Abstract
The CRISPR-associated protein Cas9 from Streptococcus pyogenes is an RNA-guided DNA endonuclease with widespread utility for genome modification. However, the structural constraints limiting the engineering of Cas9 have not been determined. Here we experimentally profile Cas9 using randomized insertional mutagenesis and delineate hotspots in the structure capable of tolerating insertions of a PDZ domain without disrupting the enzyme’s binding and cleavage functions. Orthogonal domains or combinations of domains can be inserted into the identified sites with minimal functional consequence. To illustrate the utility of the identified sites, we construct an allosterically regulated Cas9 by insertion of the Estrogen Receptor α Ligand Binding Domain. This protein displayed robust, ligand-dependent activation in prokaryotic and eukaryotic cells, establishing a versatile one-component system for inducible and reversible Cas9 activation. Thus, domain insertion profiling facilitates the rapid generation of new Cas9 functionalities and provides useful data for future engineering of Cas9.
Domain insertions—the transfer of coding sequences for unique folds from one open reading frame (ORF) to another—are common occurrences throughout protein evolution1,2. These events are known to facilitate the diversification and rapid functionalization of new types of protein modules that fill needed roles in both eukaryotic and prokaryotic genomes3. In this study we investigated how synthetic domain insertions can be used to functionalize the programmable endonuclease Cas9 from S. pyogenes (hereafter Cas9).
Cas9 is an RNA-guided, DNA-binding and cleaving protein that has been adapted to enable the facile modification or perturbation of genes and regulatory and non-coding genomic elements in a wide variety of organisms4–6. Recently, there have been numerous attempts to develop Cas9 variants with additional functions by fusing protein domains directly to its N- or C-terminus7–9. However, the N- and C-termini of Cas9 are within ~40 Å of each other, leaving a large fraction of the protein structure unexplored by termini fusions10,11. This close spatial proximity can lead to steric incompatibility and may explain a relative lack of activity for many fusions, such as with VP6412. Another solution would be to identify insertion points within Cas9 that are capable of accepting functional domains, as occurs naturally in evolution1–3. This strategy would allow the engineering of complex functionalities. For example, an allosterically regulated Cas9 would permit conditional control of activity and allow precise interrogation of development, disease progression and differentiation5,6,13,14.
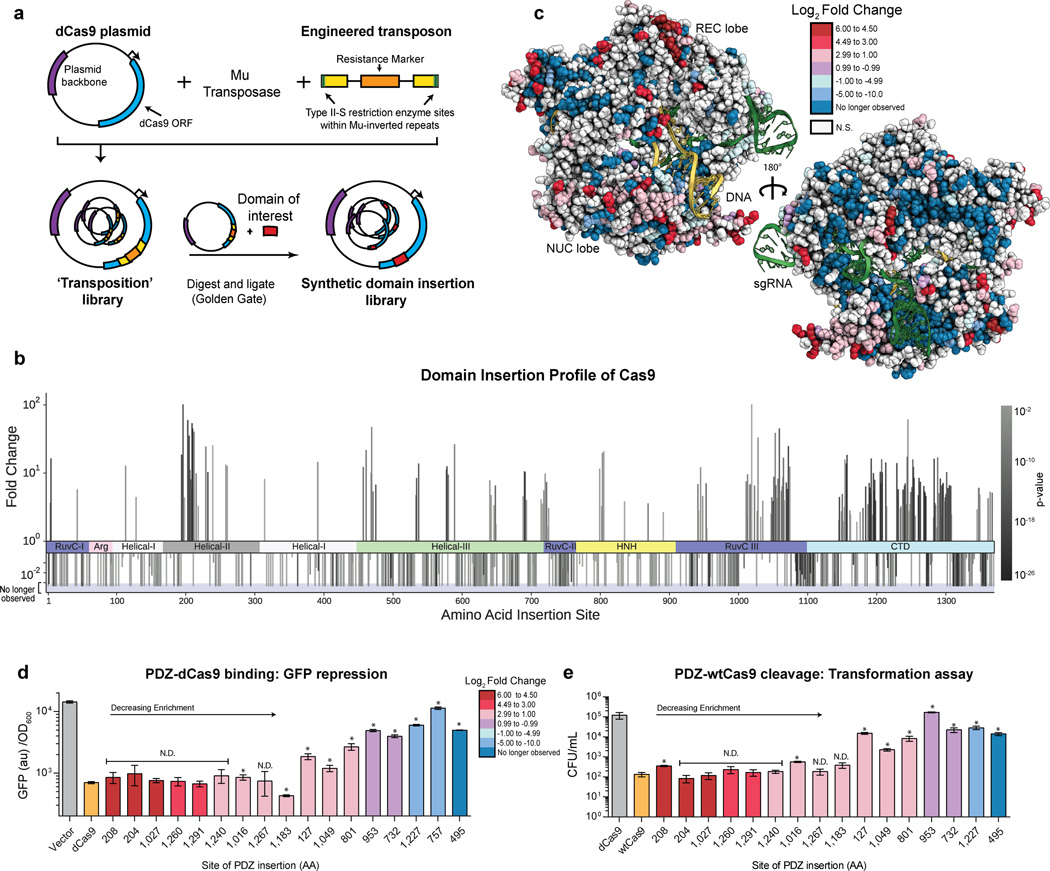
