Abstract
The balance between reactive oxygen species and antioxidants plays an important role in periodontal health. We previously demonstrated that high reactive oxygen species production by oral polymorphonuclear neutrophils (oPMNs) in chronic periodontitis (CP) refractory to conventional therapy is associated with severe destruction of periodontium. Herein, we show that inhibition of antioxidant production through down-regulation of nuclear factor erythroid 2-related factor 2 (Nrf2) pathway in oPMN, despite enhanced recruitment in the oral cavity, is associated with severe CP. Twenty-four genes in the Nrf2-mediated oxidative stress response pathway were down-regulated in PMNs of diseased patients. Downstream of Nrf2, levels of oPMN superoxide dismutase 1 and catalase were decreased in severe CP, despite increased recruitment. Nrf2−/− mice had more severe loss of periodontium in response to periodontitis-inducing subgingival ligatures compared with wild-types. Levels of 8-hydroxy-deoxyguanosine were increased in periodontal lesions of Nrf2−/− mice, indicating high oxidative damage. We report, for the first time, Nrf2 pathway down-regulation in oPMNs of patients with severe CP. PMNs of CP patients may be primed for low antioxidant response in the context of high recruitment in the oral cavity, resulting in increased oxidative tissue damage.
Periodontal diseases are a group of highly prevalent inflammatory diseases affecting up to 70% of the US population.1 Chronic periodontitis (CP), the most common form of periodontal disease, is characterized by progressive destruction of tooth-supporting structures as a result of altered host-biofilm interactions in the gingival crevice and unresolved inflammation. Although pathogenic biofilms are required for disease onset, host factors drive inflammation-mediated resorption of alveolar bone and damage to the connective tissue attachment. Neutropenia, altered PMN recruitment into periodontium, and impaired bacterial killing lead to severe forms of periodontitis.2 On the other hand, hyperresponsiveness and excess reactive oxygen species (ROS) production likely contribute to relapse after CP treatment. Untreated CP is associated with considerable morbidity and represents a risk factor for numerous systemic conditions with underlying low-grade inflammation. In addition to persistent inflammation, oxidative stress is believed to be a link between CP and metabolic syndrome.3
One mechanism suggested to contribute to periodontal tissue breakdown in CP is the increase in endogenous-produced oxidative stress observed in periodontitis lesions.4, 5 We have previously shown that CP patients refractory to conventional therapy present with more severe loss of tooth attachment when their oral polymorphonuclear neutrophils (oPMNs) produce high levels of ROS, suggesting increased oxidative stress in periodontal tissues through excess PMN-derived ROS in these patients.6 On the other hand, the protective role of antioxidants (AOs) in CP has been investigated by total antioxidant capacity measurements in whole saliva and gingival crevicular fluid (GCF) samples. Total antioxidant capacity levels were found to be significantly lower in CP patients when compared with healthy controls, although the mechanism responsible for oral AO decrease in CP has yet to be investigated.7, 8 Nonetheless, adjuvant AO therapy for CP has a potential to improve periodontal clinical parameters.9, 10
Nuclear factor erythroid 2-related factor 2 (Nrf2) regulates gene transcription of a large group of antioxidant and phase 2 detoxifying enzymes playing a central role in cellular defense against oxidative and electrophilic insults.11, 12 In response to oxidative stress, cytoplasmic Nrf2 is released from Kelch-like ECH-associated protein 1 and binds to antioxidant response elements in the promoter region of many antioxidant enzymes, including catalase (CAT), superoxide dismutase (SOD), NAD(P)H:quinone oxidoreductase, glutathione S-transferase, heme oxygenase-1, glutathione peroxidase, glutamate cysteine ligase, and peroxiredoxin I.12 In addition, because NF-κB–mediated production of cytokines and chemokines, such as tumor necrosis factor-α, IL-6, IL-8 (CXCL8), and chemokine ligand 2, is likely to be reduction-oxidation regulated, Nrf2 may play critical roles in control of inflammatory injury. Nrf2 was shown to play important roles in rheumatoid arthritis, gastrointestinal, renal, brain, skin, and pulmonary inflammation, and atherosclerosis.13
Herein, we investigated the oxidative state of local and systemic PMNs from patients with severe CP. It was found that down-regulation of the Nrf2 pathway in oPMNs is associated with increased recruitment in the oral cavity and severe CP. Nrf2−/− mice had increased local oxidative damage and alveolar bone and attachment loss at sites with CP.
Materials and Methods
Subject Population
Patients diagnosed with severe CP (eight subjects, two males and six females, aged 37 to 74 years) and periodontally healthy controls (six subjects, four males and two females, aged 24 to 38 years) were recruited in the periodontics graduate clinic at the Faculty of Dentistry, University of Toronto (Toronto, ON, Canada). Severe CP was defined using the American Association of Periodontology and the Eke et al1 criteria as the presence of two or more interproximal sites with ≥6-mm attachment loss and one or more interproximal site(s) with ≥5-mm probing depth.14 All subjects were systemically healthy and nonsmokers. Sample collection was completed before periodontal therapy. Subjects provided a written informed consent to participate, and the study was approved by the Research Ethics Board at the University of Toronto (protocol 24567).
PMN Isolation
Oral rinse and venous blood samples were collected from each subject. Oral rinse samples were collected as previously described.15 Rinses were collected from subjects who had not eaten or performed oral hygiene for the previous 1 hour to avoid clearance of PMNs before donations.16 Subjects were asked to rinse with 5 mL 0.9% isotonic sodium chloride solution (Baxter, Toronto, ON, Canada) for 30 seconds (six times with 3-minute breaks) and then asked to expectorate into a sterile 50-mL falcon tube. The same participants provided 10 mL of nonfasting venous blood. PMN isolation procedures from both oral rinse and blood samples were previously described.15 Isolation procedures were initiated within 2 hours of sample collection. In brief, blood PMNs were isolated using a one-step PMN isolation solution (one-step polymorphs; Axis-Shield PoC, Oslo, Norway) and oPMNs were isolated through a series of filtrations through 40-μm (Fisher Scientific, Loughborough, UK), 20-μm, and 11-μm nylon mesh (EMD Millipore, Billerica, MA). Both isolation procedures resulted in >95% PMNs with >95% viability.
RNA Isolation
Total PMN RNA isolation was completed on the day of sample collection, using mirVana isolation kit (Ambion, Austin, TX) using the manufacturer-suggested protocol, as was previously described by the authors.15 RNA samples were kept at −80°C until analyzed.
