Abstract
Like most positive-strand RNA viruses, hepatitis C virus (HCV) forms a membrane-associated replication complex consisting of replicating RNA, viral and host proteins anchored to altered cell membranes. We used a combination of qualitative and quantitative electron microscopy (EM), immuno-EM, and the 3D reconstruction of serial EM sections to analyze the host cell membrane alterations induced by HCV. Three different types of membrane alteration were observed: vesicles in clusters (ViCs), contiguous vesicles (CVs), and double-membrane vesicles (DMVs). The main ultrastructural change observed early in infection was the formation of a network of CVs surrounding the lipid droplets. Later stages in the infectious cycle were characterized by a large increase in the number of DMVs, which may be derived from the CVs. These DMVs are thought to constitute the membranous structures harboring the viral replication complexes in which viral replication is firmly and permanently established and to protect the virus against double-stranded RNA-triggered host antiviral responses.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1213-0) contains supplementary material, which is available to authorized users.
Keywords: HCV, Host cell membranes, Membranous web, Electron microscopy, 3D reconstruction
Introduction
Hepatitis C virus (HCV), a member of the Flaviviridae family, is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide [1]. It has a single positive-strand RNA genome encoding a large polyprotein precursor that is processed co- and post-translationally, by cellular and viral proteases, to yield 10 different proteins [2, 3]. The structural proteins core, E1, and E2 are the main constituents of the virus particle [4]. The p7 and NS2 proteins are required for virion assembly [5, 6], whereas NS3 to NS5B constitute the minimal viral replicase [7, 8]. This replicase includes the NS3/4A protein, which has serine-type protease, NTPase, and helicase activities, the NS5B RNA-dependent RNA polymerase, and the NS5A replicase factor. NS4B is a highly hydrophobic protein that plays a major role in triggering the rearrangement of intracellular membranes [9, 10]. Like most positive-strand RNA viruses, HCV forms a membrane-associated replication complex, consisting of viral proteins, replicating RNA, altered cellular membranes, and other host factors [11, 12]. These specific membrane alterations, known as the “membranous web”, were first identified with the HCV replicase construct model (subgenomic replicon) and were described as a cluster of vesicles measuring 80–100 nm in diameter [9, 13].
We recently studied these membrane alterations in Huh7.5 cells stably carrying a highly replicative subgenomic replicon of the JFH-1 strain [14]. We discovered that these HCV-induced membranous structures were much more complex than previously thought, including the formation of numerous double-membrane vesicles (DMVs), resembling those observed for other RNA viruses, such as poliovirus [15] and coronavirus [16]. Our observations were subsequently confirmed by other studies [17, 18]. In this study, we used the Huh7.5/JFH-1 HCVcc model reproducing the complete infectious HCV cycle [19–21] to analyze, by electron microscopy (EM), the kinetics of virus-induced host cell membrane alterations during the course of HCV infection.
Materials and methods
Cell culture
The Huh7.5 cell line (a gift from Dr Rice, Rockefeller University, New York, USA) is a clone of the human hepatoma cell line Huh7 obtained by curing stably selected replicon-containing cells with interferon and thus supporting efficient HCV replication [22]. This cell line was grown in Dulbecco’s modified Eagle medium (DMEM; Invitrogen) supplemented with 10 % fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen).
Antibodies
Mouse monoclonal antibody (mAb) against HCV core protein (clone C7-50) was obtained from Abcam. The mouse anti-NS5A mAb (clone 2F6) was obtained from BioFront Technologies. The mouse anti-dsRNA mAb (clone J2) was obtained from Scicons.
HCVcc and infection of Huh7.5 cells
Cells were infected with the JFH-1 strain [19], optimized by 10 cycles of infection in naive Huh7.5 cells, resulting in cytopathic effect 6 days after infection. This optimized JFH-1 virus has adaptive T414I (E2), V1048M (NS3), E1703K (NS4A), D2437N, T2439P, V2440L, C2441G (NS5A), E2459K, E2460K, V2479G, P2571S, L2804P, and A2961T (NS5B) mutations (number of amino acids in accordance with the JFH1 polyprotein sequence—data not shown). In these infection conditions, this optimized virus with titer-increasing mutations infected 100 % of the cells within 3 days of infection (Fig. S1 in the supplemental material). Infectivity titers were calculated in focus forming units (FFU), by counting the foci after core immunostaining. Huh7.5 cells were used to seed 75-cm2 flasks (2 × 106 cells per flask), 24 h before infection. The cells were incubated for 16 h with 2 × 105 FFU of the adapted JFH-1 virus. The inoculum was removed and cells were washed with medium and cultured for a further 3 days. One flask was treated with trypsin on day 2 to initiate a subculture, which was continued until day 6. Each day, a cell pellet was collected for qualitative and quantitative EM analysis and for intracellular HCV RNA quantification. Intracellular HCV RNA and extracellular HCV RNA present in the supernatant were quantified as previously described [14].