Here, we profiled the inherent plasticity of the Cas9 structure by examining its ability to tolerate a synthetic domain insertion while retaining RNA-guided DNA binding activity. An unbiased Cas9 insertion library was created using randomized transposition (Fig. 1A). Briefly, an engineered Mu transposon possessing an antibacterial selection marker flanked by BsaI endonuclease sites was inserted randomly throughout a catalytically inactive Cas9 (dCas9)-containing plasmid by in vitro transposition (Fig. 1A, Supplementary Fig. 1)15. After selection and sub-cloning to isolate plasmids with single transposition events within the dCas9 ORF, the library was characterized by deep sequencing. This analysis revealed that the transposition library possessed good insertion coverage, with insertions at >70% of all possible amino acid (AA) sites observed at least once (Supplementary Fig. 2). Once isolated, this library was used to construct specific domain insertion libraries by cleavage with BsaI and re-ligation with DNA fragments containing an ORF of interest. (Fig. 1A, Supplementary Fig. 1, Supplementary Fig. 2).
Figure 1. Mapping the insertion potential of Cas9 with the Alpha-Syntrophin PDZ protein interaction domain.
(a) Generation of the transposon-based domain insertion library. (b) Fold change values for insertions at specific amino acid sites derived from sequencing data over two rounds of screening. A positive value indicates the preference of the domain insertion at a site to remain in the library after screening for function. A negative value indicate a loss of the clone with an insertion at the site. More significant P values (DESeq, multiple hypothesis testing corrected) are represented as darker color bars. Positive values which attain 102 represent sites that were not sequenced before screening. Negative bars that extend into the shaded region represent clones which have been cleared from the library (i.e. not observed after screening). (c) Log2 fold change values from (b) mapped onto the structure of Cas9 (PDB ID:4UN3)11. (d) GFP repression activity of individual PDZ insertion sites. Values represent biological replicates with standard deviation (n=3), constructs are in order of decreasing fold change from sequencing. Positive and negative controls dCas9 and vector only are colored orange and grey, respectively. N.D. stands for no difference detected from dCas9, t-test (p > 0.01), (* = p < 0.01) (e) Cleavage activity of clones via an E. coli based transformation assay. Cas9 activity results in genomic cleavage and lower CFU/mL, values represent biological triplicates with standard deviation (n=3). Positive and negative controls Cas9 and dCas9 are colored orange and grey, respectively. N.D. stands for no difference detected from WT Cas9, t-test (p > 0.01), (* = p < 0.01)
The mouse alpha-1-syntrophin, Snta1, PDZ domain (PDZ) was chosen as a proof of concept insertion domain due to its small size (86 amino acids), well-folded nature and adjacent N- and C- termini (Supplementary Fig 3A)16,17. We hypothesized that this domain would be minimally perturbative and act as a molecular ‘potentiometer’— where the capacity to accommodate a PDZ insertion is indicative of the general insertion potential of a given amino acid site within Cas9. Moreover, as the PDZs are known protein-protein interaction domains, PDZ-Cas9s identified here may be further used as protein scaffolds to recruit other domains for editing, epigenetic modification and activation or repression purposes17 (Supplementary Fig 3B).
