Summary
The ongoing epidemic of Zika virus (ZIKV) illustrates the importance of flaviviruses as emerging human pathogens. All vector-borne flaviviruses studied thus far have to overcome type I interferon (IFN) to replicate and cause disease in vertebrates. The mechanism(s) by which ZIKV antagonizes IFN signaling is unknown. Here, we report that the nonstructural protein NS5 of ZIKV and other flaviviruses examined could suppress IFN signaling, but through different mechanisms. ZIKV NS5 expression resulted in proteasomal degradation of the IFN-regulated transcriptional activator STAT2 from humans but not mice, which may explain the requirement for IFN-deficiency to observe ZIKV-induced disease in mice. The mechanism of ZIKV NS5 resembles dengue virus (DENV) NS5 and not its closer relative, Spondweni virus (SPOV). However, unlike DENV, ZIKV did not require the E3 ubiquitin ligase UBR4 to induce STAT2 degradation. Hence, flavivirus NS5 proteins exhibit a remarkable functional convergence in IFN antagonism, albeit by virus-specific mechanisms.
Introduction
Flaviviruses constitute the most important and diverse group of arthropod transmitted viruses causing disease in humans. They include DENV, tick-borne encephalitis viruses (TBEV), yellow fever virus (YFV), and West Nile virus (WNV), among other human pathogens. In the last decades, mosquito-borne flaviviruses have caused significant epidemic outbreaks including DENV, WNV, and more recently ZIKV, associated with dramatic expansion of their geographical distribution (Coffey et al., 2013; Lazear and Diamond, 2016). ZIKV is a mosquito-borne member of the Spondweni virus (SPOV) group. Both ZIKV and SPOV were known to cause limited, sporadic outbreaks in Africa and South East Asia (Haddow et al., 2012; Wolfe et al., 1982). However, ZIKV has emerged in the South Pacific and South and Central America associated with sustained transmission affecting an estimated 0.5 to 1.5 million people. The ZIKV epidemic is associated with severe fetal abnormalities including stillbirth and microcephaly (Rasmussen et al., 2016), or with the Guillain-Barré syndrome in infected adults (Cao-Lormeau et al., 2016).
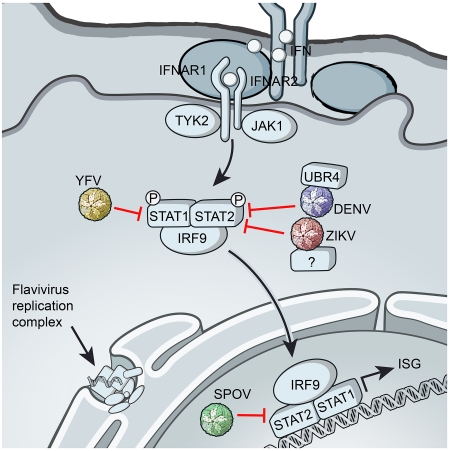
Type I IFN invokes a potent antiviral state in the cell following stimulation of two IFN receptor subunits (IFNAR1 and IFNAR2) to activate the Janus kinases (Jak1 and Tyk2) and signal transducers of transcription (STAT1 and STAT2) leading to the upregulation of hundreds of IFN-stimulated genes (ISGs) (MacMicking, 2012). The importance of IFN in host antiviral innate immunity is highlighted by the diversity of viral IFN antagonists found in vertebrate viruses (Versteeg and Garcia-Sastre, 2010). Efficient IFN antagonism might be particularly critical for mosquito-borne arboviruses such as flaviviruses that require viral loads in blood (viremia) to maintain their vector-host cycles. Various strategies of IFN antagonism have been demonstrated for well-known flaviviruses, such as DENV, WNV, YFV, and TBEV (Versteeg and Garcia-Sastre, 2010). However, the flaviviruses encompass more than 70 diverse but phylogenetically related viruses suggesting that the full spectrum of flavivirus-encoded strategies to suppress this critical host response are only beginning to be explored.
Flaviviruses share a replication strategy based on the generation of a polyprotein from the single stranded positive RNA genome. The polyprotein is proteolytically processed by viral and host proteases to produce three structural (C, prM and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5), the latter of which are involved in viral RNA synthesis and replication. While multiple viral proteins have been described for WNV and DENV to either prevent IFN induction or to inhibit IFN signaling (Aguirre et al., 2012; Liu et al., 2004; Liu et al., 2005; Munoz-Jordan et al., 2003), the NS5 protein is a potent and specific antagonist of IFN signaling shown for all human flaviviruses tested so far (Ashour et al., 2009; Best et al., 2005; Laurent-Rolle et al., 2010; Laurent-Rolle et al., 2014; Lin et al., 2006; Lubick et al., 2015; Mazzon et al., 2009; Morrison et al., 2013). In addition, the flavivirus NS5 encodes both the viral methyltransferase and the RNA-dependent RNA polymerase required for viral RNA synthesis. Intriguingly, NS5-mediated inhibition of IFN signaling is exerted by different mechanisms depending on the specific flavivirus. WNV NS5 targets the host protein prolidase to prevent surface expression of IFNAR1 (Lubick et al., 2015); by contrast DENV NS5 recruits the host E3 ubiquitin ligase UBR4 to degrade STAT2 (Morrison et al., 2013). In addition, YFV NS5 binds to STAT2, but only after IFN treatment, and this prevents STAT2 binding to ISRE (IFN stimulated responsive element) promoter elements present upstream of ISGs (Laurent-Rolle et al., 2014). NS5-mediated IFN antagonism has been shown to be essential for resistance to IFN in cell culture (Laurent-Rolle et al., 2010; Laurent-Rolle et al., 2014) and for virulence in mouse models of infection (Lubick et al., 2015).