Electron microscopy
For standard ultrastructural analysis by EM, cell pellets were collected daily and treated as previously described [14, 23]. Briefly, cells were fixed by incubation for 48 h in 4 % paraformaldehyde and 1 % glutaraldehyde in 0.1 M phosphate buffer (pH 7.2), washed in PBS, postfixed by incubation for 1 h with 2 % osmium tetroxide and dehydrated in a graded series of ethanol solutions. Cell pellets were embedded in Epon resin (Sigma), which was allowed to polymerize for 48 h at 60 °C. Ultrathin sections were cut, stained with 5 % uranyl acetate and 5 % lead citrate, and deposited on EM grids coated with Formvar (polyvinyl formal resin) membrane for examination under a JEOL 1230 transmission electron microscope (TEM). For quantitative analysis, the various membrane rearrangements were monitored and counted in 60 cells (60 consecutive sections on the EM grid) from each cell pellet. Uninfected Huh7.5 cells were used as a negative control.
Immuno-electron microscopy
Huh7.5 cells infected with our adapted JFH-1 virus or Huh7.5 control cells were fixed on day 6 post infection. Cells were incubated in a solution of 1.5 % paraformaldehyde and 0.025 % glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 1 h. They were collected by centrifugation and the cell pellet was dehydrated in a graded series of ethanol solutions at −20 °C, with an automatic freezing substitution system (AFS; Leica), and embedded in London Resin Gold (LR Gold; Electron Microscopy Science). The resin was allowed to polymerize at −25 °C under UV light for 72 h. Ultrathin sections were cut and placed on collodion-coated EM grids. These sections were then blocked by incubation in PBS supplemented with 2 % BSA (Sigma). They were then incubated with the monoclonal antibody against HCV core protein, the monoclonal antibody against NS5A, or the monoclonal antibody against dsRNA, diluted 1:50 in PBS supplemented with 1 % BSA. Sections were then washed and incubated with an appropriate 15-nm gold-particle-conjugated secondary antibody (TebuBio) diluted 1:30 in PBS supplemented with 1 % BSA. Ultrathin sections were stained with 5 % uranyl acetate and 5 % lead citrate and observed as describe above. For quantitative analysis, 10 cells were monitored, counting all the gold particles present in these cells, for each type of immunogold labeling and for both infected and uninfected Huh7.5 cells (analysis of 10 consecutive cell sections on the EM grid for each pellet).
Three-dimensional reconstruction
We recently described the use of serial EM sections for the three-dimensional (3D) reconstruction of entire cells producing the HCV core protein [24]. We used a similar approach here, to generate 3D reconstructions of several whole Huh7.5 cells 2 days after infection with our JFH-1 adapted virus. Briefly, standard Epon EM blocks were resized for the cutting of ribbons of serial ultrathin sections (70 nm thick), which were placed on EM grids. These grids were then stained as described above, and electron micrographs were acquired with a digital camera for each of the sections of the series. Photoshop software was used to align image stacks for these serial EM sections. Contours were drawn with IMOD software, through specific cellular structures, including the host cell membrane rearrangements induced by HCV, on the various serial sections. The contours from a stack of serial sections were then arranged into objects with IMOD. The contours of each object were then joined, using the IMODmesh feature of IMOD, to form a 3D model.
