Abstract
We compare the ability of shRNA and CRISPR/Cas9 screens to identify essential genes in the human chronic myelogenous leukemia cell line K562. We find that the precision of the two libraries in detecting essential genes is similar and that combining data from both screens improves performance. Notably, results from the two screens show little correlation, which can be partially explained by identification of distinct essential biological processes with each technology.
Efficient gene knockdown and knockout using RNAi1–3 and CRISPR/Cas9 systems4–8 allow for systematic evaluation of gene function, but it is unclear how the choice of technology can affect results. For example, heterogeneity of reagents has historically been associated with poor performance in RNAi-based screens9,10 and may also influence CRISPR/Cas9 deletion screens4,10 (Supplementary Fig. 1–3). Whereas the variability of shRNAs in RNAi screens stems from differences in knockdown efficiency10, variability of sgRNAs in CRISPR/Cas9 screens likely stems from the array of genotypes (true knockouts, heterozygotes, and wild-type cells) created4,10. Notably this depends on the efficiency of guide cutting as well as the relative fitness between these subpopulations. Other possible differences include interference by non-specific effects such as miRNA deregulation during RNAi11,12 or intrinsic differences between knockouts and knockdowns. These concerns, as well as others, necessitate a careful comparison between these techniques.
To directly compare the phenotypes obtained using CRISPR/Cas9 and shRNA-based screening technologies, we performed parallel screens in duplicate for genes affecting growth rates in K562 using both a 25 hairpin/gene shRNA library13 and a 4 sgRNA/gene CRISPR/Cas9 library14. Briefly, sgRNA and shRNA libraries were lentivirally infected into cells, replicate populations were split at time zero, and the composition of these populations was investigated after two weeks of unperturbed growth by comparison to the starting plasmid library (Fig. 1a). The screens were conducted in parallel for minimal technical variation, allowing a quantitative assessment of performance. A previously established gold standard of 217 genes expected to have growth phenotypes in all cell types (essential) and 947 genes (Supplementary Data 1) expected to have growth phenotypes in no cell type (nonessential)15 was used to estimate true positive and false positive rates.
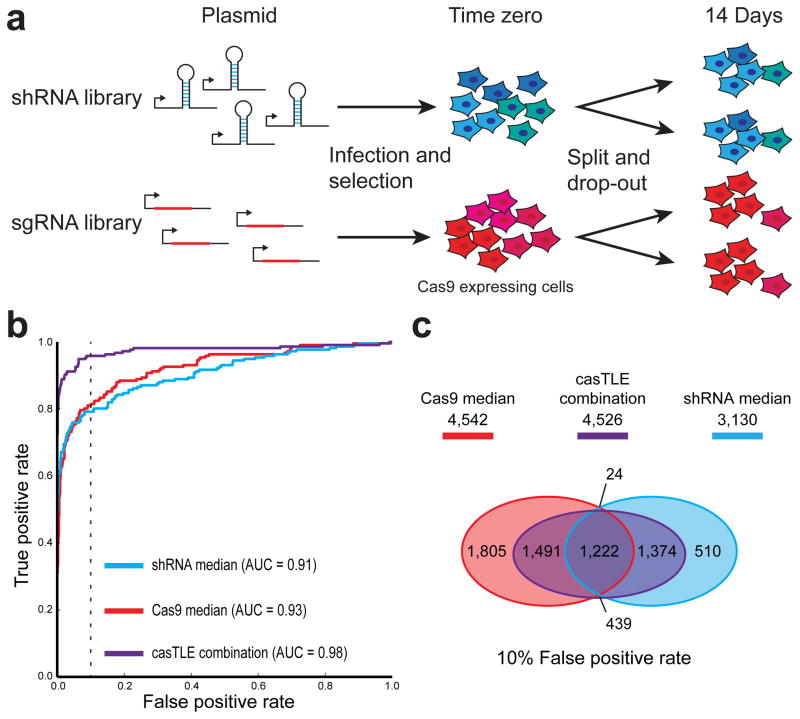
Figure 1. Parallel shRNA and CRISPR/Cas9 deletion screens to identify essential genes in K562.
(a) Schematic of screen. shRNA and Cas9 libraries were lentivirally infected into K562 and selected via puromycin treatment. After this time zero, replicate cell populations were maintained in log phase growth for two weeks. Library representation at each time point was monitored by deep-sequencing of the inserted locus. (b) ROC curves indicate screen performance in identifying essential genes by comparing the library composition between the plasmid library and cells after two weeks growth. True positive rates and false positive rates were calculated using a previously established gold standard set of essential and nonessential genes15. ROC curves for Cas9 (red) and shRNA (blue) screens based on the median score averaged over two replicates. Alternatively, data from single replicates of both Cas9 and shRNA screens were combined using casTLE (purple). (c) The number of essential genes at 10% false positive rate and their overlap based on the average median data from Cas9 and shRNA screens, as well as combination of a single replicate from both screens using casTLE. False positive rate was estimated using gold standard nonessential genes.