A PDZ domain with flanking amino acid linkers was cloned into the naïve library (Supplementary Fig 1, Supplementary Fig. 2) and passaged through two rounds of a CRISPRi screen18,19. Briefly, cells expressing Red Fluorescent Protein (RFP) and Green Fluorescent Protein (GFP) were assayed using fluorescence activated cell-sorting (FACS) to identify Cas9 variants capable of repressing RFP in a single guide RNA (sgRNA)-dependent fashion (Supplementary Fig 4)18,19. After sorting, the dCas9-PDZ libraries were subjected to deep sequencing to identify insertion sites. Comparing the transposition and PDZ libraries revealed that while the unscreened PDZ library was enriched in out-of-frame and reverse insertions, screening for dCas9-mediated gene repression increased the fraction of in-frame insertions by ~20 fold (Supplementary Fig 5). We refer to such insertions as ‘productive’ because they produce a full-length insertion protein. Thus, the screen enriched for productive PDZ-dCas9 insertion constructs.
Calculation of the log2-fold enrichment of insertion sites between the unscreened and final PDZ insertion library revealed statistically significant (p < 0.1) changes for roughly half of the amino acid (AA) sites in Cas9 (Fig 1B, C Supplementary Table 1). Domain insertions at the majority of residues within Cas9 are strongly selected against, with the bulk of clones falling out of the pool by the second round of screening (Fig 1B, C Supplementary Table 1). Sites with negative fold changes were highly overrepresented in critical motifs such as the globular core of Cas9, sgRNA-binding grooves, the bridge helix, the PAM-binding pocket and the DNA:RNA heteroduplex annealing channel (Fig 1C, Supplementary Fig. 6). Nevertheless, we identified small local clusters of amino acids that are tolerant to insertions and recovered a total of 175 statistically significant (p < 0.1) sites enriched ≥ 2-fold (Fig. 1B, C). When mapped onto the holo Cas9-DNA-RNA crystal structure11, the enriched insertion sites tend to cluster in discrete regions, often around flexible loops, the ends of helices, and at solvent-exposed residues. Specifically, hotspots are found in an abundance of Cas9’s domains: at six clusters within the helical recognition (REC) lobe, the linker between the REC and nuclease lobes, the HNH domain, three extended sites in the RuvCIII region and throughout the PAM interacting (C-terminal) domain (Fig 1B,C, Supplementary Fig. 7). These insertion sites provide access to both 5’ and 3’ ends of the bound DNA (~10 Å) as well as the groove hypothesized to hold the non-targeted DNA strand10,11. Consequently, these insertions might allow engineering of specific and functional interactions with bound DNA. Although insertions are enriched in undefined regions from three different Cas9 x-ray crystal structures, they also occur in nearly every secondary structure element of Cas9 (Supplementary Fig. 8). This poses the possibility that the oft-used rational design strategy of inserting domains into flexible loops underestimates the protein space available for manipulation. Moreover, as the insertions into an alpha-helix or beta sheet presumably disrupt the secondary structure in these areas, such insertions may be informative in future efforts to dissect Cas9 structure and function.
In order to confirm the binding activity of dCas9 proteins with PDZ domain insertions, plasmid DNA for individual clones were isolated, at random, from all stages of the selection and used to co-transform E. coli with a GFP-targeting sgRNA-expression plasmid. We found that effective GFP repression corresponds well with the calculated fold change of the insertion site; highly enriched clones perform at near wild-type (WT) dCas9 levels (Fig. 1D) (Supplementary Fig. 9)18. To determine whether the PDZ-Cas9 clones also possessed nuclease activity, the catalytic residues (D10 & H840) were reintroduced and tested in an E. coli based transformation assay4,20. In this assay, nuclease activity leads to genomic cleavage and cell death. Once again, cleavage activity correlated well with fold change, the most highly enriched PDZ-Cas9 clones, except for the insertion at amino acid 208, maintained levels of cleavage-induced cell death similar to WT Cas9 (Fig. 1E) (Supplementary Fig. 9). Thus, it is possible to insert an entire exogenous domain at numerous sites within Cas9’s primary sequence while maintaining near-native levels of activity.
To determine the effect of inserting an orthogonal domain at the sites recovered with the PDZ screen, we created eight synthetic domain insertions using the mouse SRC Homology 3 domain of adapter protein Crk (SH3)17. The SH3 domain was inserted into highly-enriched sites (> 10 fold) chosen to represent a diverse sampling of the Cas9 primary sequence. We found that all clones were functional for GFP repression and seven out of eight were able to obtain near-native activity levels (Supplementary Fig. 10). Finally, in order to examine the potential restrictions for domain insertion into Cas9 we surveyed the effect of multiple domain insertions on Cas9 function. We selected a subset of the PDZ-dCas9 and SH3-dCas9 insertions previously validated, 3 PDZ insertions, a PDZ C-termini fusion and 2 of the SH3 insertions, and created stacked constructs with up to three separate domain insertions and a terminal fusion. Although greater numbers of insertions and fusions have a perturbative effect on binding and repression activity, many of the stacked constructs are capable of repressing GFP to a degree comparable with dCas9 (Supplementary Fig. 10).