Results
Ultrastructure of host cell membrane rearrangements during the course of HCV infection
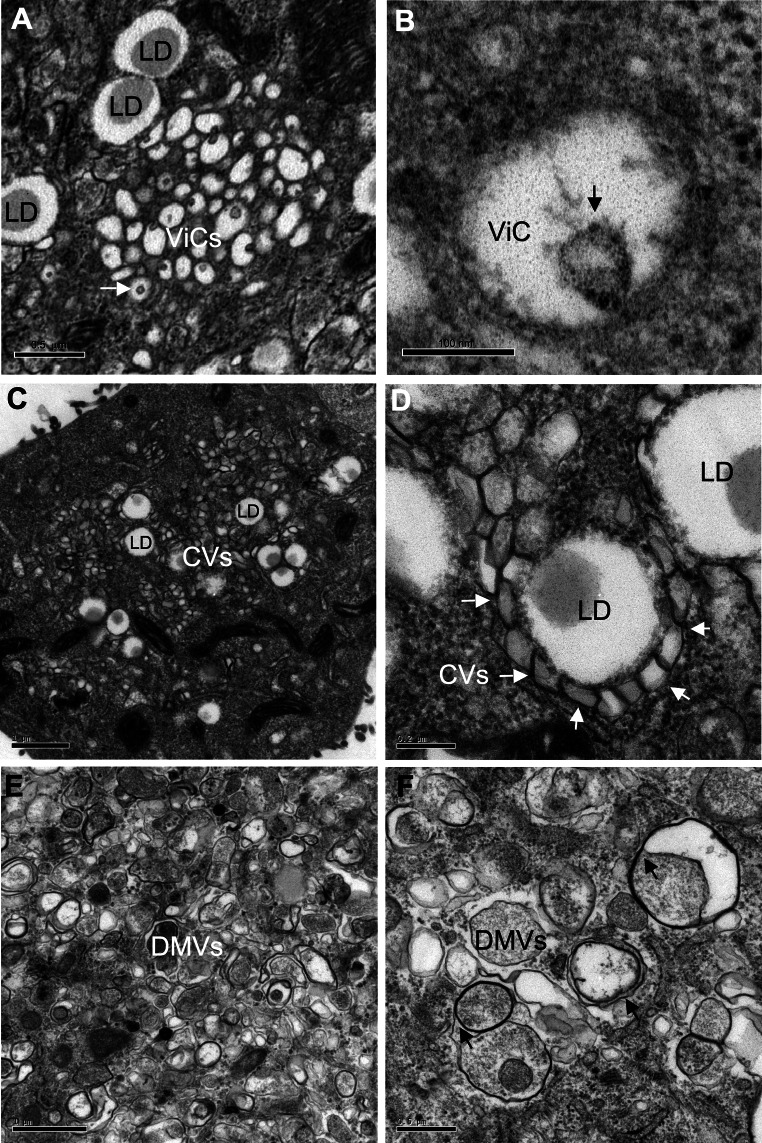
Three different types of membrane alteration were observed (Fig. 1): vesicles in clusters (ViCs), contiguous vesicles (CVs), and double-membrane vesicles (DMVs). The ViCs were small single-membrane vesicles grouped together in well-delimited areas (Fig. 1a). Most of these ViCs had an internal invagination (high magnification in Fig. 1b). The CVs were small single-membrane vesicles, present in large number and widely distributed through the cytoplasm (Fig. 1c). They were tightly associated and tended to form a collar around the lipid droplets (LD, high magnification in Fig. 1d). Finally, DMVs were heterogeneous in size and had a thick, electron-dense membrane (Fig. 1e). Examination at higher magnification (Fig. 1f) revealed the presence of a double membrane in some areas, suggesting the close association of two membranes in these vesicles, as previously reported in cells stably carrying a subgenomic JFH-1 replicon [14] and in cells infected with coronaviruses [16]. Some very large DMVs presented more complex structures forming multimembrane vesicles (MMV) or multivesicular bodies (MVB) (data not shown), as previously reported in the subgenomic replicon model [14].
Fig. 1.
Main ultrastructural changes encountered in Huh7.5 cells during the course of HCVcc infection (adapted JFH-1 strain). Cells displayed three types of membrane alterations: a, b vesicles in cluster (ViC), corresponding to a small number (10–40) of single-membrane vesicles of variable size (100–200 nm) grouped together in a well-delimited area; most, if not all, of these vesicles had an internal invagination (white arrow in the low magnification image in a; black arrow in an individual