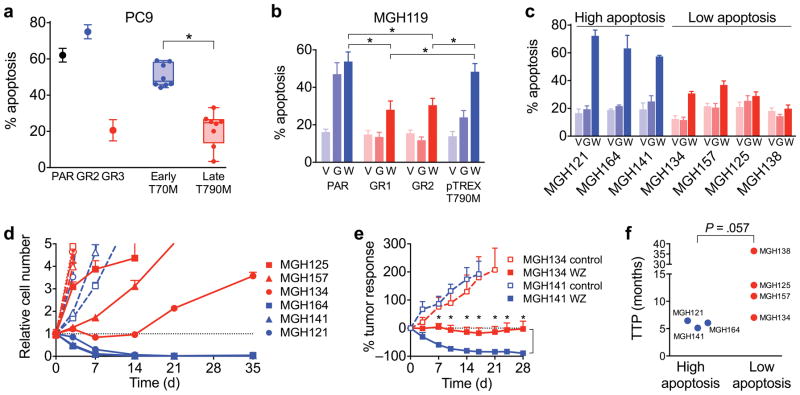
By definition, evasion of drug-induced apoptosis is a characteristic feature of drug tolerant cells (Supplementary Fig. 12a,b). Since late resistant T790M cells evolved from drug tolerant cells and continue to share a similar mRNA expression profile (Fig. 3e), we reasoned that they might also exhibit reduced apoptotic sensitivity to subsequent EGFR inhibition. Similar to PC9-GR3 cells (Fig. 1b), late T790M PC9 clones that emerged from drug tolerant cells had a reduced apoptotic response to WZ4002 treatment compared to early resistant T790M clones that were derived from pre-existing cells (Fig. 4a). Moreover, when we induced expression of T790M EGFR in drug tolerant cells, the T790M clones that grew out had reduced apoptotic sensitivity to WZ4002 (Supplementary Fig. 13a,b). MGH119-GR1 and GR2 cells, which developed T790M late consistent with evolution from drug tolerant cells, also exhibited reduced apoptotic response to WZ4002 relative to MGH119 parental cells and MGH119 cells in which we introduced T790M EGFR by lentiviral pTREX expression (Fig. 4b and Supplementary Fig. 13c). These results demonstrate that T790M cells that evolve from drug tolerant cells have reduced dependence on EGFR activation for survival compared to pre-existing T790M cells. This is consistent with the notion that drug tolerant cells are, by definition, the cells that do not undergo apoptosis following initial EGFR inhibition.
Figure 4. Late evolving T790M acquired resistance is associated with decreased apoptotic response to EGFR inhibition.
(a) Late T790M PC9 resistant clones derived from parental and single cell-derived clones and early resistant T790M clones were treated with 1 μM WZ4002 and apoptosis determined. Each dot represents % apoptosis over vehicle control for an individual clone (mean of 4 independent experiments; *P < 0.05, two-tailed t-test). PAR = PC9 parental cells.
(b) Gefitinib-resistant T790M MGH119 (GR1, GR2), MGH119-pTREX-T790M and parental MGH119 cells were treated with 1 μM gefitinib (G), WZ4002 (W) or vehicle (V) and apoptosis determined (mean and s.e.m. of 3 independent experiments; *P < 0.05, two-tailed t-test).
(c) Patient-derived T790M EGFR mutant NSCLC cell lines were treated with gefitinib (G), WZ4002 (W) or vehicle (V) and apoptosis determined (mean and s.e.m. of 4 independent experiments).
(d) Patient-derived cell lines with decreased apoptotic response were treated with 1 μM WZ4002 (closed symbols, solid lines) or vehicle (open symbols, dashed lines) and cell number determined by nuclear counting by high content imaging (mean and s.e.m. of four replicates).
(e) Mice bearing MGH134 and MGH141 subcutaneous xenograft tumors were treated with 50 mg/kg/day WZ4002. (MGH134 - control (N=8), WZ (N=10); MGH141 - control (N=7), WZ (N=8)) P < 0.01 (*) comparing WZ treatment arms at indicated time points by multiple t-tests with Sidak-Bonferroni multiple comparison.
(f) Time to progression (TTP) of patients from whom T790M patient-derived cell lines were established. TTP was determined by time from initiation of first line EGFR inhibitor therapy to first restaging scan showing definitive clinical progression of disease. Grouping of cell lines corresponds to Fig. 4c. Statistical significance was calculated using the Mann-Whitney U test.
To investigate the clinical implications of the laboratory-derived acquired resistance models, we examined cell lines derived from EGFR mutant NSCLC patient tumors at the time of clinical progression due to T790M acquired resistance14 (Supplementary Table 1). Three cells lines were exquisitely sensitive to WZ4002, whereas four had a diminished apoptotic response and increased in vitro survival (Fig. 4c,d). In vivo sensitivity of MGH141 and MGH134 xenograft tumors mirrored the in vitro response, with the low apoptosis MGH134 cell line xenografts failing to undergo tumor regression (Fig. 4e) despite suppression of EGFR and ERK signaling (Supplementary Fig. 3b). Intriguingly, the least sensitive cell lines corresponded to patients with the longest duration of response to first line EGFR inhibitor therapy prior to development of drug resistance (Fig. 4f), raising the possibility that these low-apoptotic resistant cells evolved from drug tolerant cells.