The design of synthetic protein switches and sensors is limited by the difficulty of predicting insertion sites that have the potential to confer allostery21. Therefore we next explored whether domain insertion profiling could be used to reveal sites in Cas9 amenable to allosteric coupling14,21. As a proof of concept, we chose to use the well-studied Human Estrogen Receptor α Ligand Binding Domain (ER-LBD; residues 302–552 of ESR1)22–24. Based on crystallographic data, this domain is known to adopt distinct conformations dependent on ligand binding; the antagonist-bound conformation places the N- and C-termini of ER-LBD substantially closer together (~20 Å) than either the apo or agonist bound forms (Supplementary Fig. 11). This conformational switch can serve as a mechanism by which an allosteric signal is transduced25.
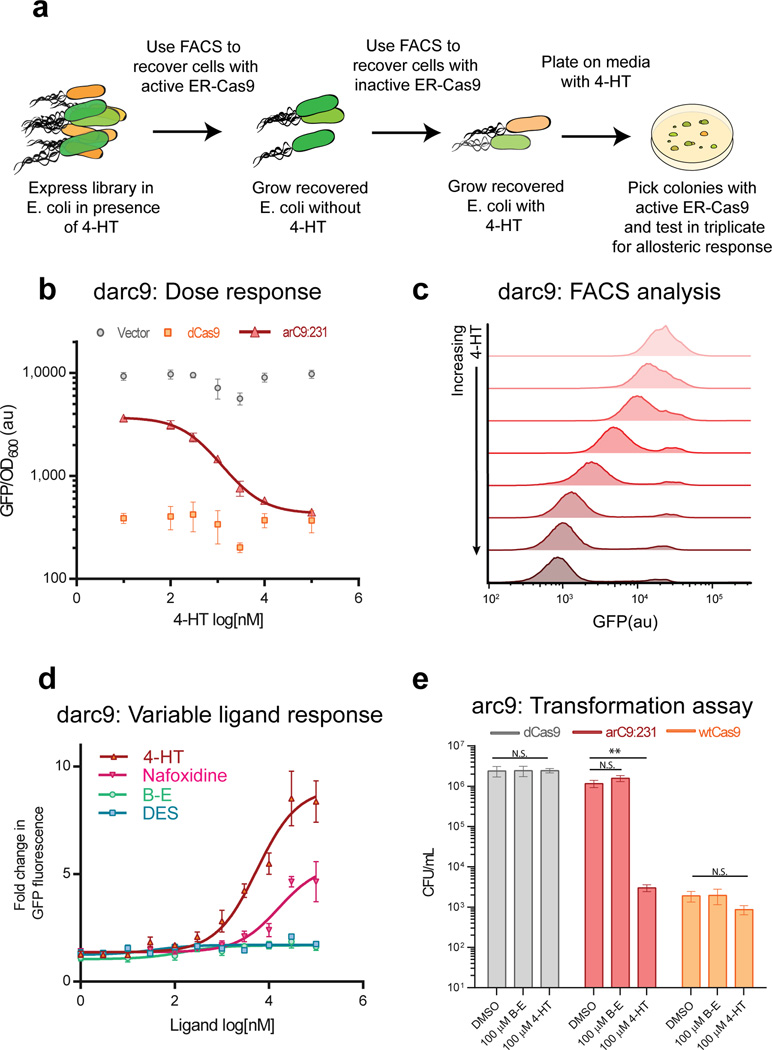
In order to create an allosterically-regulated Cas9 (arC9), the ER-LBD was introduced into the naïve dCas9 transposition library in the manner previously described and passaged through a modified version of the CRISPRi-based screen. Briefly, a positive screen in the presence of ligand 4-hydroxytamoxifen (4-HT; antagonist) was carried out, followed by a negative screen for loss of activity in the absence of ligand (Fig 2A). Clones were recovered by plating, re-transformed with a sgRNA targeting GFP, and assayed for repression in E. coli. We identified an insertion site at AA 231 that demonstrated a 4-HT-dependent decrease in GFP fluorescence, indicating switch-like behavior (Supplementary Fig. 12). Notably, this site was also enriched during the PDZ profile of Cas9 (Supplementary Table 1).