vesicle shown at high magnification in b). c, d Contiguous vesicles (CV), corresponding to single-membrane vesicles of a more homogeneous size (around 100 nm) present in large numbers; these vesicles clustered together and frequently formed a collar around lipid droplets (LD, see the white arrows on the high magnification image in d). e, f Double-membrane vesicles (DMV) of extremely variable size (150–1,000 nm); these vesicles were characterized by a thick, electron-dense membrane consisting of two closely apposed membranes (black arrows on the high magnification image in f). All these structures were highly specific to HCV infection and were not observed in uninfected Huh7.5 cells (not shown)
Sequential biogenesis of the various membranous structures induced by HCV infection
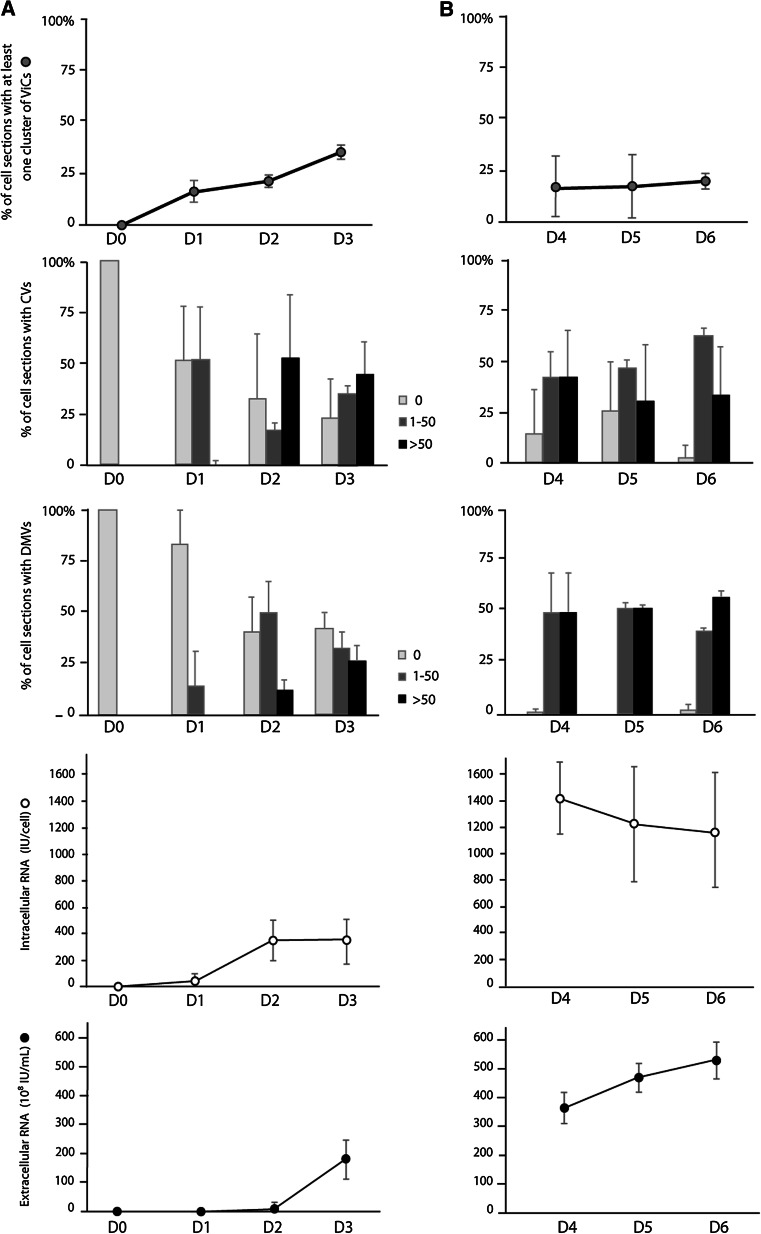
We analyzed 60 consecutive standard EM sections of Huh7.5 cells collected daily from initial infection (day 0) to day 6, in three independent infection experiments (Fig. 2). All types of vesicles (ViCs, CVs, DMVs) were observed on day 1. The percentage of cell sections containing at least one ViC cluster (composed of 10–40 vesicles) increased between days 0 and 3, subsequently decreasing after cell passage (Fig. 2). Cell sections containing more than 50 CVs appeared on day 2, and their percentage was variable over time in the different experiments. The percentage of cell sections containing more than 50 DMVs was low on day 2, but gradually increased thereafter, reaching levels of more than 50 % on day 6 (Fig. 2). The quantification of viral RNA by real-time PCR showed that intracellular HCV RNA levels increased steadily from day 1 to day 4, and then decreased slightly, whereas extracellular HCV RNA levels increased steadily from day 3 to day 6 (Fig. 2).
Fig. 2.