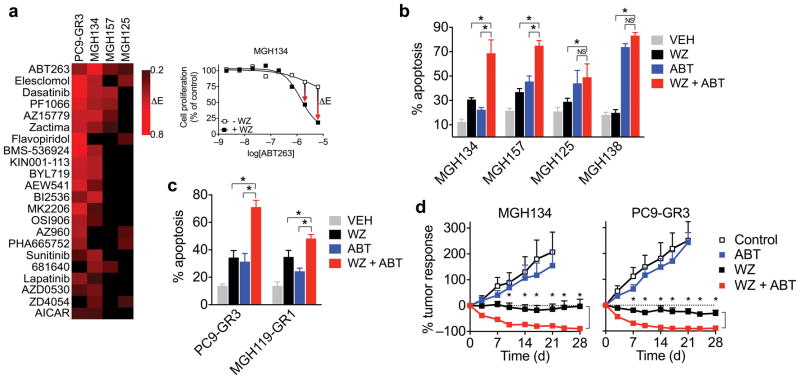
These results suggest that resistant cancers in which T790M arises during therapy may have a reduced apoptotic sensitivity to third generation EGFR inhibitors that are currently being evaluated in the clinic27–29. Therefore, we sought to identify therapeutic strategies that could enhance the apoptotic response of these cells to third generation EGFR inhibitors. Recently, we reported a drug screening strategy for identifying effective drug combinations for cancers with acquired resistance to targeted therapies14. Because this screen was focused on overcoming bypass resistance mechanisms, effective drug combinations were identified based on a shift in GI50 (drug concentration achieving 50% growth inhibition). However, we noted that the decrease in apoptosis in our models was most clearly reflected by a change in Emax (maximum growth inhibition achieved) (Supplementary Fig. 14). Therefore we reanalyzed the screen data for three T790M patient-derived cell lines (MGH134, MGH157, MGH125) to identify drugs that increased Emax when combined with WZ4002. Additionally, we performed the identical drug screen on PC9-GR3 cells also using WZ4002 as the anchor. Of the top scoring compounds for each cell line, the only one common to all four was the dual BCL-XL and BCL-2 inhibitor ABT-263 (also called navitoclax) (Fig. 5a). Notably, prior laboratory studies have demonstrated that dual BCL-XL and BCL-2 inhibitors can enhance the apoptotic activity of targeted therapies that induce apoptosis by inducing BIM17,30,31, presumably by decreasing the capacity of BCL-XL to neutralize BIM. The combination of WZ4002 and navitoclax induced significantly more apoptosis compared to WZ4002 alone in low T790M patient-derived cell lines (Fig. 5b) and in vitro generated resistant T790M cell lines (Fig. 5c). Combined treatment with WZ4002 and navitoclax also induced regression of MGH134 and PC9-GR3 xenograft tumors in mice (Fig. 5d). Thus, combining navitoclax with third generation EGFR inhibitors may be an effective strategy for treating T790M cancers with decreased apoptotic response to EGFR inhibition. Indeed, there are plans to examine the combination of AZD9291 and navitoclax in an upcoming clinical trial of EGFR NSCLC with acquired resistance to first line EGFR inhibitor therapy (NCT02520778).
Figure 5. Navitoclax enhances the apoptotic response of late resistant T790M cells with decreased sensitivity to EGFR inhibition.
(a) Top drug hits from a combination drug screen14 that led to an enhancement of Emax when combined with WZ4002 in at least two of four T790M cell lines screened. The response of MGH134 cells to ABT-263 is shown as an example.
(b) Patient-derived T790M cell lines were treated with 1μM WZ4002, ABT-263 or combination and apoptosis determined (mean and s.e.m. of 4 independent experiments; *P < 0.05, two-tailed t-test).
(c) In vitro derived late T790M gefitinib resistant cell lines were treated with 1 μM WZ4002, ABT-263 or combination and apoptosis determined (mean and s.e.m. of 4 independent experiments; *P < 0.05, two-tailed t-test;).
(d) Mice bearing MGH134 and PC9-GR3 subcutaneous xenograft tumors were treated with 100 mg/kg/day navitoclax and/or 50 mg/kg/day WZ4002. (MGH134 N=8 control, 10 WZ, 8 ABT, 7 WZ+ABT; PC9-GR3 N=8 control, 8 WZ, 7 ABT, 7 WZ+ABT). Control and WZ data from Figs. 1d and 4e are shown for comparison. P < 0.01 (*) comparing WZ and WZ + ABT treatment arms at indicated time points by multiple t-tests with Sidak-Bonferroni multiple comparison.
Discussion
It is now well appreciated that substantial genetic heterogeneity exists between cancer cell clones within a tumor32,33. Pre-existing intertumoral heterogeneity has been shown to influence treatment response and the development of acquired resistance to targeted therapy in some contexts34, but there has been little direct clinical evidence supporting either emergence of pre-existing resistant clones or de novo evolution as the predominant mechanisms of acquired resistance in lung cancer. Several studies have reported the detection of low frequency T790M-positive clones in pre-treatment clinical specimen9,35, although doubts have been raised as to whether the majority of these may be sequencing artifacts related to tissue fixation10.