Figure 2. Creation of a switch-like Cas9 though insertion of the Estrogen Receptor ligand binding domain.
(a) Schematic of the screen / counter-screen procedure to select for ligand responsive Estrogen Receptor ligand binding domain (ER-Cas9) insertions. (b) Dose-response curve to 4-HT. darC9:231 has an IC50 of 440 ± 70 nM (S.D) and a Hill coefficient of 1.04 as expected for non-cooperative binding of 4-HT to ER-LBD. (c) Single cell analysis of darC9:231 binding in response to increasing concentrations of 4-HT. Flow cytometry data tracks ensemble data demonstrating a > 9 fold switch between darC9 based GFP repression plus and minus 4-HT (darC9 GFP signal without 4-HT mean: 24,310 (au) and with 100 µM 4-HT: 2,631 (au) (d) Dose response of darC9:231 binding to various ligands (B–E and DES are beta-estradiol & diethylstilbestrol). Response is normalized to vector control fluorescence under the same conditions. (e) Switching of arC9:231 cleavage activity. Transformation assays demonstrate that ligand dependent arC9 switching also extends to cleavage activity, t-test (** p-value <0.01). All experiments performed in E. coli.
Insertion of ER-LBD at AA 231 (arC9:231) was first characterized in E. coli. A catalytically dead arC9:231 (darC9) exhibited 4-HT dose-dependent repression of GFP with a ~10 fold change in activity in pooled and single cell experiments (Fig. 2B, C). Therefore darC9 represents a tunable CRISPRi effector. darC9 also exhibited clear ligand discrimination. CRISPRi-based GFP repression increased with ligands that encourage the ER-LBD to enter an antagonist conformation, 4-HT and nafoxidine, as opposed to ligands that promote the agonist conformation, beta-estradiol and diethylstilbestrol (Fig. 2D). This further supports the argument that the ER-LBD insertion at AA 231 is able to transduce ligand specific binding of 4-HT, through induction of the antagonist ER-LBD conformation, into Cas9 activity. To determine if arC9:231 also exhibited allosteric control over cleavage activity, the catalytic residues were reintroduced (D10 & H840) and tested in the E. coli transformation assay described above (Fig. 2E). We found that arC9 increased chromosomal cleavage and death at least 100-fold more in the presence of 4-HT than with a DMSO vehicle control (p value < 0.01), indicating that allosteric modulation of arC9 also extends to cleavage activity.
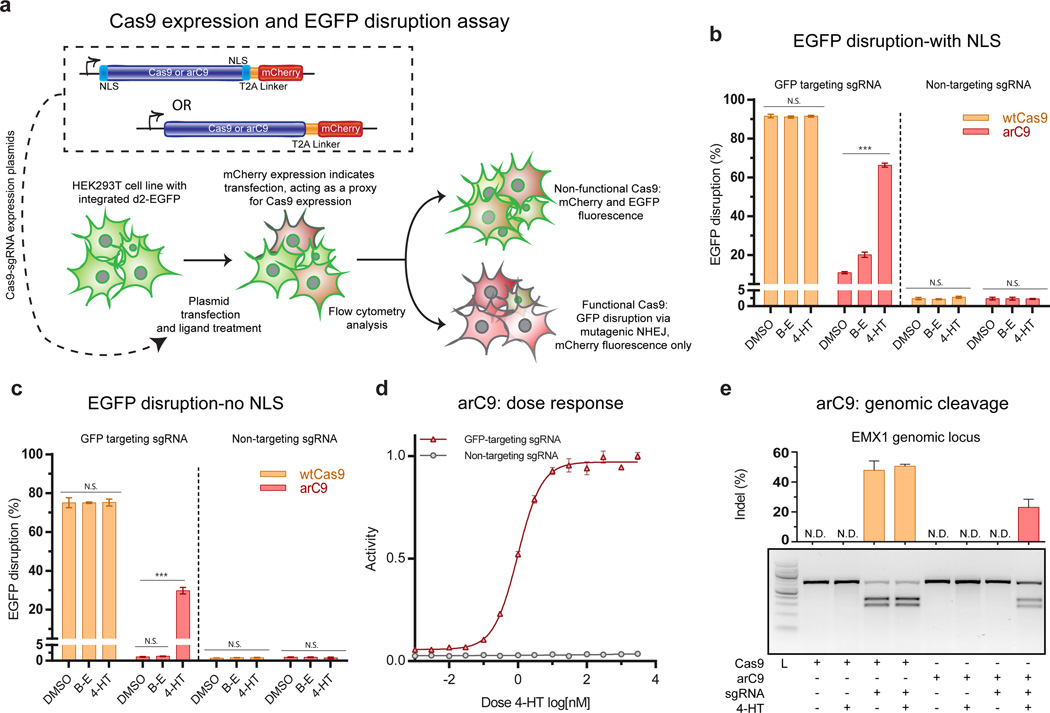
W next tested arC9:231 function in eukaryotic cells. Cas9 and arC9:231 constructs flanked by nuclear localization sequences (NLS) were transfected into a Human Embryonic Kidney (HEK293T) cell line expressing a stably integrated EGFP-PEST, and assayed for GFP disruption (Fig. 3A)26. After 72 hours, WT Cas9 was found to disrupt 91% of the EGFP signal regardless of treatment condition (Fig. 3B, Supplementary fig 12). A 6-fold induction of arC9-mediated GFP disruption upon the addition of 4-HT (10.9 ± 0.5 % to 66 ± 1%) (Fig. 3B) was observed over the same time period. We also found that higher expression conditions led to an increase of GFP disruption even in the absence of 4-HT (Supplemental Figure 13).
Figure 3. Validating arC9 in eukaryotic cells.