Quantitative analysis of the various types of vesicle observed in Huh7.5 cells in the 6 days following HCVcc infection, and quantification of intracellular/extracellular HCV RNA. a Early in the time course, cells were infected on day 0 by incubation for 16 h with an optimized JFH-1 virus strain, then washed with medium and cultured for a further 3 days. b One flask was then treated with trypsin on day 2 to initiate a subculture which was continued until day 6. Each day, a cell pellet was collected for the qualitative and quantitative analysis, by EM, of 60 consecutive cell sections. ViCs were the least abundant structures and their frequency is indicated as the percentage (gray circles) of cell sections with at least one vesicle cluster. CVs and DMVs were extremely abundant in infected cells and their frequency is indicated as the percentage of cell sections containing no (0) vesicles, 1–50 vesicles, or more than 50 vesicles (light gray, dark grey, and black bars, respectively). Intracellular (white circles) and extracellular (black circles) HCV RNA were quantified at each time point, with the Abbott m2000sp–m2000rt real-time PCR assay. In all experiments, uninfected Huh7.5 cells were used as a negative control. The data presented are the mean values and standard deviations for three independent experiments
Association of the HCV proteins and RNA with the various membranous structures
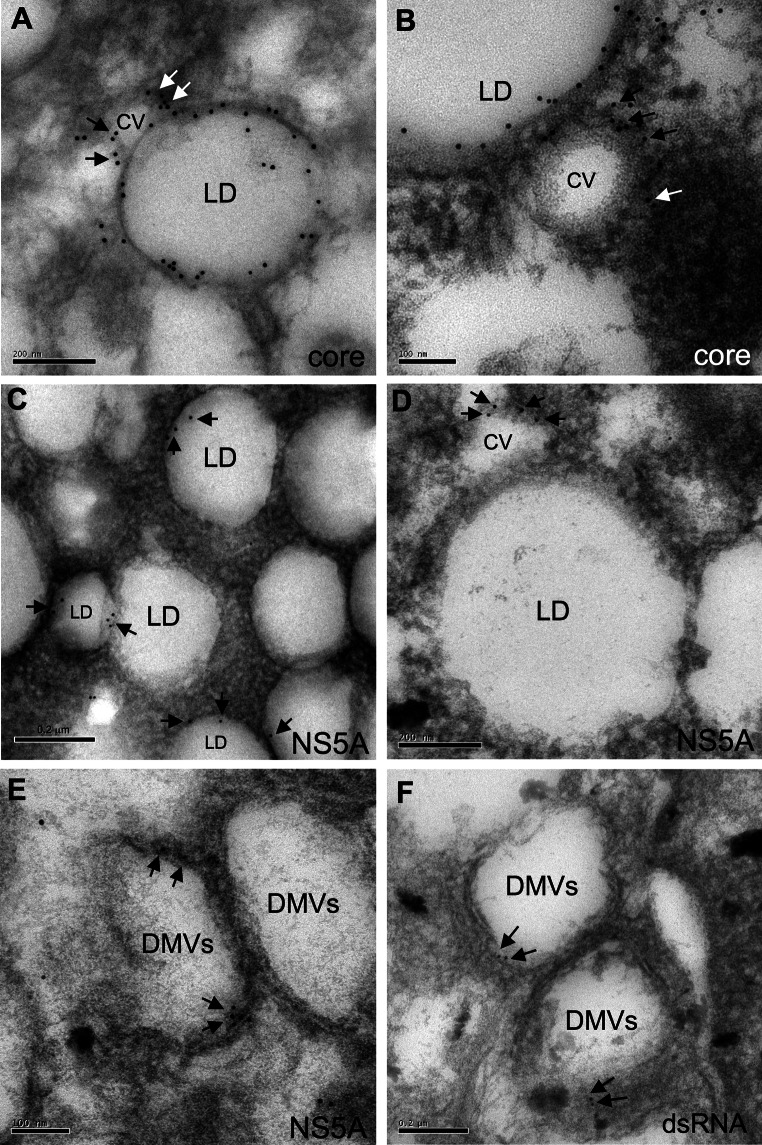
We investigated the presence of HCV proteins and RNA potentially associated with these structures by performing immuno-EM on control and infected cells prepared by freeze substitution on days 2 and 6 post infection. We also studied these cells by confocal microscopy (Fig. S2 in the supplemental material), to allow comparison of the overall distribution of these HCV components by immuno-EM and immunofluorescence microscopy. The core protein was observed in discrete foci on day 2 but strongly associated with the LD on day 6 post infection (Fig. S2). The NS5A protein remained dispersed throughout the cytoplasm throughout infection but displayed partial colocalization with LD on day 6 post infection. The labeling of dsRNA remained highly punctate from day 2 to day 6 post infection (Fig. S2). For the HCV RNA, core, and NS5A proteins, the signal was much more intense on day 6 post infection than on day 2 post infection (Fig. S2). This may account for the lack of significant immuno-EM labeling on immuno-EM of ultrathin cell sections on day 2 post infection (data not shown). We therefore focused our immuno-EM analysis on cells studied 6 days after infection (Fig. 3). For these cells, analyzed by freeze substitution, the fixation and embedding procedure resulted in a poorer preservation of cell structure than observed with standard EM methods, as previously reported by other groups [25]. However, cell structure was sufficiently well preserved for the observation of HCV core and NS5A proteins at the LD surface (Fig. 3a–c), as reported by others in Huh7.5 cells infected with the HCV JFH-1 strain [26]. Both proteins were also detected in the CVs, particularly in those surrounding the LDs (Fig. 3a, b, d). NS5A was also associated with the membranes of the DMVs (Fig. 3e), and dsRNA was detected within the DMVs or associated with their membranes (Fig. 3f), as previously observed with the subgenomic JFH-1 replicon model [14]. By monitoring and counting the gold particles in 10 consecutive cell sections for each type of immunogold labeling, we determined that 81 % of the core protein signal was associated with the LD surface, whereas 16 % of this signal was associated with the CVs (Table 1). For NS5A, we estimated that 60 % of the signal was associated with the DMVs, 27 % with the CVs, and 13 % with the LD surface (Table 1). Finally, the dsRNA signal was more heterogeneously distributed, with some labeling of the nucleus and mitochondria in some cells, but it was nonetheless predominantly associated with the DMVs (70 % of the signal, Table 1).
