Abstract
Zellweger spectrum disorder (ZSD) is a disease continuum that results from inherited defects in PEX genes essential for normal peroxisome assembly. These autosomal recessive disorders impact brain development and also cause postnatal liver, adrenal, and kidney dysfunction, as well as loss of vision and hearing. The hypomorphic PEX1-G843D missense allele, observed in approximately 30% of ZSD patients, is associated with milder clinical and biochemical phenotypes, with some homozygous individuals surviving into early adulthood. Nonetheless, affected children with the PEX1-G843D allele have intellectual disability, failure to thrive, and significant sensory deficits. To enhance our ability to test candidate therapies that improve human PEX1-G843D function, we created the novel Pex1-G844D knock-in mouse model that represents the murine equivalent of the common human mutation. We show that Pex1-G844D homozygous mice recapitulate many classic features of mild ZSD cases, including growth retardation and fatty livers with cholestasis. In addition, electrophysiology, histology, and gene expression studies provide evidence that these animals develop a retinopathy similar to that observed in human patients, with evidence of cone photoreceptor cell death. Similar to skin fibroblasts obtained from ZSD patients with a PEX1-G843D allele, we demonstrate that murine cells homozygous for the Pex1-G844D allele respond to chaperone-like compounds, which normalizes peroxisomal β-oxidation. Thus, the Pex1-G844D mouse provides a powerful model system for testing candidate therapies that address the most common genetic cause of ZSD. In addition, this murine model will enhance studies focused on mechanisms of pathogenesis.
Keywords: Bile Acids, Photoreceptor Degeneration, Peroxisome, PEX1, Retinopathy, Zellweger Spectrum Disorder
1. INTRODUCTION
Peroxisomes are subcellular organelles that play a vital role in the metabolism of lipids, amino acids, and other substrates crucial to the development and maintenance of the central nervous system and other organ systems [1]. PEX proteins (peroxins) cooperatively function to assemble peroxisomes, including the import of over 50 different enzymes with PTS1 or PTS2 peroxisome targeting sequences [2]. Sixteen PEX genes are known to be involved in the formation of functional human peroxisomes and fourteen of these genes are associated with human disease [3-5]. An inherited defect in any one of twelve PEX genes is associated with Zellweger Spectrum Disorder (ZSD).
ZSD is a preferred umbrella term that encompasses three autosomal recessive conditions described prior to discovery of their shared peroxisomal etiology was known: Zellweger syndrome (ZS), neonatal adrenoleukodystrophy (NALD) and infantile Refsum disease (IRD) [6-9]. Zellweger syndrome is associated with null PEX gene mutations and is distinguished by congenital developmental abnormalities including neuronal migration defects, renal cortical cysts, eye malformations and chondrodysplasia punctate [10, 11]. These developmental abnormalities highlight the role of peroxisome functions in fetal development. These patients rarely survive the first year of life due to cerebral dysgenesis. In contrast, the majority of ZSD patients have intermediate and milder phenotypes associated with hypomorphic PEX gene mutations that encode peroxins with residual function. Generally NALD and IRD patients are born without major malformations, but have a progressive disease due to ongoing peroxisome dysfunction. This progression includes the development of leukodystrophy, adrenal insufficiency, retinal pigmentary changes leading to blindness, sensorineural hearing loss, enamel dysplasia in secondary teeth and osteopenia leading to pathological fractures [12, 13]. Liver dysfunction, common in the severe form of ZSD, is a postnatal event [14]. Although failure to thrive and developmental delays are frequently observed, some patients have been reported with normal or near normal cognition that extends into adulthood [15, 16].
Approximately 60-70% of ZSD patients have PEX1 defects due to two common PEX1 mutations found in individuals of European descent [17]: (1) PEX1 c.2098insT (p.700fs) which is a null allele [18], and (2) PEX1 c.2528G>A (p.Gly843Asp or p.G843D) which encodes an unstable, misfolded protein that has residual activity and is associated with a milder clinical phenotype [19, 20]. Homozygosity for PEX1-700fs is associated with a severe clinical and biochemical phenotype. In contrast, heterozygosity for PEX1-G843D is associated with an intermediate phenotype and homozygosity for this allele is associated with some of the mildest phenotypes reported [15, 19]. Overall about 20-30% of ZSD patients carry at least one PEX1-G843D allele [21].
Most ZSD patients have progressive disorders in the intermediate and milder end of the spectrum and should benefit from therapeutic interventions that can halt or ameliorate further deterioration [2]. Improving the function of a mildly impaired PEX protein could help protect against development of leukodystrophy and blindness. Previous reports have shown that residual PEX1-G843D protein function can be boosted by treatment with small molecules with chaperone-like properties [22]. We have shown that ZSD patient cultured fibroblasts with at least one PEX1-G843D allele show improved peroxisome assembly in response to chemical chaperones such as dimethylsulfoxide (DMSO), glycerol, trimethylamine N-oxide (TMAO), and betaine. Furthermore, we have performed small molecule library screens that identified other drugs that improve PEX1-G843D function and may act as pharmacological chaperones [23].
Here, we report the generation of the murine equivalent model of mild ZSD caused by the common human PEX1-G843D allele. This represents the first knock-in mouse model of a peroxisomal disorder, which complements existing Pex gene knock-out mouse models that mimic the severe ZS phenotype [24-26]. We describe the initial characterization of this model that faithfully recapitulates multiple aspects of more mildly affected ZSD patients. In addition, we demonstrate the responsiveness of cultured cells, derived from these mice, to general chemical chaperones, the result of which are similar to observations made in human PEX1-G843D cells. Overall, the Pex1-G844D mouse provides a valuable new animal model for testing candidate drugs and gene therapies, as well as a platform for exploring pathogenetic mechanisms of peroxisomal dysfunction over postnatal life.
2. MATERIAL AND METHODS
2.1. Mouse Care
Mice were maintained in animal care facilities at Johns Hopkins School of Medicine. The Johns Hopkins Animal Care and Use Program was in compliance with the Animal Welfare Act and Public Health Service policies and accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. The Johns Hopkins Animal Care and Use Committee approved the protocol M012M107 and the competency of the study participants directly involved in the care of the mice used in this study. The mice had unrestricted access to food and water. Weaning was initiated at 21 days postnatal with DietGel and HydroGel supplements (Clear H2O, Portland, ME) during the transition to assure that animals were sufficiently hydrated and fortified.
2.2. Generation of Pex1-G844D Mice
The inGenious Targeting Laboratories (Stony Brook, New York) was contracted to generate Pex1-G844D heterozygous mice. We designed the construct to substitute c.2531G>A within exon 15, introducing the amino acid substitution p.G844D (Supplemental Figure S1). We also introduced loxP sites in introns 11 and 13 for future crosses with Cre recombinase mice for the creation of Pex1 conditional knock out animals The construct was electroporated into hybrid (129xC57BL/6) embryonic stem (ES) cells and selected with neomycin. ES clones were screened for homologous recombination by Southern blot analysis. Recombinant clones were injected into blastocysts for implantation. Chimeras were bred with LP (C57BL/6) transgenic mice to eliminate the neomycin cassette. Germ line transmission of the knock-in mutation was confirmed in one male mouse and one female mouse and was the basis of establishing a breeding colony. InGenious Targeting Laboratory performed these steps. Heterozygotes were bred together and with C57BL/6N wild type mice. All of the results reported in section 3 are based on mice bred on this mixed background.
2.3. Genotyping Methods
Genomic DNA isolated from tail snips of the initial heterozygous mice was PCR amplified and sequenced to confirm the presence of the correct nucleotide transition (results not shown). For routine genotyping genomic DNA was isolated from 3mm sterilely collected tail snips between 7-21 days postnatal using Direct PCR Lysis reagent (Viagen Biotech) spiked with Proteinase K following manufacturer instructions. Genotypes were ascertained by PCR amplification (forward primer 5’-TCCCTGCTCACTTCRGGAC-3’; reverse primer 5’-TAGGCAAGCTCTTTATCACC-3’) that produce products with a size difference associated with a small residual Neo cassette fragment (177bp) in an intron near the Pex1-G844D mutation. The Pex1-G844Dallele yields a 407-base pair product and a wild type allele yields a 230-base pair product.
