Abstract
Commercially available, highly passaged pancreatic cancer (PC) cell lines are of limited translational value. Attempts to overcome this limitation have primarily consisted of cancer cell isolation and culture directly from human PC specimens. However, these techniques are associated with exceedingly low success rates. Here, we demonstrate a highly reproducible culture of primary PC cell lines (PPCLs) from patient-derived xenografts, which preserve, in part, the intratumoral heterogeneity known to exist in PC. PPCL expansion from patient-derived xenografts was successful in 100% of attempts (5 of 5). Phenotypic analysis was evaluated with flow cytometry, immunofluorescence microscopy, and short tandem repeat profiling. Importantly, tumorigenicity of PPCLs expanded from patient-derived xenografts was assessed by subcutaneous injection into nonobese diabeteic.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice. Morphologically, subcutaneous injection of all PPCLs into mice yielded tumors with similar characteristics to the parent xenograft. PPCLs uniformly expressed class I human leukocyte antigen, epithelial cell adhesion molecule, and cytokeratin-19. Heterogeneity within each PPCL persisted in culture for the frequency of cells expressing the cancer stem cell markers CD44, CD133, and c-Met and the immunologic markers human leukocyte antigen class II and programmed death ligand 1. This work therefore presents a reliable method for the rapid expansion of primary human PC cells and, thereby, provides a platform for translational investigation and, importantly, potential personalized therapeutic approaches.
Pancreatic cancer (PC) is projected to be the second leading cause of cancer deaths by 2030.1 Systemic cytotoxic and kinase-targeted regimens represent the standard of care for most patients presenting with PC. Most tumors, however, will develop rapid resistance to these regimens and will continue to progress by unknown mechanisms.2, 3 As a result, both the median survival and annual death rate for patients with PC have remained unchanged over the past 20 years.4 To this end, an analysis of phase 1 cancer trials, which used agents with demonstrated efficacy in models derived from established commercially available cancer cell lines, revealed an overall objective response rate in only 3.8% of patients. For the past 40 years, commercially available PC cells (PCCs) such as MIA-PaCa2 (established in 1975),5 CFPAC-1 (liver metastases established from a cystic fibrosis patient in 1990),6 PANC-1 (established in 1975),7 and BxPC-3 (K-ras wild-type established in 1986)8 have been widely used in PC models and derived from an extremely small set of PC patients from >30 years ago. Thus, the poor predictive value of studies with the use of established cancer cell lines is a major barrier to the development of new interventions.9, 10 Our understanding of PC responses to therapy is also complicated by the marked molecular heterogeneity that exists among primary tumors from different patients and intrapatient tumor cell heterogeneity, neither of which can be captured with currently available commercial cell lines.11
To begin to address some of these barriers, groups have attempted direct isolation and culture of patient-derived primary human PCCs from viable surgical specimens. The success rate of this method was reported to be approximately 10% in experienced hands,12 although most groups do not publish failed attempts.13, 14, 15, 16 Conversely, in other cancer types, cell line derivation from patient-derived xenotransplantation demonstrates up to 10 times the success rate compared with that from cancer specimens at the time of resection.17, 18, 19, 20 Thus, to address these limitations, we leveraged our recently developed PC–patient-derived xenograft (PDX) model, whereby we have demonstrated that the early PDX is morphologically similar to the original cancer and retains both interpatient and intrapatient heterogeneity of the human disease.17, 21
Specifically, here, we demonstrate the expansion of human PCCs through a PDX model that preserves the tumor heterogeneity with a 100% success rate. These established PDX-derived primary cell lines display uniform markers associated with a human PC origin with retained tumorigenicity. In addition, after several passages, these PCCs continue to exhibit significant heterogeneity in the expression of markers associated with their initial molecular phenotype and PC-immune cell interactions. In summary, this method of primary PCC isolation may provide a critical model that enhances clinical relevance by augmenting preclinical investigations and affording a personalized therapeutic examination.
Materials and Methods
Ethical Statement
Informed written consent was obtained from all patients, and the collection of all patient material was approved by the University of Florida Institutional Review Board. All animal studies were performed in accordance with the guidelines of the University of Florida Institutional Animal Care and Use Committee.
Histologic Analysis
Human PC specimens and all xenografts were evaluated by expert gastrointestinal pathologists, in accordance with the World Health Organization Classification of Tumors of the Digestive System. Tumors were classified according to site of origin and tumor stage.
Tumor Transplantation
The implantation of surgical tumor tissue into immunocompromised mice was described previously.21 Briefly, a viable portion of resected tissue 2 × 2 mm in size was isolated immediately from surgically resected primary PC specimens with care to minimize critical ischemia time. PC tissue was then implanted subcutaneously into an 8-week-old female nonobese diabetic.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mouse (The Jackson Laboratory, Bar Harbor, ME). Xenografts were allowed to grow to a maximum diameter of 1.5 cm before passage and/or in vitro culture.
Isolation and Propagation of Patient-Derived PCC Populations
Tissue collected from xenograft tumors was minced into small pieces and enzymatically dissociated into single cells with 2 mg/mL STEMxyme 1 Collagenase/Neutral Protease solution (Worthington Biochemical Corporation, Lakewood, NJ) for 30 minutes at 37°C. After washing with Hank's balanced salt solution that contained 2% of fetal bovine serum, cells were filtered through a 100-μm filter to remove nondissociated cell clumps. The single cell suspension was then cultured in advanced Dulbecco's Modified Eagle Medium with nutrient mixture F12, 10% fetal bovine serum, 6 mmol/L glutamine, 1% of penicillin/streptomycin, and 40 ng/mL dexamethasone, at a density of 105 cells/mL on culture plates coated with rat tail collagen I. Media were changed every second or third day, and cells were passaged with Accutase (Innovative Cell Technologies, San Diego, CA) at 80% confluence. To avoid the outgrowth of contaminating fibroblasts, differential trypsinization was performed until a homogenous population of cytokeratin (CK)19/class I human leukocyte antigen (HLA)-expressing human PCCs was achieved by flow cytometry analysis.