Gene Expression Microarray
Gene expression microarray was completed at the Center for Applied Genomics, Hospital for Sick Children Research Institute (Toronto, ON, Canada). Before gene expression microarray analysis, RNA samples were analyzed on a bioanalyzer using an RNA kit (2100 Bioanalyzer using an RNA 6000 PicoLabChip Kit; Agilent Technologies, Santa Clara, CA). All samples subjected to further analysis presented with an RNA integrity number >8. Microarray analysis was completed for oral and blood PMNs of four CP and four periodontally healthy subjects using the Illumina Human 12WG Expression BeadChip (48,000 gene transcripts; Illumina Inc., San Diego, CA). The microarray data comply with Minimum Information About a Microarray Experiment guidelines.17 The data set was deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE435525).
Microarray Data Analysis
The raw microarray data were preprocessed using R loaded with Lumi R package (http://www.r-project.org, last accessed January 11, 2016), followed by background correction, completed with BeadStudio Gene Expression Module (Illumina). Data were normalized by quantile normalization implemented with R loaded with Lumi R package. Differentially expressed genes were identified using linear models for microarray data. Genes were selected controlling for false discovery rate at the level of 0.01 and by a minimum fold change of 2 (at P < 0.05). Pathway analyses were performed using the Ingenuity Pathway Analysis software version 01-07 (Ingenuity Systems, Redwood City, CA). Results from analysis of the same data set were previously reported.18
RT-qPCR
Quantitative RT-PCR (RT-qPCR) was completed for two subjects from each of the study groups, on the same RNA samples used for microarray analysis. RT-qPCR was performed in triplicate (CFX96 Real-Time System; Bio-Rad, Hercules, CA), as previously described.15 Total RNA was reverse transcribed into cDNA using murine leukemia virus reverse transcription (Superscript II; Invitrogen, Thermo Fisher Scientific, Waltham, MA), and oligo-dT18 VN primer (ACGT, Toronto, ON, Canada) in a 20-mL reaction system. A sample without template RNA, and another without reverse transcription were used as negative controls. GAPDH (forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′; reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′) was used to normalize the expression data of the tested genes (SOD1: forward, 5′-GGTGGGCCAAAGGATGAAGAG-3′; reverse, 5′-CCACAAGCCAAACGACTTCC-3′; CAT: forward, 5′-TGGAGCTGGTAACCCAGTAGG-3′; reverse, 5′-CCTTTGCCTTGGAGTATTTGGTA-3′).
WB Analysis
SOD1 and CAT protein abundance was investigated through Western blot (WB) analysis. Oral and blood PMNs were isolated from four control subjects (two males and two females, aged 32 to 38 years) and five CP subjects (two males and three females, aged 37 to 74 years). Proteins were resolved by SDS-PAGE, transferred onto nitrocellulose membranes (Amersham-GE, Baie d'Urfe, QC, Canada), and probed with appropriate dilutions of the following antibodies: rabbit anti-human SOD1, anti-human CAT, and anti–β-actin (Cell Signaling Technology, Danvers, MA; CAT: EMD Millipore, Billerica, MA). Immunoreactivity was detected using an electrochemiluminescence system (Perkin Elmer, Akron, OH), and band intensity was quantified using ImageJ software version 1.50e (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Nrf2−/− Mice
Nfe2l2 knockout mice (Nrf2−/−) are viable and have a normal phenotype under physiological conditions.19, 20 Nrf2−/− mice bred on a C57BL/6J background were purchased from Jackson Laboratories (Bar Harbor, ME). C57BL/6J mice were used as wild-type (WT) control. Mice were handled in accordance with the Guide for the Humane Use and Care of Laboratory Animals, and all of the experiments were approved by the Animal Care Committee, Faculty of Dentistry, University of Toronto. The Nrf2−/− genotype was verified before breeding using PCR, following Jackson Laboratories' instructions. Mice of both sexes, aged 6 to 10 weeks, were used.
Bone Marrow PMN Isolation
Bone marrow PMNs were harvested from mice femur and tibia and isolated with Percoll gradient separation, as previously described.21 Erythrocytes were lysed by addition of 5 mL of Pharm Lysis buffer (BD Biosciences, Mississauga, ON, Canada). Samples were washed and cells were resuspended in phosphate-buffered saline (Sigma-Aldrich, St. Louis, MO) and quantified using a Coulter counter (Beckman Coulter, Brea, CA).
Extracellular ROS Measurements
Extracellular ROS levels were measured by isoluminol-amplified chemiluminescence assay.22, 23 Bone marrow PMNs from six WT and four Nrf2−/− mice were incubated in a mixture of 3 mmol/L isoluminol and 6 U of horseradish peroxidase (Sigma-Aldrich) on preblocked 96-well plates, and stimulated with 10−6 mol/L phorbol myristate acetate (Sigma-Aldrich). Relative light units (RU) were measured using a fluorescence spectrophotometer (Fisher Scientific, Waltham, MA).
PMN Chemotaxis Assay
PMN chemotaxis was measured using a Zigmond chamber, as previously described.21 PMN movement for bone marrow PMNs from three WT and three Nrf2−/− mice in formylmethionine–leucyl-phenylalanine gradient was recorded using time-lapse video microscopy with differential interference contrast optics (Nikon Eclipse E1000 with Nikon Coolpix 995 camera). Images captured at 15-second intervals for 15 minutes were analyzed with ImageJ software.
Ligature-Induced Periodontitis
As previously described, a split-mouth experimental periodontitis model was used to investigate periodontal tissue destruction in Nrf2−/− mice with disease.24 Mice were anesthetized with ketamine/xylazine, and a 9-0 silk suture was placed in the gingival sulcus around the left second maxillary molar. The suture serves as biofilm retentive medium for subgingival biofilm maturation and induction of periodontitis. The contralateral right second molar served as the control. Mice were euthanized 21 days after ligature placements. Maxillae were harvested and defleshed in a colony of dermestid beetles at Royal Ontario Museum (Toronto, ON, Canada), and then freeze fumigated for 7 days at −20°C. Dry skulls were stained with methylene blue (1% in water) for 1 minute. Images of the buccal aspects of second molars (right, healthy; left, periodontitis) were taken in ×5 magnification using a video camera (PixeLINK, Ottawa, ON, Canada) mounted onto a stereomicroscope (Nikon Eclipse E1000). Alveolar bone loss (ABL) was measured by morphometry from the cemento-enamel junction to the alveolar bone crest in three locations per tooth: midbuccal, mesiobuccal, and distobuccal (using ImageJ software).