2.4. Biochemical Assays
C26:0-lysophosphatidylethanolamine (C26:0-LPC) and phosphatidylethanolamine (PE) plasmalogen species were extracted and measured by liquid chromatography tandem mass spectrometry (LC-MS/MS) as previously described [27-29]. Bile acid intermediates were also measured by LC-MS/MS [30]. Plasma total lipid fatty acids were measured by capillary gas chromatography mass spectrometry [31]. Assays in cultured dermal fibroblasts for total lipid very long chain fatty acids (VLCFA), plasmalogen biosynthesis, phytanic acid oxidation, pristanic acid oxidation, and immunocytochemistry were performed as previously described [16, 32-34]. Catalase latency [34] was performed by treating harvested fibroblasts with digitonin (10ng/μl in 0.25M sucrose, 10mM TRIS, pH 7.0)- this concentration made the cytoplasmic membrane leaky, but did not affect the permeability of the peroxisomal membrane. Cells were separated into a supernatant fraction which contained cytosolic enzymes and a pellet fraction which contained organelle-associated enzymes for the spectrophotometric determination of catalase activity. Titanium oxylulfate was used as an indicator for H2O2 and results were reported as the percentage cytosolic catalase i.e. [cytosolic catalase activity ÷ (cytosolic + membrane catalase activity)]. VLCFA measurement in total lipid and phospholipid fractions was performed as previously described [35].
2.5. Chaperone Testing
Dermal fibroblast cultures were established from seven-day-old pups using hairless abdominal skin. Cells were sub-cultured into T25 flasks with 10% fetal bovine serum-Eagle's minimum essential media (FBS-EMEM). The next day, the media was replaced with FBS-EMEM, FBS-EMEM supplemented with 1% DMSO or FBS-EMEM supplemented with 200mM TMAO. Cells were cultured until the untreated cells reached 100% confluence, which occurred at 7-10 days of growth. Fibroblasts were harvested and processed for VLCFA analysis [16].
2.6. Liver Histology
Liver was obtained from 15-day-old postnatal mice cardiac perfused with 4% paraformaldehyde in phosphate buffered saline (pH 7.4). The liver was post-fixed overnight at 4°C in the same fixative, paraffin embedded, sectioned at 10μm, and stained with hematoxylin-eosin following standard procedures.
2.7. Retinal Histology
Three-week-old and 22-week-old mouse eyes were fixed with 1% paraformaldehyde (PFA) in 0.1M PB at room temperature (RT) for 1 hour (h). Eyecups were then prepared, cryoprotected in increasing concentrations of sucrose from 5% to 20% in 0.1 M PB, embedded in a 20% sucrose/OCT compound (Sakura Finetek USA, Torrance, CA) mix at 2:1, and quick frozen. Retinal sections were cut at 10μm and placed on Superfrost slides. The retinal sections of 3-week mice were pre-incubated in 10% horse serum for 1 h at RT, and then incubated with rhodamine labeled peanut agglutinin (PNA) (1:1,000; Vector Laboratory, Burlingame, CA) in 3% horse serum for 1 h at RT. After the incubation, sections were mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratory). The retinal sections of 22-week mice were preincubated in 10% horse serum for 1 h at RT, and then incubated with an antibody against rhodopsin (1:200; Gene Tex, Irvine, CA) overnight at 4°C. After incubation with Alexa Fluor 488-conjugated anti-mouse IgG (1:500; Invitrogen, Carlsbad, CA) and rhodamine labeled PNA (1:1,000) for 1 h at RT, sections were mounted with VECTASHIELD Mounting Medium with DAPI. Images were taken on the LSM 510 Meta laser-scanning microscope (Carl Zeiss, Thornwood, NY).
2.8. Hearing Assessment
Auditory brainstem response (ABR) thresholds were used to screen mice for hearing loss. Mice were tested using established protocols [36, 37]. Briefly, mice were anesthetized with 100 mg/kg ketamine and 20 mg/kg xylazine (intraperitoneal) and placed inside a small sound-attenuating chamber. Core temperature was maintained at 37±1°C. ABRs were recorded differentially from the scalp using subcutaneous platinum needle electrodes placed over the left bulla and at the vertex of the skull. A ground electrode was inserted into the leg muscle. The animals were placed 30 cm from a speaker. Responses were amplified, filtered, and averaged over 300 stimulus presentations. Thresholds to broadband clicks and 8, 16, and 32 kHz pure tones were measured by playing sounds in a descending level series until no response emerged from the baseline noise.
2.9. Electroretinogram (ERG) Measurement
After overnight dark adaptation, mice were anesthetized with 100 mg/kg ketamine and 5 mg/kg xylazine under dim red light and placed on a heating pad during the ERG recordings. Pupils were dilated with 1% tropicamide and 2.5% phenylephrine. A corneal contact lens electrode with a gold ring on the inner surface was placed on the cornea with 2.5% hydrocellullose. The reference and ground electrodes were placed subcutaneously under the scalp and the tail, respectively. The mice were placed in a Ganzfeld bowl and stimulated by 10-20 stroboscopic LED flashes. Signals were detected, amplified, band pass filtered between 0.3 and 500 Hz, and analyzed using a UTAS Visual Electrodiagnostic Testing System (LKC Technologies, Gaithersburg, MD). Scotopic ERGs were recorded with increasing stimulus from −5.4 to 1.8 log cd-s/m2. The inter-stimulus interval was set to 3-60 seconds to avoid light adaptation. Photopic ERGs were recorded under 40 cd/m2 white background light to desensitize rod responses. White light stimulus ranging from −0.2 to 1.8 log cd-s/m2 was presented to elicit cone responses.
2.10. General Statistical Analysis
Student's T-test was used for statistical comparison between controls and Pex1-G844D homozygous mice. In all cases, comparisons were processed as unpaired analyses and one-tailed tests, because we expected the homozygote value to be higher or lower than the control group based on the known functional role of Pex1 in peroxisomal metabolism. The F-test was used to determine whether the two arrays to be compared were of equal or unequal variance. Chi square analyses were performed to assess whether genotypes and sex ratios were within expected ranges.
2.11. Gene Expression Profiling
Total RNA was isolated from mouse retina dissected at 124 days of age and subjected to global gene expression analysis on GeneChip® Mouse Genome 430a2.0 Arrays (Affymetrix, Santa Clara CA, USA) designed to interrogate over 14,000 transcripts. As previously described [38], the resulting .CEL files were preprocessed using the WebArray software [39] that uses the RMA (Robust Multi-array Average) algorithm [40] to generate log2-scaled expression values for each transcript. Using the LIMMA (Linear Models for Microarray) package [41], we selected probe sets showing absolute fold change greater than 1.2, and a false discovery rate (FDR) value less than 0.05. The FDR values were calculated by adjusting raw P-values using the SPLOSH (Spacing LOESS Histogram) method [42]. Original .CEL data files are available for download from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under GEO accession number 52348 and processed gene expression scores are available in Supplemental Table S1.
3. RESULTS
In the following results sections, unless stated otherwise, data is reported for Pex1-G844D homozygotes and wild type mice. In all instances we also performed the same studies in Pex1-G844D heterozygotes. The results from wild type and Pex1-G844D heterozygotes were similar and, thus, in most cases the results from the heterozygous mice are not reported.
3.1. Mouse Pex1 versus Human PEX1 Proteins
The murine Pex1 protein (Q5BL07.2) is 1,284 amino acids in length in contrast to the orthologous human PEX1 protein (NP_000457.1) that is one amino acid shorter. Due to this size difference, the murine Pex1-G844D amino acid residue is orthologous to the human PEX1-G843D residue. Overall, these proteins share 82% identity. The orthologous human PEX1-G843 and mouse Pex1-G844 residues reside in a region of 65 consecutive amino acids that are identical in both species (Supplemental Figure S2).
3.2. G844D Heterozygote Paired Mating Outcomes
In the first six months of breeding Pex1-G844D heterozygotes, 91 pups had genotypes that matched the Mendelian expected ratios for an autosomal recessive disorder: 23 wild type / wild type, 44 Pex1-G844D / wild type, and 24 Pex1-G844D / Pex1-G844D (Q=0.7281, P>0.25). This indicates that all genotypes survived until birth. The male to female sex ratio was 0.82:1 and is within the 1:1 expectation (Q=0.3454, P>0.5).
3.3. Survival
We assessed survival rates from 33 litters of mice conceived in the first 14 months of breeding (Table 1), excluding mice sacrificed for use in other experiments or to cull litters. The longest surviving Pex1-G844D homozygotes were over 1 year of age with no difference in long-term survival between males and females. The Pex1-G844D homozygous mice fell into shorter (n=20, median age of death was 22 days) and longer-term (n=29, current median age is 271 days) survival groups. Amongst the wild type and Pex1-G844D carrier mice, there were only a few cases of premature death (i.e. survival less than 404 days) due to natural causes, whereas 20 of 49 (41%) homozygotes died prematurely. No specific cause of premature death was evident.
Table 1.
Survival in 33 litters of Pex1-G844D heterozygote matings
| Died of Natural Causes |
Still Alive |
|||||
|---|---|---|---|---|---|---|
| Number of Micea | Ageb |
Number of Mice | Ageb |
|||
| Genotype | Range | Median | Range | Median | ||
| Pex1-G844D Homozygote | 20 | 12-117 | 22 | 29 | 174-404 | 271 |
| Pex1-G844D Heterozygote | 3 | 0-266 | 117 | 30 | 219-404 | 286 |
| Wild type | 1 | 0 | n/ac | 21 | 223-404 | 253 |
Some mice were euthanized to reduce the number of pups in a given litter or were not needed at the time for further study. These euthanized mice are not included in these numbers. n/a = not applicable.