Our results provide proof of principle that T790M clones not only emerge from selection of pre-existing clones, but also can emerge from initially T790M-negative drug tolerant cells. Survival of drug tolerant cancer cells may be facilitated by cell autonomous features such as altered epigenetic states11,36 and feedback activation of alternate survival pathways12 as well as external stimuli from the microenvironment13,37. While these mechanisms may be sufficient to prevent apoptosis and promote survival of drug tolerant cells, they may not fully recapitulate the optimal oncogenic signaling provided by EGFR. Subsequent acquisition of T790M likely provides an additional fitness advantage in the presence of drug. Thus drug tolerant cells that are capable of surviving initial drug therapy may provide a reservoir of cells from which genetic mechanisms of acquired resistance can evolve. Such resistant clones may continue to share molecular features of the drug tolerant cells (Fig. 3e,f), leading to a diminished apoptotic response to subsequent therapies. Our finding that T790M cell lines derived from tumors of patients who had experienced prolonged responses to first generation EGFR inhibitors had decreased apoptotic response to third generation EGFR inhibitors suggests that tumors in which T790M evolves during the course of therapy may be less responsive to third generation EGFR inhibitors (e.g. AZD9291, CO-1686)27,28 in the clinic. However, given the limited number of patient-derived cell lines available for this study, this hypothesis warrants further investigation with additional laboratory models and validation with clinical response data.
We believe that these findings have important clinical implications. These results provide a rationale for investigation of novel therapeutic strategies to target drug tolerant cells before acquisition of genetic mechanisms of resistance in order to prevent or delay the evolution of acquired resistance. We speculate that the majority of cancer cells that initially survive following therapy likely do not have bona fide resistant mechanisms (e.g., T790M), but more closely resemble a drug tolerant state. Moreover, in light of recent reports that some T790M EGFR mutant NSCLCs “lose” T790M upon developing resistance to AZD9291 or CO-168626,38, our studies raise the possibility that T790M-negative drug tolerant cells may persist in patients with resistant T790M tumors and undergo further evolution to acquire resistance to subsequent therapies. Since we now know that these drug tolerant cells can develop genetic mechanisms of resistance over time, we believe that it is imperative to develop therapeutic approaches that eliminate this reservoir of drug tolerant cells that fail to undergo apoptosis. Such treatment strategies will also need to integrate approaches that target the rare pre-existing bona fide resistant clones. We are hopeful that innovative approaches such as these will lead to more complete clinical remissions and improved patient outcomes.
Online Methods
Cell lines
The EGFR mutant NSCLC PC9 cell line (EGFR exon 19 del E746_A750, EGFR amplified) was obtained from the MGH Center for Molecular Therapeutics. The identity of PC9 parental, GR2 and GR3 cell lines were verified by STR analysis (Bio-synthesis, Inc.) at the time that these studies were performed. EGFR mutant NSCLC patient-derived cell lines were established in our laboratory from core biopsy or pleural effusion samples as previously described14,39. The MGH1075 stromal fibroblast line was established in our laboratory from a surgical resection specimen of an early stage primary lung adenocarcinoma. All patients signed informed consent to participate in a Dana Farber/Harvard Cancer Center Institutional Review Board-approved protocol giving permission for research to be performed on their samples. The HCC827-GR6 line (Exon 19 deletion, MET amplified) was previously described6. H358 cells were obtained from the Center for Molecular Therapeutics at MGH. PC9-T790M-RFP and MGH119-GR1-RFP cells were generated by infecting the PC9 Early T-1 clone and MGH119-GR1 cells, respectively, with Cignal Lenti RFP (Qiagen) lentivirus followed by puromycin selection. All cell lines were maintained in RPMI supplemented with 10% FBS, except MGH125 and MGH138 cells which were maintained in ACL4 with 10% FBS and TCM with 10% FBS, respectively. All experiments were performed in RPMI with 10% FBS. All cells were routinely tested and verified to be free of mycoplasma contamination.
Antibodies and reagents
For western blotting, the following antibodies were used: phospho-EGFR Y1068 (Abcam); EGFR, pERK1/2 T202/204, ERK1/2, p-AKT S473, BIM, Actin (Cell Signaling); AKT1/2/3 (Santa Cruz). For cell culture studies, gefitinib, WZ4002, NVP-AEW541, ABT-263 (all from Selleck), were dissolved in DMSO to a final concentration of 10 mmol/l and stored at −20°C. Unless otherwise specified, 1 μM concentration was used for in vitro cell culture experiments.
In vitro generation of gefitinib resistant cell lines
Gefitinib resistant PC9-GR2, PC9-GR3 and MGH119-GR1 cells were established by culturing parental cells in escalating concentrations of gefitinib (10 nM − 1 μM) as tolerated as previously described 14. MGH119-GR2 cells were established by culturing parental cells in full dose gefitinib (1 μM). Early, drug tolerant, late PC9 resistant clones were established by culturing in 300 nM gefitinib until resistant, at which point they were maintained in 1 μM gefitinib. During generation of resistance, media and drug were replaced twice per week.
Generation of PC9 single cell sub-clones
PC9 parental cells were seeded into 96-well plates at a density of 0.5 cells/well. After 2 weeks, approximately 25–40% of wells (50–80% of theoretical yield) contained colonies of about 10,000 cells. A minority of wells contained two colonies, which were easily distinguishable. Wells containing only single colonies were expanded an additional 6–10 doublings for use in experiments.
PI/Annexin apoptosis assay
Cells were seeded at low density 24 hours prior to drug addition. 72 hours after adding drugs, floating and adherent cells were collected and stained with propidium iodide and Cy5-Annexin V (BD Biosciences) and analyzed by flow cytometry. The annexin-positive apoptotic cell fraction was analyzed using FloJo software.