(a) Schematic of the arC9:231 expression constructs and GFP disruption assay. (b) Quantification of EGFP disruption at 72 hours for Cas9 and arC9 with a N- and C-terminal nuclear localization signal (NLS) (n=3). These data demonstrate a ~6 fold increase in EGFP disruption. Background activity of arC9 is 10.9 ± 0.5% (S.D.) while EGFP disruption in the presence of 300 nM 4-HT increases to 66 ± 1% (S.D.) Error bars represent one standard deviation of biological replicates. (c) Quantification EGFP disruption at 72 hours for Cas9 and arC9 without an NLS (n=3). Background activity of arC9 is not significantly different from a non-targeting negative control, t-test. EGFP disruption in the presence of 4-HT increases to 30 ± 2% (S.D.) this represents at least a 24-fold increase in arC9 activity in the presence of 300 nM 4-HT, t-test (*** p values < 0.001). (d) Dose response of arC9 w/o NLS normalized to maximum activity. IC50 is 1.0 ± 0.2 nM (S.D). (e) T7EI assay of Cas9 and arC9 mediated indel formation at the EMXI locus at 72 hours. Cas9 with the targeting guide causes indels regardless of treatment condition, arC9 only cleaves a genomic locus in the presence of 4-HT. Quantification and error bars represent the standard deviation of biological replicates (n=3). N.D. signifies not detected, below the detection limit of the assay.
To improve arC9 control we sought to take advantage of the known ligand-dependent nuclear localization activity of ER-LBD27. We removed the dual NLS from the arC9 construct and found that this reduced activity without 4-HT to background levels while maintaining roughly 30% of Cas9 disruption activity in the presence of 4-HT (Fig. 3C). Using the GFP disruption assay, we observed arC9’s dose-dependent response with an EC50 of 10 nM and full induction at 100–1000 nM 4-HT (Fig. 3D). The lack of background disruption suggests that arC9 can be adapted for very tight, reversible control of Cas9 cleavage activity.
In previous work, ER-LBD fusions to the termini of various DNA-binding proteins have been shown to regulate the ability of such proteins to enter the nucleus27,28 To explicitly demonstrate the necessity and utility of internal domain insertion, we directly compared the activity of our arC9 construct with that of a C-terminal ER-LBD fusion. In contrast to the switch-like response of arC9, a C-terminal fusion of ER-LBD minimally controls Cas9 EGFP disruption, providing a modest 0.21 fold increase in repression upon the addition of 4-HT, with greater than 50% of cells disrupted without activating ligand (Supplementary Figure 14). This result is consistent with our previous data demonstrating that WT Cas9 does not require an NLS to function in dividing human cells (Fig. 3C). To further validate arC9 we tested its nuclease activity on two endogenous human loci, EMX1 and DRKY1. T7 endonuclease I (T7EI) analysis showed efficient indel formation and gene editing in presence of 4-HT, while there was no detectable arC9 activity without ligand (Fig. 3E, Supplementary Figure 15, Supplementary Table 2). Thus, arC9 acts as a 4-HT-inducible nuclease in human cells demonstrating that unbiased domain insertion can be used to engineer robust Cas9 functionalities not achievable using terminal fusions.