Fig. 3.
Immuno-EM analyses of Huh7.5 cells on day 6 post infection with HCVcc, prepared by freeze substitution. a, b Immunogold labeling with a monoclonal antibody against core protein was observed over the entire surface of lipid droplets (LD) and on the membrane of the adjacent contiguous vesicles (CV). c, d Immunogold labeling with a monoclonal antibody against NS5A was more discrete than that for core protein, but was also observed in some areas of the LD surface (arrows in c), and on the membrane of the adjacent CVs (arrows in d). e Immunogold labeling with the monoclonal antibody against NS5A was also observed on double-membrane vesicle (DMV) membranes (arrows). f Immunogold labeling with the monoclonal antibody against dsRNA was observed within the DMVs or associated with DMV membranes (arrows). These observations were specific to Huh7.5-infected cells, as no immunogold labeling was detected in the uninfected Huh7.5 cells (not shown)
Table 1.
Detection of viral proteins and RNA in the various types of vesicle by immunogold labeling on day 6 post infection
| LDs (%) | CVs (%) | DMVs (%) | Nucleus (%) | Mitochondria (%) | |
|---|---|---|---|---|---|
| Core | 81 ± 10 | 16 ± 9 | 3 ± 1 | 0 | 0 |
| NS5A | 13 ± 5 | 27 ± 8 | 60 ± 17 | 0 | 0 |
| dsRNA | 12 ± 9 | 6 ± 10 | 70 ± 20 | 3 ± 5 | 9 ± 11 |
For each type of labeling, we monitored and counted all the gold particles in 10 consecutive cell sections, for both infected and uninfected cells. The nucleus and mitochondria were analyzed as controls. We removed the nonspecific background by subtracting the mean value obtained for a particular structure/compartment in the 10 uninfected cells from the values obtained with the infected cells. We then determined the percentage of gold particles associated with each structure/compartment for each infected cell. The data shown are the mean percentage and standard deviations for the 10 infected cells
Spatial organization of the membrane alterations induced by HCV infection
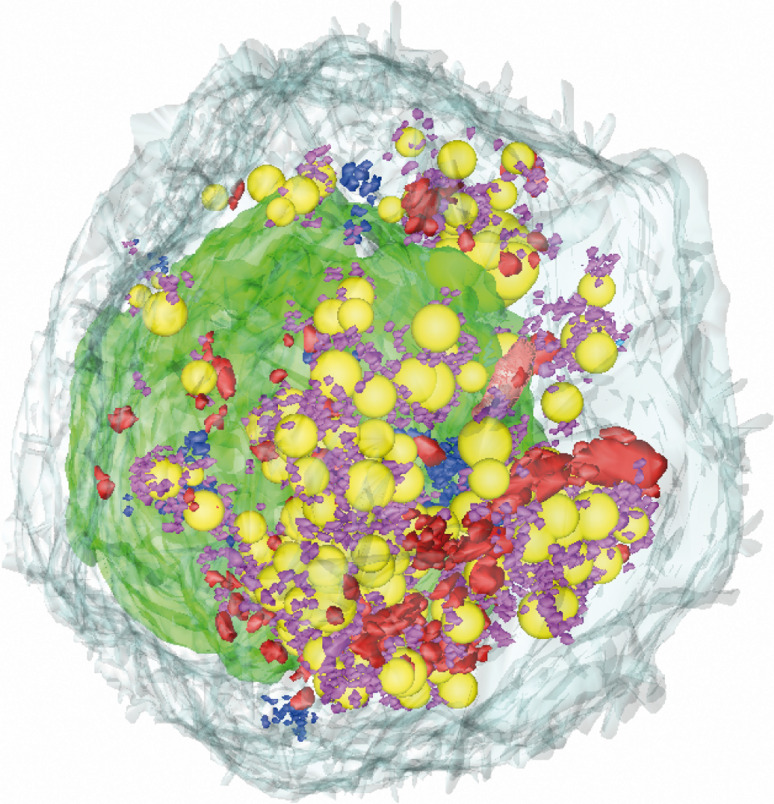
We performed 3D reconstruction on several whole infected Huh7.5 cells, using a recently developed method based on serial EM sections [24]. We found that day 2 post infection was the best time point for 3D reconstructions, because the cells contained numerous CVs and a small number of DMVs at this time. The larger numbers of DMVs at later stages of infection made it difficult to visualize intracellular organization clearly. Figure 4 shows a typical cell, with HCV-induced membranous structures closely associated with LD clusters (in yellow). Numerous small CVs (in purple) tend to surround these LDs, with the DMVs (in red) forming a massive network. ViCs (in blue) are more discrete structures, with only several small clusters identified in this cell.