Immunohistochemistry
Immunohistochemistry was completed at the Toronto Center for Phenogenomics (Mount Sinai Hospital, Toronto, ON, Canada). Maxillae collected 21 days after induction of periodontitis were fixed in 4% paraformaldehyde, decalcified in 10% formic acid for 7 days, and embedded in paraffin. Coronal sections of healthy and diseased areas were probed with primary goat anti–8-hydroxy-deoxyguanosine (8-OHdG; EMD Millipore, Billerica, MA) and secondary horse anti-goat biotinylated IgG (Vector Labs, Burlington, ON, Canada) and stained using an avidin/biotin tissue staining system (VECTASTAIN Elite; Vector Labs) and hematoxylin and eosin counterstaining. Three regions of interest were defined for each site (ligature and control): alveolar bone crest, sulcular epithelium, and gingival connective tissue. Cells positive for 8-OHdG were quantified using ImageJ software by automated particle counting after setting a uniform intensity threshold across all images analyzed.25 Numbers of positive particles per area were calculated.
Statistical Analysis
One-way analysis of variance with Tukey-Kramer honestly significant difference post hoc analysis and paired t-tests were used as specified.
Results
Nrf2 Pathway Is Down-Regulated in Oral PMNs of CP Patients
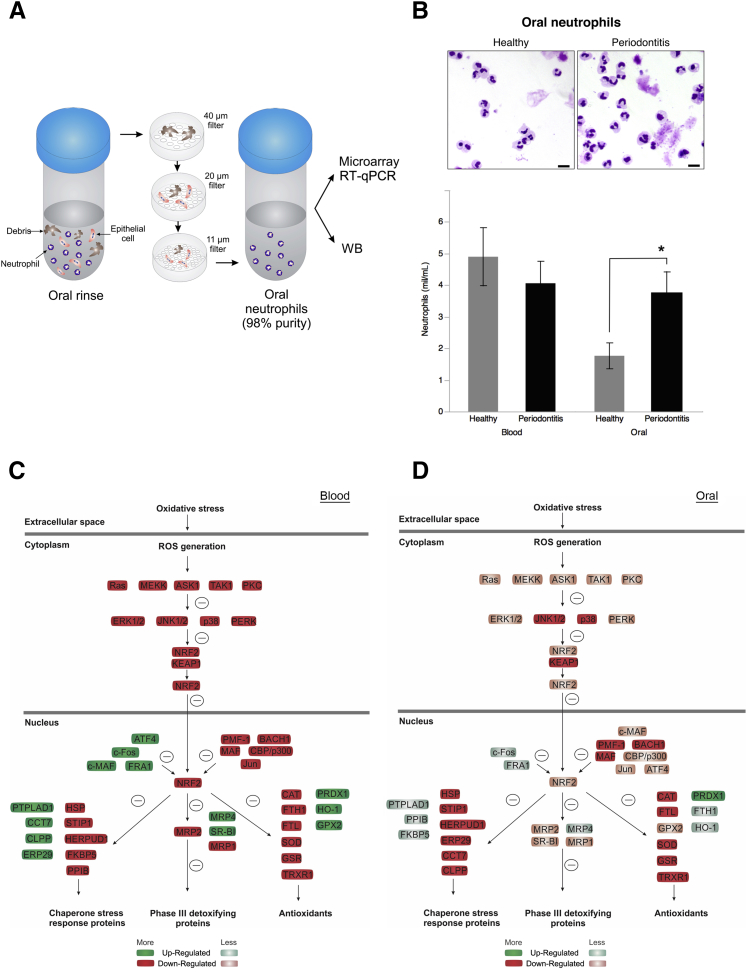
Untreated CP patients and healthy controls were assessed for periodontal clinical parameters before blood and oral PMN collection. Patients with CP had significant loss of clinical attachment and generalized bleeding on probing (Supplemental Table S1). Sequential filtering of oral rinses removed debris and epithelial cells, resulting in 98% purity of oPMN isolates. PMN RNA was further isolated for microarray and RT-qPCR assays and proteins isolated for WB analysis of AO production (Figure 1A). CP patients had significantly higher oPMN numbers compared with healthy controls, despite similar peripheral blood PMN numbers (oPMN: healthy, 1.76 ± 0.41 × 106/mL and CP, 3.75 ± 0.68 × 106/mL; blood PMN: healthy, 4.90 ± 0.92 × 106/mL and CP, 4.06 ± 0.0.69 × 106/mL) (Figure 1B).
Figure 1.
Nuclear factor erythroid 2-related factor 2 (Nrf2) antioxidant (AO) pathway is down-regulated in polymorphonuclear neutrophils (PMNs) of chronic periodontitis (CP) patients despite high oral recruitment rate. A: Oral rinses with 0.9% saline solution were used to isolate oral PMNs (oPMNs). Sequential filtering through 40-, 20-, and 11-μm filters removed debris and epithelial cells from the rinse, which resulted in 98% purity of oPMN samples. PMN RNA was further isolated for microarray and RT-qPCR assays and proteins isolated for Western blot analysis of AO production. B: Representative micrographs of Diff-Quick stained cytospins of oPMNs illustrating increased recruitment in CP patients compared with healthy controls. Oral and blood PMNs from eight CP patients and six healthy controls were quantified with a Coulter counter. C and D: PMN RNA was isolated from oPMNs of healthy and CP patients, as described in Materials and Methods. Blood PMNs were isolated using a one-step PMN isolation protocol. Regulation of Nrf2 antioxidant stress response pathway was predicted on the basis of microarray gene expression data using the Ingenuity Pathway Analysis software for blood and oPMN in CP compared with health. Data are given as means ± SD (B). ∗P < 0.05 (paired t-test). Scale bar = 10 μm (B). ROS, reactive oxygen species; RT-qPCR, quantitative RT-PCR.