Age is presented in days.
n/a = not applicable.
3.4. Growth Curves
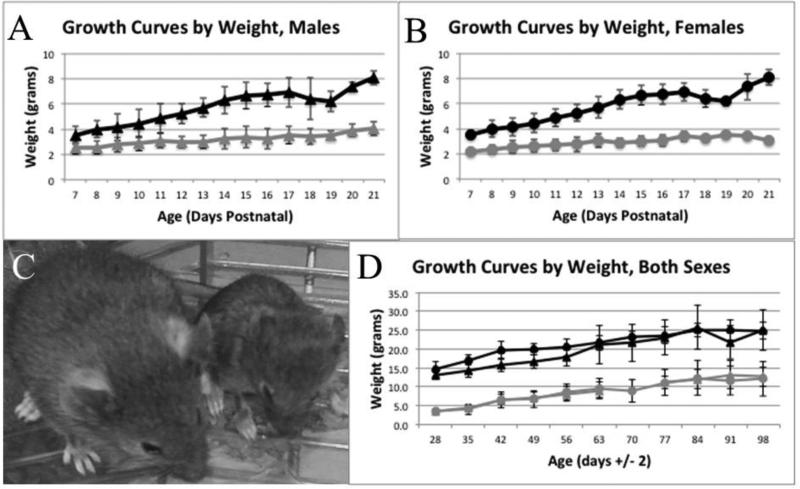
We measured total body weights on pups from postnatal day 7 through 21. On average, 23 male and 30 female controls and 10 male and 7 female homozygotes were weighed at each time point (Figure 1A and 1B). The body weight of homozygous male and female pups was significantly lower than their control littermates. Even at postnatal day 7 there was a statistically significant lower weight in the homozygotes compared to the controls. In males, the controls weighed 3.64±0.59g (n=21) and the homozygotes weighed 2.52±0.49g (n=9) [P<3×10−5]. Similarly in the female mice, the controls weighed 3.60±0.76g (n=18) and the homozygotes weighed 2.15±0.26g (n=4) [P<0.001]. In the first three postnatal weeks, Pex1-G844D homozygotes showed marked growth retardation with minimal increase in weight, in contrast to the normal steady weight increase observed in control mice. The side-by-side comparison of a Pex1-G844D heterozygote and Pex1-G844D homozygote mouse highlights the stunted growth in the homozygous animals (Figure 1C). We also recorded bodyweights on a subset of mice on a weekly basis from P28 through P98 (Figure 1D). Overall, the control mice continue to show better growth than the homozygous mice. By P98, there was more variation within each group, but the homozygotes and controls were clearly on different growth paths (at P98 males P=0.0077 and females P=8.25×10−5). Based on a two-week feeding study in adult mice, the homozygotes (n=2) and controls (n=2) consumed similar grams of chow per day per grams of body weight, 0.15 and 0.14, respectively.
Figure 1.
Growth from 7 to 98 Days Postnatal. For panels A, B and D, circles denote males, triangles denote females; black denotes control mice, grey denotes Pex1-G844D homozygotes. Panels A and B show the weight in grams of pups from 7 to 21 days postnatal for males (average n = 23 controls and 10 homozygotes) and females (average n = 30 controls and 7 homozygotes), respectively. Panel C shows 21-day-old littermates and highlights the significant growth retardation observed in homozygotes. The mouse on the left is a Pex1-G844D heterozygote and on the right is a Pex1-G844D homozygote and highlights the significant growth retardation observed in homozygotes. Panel D shows the weight in grams of mice from 28 to 98 days postnatal. Males (average n = 13 controls and 5 homozygotes) and females (average n = 14 controls and 4 homozygotes) are shown together.
3.5. Biochemistry
Total lipid fatty acid profile (total indicates fatty acids belonging to all lipid classes) analysis, including C8-C30 saturated, monounsaturated and polyunsaturated fatty acids, was performed in plasma collected from three Pex1-G844D homozygous and seven control mice from 9-28 days of age. Most of the plasma samples collected were slightly hemolyzed. Statistical analysis using unpaired Student t-test was performed on all of the parameters calculated with significant findings summarized in Table 2. The percentage of C26:0, C26:1, and C26:2 fatty acids were higher in the Pex1-G844D homozygote relative to the control group. The triene/tetraene ratio was elevated in homozygotes compared to control mice, but was not in the range (>0.2) associated with essential fatty acid deficiency [43]. Although the essential fatty acid precursors for synthesis of arachidonic acid (20:4n-6) and docosohexaenoic acid (22:6n-3 or DHA) were within normal limits, there was a trend for the homozygotes to have lower DHA levels (2.79±1.25 homozygotes, 5.95±2.75 controls, P=0.09). Interestingly, all of the C22 polyunsaturates upstream to DHA synthesis were significantly elevated in the homozygote relative to the control group. In addition, the 20:4 / 22:6 fatty acid ratio was significantly elevated in the homozygotes compared to the controls. Overall, these results are consistent with impaired DHA synthesis in the Pex1-G844D homozygote group.
Table 2.
Plasma fatty acid, blood spot lipid and skin fibroblast analyses
| Parameter | Homozygotes | Controls | P value |
|---|---|---|---|
| Plasma Total Lipid Fatty Acids |
|||
| 20:4 / 22:6 | 4.99 ± 1.30a (n=3) | 2.38 ± 0.19 (n=7) | 1.1×10−5 |
| Triene/Tetraeneb | 0.011 ± 0.006 (n=3) | 0.006 ± 0.001 (n=7) | 0.021 |
| C26:0 % | 0.033 ± 0.011 (n=3) | 0.021 ± 0.006 (n=7) | 0.013 |
| C26:1 (n-7 & n-9) % | 0.028 ± 0.005a (n=3) | 0.011 ± 0.006 (n=7) | 0.001 |
| C26:2 % | 0.018 ± 0.009a (n=3) | 0.008 ± 0.004 (n=7) | 0.019 |
| C22:2 (n-6) % | 0.109 ± 0.027a (n=3) | 0.044 ± 0.015 (n=7) | 9.3×10−5 |
| C22:4 (n-6) % | 0.841 ± 0.216a (n=3) | 0.308 ± 0.109 (n=7) | 4.8×10−5 |
| C22:5 (n-6) % | 0.944 ± 0.666a (n=3) | 0.165 ± 0.050 (n=7) | 9.6×10−4 |
| C22:5 (n-3) % | 0.721 ± 0.196 (n=3) | 0.453 ± 0.144 (n=7) | 0.020 |
| Blood Spot Lipids |
|||
| C26:0-LPC % | 0.558 ± 0.363a (n=17) | 0.052 ± 0.016 (n=42) | 9.86×10−13 |
| PE-Plasmalogensc | 2.82 ± 2.22 (n=17) | 5.97 ± 2.34 (n=42) | 1.42×10−5 |
| Skin Fibroblast Analysesd |
Δ
f
|
||
| C26:0 (μg/mg) | 0.465 | 0.023 | 20.2 |
| C26:1 (μg/mg) | 0.213 | 0.023 | 9.3 |
| C26:0/C22:0 | 1.298 | 0.053 | 24.5 |
| Phytanic Acid Oxidatione | 119.1 | 582.3 | −5.0 |
| Pristanic Acid Oxidatione | 300.0 | 487.1 | −1.67 |
| Plasmalogen Synthesis | 0.83 | 0.69 | 1.2 |
| % Cytosolic Catalase | 83.3 | 11.6 | 7.2 |
There was no overlap in the values measured in the homozygote and control groups.
The Triene to Tetraene ratio is (Mead Acid, 20:3n-9) / (Arachidonic Acid, 20:4n-6) and is a marker of essential fatty acid deficiency.
This is the sum of four phosphatidylethanolamine (PE) plasmalogens: 16:0p/20:4; 16:0p/18:1; 18:1p/20:4; 18:0p/20:4.
All results are an average of duplicate analyses.
Units of activity is reported as pmol / 48 hours / mg protein.
Δ=Fold difference.
Blood spots collected from 21-day-old mouse pups were solvent extracted for LC-MS/MS to measure the metabolites C26:0-LPC and four PE plasmalogens: 16:0p/20:4; 16:0p/18:1; 18:1p/20:4; 18:0p/20:4. More samples were available from wild type mice than homozygous mice, as it was not always possible to collect enough spotted blood from the small, growth-retarded, homozygous mice. The mean amount of C26:0-LPC was 10-fold elevated in homozygotes in comparison to the controls (Table 2); heterozygote values are not shown, but were identical to the wild type results. There was no overlap of the homozygote C26:0-LPC level compared to the control group level (P<9.9×10−13). Furthermore, the mean PE plasmalogen level in the homozygotes was 0.53-fold reduced relative to the controls (homozygote versus wild type, P=1.4×10−5; homozygote versus heterozygote, P=5.7×10−5). Nevertheless, there was some overlap in the range of PE plasmalogen levels between the Pex1-G844D homozygotes and the controls. This assay was developed for the newborn screening of X-linked adrenoleukodystrophy and ZSD [27] and could be useful for monitoring the response of mice to treatment during the course of therapeutic trials.