Based on the remarkable functional similarities but mechanistic diversities by which the NS5 of well-known flaviviruses inhibit IFN signaling, we asked whether this paradigm applies to other members of the flavivirus group, including SPOV and ZIKV. In all cases, NS5 inhibited IFN induced gene expression in human cells, indicating a functional conservation of IFN antagonism among flavivirus NS5 proteins. However, mechanistically, NS5 function was diverse. These findings highlight remarkable diversity in the function of NS5 in IFN antagonism by pathogenic flaviviruses.
Results
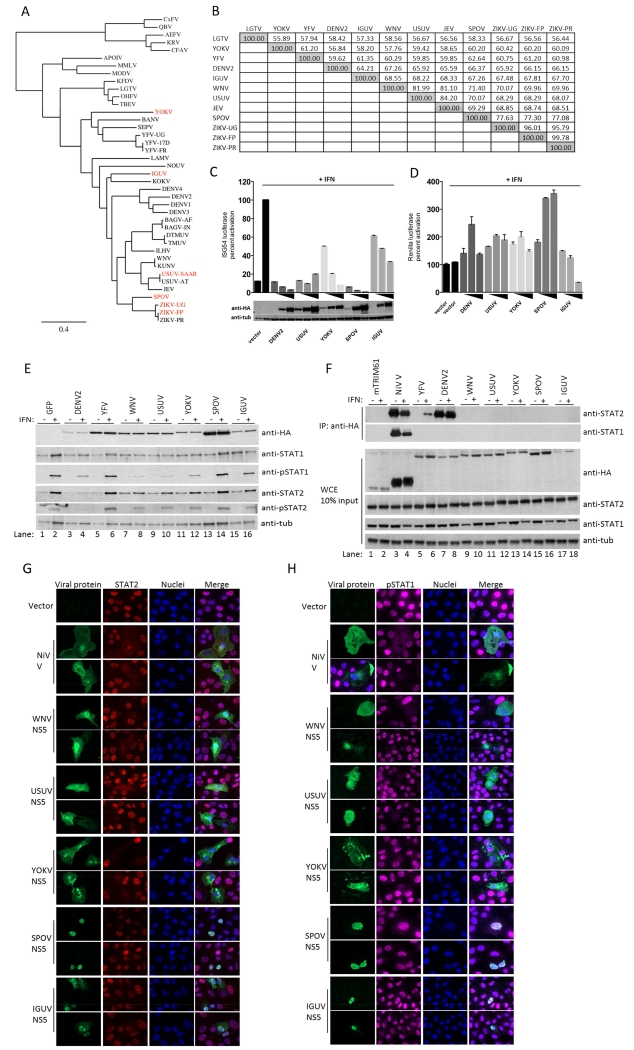
The NS5 protein is the most conserved of flavivirus proteins, although it exhibits up to 45% amino acid difference amongst vector-borne flaviviruses. A phylogenetic tree of NS5 sequences from representative flaviviruses is shown in Figure 1A. We were interested in determining if NS5 from distantly related flaviviruses with known vertebrate hosts were capable of inhibiting IFN signaling and the mechanism behind it. We initially examined NS5 function from four different evolutionary clades members within the Flaviviridae family: Yokose (YOKV), Iguape (IGUV), SPOV and Usutu (USUV) viruses. The amino acid identity of the NS5 of these flaviviruses as compared to the more studied Langat virus (LGTV), YFV, DENV, Japanese encephalitis virus (JEV) and WNV is shown in Figure 1B.
Figure 1. Phylogenetically diverse NS5 proteins antagonize Type I IFN signaling.
(A) Phylogenetic tree of NS5 proteins; red indicates proteins chosen for characterization. (B) Amino acid percent identity matrix of NS5 proteins. (C) ISG54 reporter assay performed in triplicate in 293T cells transfected with increasing amounts of NS5 plasmids. (D) Renilla luciferase values for (C). (E) Immunoblot of 293T cells transfected with GFP and flavivirus NS5 plasmids. GFP positive cells of similar intensity were sorted by flow cytometry. (F) HA-tag IP assay and immunoblot of 293T cells transfected with indicated HA-tagged plasmids. (G) Vero cell IFA for endogenous STAT2 levels post IFN treatment for 30 minutes. (H) Same as (G), except staining for endogenous pSTAT1. Error bars represent mean ±SD. Results are representative of three or more independent experiments.