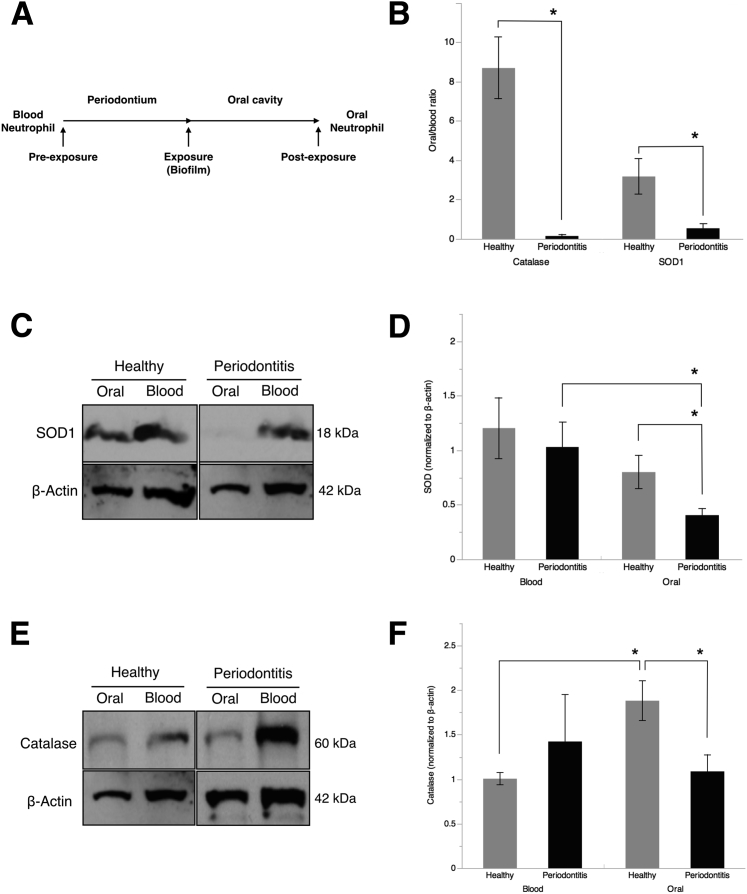
Microarray analysis of PMN gene expression revealed significantly higher CXCL8, CXCR1, and TREM1 expression by oPMN in CP compared with blood PMN (6.41-, 1.43-, and 3.1-fold increase, respectively). There was a mild down-regulation (less than twofold) of critical factors involved in ROS production in oPMNs in CP compared with health (Supplemental Table S2). Ingenuity pathway analysis of microarray data predicted significant down-regulation of AO expression downstream of Nrf2 in CP PMN (blood and oral) compared with health. Down-regulation of AO expression was predicted on the basis of down-regulation found in several upstream and downstream factors in the Nrf2 pathway (Figure 1, C and D). Analysis of oral versus blood PMN samples within each of the study groups revealed that unlike healthy controls, CP patients present a significant down-regulation of the Nrf2-mediated oxidative stress response pathway in oPMNs. Twenty-four and six Nrf2 pathway–associated genes were down-regulated or up-regulated, respectively (more than twofold) in oPMNs of CP patients when compared with blood PMNs (Supplemental Table S3). The same genes had similar expression in blood PMNs of CP and healthy subjects, with the exception of SOD1 and AKR1A1 (Supplemental Table S4). SOD1 and CAT, key regulators of superoxide and hydrogen peroxide, were used to verify the gene expression microarray results. RT-qPCR analysis confirmed the reduced expression levels of SOD1 and CAT in CP oPMN (oral/blood ratio: healthy-CAT, 8.70 ± 1.56; SOD1, 3.18 ± 0.90; CP-CAT, 0.15 ± 0.08; SOD1, 0.45 ± 0.22) (Figure 2, A and B). WB analysis of CAT and SOD1 production by oPMN of CP patients revealed significant reduction compared with health (CAT: healthy, 1.88 ± 0.22; CP, 1.09 ± 0.18; SOD1: healthy, 0.8 ± 0.15; CP, 0.41 ± 0.06) (Figure 2, C–F).
Figure 2.
Oral polymorphonuclear neutrophils (oPMNs) of chronic periodontitis (CP) patients produce low levels of catalase (CAT) and superoxide dismutase (SOD1). A: We investigated the PMN antioxidant (AO) response to subgingival biofilms by considering blood PMNs as preexposure and oPMNs as postexposure in the context of PMNs' migratory path from vasculature to the oral cavity through the gingival crevice, where periodontitis-inducing subgingival biofilms reside and induce the inflammatory response. PMN RNA was isolated from oral and blood PMN, as described in Materials and Methods, and quantitative RT-PCR performed for CAT and SOD1. B: Oral versus blood ratios for CAT and SOD1 normalized to GAPDH gene expression were compared in health and disease. C–F: CAT and SOD1 production by oral and blood PMNs was analyzed by Western blot analysis and normalized to β-actin. The oral/blood band intensity ratio was then calculated for each patient. All data were expressed as means ± SEM (C–F). n = 4 (B–F, controls); n = 5 (C–F, patients). ∗P < 0.05 [paired t-test (B) or one-way analysis of variance (C–F)].