Isolation of Nucleic Acid, DNA Fingerprinting, and Somatic Mutation Detection
DNA fingerprinting assays were performed to establish a unique genetic identification for each patient's PCC population. Total genomic DNA was extracted from cell pellets with Qiagen DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) according to the manufacturer's protocol, and sent to the Sanger Sequencing Core at the University of Florida for cell line authentication. The identity of the DNA profiles was determined by short tandem repeat profiling with the use of GenePrint 10 System (Promega, Madison, WI). This kit amplifies nine short tandem repeat human loci (TPOX, vWA, D21S11, TH01, CSF1PO, D16S539, D7S820, D13S317, and D5S818) and AMEL for sex identification, which was cross-referenced against patient sex. The samples were processed on an ABI 3130XL Genome Analyzer (Applied Biosystems, Foster City, CA), and data were analyzed with GeneMapper software version 4.0 (Applied Biosystems). Fingerprints were then compared with an established database of DNA profiles in the ATCC (Manassas, VA) to verify uniqueness.
Isolated DNA was also profiled for 38 somatic mutations within the following genes associated with PC: APC, BRAF, CDKN2A, CTNNB1, KRAS, NRAS, PIK3CA, SMAD4, and TP53 with the use of the Pancreatic Cancer qBiomarker Somatic Mutation PCR Array (Qiagen). Data were collected on the Applied Biosystems 7900HT instrument, and analyzed with the use of the Sequence Detection Systems software version 2.4.1 (Applied Biosystems). Samples were evaluated for the presence or absence of a specific somatic mutation and were normalized to gene copy reference assays for each gene present in the array.
In Vivo Tumorigenicity
To establish tumorigenicity of cultured primary PCCs, 3 × 106 cells from each patient's cell population was suspended in 200 μL of a 1:1 mixture of Dulbecco's Modified Eagle Medium with nutrient mixture F12 and Matrigel Matrix (Corning, Corning, NY) and inoculated subcutaneously into the right flank of NSG mice. Mice were inspected twice a week, and tumor size was measured with a digital caliper. Mice were euthanized 40 days after the implantation. Tumors were fixed in 10% formalin and subjected to histologic analysis with hematoxylin and eosin staining. The growth rate (%/day) was used to evaluate the growth kinetics of primary PCC.
In Vitro Cell Growth Kinetics
Short-term proliferation assays were performed to determine growth rates in culture. Cells were seeded in 24-well plates at 1 × 105 cells/well on day 0. Cell number was determined with the TC20 Automated Cell Counter (Bio-Rad, Hercules, CA) on days 1 through 6. The growth curve was fitted to Gompertz growth model with GraphPad Prism software version 6 (GraphPad Software, La Jolla, CA). Doubling time was determined from the exponential phase with the use of the equation ln(2)/k, in which k is the rate constant. Assays were performed in triplicate and repeated at least three times.
Immunohistochemistry and Immunocytochemistry
Tissue staining was performed by the University of Florida Molecular Pathology Core. Briefly, formalin fixed, paraffin-embedded patient tumors and PDX specimens were deparaffinized and cut into 5-μm sections. Sections were stained with hematoxylin and eosin. Additional 5-μm sections of PDX specimens were probed with anti–programmed death ligand 1 (PDL1; Abcam, Cambridge, MA) according to the manufacturer's recommended protocol.
Primary PCCs were cultured on collagen-coated 24-well plates and fixed in 4% paraformaldehyde for 15 minutes at room temperature. After fixation, cells were permeabilized and blocked in 3% bovine serum albumin and 0.1% Triton X-100 for 1 hour at room temperature. The cells were then incubated in 1% bovine serum albumin and 0.1% Triton X-100 overnight at 4°C with the following antibodies: Alexa Fluor 647 (AF647) anti-human CK18, AF647 anti-human epithelial cell adhesion molecule (EpCam), AF647 anti-human CK19 (BioLegend, San Diego, CA), and rabbit anti-human CD45 (Cell Signaling Technologies, Danvers, MA) followed by Alexa Fluor 568 goat anti-rabbit secondary for 2 hours at room temperature. The DNA dye DAPI was used to counterstain nuclei. Images were collected on an EVOS FL digital inverted microscope (Life Technologies, Carlsbad, CA) and processed with ImageJ software version 1.48V (NIH, Bethesda, MD; https://imagej.nih.gov/ij/download.html).
Flow Cytometric Analysis
Cells were dissociated from 24-well culture dishes with the use of Accutase, washed in Dulbecco's phosphate-buffered saline that contained 5% fetal bovine serum, 5 mmol/L EDTA, and 0.1% sodium azide (Sigma-Aldrich, St. Louis, MO), and probed for CD45, EpCam, CD44, CD133, HLA-ABC, HLA-DR/DP/DQ, CD80, CD86, and PDL1 with the use of the following antibodies: Pacific Blue-conjugated CD45, Alexa Fluor 488 (AF488)-conjugated CD44, phycoerythrin (PE)-conjugated CD133 (Miltenyi Biotec, Bergisch Gladbach, Germany), PE-cyanin 7–conjugated EpCam, allophycocyanin-cyanin 7–conjugated HLA-ABC, PE-conjugated CD86, AF647-conjugated HLA-DR/DP/DQ, PE-cyanin 7–conjugated PDL1, and allophycocyanin-H7–conjugated CD80 (BD Biosciences, Franklin Lakes, NJ). All antibodies were purchased from BioLegend unless otherwise indicated and used at a 1:100 dilution. Manufacturer's recommended isotype controls were used as negative controls for all antibodies used. A total of 10,000 events per sample were acquired with a BD LSR II (BD Biosciences), and data were analyzed with FlowJo data analysis software version 9 (FlowJo LLC, Ashland, OR).
Gemcitabine Treatment and Cell Cytotoxicity Analysis
Cell were seeded in 96-well plate at a density of 5 × 103 cells/well in a total of 100 μL of culture medium and treated with gemcitabine (Selleck Chemicals, Houston TX) at multiple concentrations of 3, 10, 30, 100, and 300 nmol/L and 1 μmol/L. After 6ix days, viability was analyzed with the Cell Counting Kit-8 (Dojindo Molecular Technologies Inc., Rockville, MD), according to the manufacturer's protocol. The absorbance of yellow formazan dye, a derivative of water-soluble [2-(2-methox y-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium] monosodium salt from dehydrogenase activity in cells was detected with colorimetric microplate reader at 450 nm. This assay was performed in triplicate and was repeated at least three times.