The discovery and application of CRISPR-Cas9 has revolutionized functional genomics. However, large-scale screens and more focused in-vivo applications that require precise timing, such as the study of development, differentiation and late onset disease, have been limited by the constitutive activity of Cas9. Tet-systems can enable Cas9 expression control29–33, but are cumbersome due to the need for additional components. We therefore tested whether arC9 could be stably integrated into the genome from a single-vector lentiviral system and enable conditional gene editing. To assess long-term leakiness, induction, and reversibility of the arC9 protein, we also generated a sensitive monoclonal reporter cell line by transducing murine BNL CL.2 cells with a retroviral vector expressing EGFP (Supplementary Figure 16a). Upon low-copy infection with the Cas9/arC9 lentiviral constructs and hygromycin B selection, we measured the reduction in the number off GFP expressing cells for arC9 over 24 days. Whereas WT Cas9 showed up to ~80% GFP disruption with GFP-targeting sgRNAs, no leakiness was observed with arC9 in DMSO (Supplementary Figure 16c). After 12 days, we treated a subpopulation of arC9-expressing cells with 4-HT (1 µM), beta-estradiol (1 µM) or vehicle control (DMSO). In the presence of 4-HT, arC9 disrupted GFP in up to ~16% of cells, whereas no measurable editing was observed in the absence of ligand (Supplementary Figure 16d). Some background GFP-negative cells were present in all samples.
To test whether the activation of arCas9 is reversible we recovered the 4-HT treated cells at the end of the treatment, cultured them for two days in regular culture medium, and then infected them with a secondary lentiviral vector expressing a sgRNA targeting Pcsk9. T7EI analysis of the endogenous Pcsk9 locus after six days of treatment with DMSO or 4-HT showed that arC9 can be turned off in the absence of ligand the or used for controlled serial genome editing by repeated addition of 4-HT (Supplementary Figure 16e). After removal of 4-HT from the media a small amount of residual arC9 activity remained. This may be due to the high affinity of arC9 for 4-HT (EC50 10 nM) and likely slow dissociation of the complex (Figure 3D). Nevertheless, arC9 can serve as a single-component system for conditional genome editing and/or be combined with other sgRNA expression platforms to render gene editing inducible and reversible.
Although arC9 functions robustly as an inducible gene repressor and endonuclease in prokaryotic and eukaryotic cells, it is unclear how arC9 is activated in the presence of 4-HT. The arC9:231 insertion is especially counterintuitive as the Helical-II (REC2) domain into which the ER-LBD is inserted has previously been shown to be dispensable for cleavage activity10. A recent crystal structure of sgRNA bound Cas934 helps rationalize the ligand-dependence of arC9, suggesting that ER-LBD may disrupt the required conformational changes of the Helical-II domain. This disruption would sterically occlude the RNA:DNA binding channel of Cas9 and prevent functional DNA unwinding and RNA:DNA hybridization (Supplementary Figure 17)34.
In this work we identify a range of potential insertion sites for Cas9 engineering and outline a methodology for the development of new Cas9 constructs with improved control over genome editing and modification13,35. Previous efforts to engineer Cas9 as a molecular scaffold have constructed systems in which the sgRNA can recruit effector proteins36,37. Others have built inducible Cas9 nucleases using intein splicing or the splitting of the protein itself13,38,39. All of these efforts have used iterative, rational design to isolate functional molecules. By contrast, we demonstrate an unbiased profiling of domain insertions across Cas9 structure. Coupled with high-throughput screening and sequencing, this profiling rapidly queries structure and informs the protein engineering process. Specifically, we have generated a host of Cas9 scaffolds, containing PDZ or SH3 interaction domains that are functional and may prove useful for the recruitment of accessory proteins in future work. Notably, we also find that all previously reported and validated split Cas9 constructs fall within or adjacent (≤2 amino acids) to a small number of the insertional hotspots identified here (Supplementary Figure 18)13,38–40. To assess the generality of domain insertion profiling, the process was repeated with ER-LBD and identified a variant, arC9, whose activity is allosterically coupled to ER ligand binding. It functions as a conditional DNA binding protein and nuclease in both prokaryotes and eukaryotes and displays substantial ligand-dependent activity with very low background.