YOKV is distantly related to, but within the clade of YFV (Figure 1A). YOKV was isolated from a bat (Miniopterus fuliginosus) in Japan in 1971 (Cook and Holmes, 2006). IGUV is in a clade together with Kokobera virus (Figure 1A). It was isolated from a sentinel forest mouse in Brazil in 1979 and is suspected to infect mice, marsupials and birds (Coimbra et al., 1993). SPOV is a mosquito-borne flavivirus closely related to ZIKV. It is the causative agent of Spondweni fever in sub-Saharan Africa and Papua New Guinea (Grard et al., 2010). USUV is within the clade of the JEV complex, which also encompasses WNV. It was first identified in South Africa in 1959, and its primary hosts include Culex mosquitoes and birds, and like WNV, it can also cause encephalitis in humans (Engel et al., 2016). In 2001 USUV was found for the first time outside Africa as the causative agent of high mortality in blackbirds in Austria (Engel et al., 2016).
Expression of all YOKV, IGUV, SPOV and USUV NS5s suppressed IFN-induced ISRE-dependent gene expression in a dose dependent manner in human 293T cells, similar to DENV2 NS5, albeit with different efficiencies (Figure 1C). Most of the inhibitory activity of the IGUV NS5, but not of any other NS5, appeared due to general inhibition of gene expression, as monitored by reporter gene expression under a constitutively active promoter (Figure 1D). In order to investigate whether NS5-mediated IFN antagonism functioned upstream or downstream STAT1/STAT2, we monitored STAT1/2 levels and phosphorylation before and after IFN treatment in 293T cells (Figure 1E). In these experiments, NS5 from DENV2, YFV and WNV were used as controls. In the case of DENV NS5, this protein was expressed as a precursor polyprotein that becomes proteolytically cleaved, as we previously demonstrated this was required for its functional activity (Ashour et al., 2009). As expected (Ashour et al., 2009), DENV2 NS5 expression degraded STAT2 (Figure 1E, lanes 3 and 4). However, no other flavivirus NS5 appeared to degrade STAT2 or STAT1. Also as expected (Laurent-Rolle et al., 2010), expression of WNV NS5 inhibited STAT1 phosphorylation after IFN treatment (Figure 1E, lane 8). A similar suppression of STAT1 phosphorylation was observed in cells expressing either USUV or YOKV NS5 (Figure 1E, lanes 10 and 12). Western blots also showed a trend of WNV, USUV and YOKV NS5 to inhibit STAT2 phosphorylation. The inability of IGUV NS5 to prevent STAT phosphorylation (Figure 1E, lane 16) is consistent with a mechanism of preventing ISRE induction based on general inhibition of gene expression (Figure 1D). Finally, SPOV NS5 did not prevent STAT phosphorylation (Figure 1E, lane 14), suggesting that it suppresses IFN signaling downstream of STAT phosphorylation, as is also the case for YFV NS5 (Laurent-Rolle et al., 2014) (Figure 1E, lane 6). We next tested whether any of these flavivirus NS5 proteins were competent to bind either STAT1 or STAT2 by immunoprecipitation (IP). YFV NS5 is known to bind and inhibit STAT2 after type I IFN treatment (Laurent-Rolle et al., 2014) (Figure 1F, lane 6). DENV2 NS5 also binds to STAT2 but this binding is independent of IFN treatment (Ashour et al., 2009) (Figure 1F, lanes 7 and 8). As a positive control for both STAT1 and STAT2 binding, we used the Nipah virus (NiV) V protein (Rodriguez et al., 2004; Shaw et al., 2004) (Figure 1F, lanes 3 and 4). The irrelevant protein mTRIM61 was used as negative control (Figure 1F, lanes 1 and 2). None of the other flavivirus NS5s were found to bind to either STAT1 or STAT2, with the exception of SPOV NS5, which showed very weak binding to STAT2 only after Western blot overexposure (data not shown).
Type I IFN-induced STAT1/2 phosphorylation triggers their translocation to the nucleus. To confirm the results on STAT1/2 phosphorylation of Figure 1E, we conducted immunofluorescence assay (IFA) in cells expressing flavivirus NS5s. Expression of USUV and YOKV NS5s prevented STAT1/2 nuclear translocation after IFN treatment in Vero cells (Figure 1G and 1H), similar to NiV V protein and WNV NS5, and consistent with their inhibition of STAT phosphorylation. However, expression of the SPOV and IGUV NS5s did not prevent IFN-mediated translocation of STAT1 or STAT2 (Figure 1G and 1H, lower panels), indicative of a mechanism of inhibition of IFN signaling downstream of STAT phosphorylation and translocation.