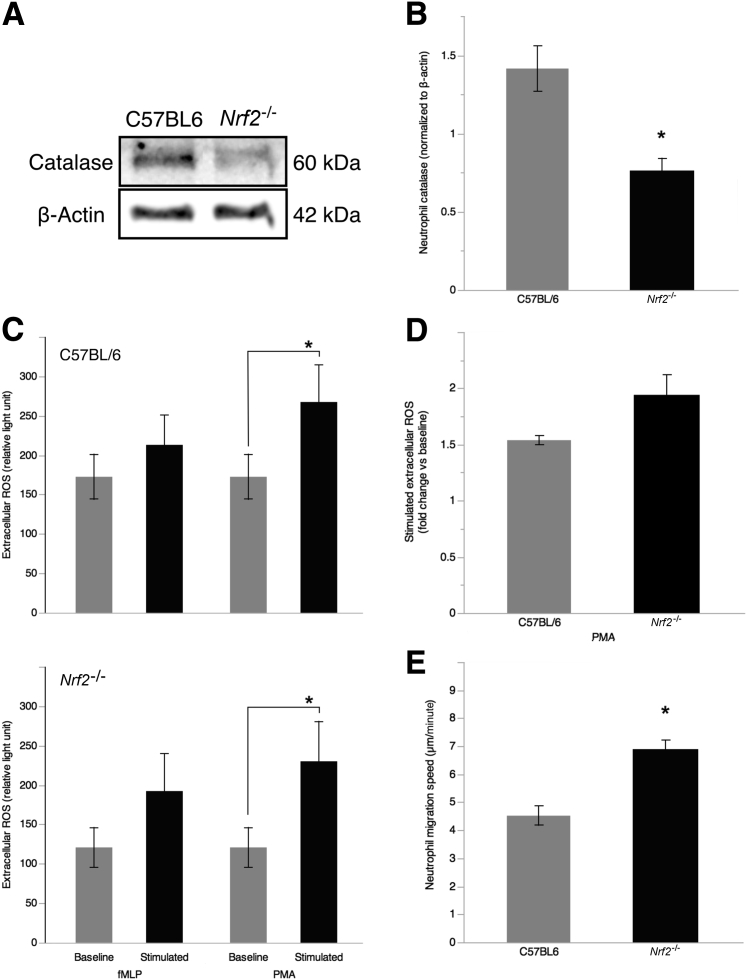
Nrf2−/− PMNs Produce Low CAT Levels
Nrf2−/− mice were used as a model of primary AO deficiency to assess the role of PMN AO deficiency on extracellular ROS production and migration toward chemoattractants. Protein extracts of bone marrow PMN assessed by WB showed lower CAT production by Nrf2−/− compared with WT (C57BL/6, 1.52 ± 0.42; Nrf2−/−, 0.76 ± 0.09 relative to β-actin) (Figure 3, A and B). Both WT and Nrf2−/− produce high ROS levels in response to phorbol myristate acetate (baseline: C57BL/6, 172.5 ± 28.7 RU; Nrf2−/−, 120.75 ± 25.3 RU; stimulated: C57BL/6, 267.3 ± 46.7 RU; Nrf2−/−, 230 ± 50.7). Extracellular ROS levels were measured as peak RUs using an isoluminol-amplified chemiluminescence assay. Extracellular ROS production in response to phorbol myristate acetate stimulation was similar in Nrf2−/− and WT PMNs (C57BL/6, 1.53 ± 0.04; Nrf2−/−, 1.93 ± 0.18) (Figure 3C). Because ROS were found to regulate migration speed and directionality of PMNs during chemotaxis, we investigated the migratory properties of Nrf2−/− PMNs.26 Nrf2−/− had higher migration speed along an formylmethionine–leucyl-phenylalanine gradient compared with WT (C57BL6, 4.52 ± 0.34 μm/second; Nrf2−/−, 6.86 ± 0.34 μm/second), despite similar overall response to the chemoattractant (migrated cells: C57BL/6, 83.32% ± 0.45%; Nrf2−/−, 84.33% ± 4.90%) (Figure 3, D and E).
Figure 3.
Polymorphonuclear neutrophils (PMNs) of nuclear factor erythroid 2-related factor 2 (Nrf2−/−) mice produce low levels of catalase (CAT) and have increased migration speed. A: CAT production by bone marrow PMNs from Nrf2−/− mice was assessed by Western blot analysis and compared with wild-type C57BL/6 mice. B: CAT levels normalized to β-actin were compared between genotypes. C: Extracellular reactive oxygen species (ROS) production by Nrf2−/− PMN in response to fMLP and 10−6 mol/L PMA (stimulation, 30 minutes) was measured by isoluminol-amplified chemiluminescence. D: Chemotaxis was measured in Zigmond chambers along an fMLP gradient (10−6 mol/L). PMN movement was recorded using time-lapse video microscopy. E: Migration speed and efficiency (percentage migrated cells) were measured by playback analysis of 15-minute recordings. All data were expressed as means ± SEM (E). n = 4 mice per group (B and E). ∗P < 0.05 (t-test). fMLP, formylmethionine–leucyl-phenylalanine; PMA, phorbol myristate acetate.
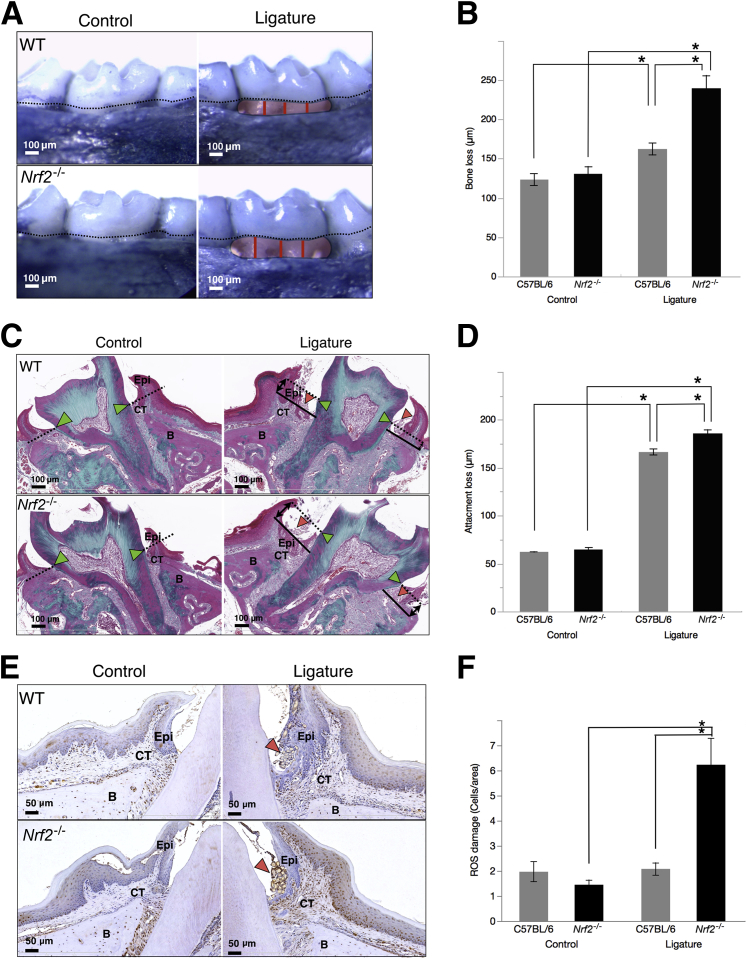
Nrf2−/− Mice Have Increased Periodontal Tissue Breakdown at Sites with Periodontitis
To get further insight into the impact of AO deficiency on periodontal tissue breakdown in periodontitis, ABL at sites with ligature-induced periodontitis was measured by morphometry in Nrf2−/− mice. Statistically significant ABL was observed in Nrf2−/− mice at sites with periodontitis compared with C57BL/6 (WT) mice (239.7 ± 16.4 μm versus 162.4 ± 7.6 μm, respectively). Both groups presented significant ABL at periodontitis sites compared with contralateral healthy molars, and no differences were found between control sites (Figure 4, A and B). Loss of attachment was measured on histological sections of diseased molars stained with Masson's trichrome. Nrf2−/− mice had more epithelial and connective tissue attachment loss compared with WT mice (172.6 ± 8.1 μm versus 166 ± 2 μm, respectively) (Figure 4, C and D). These results demonstrate a more severe tissue breakdown in Nrf2−/− mice, indicating a cytoprotective role of Nrf2 in periodontitis.