CellTiter-Glo proliferation assay
For dose response assays, cell lines were seeded into 96-well plates 24 hours before addition of drug. Cell proliferation was determined by CellTiter-Glo assay (Promega) 72 hours after adding drug using standard protocols. For time course experiments, multiple plates were seeded and drugged in identical fashion and at indicated time points plates were frozen at −80°C; all plates in an experiment were developed with CellTiter-Glo simultaneously. For early resistant/drug tolerant cell experiments, CellTiter-Glo was performed at indicated times on cells in situ.
RealTime-Glo viability assay
Cell viability was assayed in situ once weekly starting the day after seeding using the RealTime-Glo assay (Promega) according to the manufacturers protocol. Briefly, MT Cell Viability Substrate and NanoLuc Enzyme were diluted 1:500 in media and 25 μl was added to each well (1/5 total final volume). Cells were incubated for 1 hour at 37°C and luminescence measured. For experiments including RFP-labeled cells, RFP fluorescence was measured using Ex 553 nm/Em 574 nm. We did not observe any effect of the presence of RFP on RealTime-Glo luminescence or vice versa. Fresh media containing gefitinib was replaced immediately after each assay.
Long term viability assay
Long term viability assays were completed by plating 1,000–4,000 cells/well in replicate 96-well plates. Cells were treated in quadruplicate 2x/week and fixed at days 0, 3, 7, 14, 21, 28 and 35 with a mixture of formaldehyde (1.0%), PBST (0.04%), and Hoecsht dye (1μg/ml). Stained nuclei were imaged and then counted using a Molecular Devices ImageXpress Micro high content imager and MetaXpress software.
Cell cycle Analysis
Cells were seeded 24 hours prior to experiment to give a confluency of 30–50%. Drugs were added for 24 hours and cells harvested, stained with propidium iodide and analyzed by flow cytometry. Cell cycle sub-populations were calculated using the FloJo software cell cycle module.
JC-1 mitochondrial depolarization assay
Cells were treated with EGFR inhibitor in the presence of 10 μM QVD-Oph to prevent subsequent apoptosis. 48 hours after drug treatment, cells were harvested and stained with JC-1 (Life Technologies) according to the manufacturers recommendations. 10 μM CCCP was used as a positive control. Cells were analyzed by flow cytometry using excitation/emission filters for PE and FITC. Mitochondrial depolarization is indicated by a decrease in the red/green fluorescence intensity ratio.
Quantitative RT-PCR assay for gene expression
Cells were seeded 24 hours prior to give a confluency of 50%. Cells were treated with drugs for 24 hours and RNA was extracted using the RNeasy Kit (Qiagen). cDNA was prepared from 500 ng total RNA with the First Strand Synthesis Kit (Invitrogen) using oligo-dT primers. Quantitative PCR was performed using FastStart Sybr Green (Roche) on a Lightcycler 480. mRNA expression relative to ACTIN was calculated using the Delta-Delta threshold cycle method. Primers used: DUSP6 F 5′-CGACTGGAACGAGAATACGG-3′, R 5′-TTGGAACTTACTGAAGCCACCT-3′; SPRY4 F 5′-CCCCGGCTTCAGGATTTA-3′, R 5′-CTGCAAACCGCTCAATACAG-3′; HER3 F 5′-CTGATCACCGGCCTCAAT-3′, R 5′-GGAAGACATTGAGCTTCTCTGG-3′; DAPK1 F 5′-CCCTTGTCCCAGTTGAAGAA-3′, R 5′-CCGGTCGAGGAACATTCA-3′; BIM F 5′-GATCCTTCCAGTGGGTATTTCTCTT-3′, R 5′-ACTGAGATAGTGGTTGAAGGCCTGG-3; Actin F 5′-CTGTGCTATCCCTGTACGCCTC-3′, R 5′-CATGATGGAGTTGAAGGTAGTTTCGT-3′; GAPDH F 5′-AACAGCGACACCCATCCTC-3′, R 5′-CATACCAGGAAATGAGCTTGACAA-3.
T790M quantitative PCR assay
The T790M mutation was detected using allele specific primers (F) 5′-ACCATGCGAAGCCACACTGACG-3′ and (R) 5′-AGCCGAAGGGCATGAGCTGGA-3′ in conjunction with an EGFR exon 20 Taqman probe 5′-ATCACGTAGGCTTCCTGGAG-3′. A commercially available EGFR exon 19 deletion Taqman assay was used as a reference (Hs00000228_mu, Life Technologies). Quantitative PCR was performed on genomic DNA using either FastStart PCR Master or Lightcycler 480 Probes Master kits (Roche) on a Lightcycler 480. For screening assays, ΔCp(T790M-DEL19) < 8 was considered positive and ΔCp > 10 was considered negative (Probes Master), or ΔCp < 14 positive/ΔCp > 16 negative (FastStart).