Statistical Analysis
Statistical analyses were performed with GraphPad Prism software version 6 (GraphPad Software). Data are presented as means ± SEM. P values were calculated with the Student's t-test with two-tailed distribution. P < 0.05 was considered significant.
Results
Establishment of PPCLs from PDXs
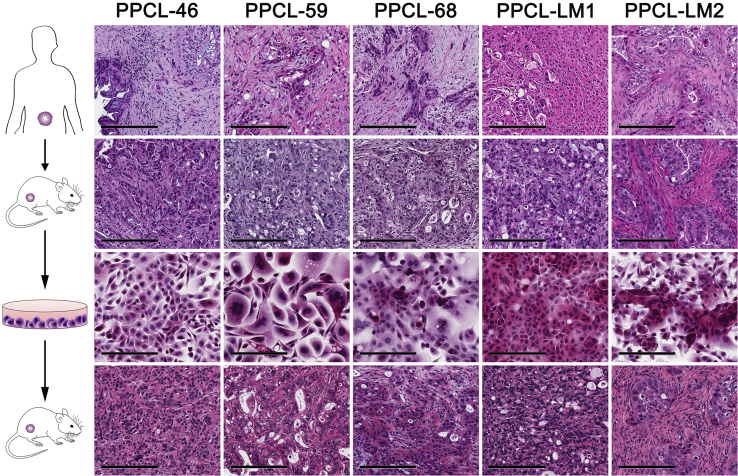
Primary pancreatic cancer cell lines (PPCLs) were isolated from PDX tumors which are enriched in patient-derived PCCs, contrary to previous attempts at direct isolation from resected PC specimens which are limited by a preponderance of myofibroblastic elements and relatively few cancer cells by volume.22 In addition, hepatic metastases from PC primaries were expanded with the PDX model and similarly isolated in culture. The isolation and expansion technique had a 100% success rate (5 of 5 lines) (Figure 1). Early depletion of murine tumor-associated stroma through differential trypsinization was critical to allow for the isolation of low-passage PCCs without the need for clonal expansion methods. All PPCLs were passaged through at least 10 generations without any sign of growth decline or morphologic change.
Figure 1.
Hematoxylin and eosin-stained micrographs are displayed as follows for (from top to bottom): human pancreatic cancer specimens, PDX tumors, PPCLs, and PPCL-derived subcutaneous xenografts. Scale bars: 200 μm. PDX, patient-derived xenograft; PPCL, primary pancreatic cancer cell line.
The patient demographic and clinicopathologic data associated with each patient from whom the PDXs and PPCLs were derived are displayed in Table 1. PPCLs presented in this study were derived from PDX established from both primary tumors (PPCL-46, PPCL-59, and PPCL-68) and hepatic metastases (PPCL-LM1, PPCL-LM2). All PPCLs demonstrated mutational profiles consistent with PC, and all identified K-Ras and TP53 mutations were conserved between primary tumor, PDX, and PPCL (Table 1), short tandem repeat profiling with 10 loci and cross-referenced against a known ATCC database, demonstrated that all PPCLs generated were unique and sex matched to the original patient. DNA fingerprinting results are displayed in Table 2.
Table 1.
Clinicopathologic Characteristics and Mutation Profiles of Five PPCLs Established from Patient-Derived Xenografts
| ID | Age, years | Sex | Tumor site | Positive lymph nodes/total | Overall survival, mo | K-Ras | TP53 |
|---|---|---|---|---|---|---|---|
| PPCL-46 | 75 | Female | Primary | 7/34 | 10 | G12V | WT |
| PPCL-59 | 73 | Female | Primary | 4/15 | Alive at 4 mo | G12V | WT |
| PPCL-68 | 64 | Female | Primary | 2/18 | Alive at 3 mo | G12D | R248W |
| PPCL-LM1 | 65 | Male | Metastasis | NA | 8 | G12V | WT |
| PPCL-LM2 | 63 | Male | Metastasis | NA | 2 | G12V | WT |
mo, month; NA, not applicable; PPCL, primary pancreatic cancer cell line; WT, wild type.
Table 2.
Short Tandem Repeat Profiling of PPCLs
| Locus | PPCL-46 | PPCL-59 | PPCL-68 | PPCL-LM1 | PPCL-LM2 |
|---|---|---|---|---|---|
| D5S818 | 12 | 11 | 9, 12 | 9, 11 | 11, 12 |
| D13S317 | 9, 14 | 9, 12 | 8, 10 | 11, 13 | 11 |
| D7S820 | 9, 11 | 9 | 11 | 10 | 8 |
| D16S539 | 11, 13 | 9, 13 | 13 | 10, 13 | 11, 12 |
| vWA | 20 | 14, 18 | 15, 17 | 15, 16 | 15, 17 |
| TH01 | 8, 9 | 9.3 | 6 | 6, 9.3 | 6 |
| TPOX | 10, 11 | 9, 10 | 8, 10 | 11 | 9 |
| CSF1PO | 12, 13 | 12 | 10 | 10, 11 | 10, 11 |
| D21S11 | 29 | 29 | 28 | 29 | 28, 32.2 |
| AMEL | X | X | X | X, Y | X, Y |
PPCL, primary pancreatic cancer cell line.
Purity of PPCLs
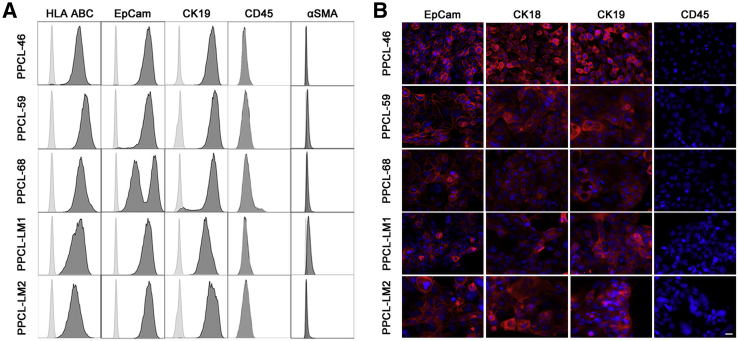
Because of the nature of the technique, contamination by immune cells and fibroblasts from both human and murine sources was evaluated. Flow cytometric analysis was used to examine the expression of HLA class I, the immune cell marker CD45, and the myofibroblast marker α-smooth muscle actin (α-SMA). All PPCLs expressed HLA class I, whereby all cultures were negative for both the leukocyte antigen CD45 and the mesenchymal antigen α-SMA, indicating the presence of human cells in the absence of immune and fibroblastic cells, respectively (Figure 2). PPCLs were further assessed for the expression of the epithelial cell markers EpCam and CK19, whereby flow cytometric analysis and immunofluorescence microscopy demonstrated that all cultures were universally positive (Figure 2). Together these data demonstrate a culture that consisted of human epithelial cells free from immune and fibroblast contamination.