Fig. 4.
Three-dimensional reconstruction of a whole Huh7.5 cell 2 days after infection with HCVcc. The standard EM block was resized for the cutting of a ribbon of 140 serial ultrathin sections (70 nm thick) to reconstruct this particular cell. Contours were drawn with IMOD software through the same specific cellular structures on different serial sections, including the plasma membrane (light gray), the nucleus (green), the lipid droplets (yellow), and the three types of membrane alterations specifically induced by HCV: ViCs (blue), CVs (purple) and DMVs (red). A QuickTime movie of the 3D reconstruction of this cell is also provided as supplemental material, to improve visualization of the spatial distribution of these structures within the cell
Discussion
Positive-strand RNA viruses are known to rearrange host cellular membranes to facilitate viral genome replication. However, the biogenesis and 3D organization of these membrane alterations and their relationship to the various stages of the infectious cycle remain unclear. Viruses from the Flaviviridae family, including the Kunjin virus (KUNV)—the Australian variant of the West Nile virus (WNV)—and Dengue virus (DENV), induce morphologically different membrane structures that probably play distinct roles in the viral infectious cycle [27, 28]. Immunolabeling studies with an anti-dsRNA antibody have suggested that membrane sacs containing small vesicles, known as vesicle packets (VPs), are the site of replication, whereas large convoluted membranes (CMs) are the site of viral polyprotein processing [29, 30]. In this study, we showed that HCV produces three morphogically different types of vesicle (i.e. ViCs, CVs, and DMVs), which appear as soon as 1 day after infection. The increase in the number of CVs observed 2 days after cell infection was correlated with an increase in intracellular HCV RNA levels. This correlation suggests a possible role for CVs in the early stages of viral replication. The presence of NS5A in these CVs, as demonstrated by immunogold staining, is consistent with this hypothesis. We also observed that the number of DMVs gradually increased over time. Further investigations are required, but this observation suggests that DMVs may be derived from CVs. Interestingly, a similar filiation is now proposed for the poliovirus and coronavirus models [31, 32]. DMVs were initially thought to carry the coronavirus replication complexes [16], but it has been shown that DMVs contain mostly dsRNA associated with several nonstructural proteins, and that their contents do not appear to be connected to the cytoplasm [31]. These previously reported findings for various viral models suggest that DMVs may help to conceal the viral RNA, enabling the virus to evade the dsRNA-triggered antiviral responses of the host, such as those mediated by the dsRNA-dependent protein kinase. In the coronavirus model, viral replication seems to occur preferentially in the membrane rearrangements known as convoluted membranes, which may be the precursors of the coronavirus-induced DMVs [31]. Moreover, a recent study in the poliovirus model demonstrated that DMVs are the most prevalent structures at late stages of the infectious cycle and that they originate from single-membrane structures that predominate during the exponential phase of viral RNA synthesis [32]. It remains unclear whether the similar ultrastructural changes observed in cells in which HCV, coronaviruses, and polioviruses are replicating reflect a similar intracellular organization. In the HCV model, DMVs may be the site of active viral replication, as most of the dsRNA signal obtained in our immuno-EM analysis of HCV-infected cells on day 6 post infection was located within the DMVs or at DMV membranes. It remains unclear whether these DMVs also help to conceal the replicated RNA from the antiviral defense system of the host cell, as suggested in other viral models. Interestingly, in our previous EM investigations of Huh7.5 cells stably carrying a highly replicative JFH-1 subgenomic replicon, ViCs and CVs were rarely observed, as these cells contained almost exclusively DMVs. However, in this particular model, cells are analyzed after the definitive establishment of HCV replication. This supports the hypothesis that DMVs are derived from other membranous alterations and constitute the main membrane alteration associated with well-established HCV replication.