EGFR T790M droplet digital PCR analysis
Isolated gDNA was amplified using ddPCR™ Supermix for Probes (Bio-Rad) with EGFR p.T790M (PrimePCR™ ddPCR™ Mutation Assay, Bio-Rad) for point mutation detection. ddPCR was performed according to manufacturer’s protocol and the results reported as percentage or fractional abundance of mutant DNA alleles to total (mutant plus wild type) DNA alleles. 8 to 10 μl of DNA template was added to 10 μl of ddPCR™ Supermix for Probes (Bio-Rad) and 2 μl of the primer/probe mixture. Droplets were generated using Auto-DG where the reaction mix was added together with Droplet Generation Oil for Probes (Bio-Rad). Droplets were then transferred to a 96 well plate (Eppendorf) and then thermal cycled with the following conditions: 5 minutes at 95°C, 40 cycles of 94°C for 30s, 55°C for 1 minute followed by 98°C for 10 minutes (Ramp Rate 2°C/sec). Droplets were analyzed with the QX200™ Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes. Gating was performed based on positive and negative controls, and mutant populations were identified. The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad) to obtain Fractional Abundance of the mutant DNA alleles in the wild-type/normal background. Fractional Abundance is calculated as follows: F.A. % = (Nmut/(Nmut+Nwt))*100), where Nmut is number of mutant events and Nwt is number of WT events per reaction. ddPCR analysis of normal control (from cell lines) and no DNA template controls were always included. Samples with low positive events were repeated at least twice in independent experiments to validate the obtained results.
Clinical cancer genotyping analysis
DNA was extracted from late non-T790M gefitinib resistant PC9 clones using the DNeasy kit (Qiagen). Genotyping was performed using the MGH NGS platform which utilizes a multiplex polymerase chain reaction (PCR) technology called Anchored Multiplex PCR (AMP) for single nucleotide variant (SNV) and insertion/deletion (indel) detection in genomic DNA using next generation sequencing (NGS). Genomic DNA was sheared with the Covaris M220 instrument, followed by end-repair, adenylation, and ligation with an adapter. A sequencing library targeting hotspots and exons in 39 genes was generated using two hemi-nested PCR reactions. Illumina MiSeq 2 × 151 base paired-end sequencing results were aligned to the hg19 human genome reference using BWA-MEM. MuTect and a laboratory-developed insertion/deletion analysis algorithm were used for SNV and indel variant detection, respectively. This assay has been validated to detect SNV and indel variants at 5% allelic frequency or higher in target regions with sufficient read coverage. Variants are reported with Human Genome Variation Society (HGVS) protein and DNA nomenclature, followed by the referenced Ensembl transcript ID. The gene targets covered by this assay are as follows (exons): AKT1 (3), ALK (22, 23, 25), APC (16), BRAF (11, 15), CDH1 (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,12, 13, 14, 15, 16), CDKN2A (1, 2, 3), CTNNB1 (3), DDR2 (12, 13, 14, 15, 16, 17, 18), EGFR (7, 15, 18, 19, 20, 21), ERBB2 (10, 20), ESR1 (8), FBXW7 (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11), FGFR1 (4, 8, 15, 17), FGFR2 (7, 9, 12,14), FGFR3 (7, 8, 9, 14, 16), FOXL2 (1), GNA11 (5), GNAQ (4, 5), GNAS (6, 7, 8, 9), HRAS (2, 3), IDH1 (3, 4), IDH2 (4), KIT (8, 9, 11, 17), KRAS (2, 3, 4, 5), MAP2K1 (2, 3), MET (14, 16, 19, 21), NOTCH (25, 26, 34), NRAS (2, 3, 4, 5), PDGFRA (12, 14, 18, 23), PIK3CA (2, 5, 8, 10, 21), PIK3R1 (1, 2, 3, 4, 5, 6, 7, 8, 9, 10), PTEN (1, 2, 3, 4, 5, 6, 7, 8, 9), RET (11, 16), ROS1 (38), SMAD4 (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12), SMO (9), STK11 (1, 2, 3, 4, 5, 6, 7, 8, 9), TP53 (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11), and VHL (1, 2, 3).
Gene amplification quantitative PCR assay
Quantitative PCR was performed on genomic DNA using FastStart Sybr Green (Roche) on a Lightcycler 480. Primers for KRAS, MET, Chromosome 12 control, Chromosome 7 control were previously described40. EGFR, BRAF and NRAS primers were adapted from Dias-Santagata et al41: EGFR F 5′-CCTCCTTCTGCATGGTATTC-3′, R 5′-GCAGCATGTCAAGATCACAG-3′; BRAF F 5′-TGCTTGCTCTGATAGGAAAATG-3′, R 5′-CTGATGGGACCCACTCCAT-3′; NRAS F 5′-CAACAGGTTCTTGCTGGTGT-3′, R 5′-GAGAGACAGGATCAGGTCAGC-3′. LINE1 primers: F 5′-AAAGCCGCTCAACTACATGG -3′, R 5′-TGCTTTGAATGCGTCCCAGAG -3′). ΔCp between gene-control (KRAS - Chr.12, LINE1; MET, EGFR, BRAF - Chr. 7, LINE1; HER2, NRAS - LINE1) were calculated and normalized to MGH1075 diploid fibroblast control. HCC827-GR6 were used as a MET amplification control 6 and H358 cells were used as KRAS amplification control.