Figure 2.
PPCL population express markers consistent with pure human PCCs. A: PPCLs were examined for staining of HLA-ABC, EpCam, CK19, and CD45 and α-SMA. Cells were gated according to forward/side scatter profiles, and isotype control histograms are shown in gray. B: PPCLs were analyzed for EpCam, CK18, CK19, and CD45 expression with the use of immunocytochemistry. Each indicated marker is displayed in red. The nuclear stain DAPI is shown in blue. Scale bar, 20 μm. CK19, cytokeratin-19; EpCam, epithelial cell adhesion molecule; HLA, human leukocyte antigen; PPCL, primary pancreatic cancer cell line; α-SMA, α-smooth muscle actin.
In Vivo and in Vitro Morphology and Growth Kinetics
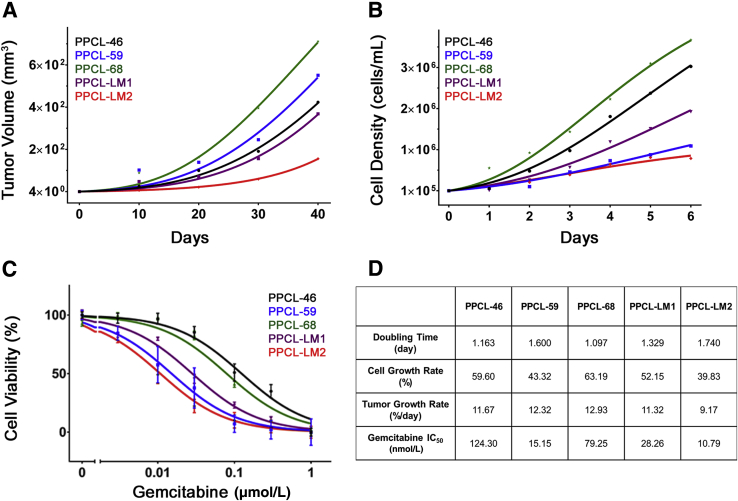
To confirm tumorigenicity of PPCL, the ability of these cultures to form tumors in NSG mice was determined. Indeed, all PPCs were able to grow in vivo, albeit at different rates (Figure 3A). Histologic analysis displayed similar architectural structure to the parental PDX and original tumors (Figure 1). All PPCLs isolated from PDX tumors formed adherent monolayers in culture and also demonstrated variable exponential growth in vitro (Figure 3B). Here, PPCLs were broadly categorized into three subgroups as a function of growth kinetics: slow (doubling time > 38 hours), intermediate (doubling time 28 to < 38 hours), and fast (doubling time < 28 hours). Interestingly, the in vitro growth kinetics were comparable with the in vivo growth dynamics of PPCLs as measured by the previously validated specific growth rate (Figure 3D).23 Collectively, these data demonstrate that all PPCL retain their tumorigenicity, regardless of the heterogeneity in the growth rates observed. These results support the tumor variability commonly observed in the clinical setting.
Figure 3.
Growth dynamics and chemosensitivity of PPCLs. A and B:In vivo (A) and in vitro (B) growth curves of PPCLs through 6 and 40 days, respectively. The growth rate of PPCLs and corresponding tumors were evaluated. Mean tumor volume is presented at each time point. C: Dose-dependent response to gemcitabine treatment of PPCLs, at concentration of 0, 3, 10, 30, 100, 300, and 1000 nmol/L. The IC50 of each line was measured. Viability at each concentration is presented. D: The doubling time, in vitro and in vivo growth rates, and IC50 for each PPCL are summarized. Data are expressed as means ± SEM. n = 3 (A and B); n = 6 (C). All experiments were performed in triplicate. IC50, half maximal inhibitory concentration; PPCL, primary pancreatic cancer cell line.