Using the median enrichment averaged over two replicates, we found that both shRNA and CRISPR/Cas9 screens have very high performance in detection of essential genes (AUC of the ROC curve > 0.90) (Fig. 1b, Supplementary Data 2–4). At a ~1% false positive rate, both screens recover >60% of gold standard essential genes. However, at a 10% false positive rate there are ~4,500 genes identified in the Cas9 screen versus ~3,100 in the shRNA screen, with ~1,200 genes identified in both (Fig. 1c). This indicates that although both our shRNA and Cas9 screens have similar levels of precision on the gold standard, both the Cas9 and shRNA screens identify numerous additional genes not in either the gold standard nor identified in the other screen.
To leverage data from both screening technologies, we developed a statistical frame work, Cas9 high-Throughput maximum Likelihood Estimator (casTLE). For each gene, casTLE combines measurements from multiple targeting reagents to estimate a maximum effect size as well as a p-value associated with that effect (Supplementary Figure 4 and see also Supplementary Methods). We validated casTLE by analyzing previous RNAi1, CRISPR deletion16, and CRISPRi/a17 screens and found consistent results (Supplementary Figure 5, Supplementary Data 5–7). casTLE performs favorably in identification of essential genes compared to previous methods1,18–21 including the median effect used here (Supplementary Figure 6 and see also Supplementary Discussion). Although casTLE performs well on single replicates from many screen types, it can also combine results from diverse data types by separately considering (a) experimental noise and (b) variability caused by heterogeneous reagents.
Using casTLE to combine data from a single replicate of the shRNA and Cas9 screens led to a noticeable improvement in performance, with an AUC of 0.98, >85% of gold standard essential genes identified at ~1% FPR, (Fig. 1b, Supplementary Fig. 7a, 8a,c, Supplementary Data 8) and the identification of ~4,500 genes with negative growth phenotypes with evidence from the combination of both screens (Fig. 1c, Supplementary Figure 8b). To test if these results depend on the number of targeting elements used, we compared the Cas9 results to a down-sampled 4 hairpin shRNA screen and found similar results (Supplementary Fig. 9a,b). The fact that the combination of both technologies can more successfully separate essential and nonessential genes suggests that the screens may be revealing different aspects of biology.
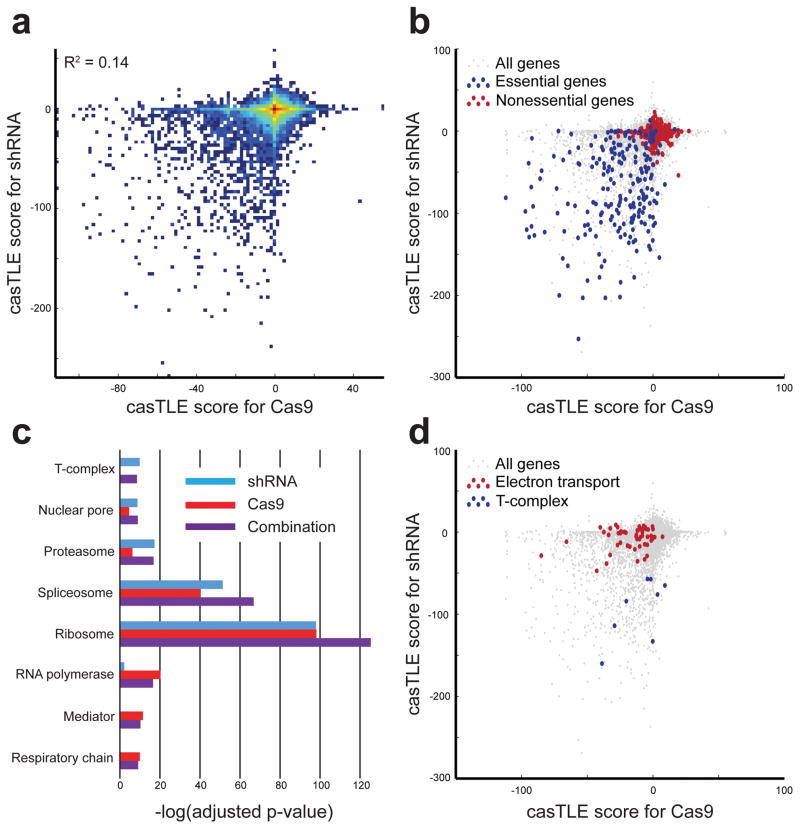
Consistent with the presence of non-redundant information, results from the Cas9 and shRNA screens show low correlation (Fig. 2a). Nonetheless, both screens effectively separate essential and nonessential genes (Fig. 1b, Fig. 2b) and are highly reproducible (Supplementary Fig. 10a,b, 7b). When we compared the enrichment of GO terms for essential complexes, we find the screens identify different biological processes. (Fig. 2c, Supplementary Data 9). For example, genes involved in the electron transport chain are enriched for essential genes in the Cas9 results, whereas all subunits of the chaperonin-containing T- complex are identified as essential by the shRNA screen (Fig. 2d). By using casTLE to combine information from each screen, we can recover each of these biological terms (Fig. 2c), further demonstrating the utility of a parallel screening approach. Again, these results did not change when we used a down-sampled 4 hairpin library (Supplementary Fig. 9c,d).