Generation of Tet-inducible EGFR pTREX cell lines
Exon 19 del and Exon 19 del/T790M plasmids were purchased from Addgene (32062, 32072). EGFR sequences were PCR amplified with the F 5′-CACCATGCGACCCTCCGGGACG-3′ and R 5′-TCATGCTCCAATAAATTCACTG-3′ primers and ligated into the pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning Kits (Invitrogen). The sequence was then introduced into the pTREX vector (kindly provided by Novartis) using Gateway LR Clonase Enzyme (Invitrogen). The pTREX-DEL19 and pTREX-DEL/T790M vectors were subsequently verified by DNA sequencing. Lentivirus was produced using standard procedures42. PC9 and MGH119 cells were infected with lentivirus followed by puromycin selection. Cells were cultured in the presence of 10 ng/ml doxycycline to induce expression of EGFR construct. For infection of PC9 drug tolerant single cells, cells were infected with a lentiviral titer that yielded a single puromycin resistant colony in ~50% of wells.
Mouse xenograft studies
All mouse studies were conducted through Institutional Animal Care and Use Committee–approved animal protocols in accordance with institutional guidelines. For xenograft studies, cell line suspensions were prepared in 1:10 matrigel and 5×106 cells were injected subcutaneously into the flanks of female athymic nude (Nu/Nu) mice (6–8 weeks). Visible tumors developed in approximately 2–4 weeks. Tumors were measured with electronic calipers and the tumor volume was calculated according to the formula V = 0.52 × L × W2. Mice with established tumors were randomized to drug treatment groups using covariate-adaptive randomization to minimize differences in baseline tumor volumes: WZ4002 50 mg/kg (10% 1-methyl-2-pyrolidone, 90% PEG300), ABT-263 100 mg/kg (30% PEG400/60% Phosal 50 PG/10% ethanol), or combinations. Drug treatments were administered by oral gavage and tumor volume measured twice weekly as above. For pharmacodynamic studies, mice were sacrificed and tumors removed 3 hours after drug administration on day 3. Tumors were snap frozen and lysates prepared for western blotting. Investigators performing tumor measurements were not blinded to treatment group. Sample size (minimum N=7 per treatment group) was chosen to verify satisfactory inter-animal reproducibility.
Estimation of frequency of pre-existing T790M clones in parental PC9 cell stock
After 2 weeks of drug exposure, 90 of 1260 wells were detected to contain resistant T790M colonies. Based on the above modeling of emergence of pre-existing RFP-labeled T790M clones, this represents approximately 72% of pre-existing T790M cells. Thus the theoretical frequency of pre-existing T790M can be estimated:
From experiments generating PC9 single cell clones, the efficiency of single PC9 cells to establish progeny is between 50–80%, thus the frequency of T790M cells in our parental PC9 line is approximately 1:25,000 – 1:50,000.
ClonTracer barcode library
Construction of the ClonTracer barcode library and generation of lentiviral particles was previously described8. Pools of 10 million PC9 cells were barcoded by lentiviral infection at a MOI of 0.1–0.2 and infected cells were selected with puromycin. Infected cell populations were expanded in culture for the minimal time period to obtain a sufficient number of cells to set up replicate experiments. 20 million cells (~10x barcode representation) were seeded in multiple replicate plates and treated with vehicle or 300 nM gefitinib (5 replicates). Vehicle treated cells reached confluency within 3 days and the entire pool was pelleted and snap frozen. Gefitinib treated plates became confluent from emergence of early resistant clones and were harvested after 3 weeks. Cell counts were approximately 20 million cells per replicate plate. Genomic DNA was extracted from the frozen cell populations with a QIAamp DNA Blood Maxi Kit (Qiagen). We used PCR to amplify the barcode sequence for NGS and introduce Illumina adaptors and index sequences8. PCR primer sequence information can be found at https://www.addgene.org/pooled-library/clontracer/. The sampling of sufficient template coverage was ensured by parallel PCR reactions.
PCR-amplified products were sequenced on an Illumina HiSeq2500 sequencer in Rapid Mode using the 50 Cycle TruSeq Rapid v2 SBS Kit, TruSeq Rapid SR v2 Cluster Kit, and HiSeq Rapid SR v2 Flow Cell (Illumina) as previously described8. Barcode-composition analysis was carried out as previously described8. A barcode was called significant if it was seen at more than 0.01% of the total population in the drug treated samples (10 times the fraction of the most enriched barcode in the vehicle treated replicates).
Cumulative density function modeling of emergence of T790M clones
Using the cumulative number (N) of RFP+ clones detected at each time point, a best fit model (without being highly sensitive to local noise) was calculated with the Matlab Curve Fitting Tool with fittype exp2, representing a model of the form a*exp(b*x) + c*exp(d*x). The 95% confidence bounds for the coefficients of equation 1 were: a = 53.56 (47.19, 59.93), b = 0.0002647 (−0.001714, 0.002243), c = −53.58 (−60.17, −47), d = −0.09483 (−0.1178, −0.07186). The equation:
was normalized by the value f(70) = 54.492, transforming it into a cumulative density function with values ranging from [0,1]:
The value of the CDF at t(14 days) = 0.7259, representing the probability that a pre-existing T790M clone would emerge within 14 days. The probability that a pre-existing T790M would not emerge within 14 days is 1−0.7259 = 0.2741. The probability of this occurring in 4 independent trials = 0.2741^4 = 0.0056.