Gemcitabine Chemosensitivity
Because chemosensitivity of PC is hypothesized to be directly related to the proliferative capacity of cancer cells, the chemosensitivity to the clinically relevant antimetabolite, gemcitabine, was evaluated for each PPCL established. Interestingly, although all PPCLs were sensitive to gemcitabine treatment (Figure 3C), an inverse correlation between proliferation rate and sensitivity was observed (Figure 3D). These findings suggest that more diverse mechanisms than rate of proliferation are responsible for antimetabolite sensitivity.24
Heterogeneity of PC Stem Cell Marker Expression
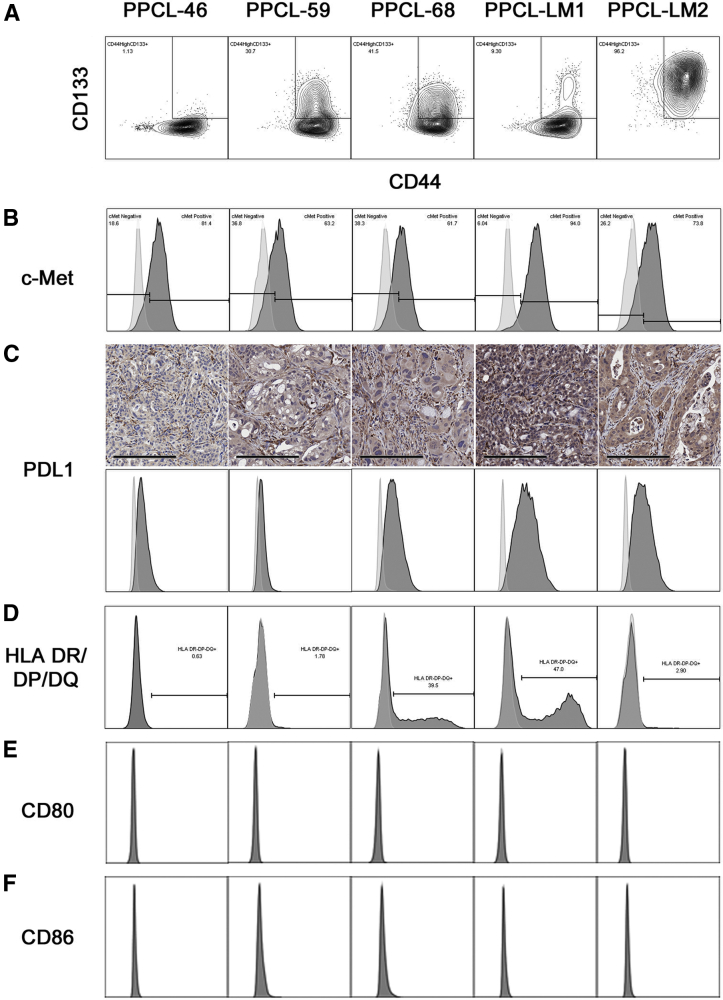
Dissociated PDX tumors have enabled careful experimental probing of intratumoral heterogeneity, critical to the discovery of PC stem cell markers.25 To assess whether the heterogeneity observed above in the PPCL cultures extends to PC stem cell markers, the established PC stem cell markers CD44, CD133, and c-Met were examined by flow cytometry.26 Interestingly, each PPCL demonstrated a unique profile of CD44 and CD133 expression (Figure 4A), exhibiting from 1% to 96% CD44highCD133+ cells. Similarly, differential c-Met expression was observed among PPCLs, with each line demonstrating between 6% and 62% c-Met− cells (Figure 4B). Taken together, these experiments demonstrate the continued presence of intratumoral cancer stem cell heterogeneity in primary culture.
Figure 4.
PPCLs demonstrate heterogeneity in markers associated with cancer stemness and PC-immune cell interactions. A and B: PPCLs were evaluated for the cancer stem cell markers CD44, CD133 (A) and c-Met (B). The percentage of cells positive for both CD144 and CD33 are indicated in the rectangular gate. C: Immunohistochemical staining for PDL1 was performed on patient-derived xenografts from each patient. C–F: In addition, cell populations were analyzed for the expression of the negative costimulatory marker PDL1 (C), antigen presentation machinery (HLA-DR/DP/DQ; D), and the positive costimulatory markers CD80 (E) and CD86 (F). Histograms are shown for isotype controls (gray) and staining for the indicated marker (black). Scale bar = 200 μm (C). HLA, human leukocyte antigen; PDL1, programmed death ligand-1; PPLC, primary pancreatic cancer cell line.
Expression of Antigen Presentation Machinery and Costimulatory Ligands
The properties of PC epithelial cells may allow for evasion of immune clearance through the down-regulation of HLA and/or T-cell–activating costimulatory molecules and/or up-regulation of T-cell inhibitory molecules.27 Thus, to evaluate the extent to which the patient-derived PPCLs express this antigen presentation machinery and costimulatory molecules, flow cytometry was performed to examine HLA class II (HLA-DR/DP/DQ), the activating costimulatory molecules CD80, CD86, and the inhibitory costimulatory molecule PDL1. Interestingly, all PPCLs expressed the inhibitory costimulatory molecule PDL1 variably, with the highest expression observed in PPCLs of metastatic origin (PPCL-LM1 and PPCL-LM2) (Figure 4C). Immunohistochemical staining of PDX tumors for PDL1 correlated with cell line expression. As evaluated by a board-certified pathologist, PDX tumors from all patients except PPCL-46 were scored as staining positive with low intensity, whereas the PDX from PPCL-46 was negative for PDL1 staining. Four of five PPCLs contained a subpopulation of HLA class II-expressing cells, which ranged from 1% to 47% of cells (Figure 4D). None of the PPCLs evaluated expressed the activating costimulatory ligands, CD80 or CD86 (Figure 4, E and F). Heterogeneity of basal PDL1 expression in unstimulated culture conditions represents an important, clinically relevant finding, given the widespread use of PD-1 inhibitors in cancer.28, 29, 30, 31 These data reinforce the intraculture and interculture heterogeneity of PPCLs and permit the retention of the immune evasive phenotype in the tumor microenvironment.
Discussion
The capacity to isolate and culture primary human cancer cells has initiated a surge of research in cancer biology that has yielded valuable insights and potential development of novel personalized therapeutic agents. Widespread use of a common pool of commercially available, highly passaged cancer cell lines, while promoting a high degree of investigative consistency, appears to experience diminishing translational returns. An analysis of phase 1 cancer trials supports this concept by demonstrating alarmingly low success rates associated with therapies that were successful in preclinical investigations.32 Both intertumor and intratumor heterogeneity contribute to these translational limitations, suggesting that not all tumors are created equal. Our work highlights the need for investigational strategies that incorporate this tumor heterogeneity to better characterize the biology of PC and disease responsiveness to conventional and experimental treatments.
To create an effective preclinical and translational model, we leveraged our PC-PDX model to rapidly and effectively generate purified populations of human PCCs in culture. Primary PPCLs isolated in this manner retain significant heterogeneity, which is commonly observed in the human disease, both between patients and within individual tumors. Importantly, we demonstrate a 100% success rate with isolation of PPCLs that are available for experimentation as early as 1 to 2 weeks after xenograft harvesting. Given the high rates of success associated with the engraftment and growth of PC-PDXs,21, 33 the establishment of PDX-derived PPCLs will provide a valuable tool to preclinical research into the biology of PC and a reliable and time-saving in vitro model for drug validation in personalized therapies.