Figure 2. Differences between Cas9 and shRNA results.
(a) Comparison of casTLE scores between single replicates of Cas9 and shRNA data. A large positive casTLE score indicates a high confidence increase in growth rate, while a highly negative casTLE score indicates a high confidence decrease in growth rate, i.e. essential. Density is in log scale. (b) casTLE scores are shown for gold standard essential and nonessential genes15. (c) Adjusted p-values for select GO terms for shRNA and Cas9 screens as well as for data from both screens combined with casTLE. (d) casTLE scores shown for genes involved in the respiratory chain complex (GO:0098803) and the chaperonin-containing T-complex (GO:0005832), which exhibit differential enrichment in Cas9 and shRNA screens.
Several technical factors could account for the observed lack of correlation and differential GO term enrichment: (1) the presence or absence of effective reagents towards a particular gene, (2) differences in off-target effects as evidenced by the distribution of phenotypes among non-targeting controls (Supplementary Fig. 1a), or (3) differences in the timing of deletion/knockdown (Supplementary Fig. 10c,d). Another possibility highlighted in a recent analysis of human essential genes22 is that RNAi is less able to perturb genes expressed at low levels. However, we find no clear signature of this in our data (Supplementary Fig. 11a), perhaps due to more effective hairpin design13 capable of targeting these genes. This increased efficacy in our shRNA library may explain the qualitatively higher performance observed here in the detection of essential genes than was seen in previous studies, which found that RNAi screens perform poorly in the identification of essential genes22,23.
Although these technical differences may account for the low overall correlation observed, it is difficult to explain differential GO term enrichment with technical reasons alone. This observation suggests that Cas9 and shRNA screens are able to detect distinct aspects of biology, although it remains unclear why particular gene sets have strong signatures using one technology but not the other. One possibility, in cases such as RNA polymerase and the Mediator complex, is the dependency of shRNA knockdown on ongoing transcription, whereas sgRNAs no longer need to be efficiently expressed after a gene is knocked out4,10. Another possibility is that for certain genes a small loss in gene product via knockdown has a completely different phenotype than a large loss via a knockout. This could reflect non-monotonic gene-dose dependence on fitness (Supplementary Fig. 4)9,24 or the necessity of a complete knockout10. Alternatively, deletion of adjacent genes by Cas9 may occur in the context of tandem duplications, as found in a previous study23. A final possibility is that distinct hits in Cas9 and shRNA screens may represent genes that interact with the nonspecific effects of RNAi and Cas9. For example, certain genes would be expected to exhibit an effect on growth in the presence of persistent DNA damage4,10,25–27 or interference with miRNA processing11,12 due to Cas9 nuclease activity or shRNA expression, respectively. The potential existence of both false positives and false negatives in each screen presents an analytical problem which combination analysis using casTLE could address.
In fact, we find that genes found uniquely in either the shRNA or Cas9 screen – but not found in the combination analysis – do not have key signatures of essential genes. Essential genes are more likely to be highly expressed15,22,23, which is clearly seen in both screens (Supplementary Fig. 11a). However, when we limit our analysis to genes found in the shRNA or Cas9 screen but not in the combination analysis, this pattern is no longer clear compared to the genes found in both the shRNA and Cas9 screens (Supplementary Fig. 11b). Similarly, hits from the shRNA and Cas9 screens show the expected enrichment for homologs of essential yeast genes15,22,23 (Supplementary Fig. 11c) in contrast to hit genes unique to one method that are depleted in essential yeast homologs (Supplementary Fig. 11d). Although there are other explanations for these observations, this suggests that the combination analysis with casTLE can limit these technology or screen-specific false positives. Given that the use of both technologies also seems to reduce technology-specific false negatives as evidenced by the more complete capture of GO terms (Fig. 2c), the use of both (1) multiple reagents per gene to control for sequence-specific off-target effects and (2) multiple technologies for perturbation to control for non-specific effects should provide a more robust determination of a gene’s phenotype. However, determining the precise sources of differences between shRNA and CRISPR/Cas9 screens will require further inquiry.
Heterogeneity is a known feature of shRNA libraries, but we have also found this phenomenon in CRISPR/Cas9-based screens (Supplementary Fig. 1–2). One likely source of this variability is the occurrence of in-frame indels, creating wildtype and heterozygous subpopulations, which has been observed previously to interfere with screening results16,28. More efficacious guide designs might help, but in some cases sgRNAs targeting conserved functional elements within genes as opposed to simply the 5′ coding region may be preferred28. Nonetheless, heterogeneity in library elements can be a useful feature for genes with a complex gene-dosage/phenotype relationship. In particular, the spectrum of expression levels generated with an shRNA library should allow the identification of non-growth phenotypes for genes where complete deletion causes severe growth effects or lethality. Indeed, in our previous work we found that an shRNA screen for drug toxicity identified a presumably obligate biosynthetic gene, DHODH, as the drug’s target, which a parallel Cas9 screen failed to identify14. This suggests that differences between CRISPR/Cas9 and shRNA technologies may be useful to fully capture relevant biology in non-growth based screens.