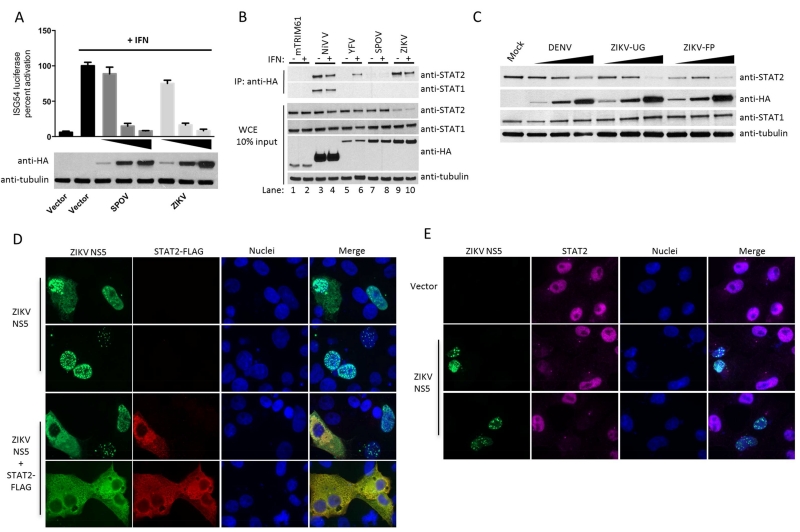
While in the process of conducting this comparative analysis of flavivirus NS5s, the emergence of the ZIKV outbreak in South America prompted us to add ZIKV NS5 to our studies. We chemically synthesized and cloned the NS5 of ZIKV based on GenBank sequence KJ776791.1 of the French Polynesia (FP) isolate, which is closely related to the ZIKV strains involved in ongoing epidemic in the Americas (Figure 1B). We then compared the IFN antagonistic properties of ZIKV NS5 to its close relative SPOV. Expression of ZIKV NS5, similar to SPOV NS5, prevented the activation of an IFN-inducible reporter gene in 293T cell treated with type I IFN (Figure 2A). However, in contrast to SPOV NS5, ZIKV NS5 strongly interacted with STAT2 (but not with STAT1) in IP experiments in 293T cells (Figure 2B). This interaction was independent of IFN treatment (Figure 2B, lanes 9 and 10) but was associated with a general reduction in STAT2 levels in ZIKV NS5 expressing cells (Figure 2B, lanes 9 and 10, WCE). This is reminiscent of DENV2 NS5, which is known to bind to STAT2 and target it for degradation (Ashour et al., 2009). In order to determine whether this observation was evolutionarily conserved amongst ZIKV strains, we cloned the NS5 from the viral RNA of the original ZIKV Uganda (UG) isolate and tested its ability to bind STAT2 and reduce STAT2 protein levels. The NS5 of ZIKV UG strain also bound to STAT2 (Figure 3F) and reduced STAT2 levels in a dose dependent manner (Figure 2C). To examine whether ZIKV NS5 colocalizes with STAT2, we co-expressed ZIKV NS5 and STAT2 in Vero cells. Ectopic expression of STAT2 was required to prevent loss of signal due to ZIKV-induced reduction of endogenous STAT2. In the absence of STAT2 overexpression, ZIKV NS5 localized mainly in the nucleus (Figure 2D, upper panels). Predominant nuclear localization has been reported for several other flavivirus NS5 proteins, such as DENV and YFV (Hannemann et al., 2013; Laurent-Rolle et al., 2014) even though the role of NS5 in viral RNA replication is in the cytosol. In the case of ZIKV NS5, nuclear localization was primarily associated with nuclear dots. We do not currently know the nature and significance of this distribution. Interestingly, following overexpression of human STAT2, ZIKV NS5 relocalized from the nucleus to the cytoplasm where it colocalized with STAT2 (Figure 2D, lower panels), indicative of a strong interaction between both proteins. ZIKV NS5 expression also prevented the translocation of endogenous STAT2 to the nucleus after IFN treatment (Figure 2E). Taken together, these results indicate that ZIKV NS5 inhibits IFN signaling by a mechanism involving STAT2 binding and most likely STAT2 degradation, which is similar to DENV NS5, yet different from its closer relative SPOV.
Figure 2. ZIKV NS5 proteins antagonize Type I IFN signaling by targeting STAT2.
(A) ISG54 reporter assay performed in triplicate in 293T cells transfected with increasing amounts of NS5 expressing plasmids. (B) HA-tag IP assay and immunoblot of 293T cells transfected with indicated HA-tagged plasmids. (C) Immunoblot of 293T cells transfected with increasing amounts of DENV2, ZIKV UG and FP NS5 plasmids. (D) Vero cell IFA for colocalization of overexpressed human STAT2-FLAG and ZIKV NS5-HA tagged proteins. (E) Vero cell IFA of endogenous STAT2 levels post IFN treatment for 30 minutes. Error bars represent mean ±SD. Results are representative of three or more independent experiments.
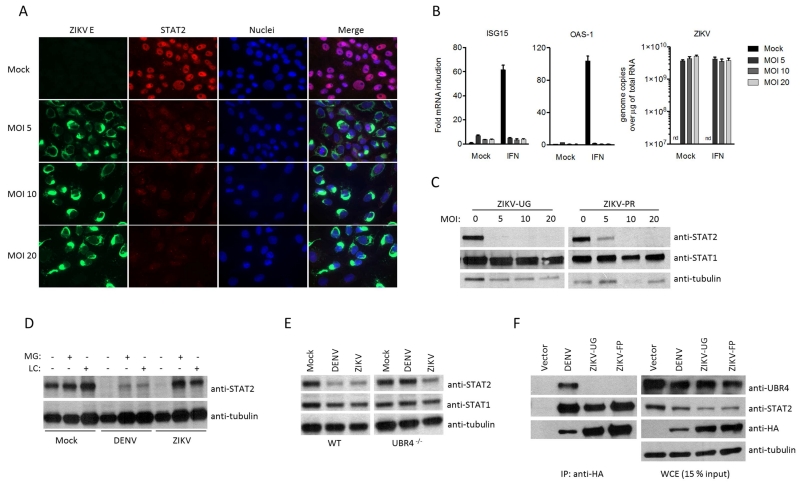
Figure 3. ZIKV degrades STAT2 in a proteasome dependent manner.