Mathematical modeling of evolution of T790M from drug tolerant cells
To model de novo acquisition of the T790M mutation during drug treatment, we assumed a branching process43 comprised of N0 drug tolerant cells that divide at a rate b, die at rate d, and upon birth, mutate at a rate μ. Acquisition of T790M mutations yields resistant cells divide at a rate of br and die at a rate dr. The range of values of the birth and death rates of drug tolerant cells were based on single cell parameters reported by Tyson et al.44 and fit to population growth rates determined by tracking the growth of pools of PC9 drug tolerant during weeks 5–11 of gefitinib treatment (Supplementary Fig. 9a). The average estimated growth rate determined by fitting the growth curves to an exponential function from the week with the smallest population, which was usually week 6 until week 11, and taking the average was b−d = 0.0015 ± [0.0012]/h. Using the inferred growth rate we back extrapolated the initial population size of the drug tolerant cells to be on average N0 = 150 ± 23. Once a cell acquired the T790M mutation, the birth and death rate of the resulting T790M cells were assumed to have similar rates as PC9 parental cells in the absence of drug (Tyson et al.44). Currently there is no agreement on the average mutation per SNV per cell division, and estimation ranges from 10−11 up to 10−7 for non-hypermutated cells45,46. More recent estimates range from 10−10 to 10−8 47, with a slight tendency towards lower values. Of note, the EGFR T790M mutation is a change of ACG to ATG generated by spontaneous deamination of 5-methylcytosine to uracil, which occurs at 18 times greater than the average rate48; furthermore the EGFR gene has a rate that is about twice the average rate47 across the genome (MutSigCV49 estimation output). Gene amplification can increase the allelic pool in which a mutation can arise, and PC9 cells have EGFR gene amplification (10 copies per cell, see Supplementary Fig. 4b) with a single T790M mutation being sufficient to confer drug resistance. Effectively, the rate of getting a T790M mutation is about 360 times more likely than an average SNV.
We modeled the process with a Monte Carlo simulation in order to estimate the fraction of drug tolerant pools predicted to acquire T790M by16 weeks after beginning drug treatment. Conversely, we simulated the fraction of drug tolerant pools predicted to acquire T790M as a function of time using fixed parameters. Additionally we derived an analytical approximation to the model
where R represent the fraction of PC9 drug tolerant pools that acquire a T790M mutation as a function of time (T). Parameters for drug tolerant cells: initial population size N0 = 150, birth rate b = 0.0162, death rate d = 0.015, mutation rate μ = 7×10−10 (estimated to be the average of many estimates by Lynch47). The birth and death rates of T790M mutant cells are br = 0.04 and dr = 0.0015, respectively44. In Supplementary Fig. 9, we compare the results of the analytical solution (black line) to the Monte Carlo simulations (red dots) based on 10,000 runs per set of values. The logic behind the solution is to estimate the number of birth events from day 0 to the end of the experiment which is
and calculate the probability of having at least one T790M mutation. One should have in mind that the mutation rate in the equation is not the average mutation per SNV per cell division, but rather 360 times larger, as explained above. It is possible that prior models may not have taken this into account, which may explain the differences in the assumed baseline mutational rates between our model and prior models (e.g., 10−8–10−7)43.
RNA-Seq analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). RNA-Seq libraries were constructed from polyA-selected RNA using NEBNext Ultra Directional RNA library prep kit for Illumina (New England Biolabs) and sequenced on Illumina HiSeq2500 instrument, resulting in approximately 34 million reads per sample on average. The data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE75602. STAR aligner50 was used to map sequencing reads to transcripts in human hg19 reference genome. Read counts for individual transcripts were produced with HTSeq-count51, followed by the estimation of expression values and detection of differentially expressed transcripts using EdgeR52. Principle component analysis (PCA) was performed on the union of differentially expressed transcripts in all samples. We used the package for gene-set enrichment analysis (GSEA)53,54 to analyze the enrichment of functional gene groups among differentially expressed transcripts. Normalized enrichment score (NES) and enrichment plots were calculated based on gene lists ranked by expression ratio between early (PC9 parental and PC9-GR2) and late (PC9-GR3 and drug tolerant) stages.
Combination drug screen
The combination drug screening platform has been described previously14. Briefly, the panel of screened compounds was comprised of 76 targeted agents directed against a broad range of biologic targets including regulators of growth factor and development signaling pathways, apoptosis, transcription and protein folding, and DNA damage. Cells were treated with vehicle or varying concentrations of screen drugs (10,000-fold range) in the absence or presence of 1 μM WZ4002 for 72 hours and cell viability determined by CellTiter-Glo assay. Each dose response curve was normalized to cell viability of corresponding vehicle treated cells (with or without WZ4002). For each screen drug dose, a ΔE was calculated:
The means of the three highest ΔE for all screen drugs were ranked, and screen drugs that scored 0.20 or higher in at least 2 of 4 cell lines were identified.
Data and Statistical Analysis
Data were analyzed using GraphPad Prism software (GraphPad Software). Unless otherwise specified, data displayed are mean and s.e.m. Pair-wise comparisons between groups (e.g., experimental versus control) were made using paired or unpaired t-tests as appropriate. For xenograft tumor measurements, individual time points were compaired using multiple t tests with Sidak-Bonferroni multiple comparison. For xenograft pharmacodynamic studies, unequal variance between groups was observed in a minority of cases, so Welch correction was performed for all comparisons. For all other experiments, variance between comparison groups was verified to be equivalent. Unless otherwise indicated, P values below 0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Cyril Benes and all members of the Engelman and Benes Lab for helpful discussions and feedback. This study was funded by support from the NIH R01CA137008 (J.A.E.), the Department of Defense (L.V.S. and J.A.E.), LunGevity (L.V.S. and J.A.E.), Uniting Against Lung Cancer (A.N.H. and M.J.N.), Conquer Cancer Foundation of ASCO (A.N.H.), Lung Cancer Research Foundation (M.J.N), Targeting a Cure for Lung Cancer, and Be a Piece of the Solution.