Skin fibroblast cell lines were established from genotyped mouse pups and tested using the standard clinical biochemical assays used to characterize human patients suspected to have a ZSD (Table 2). VLCFA were significantly elevated and the oxidation of the branched chain fatty acids phytanic acid and pristanic acid were reduced in cultured fibroblasts from Pex1-G844D homozygotes compared to controls. In contrast, plasmalogen synthesis was normal. In addition, immunocytochemical analysis with an antibody against the PEX14 peroxisomal membrane protein showed peroxisomes of normal size and number in the homozygote fibroblasts (supplemental Figure S3). However, these peroxisomes clearly had an import defect because the percentage of cytosolic catalase, a measure of import of this peroxisomal matrix protein, was significantly elevated in the homozygote cell line.
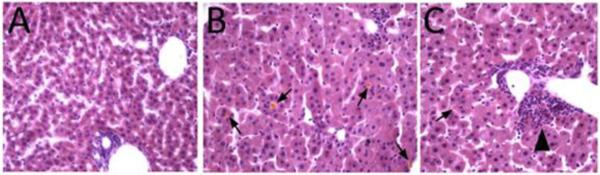
Consistent with liver peroxisome dysfunction and observations in human ZSD patients, the bile acid intermediates dihydroxycholestanoic acid (DHCA) and trihydroxycholestanoic acid (THCA) were significantly elevated in all tissues tested-feces, plasma and liver homogenate (Table 3). Early postnatal homozygous mice had evidence of severe fat malabsorption, with bulky yellow-colored large intestinal contents, similar to that observed in Pex2−/− mice. In addition, histologic sections of liver of 15-day postnatal mice showed increased lipid droplets in hepatocyte cytoplasm versus control mice, and many cholestatic deposits and foci of bile ductular proliferation in the homozygote livers (Figure 2).
Table 3.
Relative bile acid levels in affected mutant and healthy mice
| Intermediates: | THCA / Cholic Acid |
DHCA / Deoxycholic Acid |
||
|---|---|---|---|---|
| Genotype: | Normala | G844D / G844D | Normala | G844D / G844D |
| Liver Homogenates | 0.005c | 2.1-65.3d | 0.34c | 6.5-37.3d |
| Feces | 0.0008-0.03e | 0.19-1.74f | 0.01-0.90e | 4.15-38.59f |
| Plasmab | 0.02-0.25g | 1.41h | 0.02-0.21g | 8.7h |
Normal includes completely wildtype and Pex1-G844D heterozygous mice.
Reported as THCA or DHCA per 10μl plasma and NOT as a ratio to the corresponding primary bile acid.
n=1
n=2
n=11
n=4
n=8
n=1.
Figure 2.
Liver Histology at 15 Days Postnatal. Hematoxylin-eosin staining. (A) Control mouse. (B, C) Pex1-G844D homozygous mouse. Hepatocyte cytoplasmic volume is increased, associated with microvesicular fat deposition in mutant mice. Yellowish cholestatic deposits (arrows in B, C) are observed in mutant livers, and bile ductular proliferation is seen (arrowhead in C) indicating response to bile duct damage.
Total lipids were extracted from brain, spleen, lung, and kidney in at least three mice each. The mean C26:0% was elevated in spleen, lung and kidney; however, none of these differences were statistically significant. We also measured VLCFA in the phospholipid fraction isolated from these same extracts. Only spleen had a statistically significant elevation in homozygotes (0.297 ± 0.049 homozygote, 0.029 ± 0.012 controls; P=0.007). In both of these assays, the C26:0% in brain was highly similar in homozygotes and controls.
3.6. Chaperone Response
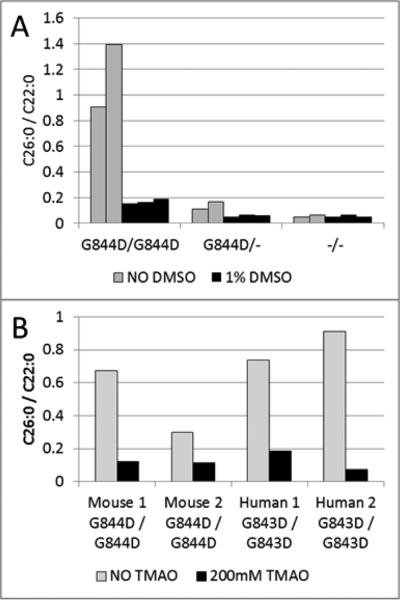
Cultured dermal fibroblasts from two different homozygotes were cultivated in the presence of 1% DMSO and 200mM TMAO for seven days to determine if chaperone-like molecules could rescue peroxisome functions. Similar to observations made in human cultured skin fibroblasts expressing the PEX1-G843D protein, the murine Pex1-G844D/Pex1-G844D cells responded to 1% DMSO with the level of C26:0/C22:0 reduced to the levels detected in the control cell lines (Figure 3A). In addition, the Pex1-G844D homozygous fibroblasts responded to TMAO similarly to human PEX1-G843D homozygous fibroblasts treated simultaneously (Figure 3B).
Figure 3.
Chaperone-molecule Response Testing in Cultured Skin Fibroblasts. Panel A shows results from three mouse fibroblast cell lines representing the three possible genotypes. Each bar in the graph represents the result from a single T25 flask. The homozygous cells treated with 1% DMSO (black bars) for seven days normalized levels of total lipid very long chain fatty acids (VLCFA) based on the C26/C22 ratio as compared to the untreated cells (grey bars). G844D homozygotes are designated G844D/G844D; G844D heterozygotes are designated G844D/-; and wild type is designated −/−. Panel B shows the results from dermal fibroblasts derived from two different Pex1-G844D homozygous mice and two human patients homozygous for PEX1-G843D. The mouse cell lines normalized VLCFA levels after treatment with 200mM TMAO (black bars); this was the same response as PEX1-G843D homozygous human fibroblasts treated simultaneously. Each bar in the graph represents the result from a single T25 flask.
3.7. Auditory Phenotype
ABR results between two Pex1-G844D homozygotes, two heterozygotes, and two wild type mice measured as a click stimulus at 8kHz, 16kHz, and 32kHz were indistinguishable. In general, all of the mice had poor hearing, in agreement with previous reports of the background strains, with a trend for the homozygous mice to have slightly better threshold responses (data not shown). Better thresholds in the Pex1-G844D homozygous mice may be attributable to their smaller head size; smaller head size likely improved the signal-to-noise ratio.