We did not detect the presence of viral components in the ViCs by immuno-EM. The lower frequency of these structures than of other membrane alterations and the difficulties involved in identifying them in cells prepared by freeze substitution may account for this finding. Alternatively, although ViCs were found specifically in HCV-infected cells, they may be free of viral proteins and dsRNA. A remarkable and intriguing feature of these ViCs was the frequent presence of an intravesicular invagination. Further investigations are required to determine whether this particular feature is also linked to the biogenesis of DMVs, acting as the source of the future double membrane. The 3D reconstruction of whole HCV-infected cells demonstrated that all these membrane rearrangements were tightly connected and closely associated with the LD clusters. It has been shown that HCV core protein accumulates on the surface of LDs, and this attachment is linked to the production of infectious viruses in HCV-infected cells [26]. NS5A is a membrane-associated RNA-binding protein involved in the HCV replication complex that has also been found on the surface of LDs and shown to be involved in HCV assembly [33]. According to recent models, viral assembly involves an interaction between HCV core and RNA-loaded NS5A [34]. However, it remains unclear whether this interaction occurs at the surface of LDs or at adjacent host cell membranes [34]. Our immuno-EM and 3D EM reconstructions of HCV-infected cells suggest that the CVs surrounding the LDs may constitute the membranous platform for viral assembly. However, we were unable to visualize unambiguously the formation of viral particles in infected cells and in CVs in particular, despite the meticulous observation of serial sections from several entire cells. Further efforts are required to improve our understanding of the HCV–host cell interactions leading to viral replication and assembly.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by a grant from the ANRS (Agence Nationale pour la Recherche sur le Sida et les hépatites virales), France. P.F. was supported by a PhD fellowship from the ANRS. We thank Dr Charles Rice and Dr Takaji Wakita for providing us with the Huh7.5 cell line and the JFH-1 virus strain, respectively. We thank Fabienne Arcanger and Juliette Rousseau for technical assistance with EM sections. We thank Catherine Gaudy and Alain Goudeau for HCV RNA quantifications and Christophe Hourioux and Jean-Christophe Meunier for helpful discussions on this work. Our data were obtained with the assistance of the RIO Electron Microscopy Facility of François Rabelais University.
Abbreviations
- BSA
Bovine serum albumin
- CM
Convoluted membranes
- CV
Contiguous vesicles
- DENV
Dengue virus
- DMEM
Dulbecco’s modified Eagle medium
- DMV
Double-membrane vesicle
- EM
Electron microscopy
- FFU
Focus forming unit
- HCV
Hepatitis C virus
- KUNV
Kunjin virus
- LD
Lipid droplet
- MAbs
Mouse monoclonal antibody
- MMV
Multimembrane vesicles
- MVB
Multivesicular bodies
- ViC
Vesicles in cluster
- VP
Vesicle packets
- WNV
West Nile virus
Footnotes
E. Blanchard and P. Roingeard are co-last authors.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 3.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard E, Brand D, Trassard S, Goudeau A, Roingeard P. Hepatitis C virus-like particle morphogenesis. J Virol. 2002;76:4073–4079. doi: 10.1128/JVI.76.8.4073-4079.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jirasko V, Montserret R, Appel N, Janvier A, Eustachi L, Brohm C, Steinmann E, Pietschmann T, Penin F, Bartenschlager R. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J Biol Chem. 2008;283:28546–28562. doi: 10.1074/jbc.M803981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 8.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 9.Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouttenoire J, Roingeard P, Penin F, Moradpour D. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J Virol. 2010;84:12529–12537. doi: 10.1128/JVI.01798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 13.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 15.Schlegel A, Giddings TH, Jr, Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;10:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J Virol. 2011;85:6963–6976. doi: 10.1128/JVI.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rorh K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 21.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hourioux C, Ait-Goughoulte M, Patient R, Fouquenet D, Arcanger-Doudet F, Brand D, Martin A, Roingeard P. Core protein domains involved in hepatitis C virus-like particle assembly at the endoplasmic reticulum membrane. Cell Microbiol. 2007;9:1014–1027. doi: 10.1111/j.1462-5822.2006.00848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depla M, Uzbekov R, Hourioux C, Blanchard E, Le Gouge A, Gillet L, Roingeard P. Ultrastructural and quantitative analysis of the lipid droplet clustering induced by hepatitis C virus core protein. Cell Mol Life Sci. 2010;67:3151–3161. doi: 10.1007/s00018-010-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoops K, Swett-Tapia C, van den Worm SHE, te Velthuis AJW, Koster AJ, Mommaas AM, Snijder EJ, Kikkert M. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J Virol. 2010;84:833–846. doi: 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikate M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 27.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol. 2010;84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly site. Cell Host Microbe. 2010;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 30.Westaway EG, Mackenzie JM, Khromykh AA. Kunjin RNA replication and applications of Kunjin replicons. Adv Virus Res. 2003;59:99–140. doi: 10.1016/S0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- 31.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belov GA, Nair V, Hansen BT, Hoyt FH, Fisher ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:303–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.