(A) Vero cell IFA of endogenous STAT2 upon ZIKV UG infection and IFN treatment for 30 minutes. (B) qRT-PCR analysis of ISGs and ZIKV UG RNA upon infection in Vero cells for 24h at indicated MOI, following mock treatment or treatment with IFN for 14h. nd=not detected, error bars represent mean ±SD of triplicates. (C) Immunoblot of Vero cells infected with ZIKV UG or ZIKV PR for 24h at indicated MOI. (D) Immunoblot of 293T cells infected with DENV2 and ZIKV UG followed by mock, MG132 (MG) or Lactacystin (LC) proteasome inhibitor treatment. (E) Immunoblot of DENV2 and ZIKV UG infected WT 293T or Ubr4 deficient 293T cells, 24h post infection at MOI 5. (F) HA-tag IP assay and immunoblot of 293T cells transfected with indicated HA-tagged plasmids. Results are representative of three or more independent experiments.
Next we investigated type I IFN signaling in ZIKV infected cells. For this purpose, we infected Vero cells with ZIKV (UG MR766 strain), treated them with IFN at 24h post-infection, and visualized STAT2 translocation 30 min post IFN treatment by IFA. As seen in Figure 3A, ZIKV infection prevented STAT2 translocation to the nucleus. In addition, ZIKV infection inhibited the IFN induction of the ISGs ISG15 and OAS1, as measured by qRT-PCR (Figure 3B). Next, we tested STAT2 degradation upon ZIKV infection with both UG and Puerto Rico (PR) strain, which is currently circulating and evolutionarily closely related to the FP strain (Lanciotti et al., 2016). Vero cells infected with both ZIKV strains exhibited dramatically reduced STAT2 levels, suggesting a mechanistic conservation of STAT2 degradation amongst ZIKV isolates (Figure 3C). Reduced levels of STAT2 during ZIKV infection were due to proteasomal degradation, as STAT2 expression was rescued in ZIKV-infected cells by inhibitors of the proteasome (Figure 3D). This finding was similar to the mechanism of STAT2 antagonism by DENV NS5, which was included as a positive control. Overall, ZIKV infection recapitulates the inhibition of IFN signaling and reduction of STAT2 levels observed in cells exclusively expressing ZIKV NS5.
The only other flavivirus known to target STAT2 for degradation in infected cells is DENV (Ashour et al., 2009; Morrison et al., 2013). In the case of DENV NS5, we have shown that it binds to STAT2 and co-opts the host E3 ubiquitin ligase UBR4 to mediate STAT2 degradation (Ashour et al., 2009; Morrison et al., 2013). To investigate whether ZIKV induced degradation of STAT2 has a similar requirement for UBR4, we infected 293T cells that have been genetically edited by CRISPR/Cas9 to knock out UBR4 expression (Tripathi et al., 2015), and determined STAT2 levels. As shown in Figure 3E, and in contrast to DENV infection, ZIKV infection resulted in reduced STAT2 levels independently of the presence or absence of UBR4 expression. Consistent with these data, NS5 of DENV, but not of ZIKV, interacts with UBR4 in IP experiments (Figure 3F). Thus, even though both DENV and ZIKV infection result in STAT2 degradation and inhibition of IFN signaling, they utilize divergent mechanisms.
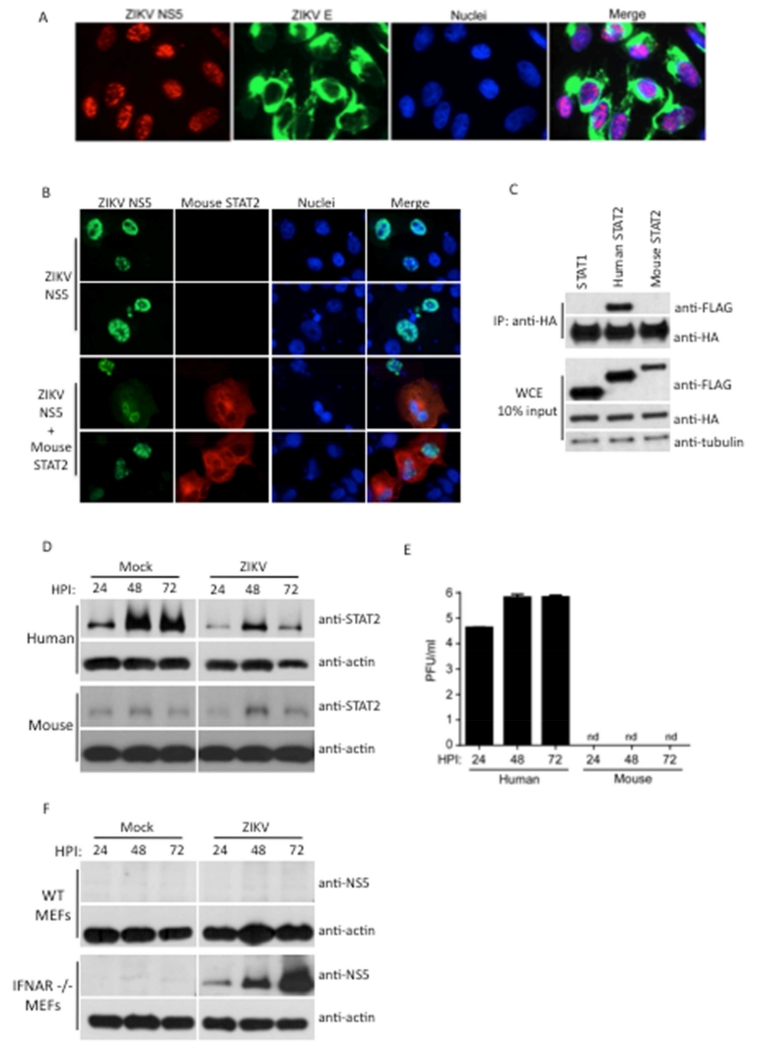