3.8. Vision Phenotypes
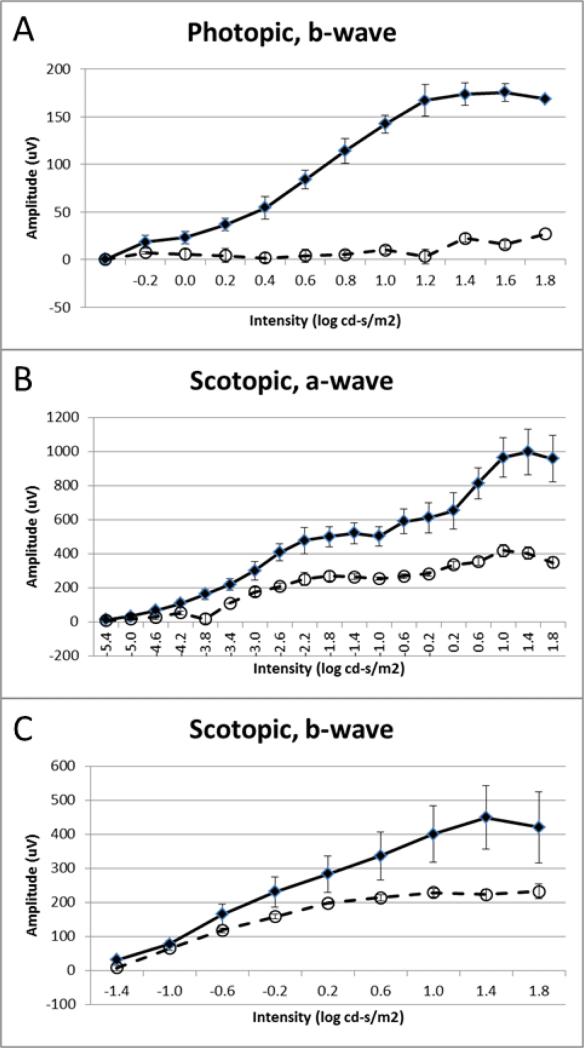
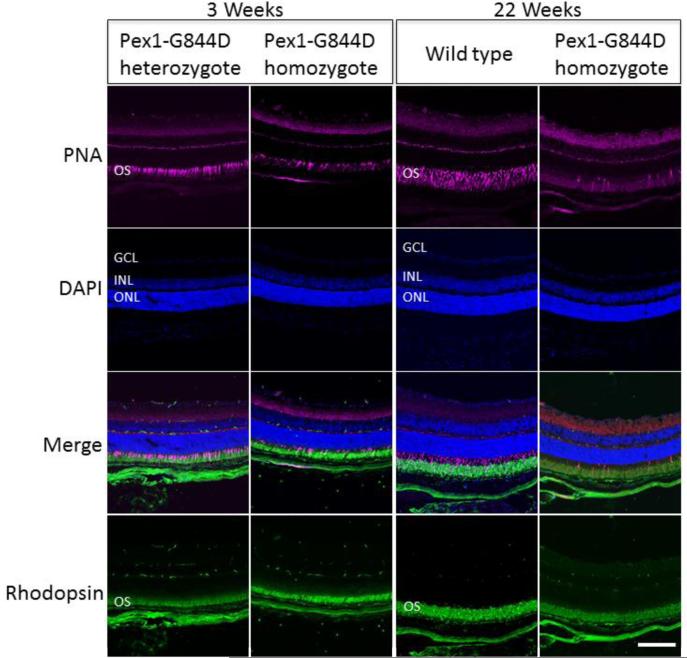
Functional analysis of cone and rod visual systems was conducted by recording ERGs under photopic and scotopic conditions, respectively. Four Pex1-G844D homozygous mice paired with wild type or heterozygous littermates between the ages of 2-4 months were tested. In the photopic ERG analysis, b-wave amplitudes were almost absent in the Pex1-G844D homozygotes (Figure 4A) indicating severe impairment of the cone visual pathway. In contrast, in the scotopic ERG analysis (Figure 4B) the amplitudes of the a-waves, which reflect rod photoreceptor activity, were not significantly reduced, although the amplitudes of the b-waves, which primarily originate from rod bipolar cells, were significantly reduced (Figure 4C). Thus, in contrast to the cone visual system, the rod visual system was relatively preserved. Consistent with these results, staining of the retinal sections with peanut agglutinin, which specifically binds to the cone inner and outer segments and the cone synaptic terminals, showed that some cone photoreceptors were retained in the Pex1-G844D homozygote retina at 3 weeks, but were completely degenerated in the adult (Figure 5). Immunostaining of the adult retina using an antibody against rhodopsin, which stains the rod outer segments, showed weaker immunoreactivity in the Pex1-G844D homozygote; this also suggested rod photoreceptor degeneration but their histology was relatively preserved compared to the cone photoreceptors.
Figure 4.
Electrophysiological Functional Analysis of Cone and Rod Visual Systems. ERG measurements were recorded in Pex1-G844D homozygous (open circles, dashed line; n=4) and wild type (black diamonds, solid line; n=4) mice at about two-months of age. A. The amplitudes of photopic b-waves. B. The amplitudes of scotopic a-waves. C. The amplitudes of scotopic b-waves. All data represented as mean ± standard error of the mean. Overall, these data indicate that the cone visual system is significantly impaired in Pex1-G844D homozygotes, whereas the rod photoreceptors are relatively preserved.
Figure 5.
Retinal Histology. Immunostaining of photoreceptor outer segments was performed for the Pex1-G884D homozygote and control mouse retinas at three-weeks and 22-weeks. Peanut agglutinin (PNA; magenta) and an antibody against rhodopsin (green) were used to stain cone and rod outer segments, respectively. Nuclei were counter-stained with DAPI (blue). Retinal layers are marked by these abbreviations: GCL, ganglion cells layer; INL, inner nuclear layer; ONL, outer nuclear layer; and, OS, outer segment. Scale bar equals100 mm. Immunostaining with the cone specific PNA in 3-weeks-old mice shows that some cone photoreceptors are preserved in Pex1-G844D homozygotes. In contrast, at 22-weeks of age PNA staining in the Pex1-G844D homozygotes is consistent with a loss of cone photoreceptors. The rhodopsin staining was not as robust in the Pex1-G844D homozygotes as the control indicating that the retinal degeneration may not be limited to the cone photoreceptors.
We conducted global gene expression analysis of retinas obtained from four homozygous mice and four heterozygous littermates. Normal retinal histology, peroxisomal biochemical profile, growth and survival in Pex1-G844D heterozygotes provided a valid basis for using them as the control group. A group of eight probe sets indicated differential expressed genes (DEGs) between these two groups (Table 4). This included six probe sets representing four unique genes (Arr3, Pde6h, Gnat2, and Opn1mw) that showed lower expression in the Pex1-G844D homozygous mice relative to the heterozygous mice. All four of these genes have been reported as being photoreceptor cone-specific genes and having reduced expression in a mouse model of retinal degeneration [44]. The decreased expression of these genes is consistent with the specific loss of photoreceptor cone cells in the retinas of Pex1-G844D homozygous mice relative to those of the controls. The two probe sets indicating increased expression in Pex1-G844D homozygous relative to heterozygous mice represented the Cyp4a14 and Ufd1l genes. Investigators reported the Cyp4 gene family is transcriptionally regulated in a PPARα/PPARγ-dependent manner with the Cyp4a14 gene product being involved in omega-oxidation of fatty acids [45]. The Ufd1l (aka Ufd1) gene plays a critical role in endoplasmic reticulum (ER)-associated degradation (ERAD) and cholesterol metabolism [46].
Table 4.
DEGs in retina tissues obtained from Pex1-G844D homozygote and heterozygote mice
| Probe ID | Symbol | Gene Description | Gene IDa | Ho Meanb | Het Meanc | Fold Changed | FDRe |
|---|---|---|---|---|---|---|---|
| 1423257_at | Cyp4a14 | cytochrome P450, | 13119 | 3.9 | 13.2 | 3.38 | 0.033 |
| family 4, subfamily a, | |||||||
| polypeptide 14 | |||||||
| 1432367_a_at | Ufd1l | ubiquitin fusion | 22230 | 28.8 | 43.2 | 1.50 | 0.075 |
| degradation 1 like | |||||||
| 1425232_x_at | Arr3 | arrestin 3, retinal | 170735 | 43.3 | 14.9 | −2.91 | 0.045 |
| 1450329_a_at | Arr3 | arrestin 3, retinal | 170735 | 124.2 | 45.5 | −2.73 | 0.005 |
| 1450765_a_at | Pde6h | phosphodiesterase 6H, | 78600 | 781.2 | 327.7 | −2.38 | 0.009 |
| cGMP-specific, cone, gamma | |||||||
| 1450766_at | Pde6h | phosphodiesterase 6H, | 78600 | 928.9 | 416.2 | −2.23 | 0.015 |
| cGMP-specific, cone, gamma | |||||||
| 1422907_at | Gnat2 | guanine nucleotide | 14686 | 417.0 | 212.0 | −1.97 | 0.061 |
| binding protein, alpha transducing 2 | |||||||
| 1419723_at | Opn1mw | opsin 1 (cone Pigments) | 14539 | 93.3 | 50.1 | −1.86 | 0.003 |
NCBI Gene ID.
Geometric mean gene expression score for Pex1-G844D homozygote retinal tissue.
Geometric mean gene expression score for Pex1-G844D heterozygote retinal tissue.
Fold change = Ho Mean / Het Mean.
False discovery rate.
4. DISCUSSION
ZSD is a disease continuum wherein patients manifest a spectrum of clinical presentations with survival ranging from early infancy (ZS), to school age (NALD), to adulthood (IRD). About 50% of patients in the Zellweger spectrum have at least one hypomorphic PEX gene mutation that yields a protein of residual function which is associated with milder clinical, biochemical and histological phenotypes [4, 47]. The PEX1-G843D missense allele is the most common hypomorphic mutation observed in ZSD patients and produces a misfolded and unstable PEX1 protein with partial activity. Although most patients with PEX1-G843D missense allele(s) survive beyond infancy, they still show progressive psychomotor retardation and hypotonia as well as visual and hearing loss. These individuals in particular may benefit from early medical interventions that either slow or halt disease progression prior to irreversible damage to multiple organ systems.
Mouse models with a complete deficiency of Pex2, Pex5, Pex13, or Pex11β gene functions have been developed. The first three models recapitulate multiple aspects of the most severe end of the ZSD disease spectrum, including absent peroxisome function, severe hypotonia, and neonatal lethality [24-26, 48]. Similar to humans, severe peroxisome deficiency is compatible with embryonic, but not postnatal, life in mice. To overcome the limitations neonatal lethality placed on testing therapies and investigating disease mechanism, mice with conditional null alleles of the Pex5 gene have been engineered and used to evaluate downstream consequences of peroxisome dysfunction in specific organs and cell types [49, 50]. Such animals can survive beyond the postnatal period and have yielded mechanistic insights into the role of peroxisomes in the development and maintenance of brain and liver cell types and functions. Nevertheless, to date, no mouse models have been reported that reflect the hypomorphic PEX alleles that are commonly found in ZSD patients on the intermediate and milder end of the Zellweger spectrum.