Here we present an experimental side-by-side comparison of CRISPR/Cas9 and RNAi screens for essential genes. Using median enrichment analysis and our casTLE analysis tool (Supplementary Fig. 2–4), we demonstrate that both a recently developed shRNA library13 and a Cas9 sgRNA library have high precision (Fig. 1b, Supplementary Fig. 8a,c), but that the Cas9 library identifies many more essential genes (Fig. 1c, Supplementary Fig. 8b). The two screening technologies identify different biological categories of genes (Fig. 2a, c). These differences can be exploited to obtain a more complete picture of genes regulating growth by combining information using casTLE (Fig. 1b, 2c, Supplementary Fig. 8a,c) and may be important when considering which technology to use to probe a given process.
Methods
Cell Culture
Cell culture performed as previously described14. Briefly, K562 cells (ATCC) were cultured in RPMI 1640 (Gibco) media and supplemented with 10% FBS (Hyclone), penicillin (10,000 I.U./mL), streptomycin (10,000 ug/mL), and L-glutamine (2 mM). Cells were grown in log phase during all biological assays by returning the population to 500,000 cells/mL each day. K562 cells were maintained in a controlled humidified incubator at 37° C, with 5% CO2.
Genome-wide shRNA screen
A previously designed 25 shRNA/gene RNAi library was used13. Library infections and shRNA prep/sequencing were performed as previously described1. Briefly, to generate sufficient lentivirus to infect the genome-wide shRNA library13 into K562 cells, we plated 293T cells on 15 cm tissue culture plates. 293T cells were transfected with third generation packaging plasmids and shRNA encoding vectors. After 48 hrs and 72 hrs of incubation, lentivirus was harvested. We filtered the pooled lentivirus through a 0.45 μm PVDF filter (Millipore) to remove any cellular debris. Approximately 560 million K562 cells were infected with our next-generation genome-wide lentiviral shRNA library to maintain roughly 1,000-fold coverage of the shRNA library after selection. Infected cells grew for 3 days before selecting the cells with puromycin (0.7 μg/mL, Sigma). After three days of selection, infection efficiency was monitored using flow cytometry (BD Accuri C6). Once the cells reached 90–100% mCherry positive cells, they were spun out of selection and allowed to recover in normal RPMI 1640 media. At T0, 500 million cells were pelleted by centrifugation (300g for 5 min). Cells were then split into two populations and maintained at logarithmic growth (500,000 cells/mL) each day for 14 days. After 14 days of growth, cells were spun down (300g for 5 min). Genomic DNA was extracted for all 3 time points separately following Qiagen’s Blood Maxi Kit protocol. shRNA encoding constructs were measured by deep sequencing.
Genome-wide CRISPR-Cas9 screen
A previously designed 4 sgRNA/gene CRISPR-Cas9 library was used targeting 5′ ends of conserved exons with sgRNAs varying in length between 19 and 25 base-pairs14. The library was generated first by infecting K562 cells with a SFFV-Cas9-BFP vector to create a stably expressed Cas9 cell line. We then infected the lentiviral genome-wide sgRNA library into approximately 120 million cells following the same protocol as the genome-wide shRNA library to maintain at least 1,000-fold representation in cells. Infected cells were selected with puromycin (0.7 μg/mL, Sigma) for 3 days. Percentage of mCherry positive cells was measured by flow cytometry (BD Accuri C6). Selected cells were spun out of selection and into normal RPMI 1640 media. At T0, 120 million cells were spun down (300g for 5 min). Cells were then split into two populations and grown for 14 days, maintaining logarithmic growth (500,000 cells/mL) each day. After 14 days of growth, cells were pelleted by centrifugation, and genomic DNA was extracted for all three time samples separately following Qiagen’s Blood Maxi Kit instructions. sgRNA encoding constructs were analyzed by deep sequencing.
Analysis of previous screens
Data from previous shRNA screen for modifiers of ricin toxicity1 were obtained as pre- computed hairpin-level enrichments averaged from two replicates, pooled data was analyzed with casTLE, and compared to signed, log-transformed published results (Supplementary Data 5). Count data for two replicates from a previous CRISPR/Cas9 cutting screen for LPS-induced TNF expression16 were obtained from the authors, analyzed with casTLE, and compared to signed, log-transformed published DESeq results (Supplementary Data 6). Data from previous CRISPRi and CRISPRa screens17 were obtained as pre-computed guide-level enrichment averaged from two replicates, analyzed with casTLE, and compared to published results (Supplementary Data 7). Where available, positive and negative results from published low-throughput validations are also presented.
Analysis of screen results
Deep sequencing on an Illumina Nextseq was used to monitor composition of libraries. Trimmed sequences were aligned to the library using Bowtie with zero mismatches tolerated. All alignment from multi-mapped reads were used. Enrichment of individual hairpins was calculated as a median-normalized log-ratio of fraction of counts, as previously described1. Raw count files are available (Supplementary Data 2), and raw FASTQ files have been deposited at the Sequence Read Archive (Accession SRP072806) and BioProject (Accession PRJNA317269; http://www.ncbi.nlm.nih.gov/bioproject/PRJNA317269).