To determine if the findings associated with ectopic expression of ZIKV are relevant to ZIKV biology, we generated an anti-peptide antibody that recognizes ZIKV NS5, which was confirmed by Western blot on virus infected cells (Figure 4F). IFA revealed predominant nuclear localization of NS5 in ZIKV-infected cells in distinct bodies (Figure 4A), similar to ectopic expression. Three recent publications demonstrate that mouse susceptibility to ZIKV requires complete genetic deletion of type I IFN signaling through the use of IFNAR-deficient mouse strains (Aliota et al., 2016; Lazear et al., 2016; Rossi et al., 2016). This suggests that ZIKV IFN antagonism may be species-dependent. In support of this, overexpression of ZIKV NS5 along with mouse STAT2 did not result in re-localization of ZIKV NS5 to the cytoplasm (Figure 4B), unlike that observed with human STAT2 (Figure 2D), indicating a lack of interaction between ZIKV NS5 and mouse STAT2. This observation was confirmed by IP assays (Figure 4C). Furthermore, virus replication in primary human fibroblasts, previously shown to be permissive to ZIKV replication (Hamel et al., 2015), resulted in STAT2 degradation. In contrast, STAT2 levels were unaffected in ZIKV infected primary mouse embryonic fibroblasts (MEFs) (Figure 4D), associated with no detectable virus production in these cells (Figure 4E). Lastly, IFNAR deficient MEFs were able to support ZIKV replication (Figure 4F), in contrast to wild type MEFs, demonstrating viral restriction is dependent on an intact IFN signaling response and further exemplifying the requirement of IFN antagonism for ZIKV replication. In summation, these results suggest that our findings observed by ectopic NS5 expression recapitulate the major features of IFN antagonism exhibited during ZIKV infection. Furthermore, with the possibility that additional restriction factors might be responsible for the lack of replication of ZIKV in MEFs, these results imply that species-restricted IFN antagonism may contribute to the efficient control of ZIKA virus replication and lack of pathology in IFN-competent mouse models.
Figure 4. ZIKV exhibits species-specific antagonism of STAT2.
(A) Vero cell IFA of viral NS5 and E proteins upon ZIKV Fortaleza infection at MOI 5 for 24h (B) Vero cell IFA for colocalization between overexpressed mouse STAT2-FLAG and ZIKV UG NS5-HA tagged proteins. (C) HA-tag IP assay and immunoblot analysis of 293T cells transfected with ZIKV UG NS5-HA, human STAT1-FLAG, human STAT2-FLAG and mouse STAT2-FLAG plasmids. (D) Immunoblot of primary human and mouse fibroblasts infected with ZIKV Fortaleza strain at MOI 5 for indicated duration. (E) Measurement of ZIKV Fortaleza viral replication by plaque assay in primary human and mouse fibroblasts infected at MOI 0.1 for indicated duration. PFU (plaque forming units). (F) Immunoblot of wild type (WT) and IFNAR −/− MEFs infected with ZIKV Fortaleza at MOI 1, probed with anti-NS5 sera. Results are representative of three or more independent experiments.
DISCUSSION
It is known that previously studied flaviviruses have evolved independent and diverse mechanisms to prevent type I IFN signaling in vertebrate hosts, despite using the same viral protein, NS5, to achieve this goal. Here, we have shown that two closely related flaviviruses, ZIKV and SPOV, encode an NS5 that inhibits type I IFN signaling in human cells. However, these two viruses utilize different mechanisms. ZIKV NS5 binds and degrades human STAT2, whereas SPOV only weakly binds STAT2, does not degrade STAT2 nor affect STAT2 phosphorylation and nuclear localization. Therefore, SPOV NS5 functions downstream of these events and could conceivably interfere with ISGF3 binding to ISRE promoter elements. However, SPOV may affect other events necessary for ISG expression, such as epigenetic histone modification or long non-coding RNA expression (Kroczynska et al., 2014; Ouyang et al., 2014). Future studies will be needed to determine the precise mechanism of action. The ability of SPOV NS5 to antagonize human IFN signaling, combined with the fact that SPOV is mosquito-borne, suggests that this virus may have the potential to emerge more broadly in human populations.