Figure 4.
In response to ligature-biofilms, nuclear factor erythroid 2-related factor 2 (Nrf2−/−) mice develop severe bone and attachment loss associated with high oxidative stress. A: Periodontitis was induced in Nrf2−/− mice using a ligature-biofilm model. Representative micrographs of buccal aspects of ligated and contralateral healthy control molars demonstrate the bone loss 21 days after induction of periodontitis. Dotted lines represent the cement-enamel junction. Red lines represent the measurement of alveolar bone loss along the exposed roots. B: Alveolar bone loss was measured by morphometry from the cement-enamel junction (dotted lines in A) to the bone crest along the exposed roots (red lines in A) on defleshed methynene blue–stained maxillae. C and D: Loss of connective tissue attachment was measured on Masson trichrome–stained sections from the cement-enamel junction as reference for baseline base of gingival sulcus (green arrowheads, dotted lines) to the base of the periodontal pocket (solid line) on buccal and palatal surfaces of ligated molars. Red arrowheads indicate the ligature. E: Reactive oxygen species damage in periodontitis lesions surrounding the ligature (red arrowheads) was assessed by immunohistochemistry for 8-hydroxydeoxyguanosine (8-OHdG), a specific marker for oxidative damage to DNA. F: Oxidative damage was quantified in epithelium (Epi), connective tissue (CT), and at bone level (B) by counting 8-OHdG+ cells per surface area. All data were expressed as means ± SEM (F). n = 12 (B, WT); n = 11 (B, Nrf2−/−); n = 3 mice per group (C, D, and F). ∗P < 0.05 (one-way analysis of variance). WT, wild type.
Nrf2−/− Mice Have High 8-OHdG Levels at Sites with Periodontitis
To assess ROS damage at sites with periodontitis, we quantified 8-OHdG–positive cells and found increased numbers around ligated molars of Nrf2−/− mice compared with healthy molars and with WT control mice (control: C57BL/6, 1.97 ± 0.39; Nrf2−/−, 1.49 ± 0.19; ligature: C57BL/6, 2.08 ± 0.25; Nrf2−/−, 6.23 ± 1.05 cells/square pixels) (Figure 4, E and F). Positive cells identified at the control sites are considered as physiological background level of DNA oxidation.27 Analysis of 8-OHdG–positive cell levels in different regions of interest in the soft tissues and along the alveolar bone surface measured cells/area found significant ROS-mediated damage in the epithelium and connective tissue compared with bone.
Discussion
The current study demonstrates, for the first time, that the Nrf2 pathway is down-regulated in oPMNs of patients with severe CP compared with periodontally healthy controls. Twenty-four upstream and downstream Nrf2 pathway factors were down-regulated twofold to fivefold in oral compared with blood PMNs of patients with CP who also recruited oPMNs in higher numbers. In addition, we showed that oxidative damage as a result of Nrf2 knockout results in more severe loss of periodontal tissues in an animal model of periodontitis. The Nrf2 pathway controls the expression of a wide array of genes essential for production of cytoprotective factors, its activation leading to cell survival and tissue protection during oxidative stress conditions.11, 12 It has also been shown that osteoclastic bone resorption is substantially promoted in Nrf2-deficient osteoclast precursor cells compared with WT cells.28 This must be kept in mind when interpreting our results because the lack of AO also affects the osteoclasts in addition to the PMNs. The role of CAT in maintaining periodontal health was previously demonstrated in acatalasemia patients who are systemically healthy, but have an increased incidence of CP.29 Similarly, SOD1 levels were decreased in whole saliva and serum samples from CP patients.30, 31, 32 Therefore, our results indicate that PMN Nrf2 down-regulation in the context of increased periodontal recruitment aggravates tissue destruction. ROS neutralization may play a role in preventing a continuous PMN influx to allow for resolution of inflammation to proceed because PMN chemotaxis is a ROS-dependent process.33, 34
Nrf2 down-regulation in oPMNs of CP patients as cause for reduced PMN ability to neutralize ROS and withstand oxidative stress supports previous reports of increased oxidative damage in CP.4, 10 Interestingly, ingenuity pathway analysis predicted a more significant Nrf2 pathway down-regulation in blood PMNs of patients with CP compared with healthy controls. This seems to suggest that constitutive Nrf2 expression may be required for adequate PMN responses to subgingival biofilms, which is associated with periodontal health. The difference in AO production we observed may also be a reflection of age and sex differences between CP patients and healthy controls in our study. Several studies demonstrated a significant increase in intracellular ROS production by peripheral blood PMNs in CP patients within a similar age range compared with sex- and age-matched healthy controls.21, 35 However, the investigation of peripheral blood PMNs may not provide a full understanding of the mechanisms underlying PMNs' role in CP pathogenesis.
Recently, our group demonstrated specific gene expression in oPMNs obtained from healthy subjects, when compared with peripheral blood PMNs.15 We further found that in a subset of patients with CP, who are refractory to conventional therapy, oPMNs are hyperactive and produce high ROS levels associated with more severe loss of periodontal tissues.6 One study showed increased stimulated levels of ROS in GCF of CP patients compared with healthy controls but no difference in superoxide generation by blood PMNs or in AO capacity of cell-free GCF in disease versus health.36 Malodialdehyde (MDA), a thiobarbituric acid reactive substance used as a marker of lipid peroxidation to assess oxidative damage, was found increased in GCF and saliva of patients with CP.10, 37, 38 Tsai et al37 reported 200- to 400-fold higher MDA levels in GCF compared with saliva but similar salivary AO capacity. However, salivary AO capacity may have a lower prediction value for CP activity than oPMN AO capacity, considering that most oPMNs reach the oral cavity through the gingival crevices.39, 40, 41 To our knowledge, the current study is the first to report an association between reduced oPMN AO production and severe CP, and opens the question of whether blood PMNs in patients with CP are primed to a low response to specific subgingival bacteria. Whether low AO production by oPMNs in severe CP is because of systemic priming or because of local inflammatory milieu remains unknown.