In our initial characterization of mice homozygous for the Pex1-G844D allele (the murine ortholog of the common human PEX1-G843D mutation) we noted growth retardation and numerous metabolic phenotypes that reflect those found in patients on the mild end of the Zellweger spectrum. The significant growth retardation in the Pex1-G844D homozygotes correlates with the poor linear growth and weight gain reported by Poll-The et al [47] in ZSD patients surviving more than one year, many of whom have at least one PEX1-G843D allele. A metabolic phenotype includes biochemical and histological evidence of a bile acid defect that is associated with intestinal fat malabsorption and cholestasis in the young Pex1-G844D homozygous mice. However, further studies on bile acids and other hepatic metabolic and morphologic defects in these more mildly affected Pex1-G844D mice are needed, as effectiveness of bile acid supplementation in Pex2−/− mice was limited by their severe hepatic metabolic defects [51].
In addition, the Pex1-G844D homozygous mice both recapitulated and informed our understanding of the mechanisms underlying vision loss in ZSD patients. Importantly, we have demonstrated that Pex1-G844D homozygotes have abnormal retinal function, as measured by ERG, associated with a loss of cone photoreceptors that were partially intact at 3 weeks of life. This indicates that, at least in mice, there is a window of opportunity where therapies that improve peroxisome assembly may be able to preserve cone cell survival and function. Although much is known about vision defects in human ZSD patients, a detailed examination of retinal photoreceptor cell loss in these individuals has not been reported [15, 52-55]. This provides an opportunity for applying the Pex1-G844D homozygous mouse model to evaluate small molecule interventions and retinal gene therapy approaches to slow down or potentially halt vision loss, approaches that could be tested in humans with milder forms of ZSD if shown to be effective in mice.
In addition to recapitulating key clinical phenotypes observed in patients with milder forms of ZSD, we have demonstrated in principle that the Pex1-G844D homozygous mice will be valuable for testing targeted therapies tailored towards the genetic basis of disease. Treatment of cultured fibroblasts from Pex1-G844D homozygotes, heterozygotes, and littermate controls with DMSO and TMAO demonstrated that the Pex1-G844D protein responded similarly to the orthologous human Pex1-G843D protein. These chemical chaperones are believed to act by stabilizing the microenvironment around the misfolded protein and enabling it to improve folding [56]. By demonstrating that the mouse mutant protein was also responsive to chaperone molecules, this model should provide a unique resource for investigating classes of compounds that enhance residual activity through chaperone actions, such as betaine that is currently in clinical trials for ZSD patients with Pex1-G843D mutations (ClinicalTrials.gov Identifier: NCT01838941). In addition, our ability to monitor key biomarkers of peroxisome function in blood spots and feces will allow us to monitor the efficacy of therapies during treatment trials.
More comprehensive analyses on the brain, retina, liver, adrenal glands, kidneys and other organs from Pex1-G844D mice could provide valuable information to guide the development of rationally designed therapies for children and young adults affected with milder forms of ZSD. For example, the Pex1-G844D mouse provides a novel platform to test hypotheses regarding the physiological impact of VLCFA accumulation in patients with ZSD and other peroxisomal disorders. Male patients with X-linked adrenoleukodystrophy have a defect in the ABCD1 gene encoding a peroxisomal membrane protein and accumulate VLCFA, but are normal at birth [57]. Nevertheless, these individuals are vulnerable to developing a leukodystrophy starting at approximately 3 years of age or pathologic changes to the spinal cord that lead to spastic paraparesis later in life [57]. Since Abcd1 knock-out mice do not develop a leukodystrophy (Lopez-Erauskin et al, 2013), we do not expect Pex1-G844D homozygotes to model the white matter degeneration that longer surviving ZSD patients may develop. Nevertheless, the Pex1-G844D mice provide an opportunity to explore the impact of peroxisome dysfunction on mouse brain development. We present evidence that DHA synthesis is impaired in homozygous Pex1-G844D mice. The importance of DHA in mammalian brain and retinal development is well established [58-60]. DHA is actively incorporated into phospholipids in the retina based on studies in multiple species, including humans, with preferential accumulation in the photoreceptors [61, 62]. Nevertheless, it is premature to say whether DHA deficiency is playing a specific functional role in the cone dystrophy we described in these animals. We also present evidence of decreased expression of genes specifically associated with cone cells. There is a need to investigate whether this is a marker of cone cell death or a direct consequence of reduced Pex1 function.
Overall, the Pex1-G844D mouse provides multiple opportunities to expand our understanding of the pathophysiology associated with mild defects in peroxisome assembly and the requirements for peroxisome functions in organ homeostasis from the postnatal period onwards. Although we have characterized Pex1-G844D homozygous mice, our model provides an opportunity to produce an animal model compound heterozygous for the Pex1-G844D hypomorphic allele and Pex1 null allele, which reflects a genotype frequently found in the human ZSD patient population. This combination of alleles is predictive of reduced peroxisome function and more severe clinical presentation relative to the Pex1-G844D homozygous mouse with two hypomorphic alleles. Furthermore, the loxP-sites present in the targeting construct used to generate the Pex1-G844D mice allow for the development of tissue and cell type specific knockouts of the Pex1 gene in the genetic background of hypomorphic Pex1 alleles. This could allow for rigorous investigations into the role varying levels of peroxisome functionality play in the cell type specific clinical phenotypes observed in ZSD patients and the pathophysiology of postnatal peroxisome dysfunction.
Supplementary Material
Highlights.
Created a knock-in mouse model that harbors Pex1-G844D
Pex1-G844D homozygotes have biochemical phenotype of Zellweger spectrum disorder
Pex1-G844D homozygotes can survive at least 14 months
Pex1-G844D homozygotes have liver phenotype similar to human patients
Pex1-G844D homozygotes have retinal phenotype similar to human patients
ACKNOWLEDGEMENTS
BAER testing was performed in the Johns Hopkins Auditory Phenotyping Core, sponsored by P30 NIH DC005211. Creation and characterization of the Pex1-G844D mouse model was made possible by contributions from the Parks-Hopkins and Woodbury families and funds raised by Pound the Pavement for Peter (http://www.poundthepavementforpeter.com). Funding was also aided by grants from the National Institutes of Health (R01EY009769 and 5P30EY001765), the Foundation Fighting Blindness, Research to Prevent Blindness and a generous gift from the Guerrieri Family Foundation.
We thank Sabine Weller and Jutta Gartner for sharing details of their Pex1 construct that established Pex1-G844D heterozygotes, but not viable Pex1-G844D homozygotes. We extend our appreciation for Ann Snowden's technical assistance in performing skin fibroblast biochemical assays, to Dominick Reisinger's contribution to perfusing mice and sectioning livers used for histology, and to Zhangyong Wei for technical assistance in carrying out retinal staining procedures.