Briefly, we built cas9 High Throughput maximum Likelihood Estimator (casTLE) that uses an Empirical Bayesian29,30 framework to account for multiple sources of variability, including reagent efficacy and off-target effects9,11,31,32 (for more complete description, see Supplementary Methods). For each gene, we have the phenotypes of multiple targeting reagents. From this, and the phenotypes of negative controls, we obtain an effect size estimate for each gene and an associated log-likelihood ratio. In the figures we present this as the casTLE score, which is twice the log-likelihood ratio, signed to match the effect size. All screens are analyzed with the same parameters, and no optimization was performed using the gold standard or related sets. casTLE is implemented using custom Python scripts. These, along with a complete screen analysis pipeline, are available at https://bitbucket.org/dmorgens/castle.
For comparison, RIGER was implemented with default settings on precomputed element enrichments with GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/). RSA was implemented with Python scripts available from http://carrier.gnf.org/publications/RSA/. MAGeCK was implemented with default settings with software available from https://sourceforge.net/projects/mageck/. HiTSelect was implemented with default settings with software available from https://sourceforge.net/projects/hitselect/. Median and highest effect heuristics as well as the MW test were implemented with custom Python scripts. GO terms were generated using GOrilla33 available at http://cbl-gorilla.cs.technion.ac.il/ by ranking genes from highest confidence negative growth phenotypes to lowest confidence genes to highest confidence positive growth phenotypes.
For direct comparison to the 4 sgRNA per gene CRISPR/Cas9 library, the 25 hairpin per gene library RNAi library was down-sampled to 4 shRNAs per gene. Hairpins were ranked according to their original computational design13 and the top four unique shRNAs were used as well as all negative control shRNAs. Note that this is independent of the data set used here and represents the 4 shRNA library that would have been designed. Essential gene predictions were then repeated using this reduced shRNA library as above (Supplementary Fig. 9).
Analysis of gene expression data and yeast essentials
Gene sets were defined by 10% FPR cutoff for Cas9, shRNA, and combination screens. Cas9 and shRNA unique gene sets were defined as genes under the 10% FPR cutoff in the Cas9 or shRNA screen but neither the other nor the combination screen. An overlap set was defined as genes under the 10% FPR cutoff for both Cas9 and shRNA screens.
Public RNA-seq data (Accession ENCFF934YBO) for the K562 cell line was obtained from ENCODE34 experiment ENCSR000AEM. Genes were filtered for FPKM greater than one and successful mapping to genes present in our libraries, leaving expression data for ~7,000 genes. Genes were then ranked from highest to lowest expressed and binned in increments of 500. The number of genes in each bin were counted for each gene set and normalized by the total number of genes in each gene set present in the RNA-seq data. This fraction of essential genes for each gene set was then graphed versus the mean log(FPKM) value for each bin (Supplementary Figure 11a,b; Supplementary Data 10).
Yeast essential genes annotated as viable or inviable as well as their human homologs were obtained from the Saccharomyces Genome Database35. The number of homologs in each gene set were counted for both inviable and viable annotations, and the fraction inviable is presented (Supplementary Figure 11c,d; Supplementary Data 11). P-values are calculated using Fisher’s exact test. The total fraction of all homologs mapped from inviable yeast genes is also presented.
Supplementary Material
Supplementary Figure 1. Distribution of targeting and control elements. (a) Distribution of negative controls for a single replicate of Cas9 and shRNA screens. Enrichments are calculated as a median-normalized log ratio of counts. (b,c) Distribution of targeting elements is shown in meta-gene plots for the top 50 (b) enriched and (c) disenriched genes found in a single replicate of the Cas9 and shRNA screens as identified by casTLE. To normalize, the enrichment of each individual element was divided by the effect size estimate for the gene generated by casTLE. The dotted line is placed at the estimated effect size and normalized to one.
Supplementary Figure 2. Distribution of targeting sgRNAs for top disenriched genes. (a–d) Enrichment of targeting elements and estimated effect size is shown for the top four disenriched genes from Cas9 data from a single replicate. Enrichments are calculated as a median-normalized log ratio of counts. Gray lines represent the smoothed distribution of non-targeting controls. Red vertical lines represent enrichment of individual targeting guides towards indicated genes. Vertical dotted line represents effect size estimate from casTLE. Red distribution is a smoothed distribution of guides targeting the genes indicated.
Supplementary Figure 3. Distribution of targeting sgRNAs for top disenriched genes. (a–d) Enrichment of targeting elements and estimated effect size is shown for the top four disenriched genes from shRNA data from a single replicate. Enrichments are calculated as a median-normalized log ratio of counts. Gray lines represent the smoothed distribution of non-targeting controls. Blue vertical lines represent enrichment of individual targeting hairpins towards indicated genes. Vertical dotted line represents effect size estimate from casTLE. Blue distribution is a smoothed distribution of hairpins targeting the genes indicated.