The microarray data found that blood PMNs of CP patients have a more pronounced Nrf2 pathway down-regulation, indicating systemic priming. One study reported high 8-OHdG levels in whole saliva and GCF of patients with CP in a Japanese population, but systemic levels of 8-OHdG were not measured.42 However, GCF levels of 8-OHdG were suggested as a biomarker for DNA damage caused by oxidative stress and an indicator for CP severity.43 Konopka et al44 showed that 8-OHdG levels in gingival blood of patients with aggressive periodontitis were higher than those with CP, suggesting a correlation between ROS damage and disease severity. D'Aiuto et al45 have shown that severe CP patients have higher Diacron-reactive oxygen metabolites in serum and reduced total serum AO capacity compared with healthy controls. In search for salivary biomarkers for oxidative stress and alveolar bone loss in CP, a recent study has found increased salivary 8-OHdG and MDA levels, and reduced salivary total antioxidant capacity in CP patients. A correlation between salivary MDA levels and C-terminal telopeptide of type I collagen, a biomarker for bone loss, was also reported.46 These findings point to a possible systemic low AO expression in CP, but the relative contribution of oPMNs to low oral AO capacity may be critical for disease progression.
In summary, our present study indicates that Nrf2 down-regulation in blood and oPMNs of CP patients may predispose to severe periodontal tissue breakdown, likely through increased local oxidative damage. Furthermore, PMNs of CP patients may be primed for low AO response in the context of high recruitment in the oral cavity.
Acknowledgments
We thank Susan Carter for assistance with mouse surgical procedures and Kevin Seymour for providing the dermestid beetle colony for skull defleshing at the Royal Ontario Museum.
Footnotes
Supported by Canadian Institute of Health Research grant TBO-122068 (M.G.) and National Institute of Dental and Craniofacial Research Award K99DE024575 (C.S.).
C.S. and G.M.A. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.01.013.
Supplemental Data
References
- 1.Eke P.I., Dye B.A., Wei L., Thornton-Evans G.O., Genco R.J. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 2.Sima C., Glogauer M. Neutrophil dysfunction and host susceptibility to periodontal inflammation: current state of knowledge. Curr Oral Health Rep. 2014;1:95–103. [Google Scholar]
- 3.Bullon P., Morillo J.M., Ramirez-Tortosa M.C., Quiles J.L., Newman H.N., Battino M. Metabolic syndrome and periodontitis: is oxidative stress a common link? J Dent Res. 2009;88:503–518. doi: 10.1177/0022034509337479. [DOI] [PubMed] [Google Scholar]
- 4.Chapple I.L., Matthews J.B. The role of reactive oxygen and antioxidant species in periodontal tissue destruction. Periodontol 2000. 2007;43:160–232. doi: 10.1111/j.1600-0757.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 5.Battino M., Bullon P., Wilson M., Newman H. Oxidative injury and inflammatory periodontal diseases: the challenge of anti-oxidants to free radicals and reactive oxygen species. Crit Rev Oral Biol Med. 1999;10:458–476. doi: 10.1177/10454411990100040301. [DOI] [PubMed] [Google Scholar]
- 6.Aboodi G.M., Goldberg M.B., Glogauer M. Refractory periodontitis population characterized by a hyperactive oral neutrophil phenotype. J Periodontol. 2011;82:726–733. doi: 10.1902/jop.2010.100508. [DOI] [PubMed] [Google Scholar]
- 7.Chapple I.L., Brock G.R., Milward M.R., Ling N., Matthews J.B. Compromised GCF total antioxidant capacity in periodontitis: cause or effect? J Clin Periodontol. 2007;34:103–110. doi: 10.1111/j.1600-051X.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 8.Brock G.R., Butterworth C.J., Matthews J.B., Chapple I.L. Local and systemic total antioxidant capacity in periodontitis and health. J Clin Periodontol. 2004;31:515–521. doi: 10.1111/j.1600-051X.2004.00509.x. [DOI] [PubMed] [Google Scholar]
- 9.Muniz F.W., Nogueira S.B., Mendes F.L., Rösing C.K., Moreira M.M., de Andrade G.M., Carvalho Rde S. The impact of antioxidant agents complimentary to periodontal therapy on oxidative stress and periodontal outcomes: a systematic review. Arch Oral Biol. 2015;60:1203–1214. doi: 10.1016/j.archoralbio.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Levy Y. Oxidative stress, antioxidants and periodontal disease. Arch Oral Biol. 2015;60:1461–1462. doi: 10.1016/j.archoralbio.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Osburn W.O., Kensler T.W. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niture S.K., Khatri R., Jaiswal A.K. Regulation of Nrf2: an update. Free Radic Biol Med. 2014;66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J., Cha Y.N., Surh Y.J. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat Res. 2010;690:12–23. doi: 10.1016/j.mrfmmm.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Armitage G.C. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Lakschevitz F.S., Aboodi G.M., Glogauer M. Oral neutrophils display a site-specific phenotype characterized by expression of T-cell receptors. J Periodontol. 2013;84:1493–1503. doi: 10.1902/jop.2012.120477. [DOI] [PubMed] [Google Scholar]
- 16.Bender J.S., Thang H., Glogauer M. Novel rinse assay for the quantification of oral neutrophils and the monitoring of chronic periodontal disease. J Periodont Res. 2006;41:214–220. doi: 10.1111/j.1600-0765.2005.00861.x. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R., Barrett T. NCBI GEO standards and services for microarray data. Nat Biotechnol. 2006;24:1471–1472. doi: 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakschevitz F.S., Aboodi G.M., Glogauer M. Oral neutrophil transcriptome changes result in a pro-survival phenotype in periodontal diseases. PLoS One. 2013;8:e68983. doi: 10.1371/journal.pone.0068983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan K., Kan Y.W. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakschevitz F.S., Visser M.B., Sun C., Glogauer M. Neutrophil transcriptional profile changes during transit from bone marrow to sites of inflammation. Cell Mol Immunol. 2015;12:53–65. doi: 10.1038/cmi.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews J.B., Wright H.J., Roberts A., Cooper P.R., Chapple I.L.C. Hyperactivity and reactivity of peripheral blood neutrophils in chronic periodontitis. Clin Exp Immunol. 2007;147:255–264. doi: 10.1111/j.1365-2249.2006.03276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahlgren C., Karlsson A., Bylund J. Measurement of respiratory burst products generated by professional phagocytes. Methods Mol Biol. 2007;412:349–363. doi: 10.1007/978-1-59745-467-4_23. [DOI] [PubMed] [Google Scholar]