Online Methods
Comprehensive methods used in this study may be found in the Supplementary Materials.
Supplementary Material
Acknowledgments
We thank Stanly Qi and John Dueber for generously providing the E. coli strain from Qi et al. 2013 and the PDZ and SH3 domain from Dueber et al. 2009. We would like to thank M. O’Connell, S. Sternberg A. Wright and S. Higgins for productive discussions and readings of the manuscript. This work was supported by a NIH New Innovator Award (1DP2EB018658-01) and a Basil O’Connor Starter Scholar Research Award from the March of Dimes to D.F.S. and by the National Science Foundation (IQJEDMS001) (J.A.D); A.F is a National Science Foundation Graduate Research Fellow, B.T.S. is a Roche Postdoctoral Fellow, RPF 311 J.A.D. is an HHMI Investigator.
Footnotes
Accession codes:
The Domain Insertion Profile-sequencing data is available at https://github.com/SavageLab/arC9_data. Sequencing data was analyzed with a custom Python pipeline that is available online at http://github.com/SavageLab/dipseq
Contributions
B.L.O, D.C.N, C.F, J.A.D & D.F.S, designed the research. B.L.O, D.C.N, C.F. A.F, B.T.S, performed the experiments. A.F performed the computational analysis. B.L.O, A.F, C.F & D.F.S analyzed the data. B.L.O, C.F, J.A.D & D.F.S wrote the paper. J.A.D & D.F.S funded.
Competing financial interests
The authors have submitted a patent disclosure on this work. J.A.D is employed by HHMI and works at the University at California Berkeley. UC Berkeley and HHMI have patents pending for CRISPR technologies on which she is an inventor. J.A.D is the executive director of the Innovative Genomics Initiative at UC Berkeley and UCSF. J.A.D is a co-founder of Editas Medicine, Intellia Therapeutics and Caribou Biosciences and a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics and Driver.
References
- 1.Consortium IHGS, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Peisajovich SG, Garbarino JE, Wei P, Lim Wa. Rapid Diversification of Cell Signaling Phenotypes by Modular Domain Recombination. Science (80-.) 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chothia C. Evolution of the Protein Repertoire. Science (80-.) 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A Programmable Dual-RNA – Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science (80-.) 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science (80-.) 2014;346:1258096–1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 6.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis KM, Pattanayak V, Thompson DB, Zuris Ja, Liu DR. Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds KA, McLaughlin RN, Ranganathan R. Hot Spots for Allosteric Regulation on Protein Surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards WR, Busse K, Allemann RK, Jones DD. Linking the functions of unrelated proteins using a novel directed evolution domain insertion method. Nucleic Acids Res. 2008;36:e78. doi: 10.1093/nar/gkn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz J, et al. Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat. Struct. Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- 17.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 18.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oakes BL, Nadler DC, Savage DF. Protein Engineering of Cas9 for Enhanced Function. Methods Enzymol. 2014;546C:491–511. doi: 10.1016/B978-0-12-801185-0.00024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briner AEEE, et al. Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality. Mol. Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Stein V, Alexandrov K. Synthetic protein switches: design principles and applications. Trends Biotechnol. 2015;33:101–110. doi: 10.1016/j.tibtech.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 23.Tanenbaum DM, Wang Y, Williams SP, Sigler PB. Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc Natl Acad Sci U S A. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warnmark a. Interaction of Transcriptional Intermediary Factor 2 Nuclear Receptor Box Peptides with the Coactivator Binding Site of Estrogen Receptor alpha. J. Biol. Chem. 2002;277:21862–21868. doi: 10.1074/jbc.M200764200. [DOI] [PubMed] [Google Scholar]
- 25.Tucker CL, Fields S. A yeast sensor of ligand binding. Nat. Biotechnol. 2001;19:1042–1046. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- 26.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIsaac RS, et al. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013;41:e57–e57. doi: 10.1093/nar/gks1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feil R, et al. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dow LE, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015;33:390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gossen M, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 33.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang F, Zhou K, Ma L, Gressel S, Doudna Ja. A Cas9-guide RNA complex preorganized for target DNA recognition. Science (80-.) 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 35.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zalatan JG, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2014;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;9:1–8. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 39.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truong D-JJ, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.