ABBREVIATIONS
- ABR
auditory brainstem response
- C26:0-LPC
C26:0-lysophosphatidylethanolamine
- DHCA
dihydroxycholestanoic acid
- DMSO
dimethylsulfoxide
- ERG
electroretinogram
- FBS-EMEM
fetal bovine serum-Eagle's minimum essential media
- h
hour
- IRD
infantile Refsum disease
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- NALD
neonatal adrenoleukodystrophy
- PE
phosphatidylethanolamine
- PEX
peroxins
- PEX1-G843D
human PEX1 mutation c.2528G>A (p.G843D)
- Pex1-G844D
murine Pex1 mutation c.2531G>A (p.G844D)
- PFA
paraformaldehyde
- PNA
peanut agglutinin
- PTS1
peroxisome targeting signal 1
- PTS2
peroxisome targeting signal 2
- RT
room temperature
- THCA
trihydroxycholestanoic acid
- TMAO
trimethylamine N-oxide
- VLCFA
very long chain fatty acids
- ZS
Zellweger syndrome
- ZSD
Zellweger spectrum disorder
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annual review of biochemistry. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 2.Braverman NE, D'Agostino MD, Maclean GE. Peroxisome biogenesis disorders: Biological, clinical and pathophysiological perspectives. Developmental disabilities research reviews. 2013;17:187–196. doi: 10.1002/ddrr.1113. [DOI] [PubMed] [Google Scholar]
- 3.Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nature genetics. 1997;15:369–376. doi: 10.1038/ng0497-369. [DOI] [PubMed] [Google Scholar]
- 4.Ebberink MS, Mooijer PA, Gootjes J, Koster J, Wanders RJ, Waterham HR. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Human mutation. 2011;32:59–69. doi: 10.1002/humu.21388. [DOI] [PubMed] [Google Scholar]
- 5.Ebberink MS, Koster J, Visser G, Spronsen F, Stolte-Dijkstra I, Smit GP, Fock JM, Kemp S, Wanders RJ, Waterham HR. A novel defect of peroxisome division due to a homozygous non-sense mutation in the PEX11beta gene. Journal of medical genetics. 2012;49:307–313. doi: 10.1136/jmedgenet-2012-100778. [DOI] [PubMed] [Google Scholar]
- 6.Goldfischer S, Moore CL, Johnson AB, Spiro AJ, Valsamis MP, Wisniewski HK, Ritch RH, Norton WT, Rapin I, Gartner LM. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science. 1973;182:62–64. doi: 10.1126/science.182.4107.62. [DOI] [PubMed] [Google Scholar]
- 7.Poulos A, Sharp P, Whiting M. Infantile Refsum's disease (phytanic acid storage disease): a variant of Zellweger's syndrome? Clinical genetics. 1984;26:579–586. doi: 10.1111/j.1399-0004.1984.tb01107.x. [DOI] [PubMed] [Google Scholar]
- 8.Scotto JM, Hadchouel M, Odievre M, Laudat MH, Saudubray JM, Dulac O, Beucler I, Beaune P. Infantile phytanic acid storage disease, a possible variant of Refsum's disease: three cases, including ultrastructural studies of the liver. Journal of inherited metabolic disease. 1982;5:83–90. doi: 10.1007/BF01799998. [DOI] [PubMed] [Google Scholar]
- 9.Wanders RJ, Saelman D, Heymans HS, Schutgens RB, Westerveld A, Poll-The BT, Saudubray JM, Van den Bosch H, Strijland A, Schram AW, et al. Genetic relation between the Zellweger syndrome, infantile Refsum's disease, and rhizomelic chondrodysplasia punctata. The New England journal of medicine. 1986;314:787–788. doi: 10.1056/NEJM198603203141216. [DOI] [PubMed] [Google Scholar]
- 10.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y. A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science. 1992;255:1132–1134. doi: 10.1126/science.1546315. [DOI] [PubMed] [Google Scholar]
- 11.Zellweger H, Maertens P, Superneau D, Wertelecki W. History of the cerebrohepatorenal syndrome of Zellweger and other peroxisomal disorders. Southern medical journal. 1988;81:357–364. doi: 10.1097/00007611-198803000-00017. [DOI] [PubMed] [Google Scholar]
- 12.Wanders RJ, Waterham HR. Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clinical genetics. 2005;67:107–133. doi: 10.1111/j.1399-0004.2004.00329.x. [DOI] [PubMed] [Google Scholar]
- 13.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochimica et biophysica acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Powers JM, Moser HW, Moser AB, Upshur JK, Bradford BF, Pai SG, Kohn PH, Frias J, Tiffany C. Fetal cerebrohepatorenal (Zellweger) syndrome: dysmorphic, radiologic, biochemical, and pathologic findings in four affected fetuses. Human pathology. 1985;16:610–620. doi: 10.1016/s0046-8177(85)80111-8. [DOI] [PubMed] [Google Scholar]
- 15.Majewski J, Wang Z, Lopez I, Al Humaid S, Ren H, Racine J, Bazinet A, Mitchel G, Braverman N, Koenekoop RK. A new ocular phenotype associated with an unexpected but known systemic disorder and mutation: novel use of genomic diagnostics and exome sequencing. Journal of medical genetics. 2011;48:593–596. doi: 10.1136/jmedgenet-2011-100288. [DOI] [PubMed] [Google Scholar]
- 16.Steinberg SJ, Snowden A, Braverman NE, Chen L, Watkins PA, Clayton PT, Setchell KD, Heubi JE, Raymond GV, Moser AB, Moser HW. A PEX10 defect in a patient with no detectable defect in peroxisome assembly or metabolism in cultured fibroblasts. Journal of inherited metabolic disease. 2009;32:109–119. doi: 10.1007/s10545-008-0969-8. [DOI] [PubMed] [Google Scholar]
- 17.Collins CS, Gould SJ. Identification of a common PEX1 mutation in Zellweger syndrome. Human mutation. 1999;14:45–53. doi: 10.1002/(SICI)1098-1004(1999)14:1<45::AID-HUMU6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell MA, Nelson PV, Chin SJ, Paton BC, Carey WF, Crane DI. A common PEX1 frameshift mutation in patients with disorders of peroxisome biogenesis correlates with the severe Zellweger syndrome phenotype. Human genetics. 1999;105:38–44. doi: 10.1007/s004399900095. [DOI] [PubMed] [Google Scholar]
- 19.Preuss N, Brosius U, Biermanns M, Muntau AC, Conzelmann E, Gartner J. PEX1 mutations in complementation group 1 of Zellweger spectrum patients correlate with severity of disease. Pediatric research. 2002;51:706–714. doi: 10.1203/00006450-200206000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Walter C, Gootjes J, Mooijer PA, Portsteffen H, Klein C, Waterham HR, Barth PG, Epplen JT, Kunau WH, Wanders RJ, Dodt G. Disorders of peroxisome biogenesis due to mutations in PEX1: phenotypes and PEX1 protein levels. American journal of human genetics. 2001;69:35–48. doi: 10.1086/321265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinberg S, Chen L, Wei L, Moser A, Moser H, Cutting G, Braverman N. The PEX Gene Screen: molecular diagnosis of peroxisome biogenesis disorders in the Zellweger syndrome spectrum. Molecular genetics and metabolism. 2004;83:252–263. doi: 10.1016/j.ymgme.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Berendse K, Ebberink MS, Ijlst L, Poll-The BT, Wanders RJ, Waterham HR. Arginine improves peroxisome functioning in cells from patients with a mild peroxisome biogenesis disorder. Orphanet journal of rare diseases. 2013;8:138. doi: 10.1186/1750-1172-8-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R, Chen L, Jiralerspong S, Snowden A, Steinberg S, Braverman N. Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5569–5574. doi: 10.1073/pnas.0914960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baes M, Gressens P, Baumgart E, Carmeliet P, Casteels M, Fransen M, Evrard P, Fahimi D, Declercq PE, Collen D, van Veldhoven PP, Mannaerts GP. A mouse model for Zellweger syndrome. Nature genetics. 1997;17:49–57. doi: 10.1038/ng0997-49. [DOI] [PubMed] [Google Scholar]
- 25.Faust PL, Su HM, Moser A, Moser HW. The peroxisome deficient PEX2 Zellweger mouse: pathologic and biochemical correlates of lipid dysfunction. Journal of molecular neuroscience : MN. 2001;16:289–297. doi: 10.1385/JMN:16:2-3:289. discussion 317-221. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell M, Bjorkman J, Nguyen T, Sharp P, Finnie J, Paterson C, Tonks I, Paton BC, Kay GF, Crane DI. Pex13 inactivation in the mouse disrupts peroxisome biogenesis and leads to a Zellweger syndrome phenotype. Molecular and cellular biology. 2003;23:5947–5957. doi: 10.1128/MCB.23.16.5947-5957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubbard WC, Moser AB, Liu AC, Jones RO, Steinberg SJ, Lorey F, Panny SR, Vogt RF, Jr., Macaya D, Turgeon CT, Tortorelli S, Raymond GV. Newborn screening for X-linked adrenoleukodystrophy (X-ALD): validation of a combined liquid chromatography-tandem mass spectrometric (LC-MS/MS) method. Molecular genetics and metabolism. 2009;97:212–220. doi: 10.1016/j.ymgme.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Hubbard WC, Moser AB, Tortorelli S, Liu A, Jones D, Moser H. Combined liquid chromatography-tandem mass spectrometry as an analytical method for high throughput screening for X-linked adrenoleukodystrophy and other peroxisomal disorders: preliminary findings. Molecular genetics and metabolism. 2006;89:185–187. doi: 10.1016/j.ymgme.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Zemski Berry KA, Murphy RC. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. Journal of the American Society for Mass Spectrometry. 2004;15:1499–1508. doi: 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Johnson DW, ten Brink HJ, Schuit RC, Jakobs C. Rapid and quantitative analysis of unconjugated C(27) bile acids in plasma and blood samples by tandem mass spectrometry. Journal of lipid research. 2001;42:9–16. [PubMed] [Google Scholar]