Supplementary Figure 4. casTLE provides a statistical framework to account for high-throughput screens. The unknown relationship between gene dosage and measured phenotype as well as the unknown distribution of shRNA and Cas9 efficacies restricts the predicted effect size of reagents to a bounded region, marked as the blue shaded region, between 0 and the maximum effect I, marked by the dotted line. Some fraction (1–θ) of the reagents have no on-target effect at all. The phenotype observed is thus the true effect obscured by noise, which is estimated using the distribution of non-targeting controls. The likelihood of models for different values of I and θ are calculated and by marginalizing θ the most likely effect size is selected. A likelihood ratio is then calculated by comparing to a null model where I is zero.
Supplementary Figure 5. Reanalysis of previous screens. (a) Results are shown for a previously published shRNA screen for ricin sensitivity reanalyzed with casTLE and compared to published results based on a MW test1. (b) Previous CRISPR/Cas9 deletion screen for LPS-induced TNF expression in primary mouse bone-marrow derived dendritic cells, analyzed with casTLE and the published DESeq results16. (c) Previous CRISPRi screen for sensitivity to the fusion toxin CTx-DTA, analyzed with casTLE versus the average of the top three sgRNA effects17. (d) Previous CRISPRa screen for sensitivity to the fusion toxin CTx-DTA, analyzed with casTLE versus the average of the top three sgRNA effects17.
Supplementary Figure 6. Comparison of casTLE to other methods. (a–d) ROC curves indicate screen performance in identifying essential genes from changing composition between the plasmid library and two weeks growth. True positive rates and false positive rates are calculated using a previously established gold standard set of essential and nonessential genes15. Genes are ranked by likelihood to be essential using the indicated methods, including casTLE. Highest effect heuristic was calculated by ranking the genes according to their most disenriched element. Data is shown from single replicates of the (a,c) Cas9 and (b,d) shRNA screens for (a,b) replicate 1 and (c,d) replicate 2.
Supplementary Figure 7. Performance of combination of shRNA and Cas9 data. (a) ROC curves from combination of different replicates of Cas9 and shRNA using casTLE. ROC curves indicate screen performance in identifying essential genes from changing composition between the plasmid library and two weeks growth. True positive rates and false positive rates are calculated using a previously established gold standard set of essential and nonessential genes15. (b) Combination score has high reproducibility. A large positive casTLE score indicates a high confidence increase in growth rate, while a highly negative casTLE indicates a high confidence decrease in growth rate, i.e. gene essentiality. The graphs compare replicate measurements of likelihood ratio between plasmid and T14 of the combination score based on replicates 1 for Cas9 and shRNA and replicates 2 for Cas9 and shRNA. Density is in log scale.
Supplementary Figure 8. Comparison of casTLE combination to casTLE analysis of single screens. (a) ROC curves indicate screen performance in identifying essential genes by comparing the library composition between the plasmid library and cells after two weeks growth. ROC curves for Cas9 (red) and shRNA (blue) screens based on duplicate data combined using casTLE. Alternatively, data from single replicates of both Cas9 and shRNA screens were combined using casTLE (purple). (b) The number of essential genes at 10% false positive rate and their overlap based on the duplicate data from Cas9 and shRNA screens, as well as combination of a single replicate from both screens. False positive rate was estimated using gold standard nonessential genes. (c) Precision recall curve for Cas9, shRNA, and combination data using casTLE.
Supplementary Figure 9. Comparison to an in silico 4 shRNA per gene library. Results from the 25 shRNA library were downsampled by only including four hairpins per gene, selected by previous computational ranking. (a) ROC curves indicate screen performance in identifying essential genes by comparing the library composition between the plasmid library and cells after two weeks growth. (b) The number of essential genes at 10% false positive rate and their overlap based on the duplicate data from Cas9 and shRNA screens, as well as combination of a single replicate from both screens. (c) Comparison of casTLE scores derived from casTLE between single replicates of Cas9 and shRNA data. (d) Adjusted p-values for select GO terms for shRNA and Cas9 screens as well as for data from both screens combined with casTLE.
Supplementary Figure 10. Screen reproducibility and time-dependence of phenotypes. (a,b) shRNA and Cas9 screens have high reproducibility. A large positive casTLE score indicates a high confidence increase in growth rate, while a highly negative casTLE score indicates a high confidence decrease in growth rate, i.e. gene essentiality. The graphs compare replicate measurements of casTLE scores between plasmid and T14 for (a) Cas9 and (b) shRNA screens. Density is in log scale. (c,d) Time dependence of phenotypes. casTLE scores in different time-frames for (c) Cas9 and (d) shRNA screens.
Supplementary Figure 11. Analysis of gene expression and yeast essential homologs. Genesets are defined for Cas9, shRNA, and Combination by a 10% FPR cutoff. Genesets are defined for Cas9-combo and shRNA-combo by the genes present in Cas9 or shRNA set and not in the Combination set. Overlap set is defined as genes present in both the Cas9 and shRNA set (See Supplementary Fig. 8b). (a,b) ~7,000 genes with detectable expression in K562 were binned by expression. The fraction of genes identified as essential in each bin is reported versus the average expression level of the bin. (c,d) Fraction of genes that are homologs of essential yeast genes versus genes that are homologs of nonessential yeast genes. P-values calculated using Fisher’s exact test.