It is important to recognize that other groups have established variations of this technique for isolating PCCs from PDXs. Both Kang et al34 and Li et al35 have described methods to dissociate PC-PDX into suspensions of viable cells for further investigation. Kang et al34 then pursued an outgrowth strategy from macroscopic tumor pieces in culture with the use of standard RPMI-1640 media with serum supplementation. However, in our experience, this technique is associated with a high rate of stromal contamination, likely necessitating their use of cloning rings to isolate and expand pure PC cell lines. Here, we demonstrate that the complete dissociation of the PDX is readily achievable with the use of a gentle collagenase/dispase cocktail and combined with epithelial-enhanced media, which greatly reduces stromal cell contamination. The few remaining stromal cells can then be removed with differential trypsinization, effectively preserving a large fraction of PCCs from the original xenograft. Because of the high efficiency of PCC preservation and early exponential growth in culture, experimentation can typically begin within 1 to 2 weeks of xenograft harvest, allowing for a more reliable and genetically stable source of cancer cells that are more predictive of clinical responsiveness.
PC stem cells were first described as a subpopulation of PCCs with a high degree of tumorigenicity,25 and careful analysis of these subpopulations has identified multiple therapeutic targets.26, 36, 37 However, as opposed to freshly isolated primary cells, highly passaged established cancer cell lines do not exhibit the same relation between cancer stem cell markers and tumorigenicity,38 likely because of the expansion of a homogenous population over long periods of time in culture. Importantly, our model demonstrates the retention of PCC heterogeneity in culture. Thus, the method presented here may provide an experimental framework to evaluate relations between cancer stem cells, tumorigenicity, and therapeutic resistance for each individual patient.
Translation of tailored treatment regimens on the individual patient level is exemplified by the routine practice of molecular profiling in the clinical care for colorectal, lung, and breast tumors.39 PDXs provide an attractive model for personalized cancer therapy and have already shown promise in the treatment of sarcomas.40 However, freshly isolated PPCLs will enable evaluation of the heterogeneity of cancer cell biology and the identification of more appropriate targets for therapeutic intervention. In addition, it will allow for the potential high-throughput pharmacologic screening of individualized therapeutics. Thus, the availability of PPCLs coupled with the syngeneic PDX provides a much more robust and personalized preclinical model for PC.
Given the recent success and continued evolution of immune modulation in cancer,29, 31, 41, 42, 43 delineating relations between cancer cells and the immune system will be critical. A long-debated question in the field of immuno-oncology is whether cancer cells are capable of antigen presentation to CD4 T cells. Although the expression of HLA class II and PDL1 is a known consequence of interferon-γ stimulation,44, 45 here, we observed a substantial proportion of unstimulated PCCs stably expressing both HLA class II and PDL1 in culture. This pattern of expression, in the absence of positive costimulatory markers CD80 or CD86, has been associated with highly regulatory immune cell phenotypes46 and may contribute to the profoundly suppressive immune microenvironment of PC.47, 48 Importantly, histologic examination of human PC specimens revealed an identical pattern to that observed in our primary PCC phenotyping for HLA class II staining.49 Conversely, we have not observed any basal expression of HLA class II expression in commercially available, established PC cell lines PANC-1, BxPC3, and L3.6 pl (data not shown), marking a significant phenotypic departure of these highly passaged PC cell lines from our PPCLs. Although the functional consequences of basal HLA class II and PDL1 expression remain speculative at this point, these data certainly highlight that primary cancer cells may also be critical to defining PC-immune cell relations.
In summary, here we introduce an efficient, highly successful technique to isolate and expand a heterogeneous population of patient-derived PCCs. Currently, the only barrier at our institution to establishing each individual's PCCs in culture is the engraftment and growth of the PDX. Combining a successful PDX program with consistent isolation and expansion of cultured PPCLs may therefore provide an opportunity to evaluate the biology and treatment of PC on a highly representative scale. Importantly, PPCLs may be cryopreserved and efficiently distributed between institutions, ultimately focusing investigations to clinically relevant subsets of this heterogeneous disease and enabling a high degree of collaborative expertise.
Footnotes
Supported by National Cancer Institute grant 5T32CA106493-09 (D.D.) and Cracchiolo Foundation grant UF115477 (D.D.).
K.P. and D.D. contributed equally to this work.
Disclosures: None declared.
Contributor Information
Chen Liu, Email: liu@pathology.ufl.edu.
Jose G. Trevino, Email: jose.trevino@surgery.ufl.edu.
References
- 1.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Koay E.J., Truty M.J., Cristini V., Thomas R.M., Chen R., Chatterjee D., Kang Y., Bhosale P.R., Tamm E.P., Crane C.H., Javle M., Katz M.H., Gottumukkala V.N., Rozner M.A., Shen H., Lee J.E., Wang H., Chen Y., Plunkett W., Abbruzzese J.L., Wolff R.A., Varadhachary G.R., Ferrari M., Fleming J.B. Transport properties of pancreatic cancer describe gemcitabine delivery and response. J Clin Invest. 2014;124:1525–1536. doi: 10.1172/JCI73455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore M.J., Goldstein D., Hamm J., Figer A., Hecht J.R., Gallinger S., Au H.J., Murawa P., Walde D., Wolff R.A., Campos D., Lim R., Ding K., Clark G., Voskoglou-Nomikos T., Ptasynski M., Parulekar W. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 4.Baxter N.N., Whitson B.A., Tuttle T.M. Trends in the treatment and outcome of pancreatic cancer in the United States. Ann Surg Oncol. 2007;14:1320–1326. doi: 10.1245/s10434-006-9249-8. [DOI] [PubMed] [Google Scholar]