Footnotes
Competing financial interests
The authors declare competing financial interests: J.A.E. is a consultant for Novartis, Sanofi, Genentech, Clovis and Astra Zeneca; owns equity in Gatekeeper Pharmaceuticals, which has interest in T790M inhibitors; has research agreements with Novartis and Astra Zeneca. A.N.H. has provided consulting services for Amgen. M.J.N. has provided consulting services for Boehringer-Ingelheim. Z.P. has provided consulting services for Clovis Oncology and Boehringer-Ingelheim. L.V.S. has provided uncompensated consulting services to Clovis Oncology, AstraZeneca, Novartis, Boehringer-Ingelheim, Merrimack Pharmaceuticals, Genentech and Taiho Pharmaceuticals. H.C.B., V.K.R., C.L.K, D.A.R and and F.S. are employees of Novartis, Inc., as noted in the affiliations.
Accession codes:
RNA-Seq datasets from PC9 parental, drug tolerant and resistant populations have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE75602 (individual datasets GSM1960262-83).
Author Contributions
A.N.H., M.J.N. and J.A.E. designed the study, analyzed the data and wrote the paper. A.N.H, M.J.N., H.L.A, M.G.C., F.M.S, H.E.M., H.H. and L.J.D. performed cell line and biochemical studies. M.G.C. and C.C. performed tumor xenograft studies. E.L., A.K. and D.L. generated the patient derived cell lines. H.C.B, V.K.R., C.L.K., D.A.R. and F.S. performed analysis of barcode analysis. Y.E.M. and G.G. performed mathematical modeling of T790M evolution. F.J. and R.I.S. performed RNA-Seq analysis. A.S.C. performed combination drug screening. G.S., A.J.I. and A.B. performed genotyping analysis. S.R. performed mathematical modeling of emergence of RFP resistant clones. A.C.F. was involved with study design. L.V.S. and Z.P. provided EGFR mutant NSCLC patient samples. A.N.H and M.J.N. contributed equally to the study. All authors discussed the results and commented on the manuscript.
References
- 1.Mok T, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 3.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 4.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 7.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhang HE, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med. 2015 doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 9.Su KY, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 10.Ye X, et al. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: truth or artifact? Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2013;8:1118–1120. doi: 10.1097/JTO.0b013e31829f691f. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HJ, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Wilson TR, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crystal AS, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014 doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hata AN, Engelman JA, Faber AC. The BCL2 Family: Key Mediators of the Apoptotic Response to Targeted Anticancer Therapeutics. Cancer Discov. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:1681–1689. doi: 10.1371/journal.pmed.0040316. discussion 1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa DB, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. discussion 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong Y, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:1655–1668. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber AC, et al. BIM expression in treatment naïve cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robasky K, Lewis NE, Church GM. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet. 2014;15:56–62. doi: 10.1038/nrg3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Z, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 24.Ohashi K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberlein CA, et al. Acquired Resistance to the Mutant-Selective EGFR Inhibitor AZD9291 Is Associated with Increased Dependence on RAS Signaling in Preclinical Models. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piotrowska Z, et al. Heterogeneity Underlies the Emergence of EGFR T790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third Generation EGFR Inhibitor. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janne PA, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 28.Sequist LV, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2015;372:1700–1709. doi: 10.1056/NEJMoa1413654. [DOI] [PubMed] [Google Scholar]
- 29.Cross DA, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cragg MS, et al. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest. 2008;118:3651–3659. doi: 10.1172/JCI35437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corcoran RB, et al. Synthetic Lethal Interaction of Combined BCL-XL and MEK Inhibition Promotes Tumor Regressions in KRAS Mutant Cancer Models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 35.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirata E, et al. Intravital Imaging Reveals How BRAF Inhibition Generates Drug-Tolerant Microenvironments with High Integrin beta1/FAK Signaling. Cancer Cell. 2015;27:574–588. doi: 10.1016/j.ccell.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thress KS, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niederst MJ, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misale S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Science translational medicine. 2014;6:224ra226. doi: 10.1126/scitranslmed.3007947. [DOI] [PubMed] [Google Scholar]
- 41.Dias-Santagata D, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hata AN, et al. Failure to induce apoptosis via BCL-2 family proteins underlies lack of efficacy of combined MEK and PI3K inhibitors for KRAS-mutant lung cancers. Cancer Res. 2014;74:3146–3156. doi: 10.1158/0008-5472.CAN-13-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chmielecki J, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Science translational medicine. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyson DR, Garbett SP, Frick PL, Quaranta V. Fractional proliferation: a method to deconvolve cell population dynamics from single-cell data. Nat Methods. 2012;9:923–928. doi: 10.1038/nmeth.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 46.Oller AR, Rastogi P, Morgenthaler S, Thilly WG. A statistical model to estimate variance in long term-low dose mutation assays: testing of the model in a human lymphoblastoid mutation assay. Mutat Res. 1989;216:149–161. doi: 10.1016/0165-1161(89)90001-0. [DOI] [PubMed] [Google Scholar]
- 47.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.