Acknowledgments
We thank Kyuho Han, Gaelen Hess, Michael Dubreuil, Michael Haney, Kim Tsui, and the Bassik Lab for technical expertise and helpful discussions. We thank Gavin Sherlock, Nathan Boley, Anshul Kundaje, Robert Tibshirani, and Andrew Fire for their critical reading of the manuscript and suggestions. Many thanks to Aviv Regev, Oren Parnas, and Rebecca H. Herbst for providing data from their published screen. We acknowledge the ENCODE Consortium and the Gingeras lab for the generation and release of the K562 gene expression data used here. This work was funded by NIH Director’s New Innovator Award Program 1DP2HD084069-01, and a seed grant from Stanford ChEM-H.
Footnotes
Author Contributions
DWM, RMD, and MCB conceived and designed the study. RMD and AL performed screens. DWM, RMD, and AL processed and sequenced screens. DWM designed and wrote casTLE. DWM performed all analysis. DWM and MCB wrote manuscript. All authors reviewed and approved the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–22. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–20. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–91. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin WG. Use and Abuse of RNAi to Study Mammalian Gene Function. Science (80-) 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrangou R, et al. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Res. 2015;43:3407–19. doi: 10.1093/nar/gkv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 12.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 13.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc Natl Acad Sci U S A. 2015;112:E3384–3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deans RM, et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2050. Published online Mar 28th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parnas O, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015 doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.König R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–9. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 20.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz AA, Qin H, Ramalho-Santos M, Song JS. HiTSelect: a comprehensive tool for high-complexity-pooled screen analysis. Nucleic Acids Res. 2014;43:e16–e16. doi: 10.1093/nar/gku1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Wang T, et al. Identification and characterization of essential genes in the human genome. Science (80-) 2015:aac7041. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsvetkov P, et al. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. Elife. 2015;4:e08467. doi: 10.7554/eLife.08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–86. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2014;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of Zinc Finger Nuclease Toxicity by Small-Molecule Regulation of Protein Levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efron B. Two modeling strategies for empirical Bayes estimation. Stat Sci. 2014;29:285–301. doi: 10.1214/13-sts455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kass RE, Steffey D. Approximate Bayesian Inference in Conditionally Independent Hierarchical Models (Parametric Empirical Bayes Models) J Am Stat Assoc. 2012 at < http://www.tandfonline.com/doi/abs/10.1080/01621459.1989.10478825>.
- 31.Birmingham A, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 32.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 33.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae S, Kweon J, Kim HS, Kim JS. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. 2014;11:705–6. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 37.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–75. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acosta-Alvear D, et al. Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits. Elife. 2015;4:e08153. doi: 10.7554/eLife.08153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 40.Haga T, et al. Attenuation of experimental autoimmune myocarditis by blocking T cell activation through 4-1BB pathway. J Mol Cell Cardiol. 2009;46:719–27. doi: 10.1016/j.yjmcc.2009.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Distribution of targeting and control elements. (a) Distribution of negative controls for a single replicate of Cas9 and shRNA screens. Enrichments are calculated as a median-normalized log ratio of counts. (b,c) Distribution of targeting elements is shown in meta-gene plots for the top 50 (b) enriched and (c) disenriched genes found in a single replicate of the Cas9 and shRNA screens as identified by casTLE. To normalize, the enrichment of each individual element was divided by the effect size estimate for the gene generated by casTLE. The dotted line is placed at the estimated effect size and normalized to one.
Supplementary Figure 2. Distribution of targeting sgRNAs for top disenriched genes. (a–d) Enrichment of targeting elements and estimated effect size is shown for the top four disenriched genes from Cas9 data from a single replicate. Enrichments are calculated as a median-normalized log ratio of counts. Gray lines represent the smoothed distribution of non-targeting controls. Red vertical lines represent enrichment of individual targeting guides towards indicated genes. Vertical dotted line represents effect size estimate from casTLE. Red distribution is a smoothed distribution of guides targeting the genes indicated.
Supplementary Figure 3. Distribution of targeting sgRNAs for top disenriched genes. (a–d) Enrichment of targeting elements and estimated effect size is shown for the top four disenriched genes from shRNA data from a single replicate. Enrichments are calculated as a median-normalized log ratio of counts. Gray lines represent the smoothed distribution of non-targeting controls. Blue vertical lines represent enrichment of individual targeting hairpins towards indicated genes. Vertical dotted line represents effect size estimate from casTLE. Blue distribution is a smoothed distribution of hairpins targeting the genes indicated.