- 5.Yunis A.A., Arimura G.K., Russin D.J. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: sensitivity to asparaginase. Int J Cancer. 1977;19:128–135. doi: 10.1002/ijc.2910190118. [DOI] [PubMed] [Google Scholar]
- 6.McIntosh J.C., Schoumacher R.A., Tiller R.E. Pancreatic adenocarcinoma in a patient with cystic fibrosis. Am J Med. 1988;85:592. doi: 10.1016/s0002-9343(88)80116-5. [DOI] [PubMed] [Google Scholar]
- 7.Lieber M., Mazzetta J., Nelson-Rees W., Kaplan M., Todaro G. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int J Cancer. 1975;15:741–747. doi: 10.1002/ijc.2910150505. [DOI] [PubMed] [Google Scholar]
- 8.Tan M.H., Nowak N.J., Loor R., Ochi H., Sandberg A.A., Lopez C., Pickren J.W., Berjian R., Douglass H.O., Jr., Chu T.M. Characterization of a new primary human pancreatic tumor line. Cancer Invest. 1986;4:15–23. doi: 10.3109/07357908609039823. [DOI] [PubMed] [Google Scholar]
- 9.Johnson J.I., Decker S., Zaharevitz D., Rubinstein L.V., Venditti J.M., Schepartz S., Kalyandrug S., Christian M., Arbuck S., Hollingshead M., Sausville E.A. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel V.C., Marchionni L., Hierman J.S., Rhodes J.T., Devereux W.L., Rudin C.M., Yung R., Parmigiani G., Dorsch M., Peacock C.D., Watkins D.N. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassidy J.W., Caldas C., Bruna A. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75:2963–2968. doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruckert F., Aust D., Bohme I., Werner K., Brandt A., Diamandis E.P., Krautz C., Hering S., Saeger H.D., Grutzmann R., Pilarsky C. Five primary human pancreatic adenocarcinoma cell lines established by the outgrowth method. J Surg Res. 2012;172:29–39. doi: 10.1016/j.jss.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Holloway S., Davis M., Jaber R., Fleming J. A clinically relevant model of human pancreatic adenocarcinoma identifies patterns of metastasis associated with alterations of the TGF-beta/Smad4 signaling pathway. Int J Gastrointest Cancer. 2003;33:61–69. doi: 10.1385/IJGC:33:1:61. [DOI] [PubMed] [Google Scholar]
- 14.Chifenti B., Morelli M., Zavaglia M., Coviello D.A., Guerneri S., Santucci A., Paffetti A., Masetti M., Locci M.T., Bertacca G., Capodanno A., Collecchi P., Campani D., Mosca F., Bevilacqua G., Cavazzana A.O. Establishment and characterization of 4 new human pancreatic cancer cell lines: evidences of different tumor phenotypes. Pancreas. 2009;38:184–196. doi: 10.1097/MPA.0b013e31818c746a. [DOI] [PubMed] [Google Scholar]
- 15.Kyriazis A.P., McCombs W.B., 3rd, Sandberg A.A., Kyriazis A.A., Sloane N.H., Lepera R. Establishment and characterization of human pancreatic adenocarcinoma cell line SW-1990 in tissue culture and the nude mouse. Cancer Res. 1983;43:4393–4401. [PubMed] [Google Scholar]
- 16.Kato M., Shimada Y., Tanaka H., Hosotani R., Ohshio G., Ishizaki K., Imamura M. Characterization of six cell lines established from human pancreatic adenocarcinomas. Cancer. 1999;85:832–840. doi: 10.1002/(sici)1097-0142(19990215)85:4<832::aid-cncr10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Tentler J.J., Tan A.C., Weekes C.D., Jimeno A., Leong S., Pitts T.M., Arcaroli J.J., Messersmith W.A., Eckhardt S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dangles-Marie V., Pocard M., Richon S., Weiswald L.B., Assayag F., Saulnier P., Judde J.G., Janneau J.L., Auger N., Validire P., Dutrillaux B., Praz F., Bellet D., Poupon M.F. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- 19.DeRose Y.S., Wang G., Lin Y.C., Bernard P.S., Buys S.S., Ebbert M.T., Factor R., Matsen C., Milash B.A., Nelson E., Neumayer L., Randall R.L., Stijleman I.J., Welm B.E., Welm A.L. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Garcia R., Juan D., Rausell A., Munoz M., Banos N., Menendez C., Lopez-Casas P.P., Rico D., Valencia A., Hidalgo M. Transcriptional dissection of pancreatic tumors engrafted in mice. Genome Med. 2014;6:27. doi: 10.1186/gm544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delitto D., Pham K., Vlada A.C., Sarosi G.A., Thomas R.M., Behrns K.E., Liu C., Hughes S.J., Wallet S.M., Trevino J.G. Patient-derived xenograft models for pancreatic adenocarcinoma demonstrate retention of tumor morphology through incorporation of murine stromal elements. Am J Pathol. 2015;185:1297–1303. doi: 10.1016/j.ajpath.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erkan M., Michalski C.W., Rieder S., Reiser-Erkan C., Abiatari I., Kolb A., Giese N.A., Esposito I., Friess H., Kleeff J. The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol. 2008;6:1155–1161. doi: 10.1016/j.cgh.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Mehrara E., Forssell-Aronsson E., Ahlman H., Bernhardt P. Specific growth rate versus doubling time for quantitative characterization of tumor growth rate. Cancer Res. 2007;67:3970–3975. doi: 10.1158/0008-5472.CAN-06-3822. [DOI] [PubMed] [Google Scholar]
- 24.de Sousa Cavalcante L., Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 25.Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., Wicha M., Clarke M.F., Simeone D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 26.Li C., Wu J.J., Hynes M., Dosch J., Sarkar B., Welling T.H., Pasca di Magliano M., Simeone D.M. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227.e5. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Delitto D., Perez C., Han S., Gonzalo D.H., Pham K., Knowlton A.E., Graves C.L., Behrns K.E., Moldawer L.L., Thomas R.M., Liu C., George T.J., Jr., Trevino J.G., Wallet S.M., Hughes S.J. Downstream mediators of the intratumoral interferon response suppress antitumor immunity, induce gemcitabine resistance and associate with poor survival in human pancreatic cancer. Cancer Immunol Immunother. 2015;64:1553–1563. doi: 10.1007/s00262-015-1760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 29.Postow M.A., Chesney J., Pavlick A.C., Robert C., Grossmann K., McDermott D., Linette G.P., Meyer N., Giguere J.K., Agarwala S.S., Shaheen M., Ernstoff M.S., Minor D., Salama A.K., Taylor M., Ott P.A., Rollin L.M., Horak C., Gagnier P., Wolchok J.D., Hodi F.S. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motzer R.J., Rini B.I., McDermott D.F., Redman B.G., Kuzel T.M., Harrison M.R., Vaishampayan U.N., Drabkin H.A., George S., Logan T.F., Margolin K.A., Plimack E.R., Lambert A.M., Waxman I.M., Hammers H.J. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33:1430–1437. doi: 10.1200/JCO.2014.59.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., Biedrzycki B., Donehower R.C., Zaheer A., Fisher G.A., Crocenzi T.S., Lee J.J., Duffy S.M., Goldberg R.M., de la Chapelle A., Koshiji M., Bhaijee F., Huebner T., Hruban R.H., Wood L.D., Cuka N., Pardoll D.M., Papadopoulos N., Kinzler K.W., Zhou S., Cornish T.C., Taube J.M., Anders R.A., Eshleman J.R., Vogelstein B., Diaz L.A., Jr. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts T.G., Jr., Goulart B.H., Squitieri L., Stallings S.C., Halpern E.F., Chabner B.A., Gazelle G.S., Finkelstein S.N., Clark J.W. Trends in the risks and benefits to patients with cancer participating in phase 1 clinical trials. JAMA. 2004;292:2130–2140. doi: 10.1001/jama.292.17.2130. [DOI] [PubMed] [Google Scholar]
- 33.Thomas R.M., Truty M.J., Kim M., Kang Y., Zhang R., Chatterjee D., Katz M.H., Fleming J.B. The canary in the coal mine: the growth of patient-derived tumorgrafts in mice predicts clinical recurrence after surgical resection of pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2015;22:1884–1892. doi: 10.1245/s10434-014-4241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y., Zhang R., Suzuki R., Li S.Q., Roife D., Truty M.J., Chatterjee D., Thomas R.M., Cardwell J., Wang Y., Wang H., Katz M.H., Fleming J.B. Two-dimensional culture of human pancreatic adenocarcinoma cells results in an irreversible transition from epithelial to mesenchymal phenotype. Lab Invest. 2015;95:207–222. doi: 10.1038/labinvest.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C., Lee C.J., Simeone D.M. Identification of human pancreatic cancer stem cells. Methods Mol Biol. 2009;568:161–173. doi: 10.1007/978-1-59745-280-9_10. [DOI] [PubMed] [Google Scholar]
- 36.Li L., Hao X., Qin J., Tang W., He F., Smith A., Zhang M., Simeone D.M., Qiao X.T., Chen Z.N., Lawrence T.S., Xu L. Antibody against CD44s inhibits pancreatic tumor initiation and postradiation recurrence in mice. Gastroenterology. 2014;146:1108–1118. doi: 10.1053/j.gastro.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abel E.V., Simeone D.M. Biology and clinical applications of pancreatic cancer stem cells. Gastroenterology. 2013;144:1241–1248. doi: 10.1053/j.gastro.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 38.Fan F., Bellister S., Lu J., Ye X., Boulbes D.R., Tozzi F., Sceusi E., Kopetz S., Tian F., Xia L., Zhou Y., Bhattacharya R., Ellis L.M. The requirement for freshly isolated human colorectal cancer (CRC) cells in isolating CRC stem cells. Br J Cancer. 2015;112:539–546. doi: 10.1038/bjc.2014.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garraway L.A. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 40.Stebbing J., Paz K., Schwartz G.K., Wexler L.H., Maki R., Pollock R.E., Morris R., Cohen R., Shankar A., Blackman G., Harding V., Vasquez D., Krell J., Zacharoulis S., Ciznadija D., Katz A., Sidransky D. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120:2006–2015. doi: 10.1002/cncr.28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert C., Long G.V., Brady B., Dutriaux C., Maio M., Mortier L., Hassel J.C., Rutkowski P., McNeil C., Kalinka-Warzocha E., Savage K.J., Hernberg M.M., Lebbe C., Charles J., Mihalcioiu C., Chiarion-Sileni V., Mauch C., Cognetti F., Arance A., Schmidt H., Schadendorf D., Gogas H., Lundgren-Eriksson L., Horak C., Sharkey B., Waxman I.M., Atkinson V., Ascierto P.A. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 42.Rizvi N.A., Mazieres J., Planchard D., Stinchcombe T.E., Dy G.K., Antonia S.J., Horn L., Lena H., Minenza E., Mennecier B., Otterson G.A., Campos L.T., Gandara D.R., Levy B.P., Nair S.G., Zalcman G., Wolf J., Souquet P.J., Baldini E., Cappuzzo F., Chouaid C., Dowlati A., Sanborn R., Lopez-Chavez A., Grohe C., Huber R.M., Harbison C.T., Baudelet C., Lestini B.J., Ramalingam S.S. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16:257–265. doi: 10.1016/S1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran E., Turcotte S., Gros A., Robbins P.F., Lu Y.C., Dudley M.E., Wunderlich J.R., Somerville R.P., Hogan K., Hinrichs C.S., Parkhurst M.R., Yang J.C., Rosenberg S.A. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roche P.A., Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loos M., Giese N.A., Kleeff J., Giese T., Gaida M.M., Bergmann F., Laschinger M.M., W Büchler M., Friess H. Clinical significance and regulation of the costimulatory molecule B7-H1 in pancreatic cancer. Cancer Lett. 2008;268:98–109. doi: 10.1016/j.canlet.2008.03.056. [DOI] [PubMed] [Google Scholar]
- 46.Hepworth M.R., Fung T.C., Masur S.H., Kelsen J.R., McConnell F.M., Dubrot J., Withers D.R., Hugues S., Farrar M.A., Reith W., Eberl G., Baldassano R.N., Laufer T.M., Elson C.O., Sonnenberg G.F. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark C.E., Hingorani S.R., Mick R., Combs C., Tuveson D.A., Vonderheide R.H. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67:9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 48.Lutz E.R., Wu A.A., Bigelow E., Sharma R., Mo G., Soares K., Solt S., Dorman A., Wamwea A., Yager A., Laheru D., Wolfgang C.L., Wang J., Hruban R.H., Anders R.A., Jaffee E.M., Zheng L. Immunotherapy converts nonimmunogenic pancreatic tumors into immunogenic foci of immune regulation. Cancer Immunol Res. 2014;2:616–631. doi: 10.1158/2326-6066.CIR-14-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaida M.M., Welsch T., Herpel E., Tschaharganeh D.F., Fischer L., Schirmacher P., Hansch G.M., Bergmann F. MHC class II expression in pancreatic tumors: a link to intratumoral inflammation. Virchows Arch. 2012;460:47–60. doi: 10.1007/s00428-011-1175-x. [DOI] [PubMed] [Google Scholar]