Antagonism of IFN-mediated signal transduction by ZIKV NS5 shares multiple similarities with that of DENV NS5. Both NS5 proteins bind and degrade STAT2 via the proteasome and accomplish this in a highly specific, species-restricted fashion, which is exemplified by the ability to degrade human and non-human primate (as shown in Vero cells) STAT2, but not mouse STAT2 (Ashour et al., 2010). This likely reflects the similar natural history of the two viruses. ZIKV was originally isolated from sentinel rhesus monkeys in 1947 in Uganda and separately in Aedes africanus mosquitoes shortly thereafter (Dick, 1952; Dick et al., 1952), suggesting a sylvatic mosquito-non-human primate cycle of transmission similar to DENV (Haddow et al., 2012). The implication from the work here is that the inability of ZIKV to cause disease in IFN-competent mice (despite lethality in IFNAR-deficient mice) (Aliota et al., 2016; Lazear et al., 2016; Rossi et al., 2016) is at least linked to the inability of ZIKV to antagonize mouse IFN responses. Interestingly, type III IFN (IFNλ) has been shown to protect barrier cells of the human placenta called trophoblasts from ZIKV infection (Bayer et al., 2016). Type III IFN utilizes different cell surface receptors to type I IFN, but utilizes the same JAK-STAT machinery including STAT2 (Kotenko et al., 2003; Sheppard et al., 2003). Thus, it is highly likely that ZIKV can also evade type III IFN signaling through STAT2 degradation by NS5, which may contribute to virus crossing the placenta during pregnancy to cause neuronal disease in the developing fetus. In addition, the use of STAT2 deficient mice may be a useful model to examine ZIKV pathogenesis in vivo.
Despite the similarities of NS5 function amongst ZIKV and DENV, a number of interesting differences exist. DENV engages the E3 ubiquitin ligase UBR4 to degrade STAT2 (Morrison et al., 2013). Degradation of STAT2 is dependent on proteolytic processing of the DENV NS5 N-terminus in the context of the viral polyprotein (Ashour et al., 2009), although binding of STAT2 is independent of this processing (Mazzon et al., 2009). ZIKV NS5 did not require an authentic N-terminus as would be generated in the context of the viral polyprotein to degrade STAT2. This was consistent with the observation that ZIKV did not utilize UBR4 in STAT2 degradation, since UBR4 appears to function specifically in the degradation of proteins containing destabilizing N termini or N-degrons (Tasaki et al., 2005).
The function of ZIKV NS5 as an IFN antagonist is likely to play an important role in the emergence and pathogenesis of ZIKV in humans. In fact, we have shown that NS5 mutants of WNV, YFV and TBEV that are unable to inhibit type I IFN signaling are attenuated in tissue culture or mice (Laurent-Rolle et al., 2010; Laurent-Rolle et al., 2014; Lubick et al., 2015). In any case, further mapping of ZIKV NS5 amino acids involved in IFN antagonism should allow the generation of mutant ZIKVs that are attenuated due to their inability to prevent IFN signaling. Such recombinant viruses could still be grown in IFN-deficient cell lines, such as Vero cells, and used as live-attenuated vaccines if proven safe and immunogenic. Moreover, further elucidation of the mechanism and host proteins required for STAT2-mediated degradation by ZIKV might unravel targets for potential therapeutic intervention to treat ZIKV infections.
Experimental Procedures
For reporter assays 293T cells were cotransfected with the NS5-HA plasmids, the IFN-inducible firefly luciferase reporter (ISG54 promoter), and a plasmid constitutively expressing the renilla luciferase protein. Twenty-four hours post transfection cells were treated with 1,000 U of universal IFN (PBL) and analyzed for reporter activity 12h post treatment. Average firefly luciferase values were normalized to average renilla luciferase values. Empty vector treated with IFN was set to 100% reporter activity; each sample was standardized to this value. For cloning NS5 of Uganda ZIKV, Vero cells were infected with MR766 ZIKV strain (ATCC VR-84), total RNA was isolated using trizol and cDNA was RT-PCR amplified using gene specific primers and cloned into pCAGGS-COOH-HA expression vector. Upon sequencing it corresponded to Genbank: HQ234498. USUV NS5 was cloned from viral RNA. For ZIKV RNA quantification, total RNA was isolated from virus-infected cells using Trizol reagent (Invitrogen). 40ng of total RNA was used with ZIKV specific primers (5’ TTGGTCATGATACTGCTGATTGC and 5’ CCYTCCACRAAGTCYCTATTGC) and probe (5’ 6FAM-CGGCATACAGYATCAGGTGCATWGGAG-MGBNFQ) for qRT-PCR (ThermoFisher) (Lanciotti et al., 2008). For ISG mRNA quantification, we used a previously described protocol (Rajsbaum et al., 2014). Immunoprecipation assays were conducted in 293T cells transfected with the indicated plasmids, and 24h post transfection cells were treated with type I IFN for 45 minutes, or mock treated (or left untreated for the UBR4 and mouse STAT2 coIPs), followed by IP. Lysates were collected (sonication was performed on UBR4 IP lysates due to the membrane bound nature of UBR4) and centrifuged to obtain whole cell extract and the remaining lysate was incubated with anti-HA beads (Sigma). HA beads were washed and bound protein was eluted by boiling the beads in SDS buffer. SDS-PAGE followed by immunoblot analysis was performed on the IP and whole cell extracts with the indicated antibodies. Vero cells were used in IFA and 293T cells in flow cytometry.
Supplementary Material
Highlights.