Supplementary Figure 4. casTLE provides a statistical framework to account for high-throughput screens. The unknown relationship between gene dosage and measured phenotype as well as the unknown distribution of shRNA and Cas9 efficacies restricts the predicted effect size of reagents to a bounded region, marked as the blue shaded region, between 0 and the maximum effect I, marked by the dotted line. Some fraction (1–θ) of the reagents have no on-target effect at all. The phenotype observed is thus the true effect obscured by noise, which is estimated using the distribution of non-targeting controls. The likelihood of models for different values of I and θ are calculated and by marginalizing θ the most likely effect size is selected. A likelihood ratio is then calculated by comparing to a null model where I is zero.
Supplementary Figure 5. Reanalysis of previous screens. (a) Results are shown for a previously published shRNA screen for ricin sensitivity reanalyzed with casTLE and compared to published results based on a MW test1. (b) Previous CRISPR/Cas9 deletion screen for LPS-induced TNF expression in primary mouse bone-marrow derived dendritic cells, analyzed with casTLE and the published DESeq results16. (c) Previous CRISPRi screen for sensitivity to the fusion toxin CTx-DTA, analyzed with casTLE versus the average of the top three sgRNA effects17. (d) Previous CRISPRa screen for sensitivity to the fusion toxin CTx-DTA, analyzed with casTLE versus the average of the top three sgRNA effects17.
Supplementary Figure 6. Comparison of casTLE to other methods. (a–d) ROC curves indicate screen performance in identifying essential genes from changing composition between the plasmid library and two weeks growth. True positive rates and false positive rates are calculated using a previously established gold standard set of essential and nonessential genes15. Genes are ranked by likelihood to be essential using the indicated methods, including casTLE. Highest effect heuristic was calculated by ranking the genes according to their most disenriched element. Data is shown from single replicates of the (a,c) Cas9 and (b,d) shRNA screens for (a,b) replicate 1 and (c,d) replicate 2.
Supplementary Figure 7. Performance of combination of shRNA and Cas9 data. (a) ROC curves from combination of different replicates of Cas9 and shRNA using casTLE. ROC curves indicate screen performance in identifying essential genes from changing composition between the plasmid library and two weeks growth. True positive rates and false positive rates are calculated using a previously established gold standard set of essential and nonessential genes15. (b) Combination score has high reproducibility. A large positive casTLE score indicates a high confidence increase in growth rate, while a highly negative casTLE indicates a high confidence decrease in growth rate, i.e. gene essentiality. The graphs compare replicate measurements of likelihood ratio between plasmid and T14 of the combination score based on replicates 1 for Cas9 and shRNA and replicates 2 for Cas9 and shRNA. Density is in log scale.
Supplementary Figure 8. Comparison of casTLE combination to casTLE analysis of single screens. (a) ROC curves indicate screen performance in identifying essential genes by comparing the library composition between the plasmid library and cells after two weeks growth. ROC curves for Cas9 (red) and shRNA (blue) screens based on duplicate data combined using casTLE. Alternatively, data from single replicates of both Cas9 and shRNA screens were combined using casTLE (purple). (b) The number of essential genes at 10% false positive rate and their overlap based on the duplicate data from Cas9 and shRNA screens, as well as combination of a single replicate from both screens. False positive rate was estimated using gold standard nonessential genes. (c) Precision recall curve for Cas9, shRNA, and combination data using casTLE.
Supplementary Figure 9. Comparison to an in silico 4 shRNA per gene library. Results from the 25 shRNA library were downsampled by only including four hairpins per gene, selected by previous computational ranking. (a) ROC curves indicate screen performance in identifying essential genes by comparing the library composition between the plasmid library and cells after two weeks growth. (b) The number of essential genes at 10% false positive rate and their overlap based on the duplicate data from Cas9 and shRNA screens, as well as combination of a single replicate from both screens. (c) Comparison of casTLE scores derived from casTLE between single replicates of Cas9 and shRNA data. (d) Adjusted p-values for select GO terms for shRNA and Cas9 screens as well as for data from both screens combined with casTLE.
Supplementary Figure 10. Screen reproducibility and time-dependence of phenotypes. (a,b) shRNA and Cas9 screens have high reproducibility. A large positive casTLE score indicates a high confidence increase in growth rate, while a highly negative casTLE score indicates a high confidence decrease in growth rate, i.e. gene essentiality. The graphs compare replicate measurements of casTLE scores between plasmid and T14 for (a) Cas9 and (b) shRNA screens. Density is in log scale. (c,d) Time dependence of phenotypes. casTLE scores in different time-frames for (c) Cas9 and (d) shRNA screens.
Supplementary Figure 11. Analysis of gene expression and yeast essential homologs. Genesets are defined for Cas9, shRNA, and Combination by a 10% FPR cutoff. Genesets are defined for Cas9-combo and shRNA-combo by the genes present in Cas9 or shRNA set and not in the Combination set. Overlap set is defined as genes present in both the Cas9 and shRNA set (See Supplementary Fig. 8b). (a,b) ~7,000 genes with detectable expression in K562 were binned by expression. The fraction of genes identified as essential in each bin is reported versus the average expression level of the bin. (c,d) Fraction of genes that are homologs of essential yeast genes versus genes that are homologs of nonessential yeast genes. P-values calculated using Fisher’s exact test.