- 31.Steinberg S, Jones R, Tiffany C, Moser A. Investigational methods for peroxisomal disorders. Current protocols in human genetics / editorial board, Jonathan L. Haines ... [et al.] 2008 doi: 10.1002/0471142905.hg1706s58. Chapter 17 Unit 17 16. [DOI] [PubMed] [Google Scholar]
- 32.Roscher A, Molzer B, Bernheimer H, Stockler S, Mutz I, Paltauf F. The cerebrohepatorenal (Zellweger) syndrome: an improved method for the biochemical diagnosis and its potential value for prenatal detection. Pediatric research. 1985;19:930–933. doi: 10.1203/00006450-198509000-00013. [DOI] [PubMed] [Google Scholar]
- 33.Hoefler G, Hoefler S, Watkins PA, Chen WW, Moser A, Baldwin V, McGillivary B, Charrow J, Friedman JM, Rutledge L, et al. Biochemical abnormalities in rhizomelic chondrodysplasia punctata. The Journal of pediatrics. 1988;112:726–733. doi: 10.1016/s0022-3476(88)80689-9. [DOI] [PubMed] [Google Scholar]
- 34.Wanders RJ, Kos M, Roest B, Meijer AJ, Schrakamp G, Heymans HS, Tegelaers WH, van den Bosch H, Schutgens RB, Tager JM. Activity of peroxisomal enzymes and intracellular distribution of catalase in Zellweger syndrome. Biochemical and biophysical research communications. 1984;123:1054–1061. doi: 10.1016/s0006-291x(84)80240-5. [DOI] [PubMed] [Google Scholar]
- 35.Theda C, Moser AB, Powers JM, Moser HW. Phospholipids in X-linked adrenoleukodystrophy white matter: fatty acid abnormalities before the onset of demyelination. Journal of the neurological sciences. 1992;110:195–204. doi: 10.1016/0022-510x(92)90028-j. [DOI] [PubMed] [Google Scholar]
- 36.Lauer AM, May BJ. The medial olivocochlear system attenuates the developmental impact of early noise exposure. Journal of the Association for Research in Otolaryngology : JARO. 2011;12:329–343. doi: 10.1007/s10162-011-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lina IA, Lauer AM. Rapid measurement of auditory filter shape in mice using the auditory brainstem response and notched noise. Hearing research. 2013;298:73–79. doi: 10.1016/j.heares.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelikan RC, Iwata J, Suzuki A, Chai Y, Hacia JG. Identification of candidate downstream targets of TGFbeta signaling during palate development by genome-wide transcript profiling. Journal of cellular biochemistry. 2013;114:796–807. doi: 10.1002/jcb.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, McClelland M, Xia XQ. Analyzing microarray data using WebArray Cold Spring Harbor protocols 2009. 2009 doi: 10.1101/pdb.prot5260. pdb prot5260. [DOI] [PubMed] [Google Scholar]
- 40.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 42.Pounds S, Cheng C. Improving false discovery rate estimation. Bioinformatics. 2004;20:1737–1745. doi: 10.1093/bioinformatics/bth160. [DOI] [PubMed] [Google Scholar]
- 43.Le HD, Fallon EM, Kalish BT, de Meijer VE, Meisel JA, Gura KM, Nose V, Pan AH, Bistrian BR, Puder M. The effect of varying ratios of docosahexaenoic acid and arachidonic acid in the prevention and reversal of biochemical essential fatty acid deficiency in a murine model. Metabolism: clinical and experimental. 2013;62:499–508. doi: 10.1016/j.metabol.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swiderski RE, Nishimura DY, Mullins RF, Olvera MA, Ross JL, Huang J, Stone EM, Sheffield VC. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Investigative ophthalmology & visual science. 2007;48:3329–3340. doi: 10.1167/iovs.06-1477. [DOI] [PubMed] [Google Scholar]
- 45.Ding J, Loizides-Mangold U, Rando G, Zoete V, Michielin O, Reddy JK, Wahli W, Riezman H, Thorens B. The Peroxisomal Enzyme L-PBE Is Required to Prevent the Dietary Toxicity of Medium-Chain Fatty Acids. Cell reports. 2013;5:248–258. doi: 10.1016/j.celrep.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 46.Cao J, Wang J, Qi W, Miao HH, Wang J, Ge L, DeBose-Boyd RA, Tang JJ, Li BL, Song BL. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell metabolism. 2007;6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Poll-The BT, Gootjes J, Duran M, De Klerk JB, Wenniger-Prick LJ, Admiraal RJ, Waterham HR, Wanders RJ, Barth PG. Peroxisome biogenesis disorders with prolonged survival: phenotypic expression in a cohort of 31 patients. American journal of medical genetics. Part A. 2004;126A:333–338. doi: 10.1002/ajmg.a.20664. [DOI] [PubMed] [Google Scholar]
- 48.Ahlemeyer B, Gottwald M, Baumgart-Vogt E. Deletion of a single allele of the Pex11beta gene is sufficient to cause oxidative stress, delayed differentiation and neuronal death in mouse brain. Disease models & mechanisms. 2012;5:125–140. doi: 10.1242/dmm.007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baes M, Van Veldhoven PP. Mouse models for peroxisome biogenesis defects and beta-oxidation enzyme deficiencies. Biochimica et biophysica acta. 2012;1822:1489–1500. doi: 10.1016/j.bbadis.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Baes M, Van Veldhoven PP. Generalised and conditional inactivation of Pex genes in mice. Biochimica et biophysica acta. 2006;1763:1785–1793. doi: 10.1016/j.bbamcr.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 51.Keane MH, Overmars H, Wikander TM, Ferdinandusse S, Duran M, Wanders RJ, Faust PL. Bile acid treatment alters hepatic disease and bile acid transport in peroxisome-deficient PEX2 Zellweger mice. Hepatology. 2007;45:982–997. doi: 10.1002/hep.21532. [DOI] [PubMed] [Google Scholar]
- 52.Cohen SM, Brown FR, 3rd, Martyn L, Moser HW, Chen W, Kistenmacher M, Punnett H, de la Cruz ZC, Chan NR, Green WR. Ocular histopathologic and biochemical studies of the cerebrohepatorenal syndrome (Zellweger's syndrome) and its relationship to neonatal adrenoleukodystrophy. American journal of ophthalmology. 1983;96:488–501. doi: 10.1016/s0002-9394(14)77913-9. [DOI] [PubMed] [Google Scholar]
- 53.Cohen SM, Green WR, de la Cruz ZC, Brown FR, 3rd, Moser HW, Luckenbach MW, Dove DJ, Maumenee IH. Ocular histopathologic studies of neonatal and childhood adrenoleukodystrophy. American journal of ophthalmology. 1983;95:82–96. doi: 10.1016/0002-9394(83)90336-7. [DOI] [PubMed] [Google Scholar]
- 54.Folz SJ, Trobe JD. The peroxisome and the eye. Survey of ophthalmology. 1991;35:353–368. doi: 10.1016/0039-6257(91)90185-i. [DOI] [PubMed] [Google Scholar]
- 55.Noguer MT, Martinez M. Visual follow-up in peroxisomal-disorder patients treated with docosahexaenoic Acid ethyl ester. Investigative ophthalmology & visual science. 2010;51:2277–2285. doi: 10.1167/iovs.09-4020. [DOI] [PubMed] [Google Scholar]
- 56.Perlmutter DH. Chemical chaperones: a pharmacological strategy for disorders of protein folding and trafficking. Pediatric research. 2002;52:832–836. doi: 10.1203/00006450-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 57.Engelen M, Kemp S, de Visser M, van Geel BM, Wanders RJ, Aubourg P, Poll-The BT. X-linked adrenoleukodystrophy (X-ALD): clinical presentation and guidelines for diagnosis, follow-up and management. Orphanet journal of rare diseases. 2012;7:51. doi: 10.1186/1750-1172-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson GJ, Connor WE, Corliss JD. Docosahexaenoic acid is the preferred dietary n-3 fatty acid for the development of the brain and retina. Pediatric research. 1990;27:89–97. doi: 10.1203/00006450-199001000-00023. [DOI] [PubMed] [Google Scholar]
- 59.Bazan NG. The metabolism of omega-3 polyunsaturated fatty acids in the eye: the possible role of docosahexaenoic acid and docosanoids in retinal physiology and ocular pathology. Progress in clinical and biological research. 1989;312:95–112. [PubMed] [Google Scholar]
- 60.Connor WE, Neuringer M. The effects of n-3 fatty acid deficiency and repletion upon the fatty acid composition and function of the brain and retina. Progress in clinical and biological research. 1988;282:275–294. [PubMed] [Google Scholar]
- 61.Rodriguez de Turco EB, Gordon WC, Peyman GA, Bazan NG. Preferential uptake and metabolism of docosahexaenoic acid in membrane phospholipids from rod and cone photoreceptor cells of human and monkey retinas. Journal of neuroscience research. 1990;27:522–532. doi: 10.1002/jnr.490270413. [DOI] [PubMed] [Google Scholar]
- 62.Bazan NG, Rodriguez de Turco EB. Review: pharmacological manipulation of docosahexaenoic-phospholipid biosynthesis in photoreceptor cells: implications in retinal degeneration. Journal of ocular pharmacology. 1994;10:591–604. doi: 10.1089/jop.1994.10.591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.