Flavivirus nonstructural NS5 proteins antagonize type I IFN by different mechanisms
Zika virus (ZIKV) NS5 target the IFN-regulated transcriptional activator STAT2
ZIKV NS5 binds to and targets human, but not mouse, STAT2 for degradation
Unlike Dengue, ZIKV NS5 mediated STAT2 degradation does not require E3 ligase UBR4
Acknowledgements
We thank Barbara W Johnson, Ana Fernandez-Sesma and Steve Whitehead for kindly providing the 2015 PR ZIKV, 16881 DENV2 and the 2015 Fortaleza ZIKV strains respectively. We thank Genhong Cheng for IFNAR1−/− MEFs. We thank the microscopy and FACS SRF at Icahn School of Medicine for their support and Richard Cadagan for excellent technical assistance. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases. These studies were also partly supported by a supplement to NIAID grant U19AI118610.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
AG-S, SMB and MJE designed experiments. AG-S, SMB and AG wrote the paper. AG, SSP, ST, VB, LM and MS conducted the experiments. MCS and MPS-S provided reagents.
References
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathogens. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. Characterization of Lethal Zika Virus Infection in AG129 Mice. PLoS Negl Trop Dis. 2016;10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Laurent-Rolle M, Shi P, García-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. Journal of Virology. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, et al. Mouse STAT2 restricts early dengue virus replication. Cell Host & Microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET, Jr., Cherry S, Sadovsky Y, Coyne CB. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host & Microbe. 2016 doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, et al. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016 doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Forrester N, Tsetsarkin K, Vasilakis N, Weaver SC. Factors shaping the adaptive landscape for arboviruses: implications for the emergence of disease. Future Microbiol. 2013;8:155–176. doi: 10.2217/fmb.12.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra TL, Nassar ES, Nagamori AH, Ferreira IB, Pereira LE, Rocco IM, Ueda-Ito M, Romano NS. Iguape: a newly recognized flavivirus from Sao Paulo State, Brazil. Intervirology. 1993;36:144–152. doi: 10.1159/000150333. [DOI] [PubMed] [Google Scholar]
- Cook S, Holmes EC. A multigene analysis of the phylogenetic relationships among the flaviviruses (Family: Flaviviridae) and the evolution of vector transmission. Arch Virol. 2006;151:309–325. doi: 10.1007/s00705-005-0626-6. [DOI] [PubMed] [Google Scholar]
- Dick GW. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg. 1952;46:521–534. doi: 10.1016/0035-9203(52)90043-6. [DOI] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- Engel D, Jost H, Wink M, Borstler J, Bosch S, Garigliany MM, Jost A, Czajka C, Luhken R, Ziegler U, et al. Reconstruction of the Evolutionary History and Dispersal of Usutu Virus, a Neglected Emerging Arbovirus in Europe and Africa. MBio. 2016;7 doi: 10.1128/mBio.01938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard G, Moureau G, Charrel RN, Holmes EC, Gould EA, de Lamballerie X. Genomics and evolution of Aedes-borne flaviviruses. J Gen Virol. 2010;91:87–94. doi: 10.1099/vir.0.014506-0. [DOI] [PubMed] [Google Scholar]
- Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann H, Sung PY, Chiu HC, Yousuf A, Bird J, Lim SP, Davidson AD. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J Biol Chem. 2013;288:22621–22635. doi: 10.1074/jbc.M113.481382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nature immunology. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Kroczynska B, Mehrotra S, Arslan AD, Kaur S, Platanias LC. Regulation of interferon-dependent mRNA translation of target genes. J Interferon Cytokine Res. 2014;34:289–296. doi: 10.1089/jir.2013.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, del Carmen Castillo Signor L. Phylogeny of Zika virus in Western Hemisphere, 2015. Emerg. Infect. Dis. 2016 doi: 10.3201/eid2205.160065. DOI: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. Journal of virology. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JM, Pisanelli G, Pham A, Ayllon J, Miorin L, Martinez-Romero C, tenOever BR, et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell host & microbe. 2014;16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Diamond MS. Zika Virus: New Clinical Syndromes and its Emergence in the Western Hemisphere. J Virol. 2016 doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Chang B, Yu H, Liao C, Lin Y. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. Journal of virology. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J Virol. 2004;78:12225–12235. doi: 10.1128/JVI.78.22.12225-12235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol. 2005;79:1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubick KJ, Robertson SJ, McNally KL, Freedman BA, Rasmussen AL, Taylor RT, Walts AD, Tsuruda S, Sakai M, Ishizuka M, et al. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell host & microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nature reviews Immunology. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. The Journal of infectious diseases. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, García-Sastre A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathogens. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martinez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity. 2014;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects - Reviewing the Evidence for Causality. The New England journal of medicine. 2016 doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Rodriguez JJ, Cruz CD, Horvath CM. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J Virol. 2004;78:5358–5367. doi: 10.1128/JVI.78.10.5358-5367.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. Characterization of a Novel Murine Model to Study Zika Virus. Am J Trop Med Hyg. 2016 doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw ML, Garcia-Sastre A, Palese P, Basler CF. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J Virol. 2004;78:5633–5641. doi: 10.1128/JVI.78.11.5633-5641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nature Immunology. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell host & microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Garcia-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS, Calisher CH, McGuire K. Spondweni virus infection in a foreign resident of Upper Volta. Lancet. 1982;2:1306–1308. doi: 10.1016/s0140-6736(82)91511-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.