- 24.Sima C., Gastfreund S., Sun C., Glogauer M. Rac-null leukocytes are associated with increased inflammation-mediated alveolar bone loss. Am J Pathol. 2014;184:472–482. doi: 10.1016/j.ajpath.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Grishagin I.V. Automatic cell counting with ImageJ. Anal Biochem. 2015;473:63–65. doi: 10.1016/j.ab.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Hattori H., Subramanian K.K., Sakai J., Jia Y., Li Y., Porter T.F., Loison F., Sarraj B., Kasorn A., Jo H., Blanchard C., Zirkle D., McDonald D., Pai S.Y., Serhan C.N., Luo H.R. Small-molecule screen identifies reactive oxygen species as key regulators of neutrophil chemotaxis. Proc Natl Acad Sci U S A. 2010;107:3546–3551. doi: 10.1073/pnas.0914351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins A.R., Cadet J., Mőller L., Poulsen H.E., Viña J. Are we sure we know how to measure 8-oxo-7,8-dihydroguanine in DNA from human cells? Arch Biochem Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Hyeon S., Lee H., Yang Y., Jeong W. Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radic Biol Med. 2013;65:789–799. doi: 10.1016/j.freeradbiomed.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Eaton J.W., Ma M. ed 7. McGraw-Hill; New York: 1995. Acatalasemia. The Metabolic and Molecular Bases of Inherited Disease; pp. 2371–2378. [Google Scholar]
- 30.Canakci C.F., Cicek Y., Yildirim A., Sezer U., Canakci V. Increased levels of 8-hydroxydeoxyguanosine and malondialdehyde and its relationship with antioxidant enzymes in saliva of periodontitis patients. Eur J Dent. 2009;3:100–106. [PMC free article] [PubMed] [Google Scholar]
- 31.Novakovic N., Todorovic T., Rakic M., Milinkovic I., Dozic I., Jankovic S., Aleksic Z., Cakic S. Salivary antioxidants as periodontal biomarkers in evaluation of tissue status and treatment outcome. J Periodont Res. 2014;49:129–136. doi: 10.1111/jre.12088. [DOI] [PubMed] [Google Scholar]
- 32.Singh N., Chander Narula S., Kumar Sharma R., Tewari S., Kumar Sehgal P. Vitamin E supplementation, superoxide dismutase status, and outcome of scaling and root planing in patients with chronic periodontitis: a randomized clinical trial. J Periodontol. 2014;85:242–249. doi: 10.1902/jop.2013.120727. [DOI] [PubMed] [Google Scholar]
- 33.Loboda A., Stachurska A., Florczyk U., Rudnicka D., Jazwa A., Wegrzyn J., Kozakowska M., Stalinska K., Poellinger L., Levonen A.L., Yla-Herttuala S., Jozkowicz A., Dulak J. HIF-1 induction attenuates Nrf2-dependent IL-8 expression in human endothelial cells. Antioxid Redox Signal. 2009;11:1501–1517. doi: 10.1089/ars.2008.2211. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Chen X., Song H., Chen H.Z., Rovin B.H. Activation of the Nrf2/antioxidant response pathway increases IL-8 expression. Eur J Immunol. 2005;35:3258–3267. doi: 10.1002/eji.200526116. [DOI] [PubMed] [Google Scholar]
- 35.Gustafsson A., Ito H., Åsman B., Bergström K. Hyper-reactive mononuclear cells and neutrophils in chronic periodontitis. J Clin Periodontol. 2006;33:126–129. doi: 10.1111/j.1600-051X.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- 36.Guarnieri C., Zucchelli G., Bernardi F., Scheda M., Valentini A.F., Calandriello M. Enhanced superoxide production with on change of the antioxidant activity in gingival fluid of patients with chronic adult periodontitis. Free Radic Res. 1991;15:11–16. doi: 10.3109/10715769109049120. [DOI] [PubMed] [Google Scholar]
- 37.Tsai C.C., Chen H.S., Chen S.L., Ho Y.P., Ho K.Y., Wu Y.M., Hung C.C. Lipid peroxidation: a possible role in the induction and progression of chronic periodontitis. J Periodont Res. 2005;40:378–384. doi: 10.1111/j.1600-0765.2005.00818.x. [DOI] [PubMed] [Google Scholar]
- 38.Akalιn F.A., Baltacιoğlu E., Alver A., Karabulut E. Lipid peroxidation levels and total oxidant status in serum, saliva and gingival crevicular fluid in patients with chronic periodontitis. J Clin Periodontol. 2007;34:558–565. doi: 10.1111/j.1600-051X.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 39.Young I. Measurement of total antioxidant capacity. J Clin Pathol. 2001;54:339. doi: 10.1136/jcp.54.5.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raeste A.M., Tapanila T. Leukocyte migration into the healthy dentuious mouth: a study in children, adolescents and adults. J Periodont Res. 1977;12:444–449. doi: 10.1111/j.1600-0765.1977.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 41.Hase M.P., Reade P.C. The oral leukocyte migration rate index as a method of assessing periodontal disease in an individual. J Periodont Res. 1979;14:153–159. doi: 10.1111/j.1600-0765.1979.tb00785.x. [DOI] [PubMed] [Google Scholar]
- 42.Takane M., Sugano N., Ezawa T., Uchiyama T., Ito K. A marker of oxidative stress in saliva: association with periodontally-involved teeth of a hopeless prognosis. J Oral Sci. 2005;47:53–57. doi: 10.2334/josnusd.47.53. [DOI] [PubMed] [Google Scholar]
- 43.Dede F.Ö., Özden F.O., Avcı B. 8-Hydroxy-deoxyguanosine levels in gingival crevicular fluid and saliva in patients with chronic periodontitis after initial periodontal treatment. J Periodontol. 2013;84:821–828. doi: 10.1902/jop.2012.120195. [DOI] [PubMed] [Google Scholar]
- 44.Konopka T., Król K., Kopeć W., Gerber H. Total antioxidant status and 8-hydroxy-2′-deoxyguanosine levels in gingival and peripheral blood of periodontitis patients. Arch Immunol Ther Exp. 2007;55:417–425. doi: 10.1007/s00005-007-0047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Aiuto F., Nibali L., Parkar M., Patel K., Suvan J., Donos N. Oxidative stress, systemic inflammation, and severe periodontitis. J Dent Res. 2010;89:1241–1246. doi: 10.1177/0022034510375830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miricescu D., Totan A., Calenic B., Mocanu B., Didilescu A., Mohora M., Spinu T., Greabu M. Salivary biomarkers: relationship between oxidative stress and alveolar bone loss in chronic periodontitis. Acta Odontol Scand. 2014;72:42–47. doi: 10.3109/00016357.2013.795659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.