Abstract
There is an urgent need to identify new treatments for tuberculosis (TB), a major infectious disease caused by Mycobacterium tuberculosis (Mtb), which results in 1.5 million deaths each year. We have targeted two essential enzymes in this organism that are promising for antibacterial therapy and reported to be inhibited by naphthoquinones. ThyX is an essential thymidylate synthase that is mechanistically and structurally unrelated to the human enzyme. DNA gyrase is a DNA topoisomerase present in bacteria and plants but not animals. The current study set out to understand the structure-activity relationships of these targets in Mtb using a combination of cheminformatics and in vitro screening. Here, we report the identification of new Mtb ThyX inhibitors, 2-chloro-3-(4-methanesulfonylpiperazin-1-yl)-1,4-dihydronaphthalene-1,4-dione) and idebenone, which show modest whole-cell activity and appear to act, at least in part, by targeting ThyX in Mtb.
Tuberculosis (TB) is a major infectious disease that knows no geographic boundary and accounts for 9 million new cases and approximately 1.5 million deaths each year1. TB and its etiological agent Mycobacterium tuberculosis (Mtb) are the focus of intense efforts to develop new tools for the control and eventual elimination2 of this devastating disease, which is associated increasingly with resistance to first- and second-line drugs3. The discovery of new TB drug candidates with novel mechanisms of action that can overcome resistance, shorten the duration of treatment, and be co-administered with antiretrovirals, is of fundamental importance in this regard4,5,6. Over the last decade, there has been considerable investment in TB drug discovery with particular emphasis on the use of high-throughput phenotypic screening of libraries of thousands to hundreds of thousands of molecules for “hit” identification5,7,8,9. Whole-cell approaches have the advantage of allowing the high-throughput screening (HTS) assay to be conducted under conditions that mimic host infection without knowledge of mechanism of action10. Importantly, all presently used antibiotics have been developed by this approach, including the recently approved drug, bedaquiline11. Moreover, the phenotypic screening format produces a wealth of data that can be used for computational machine learning12, which has the potential to improve the screening efficiency of additional compounds13,14 and assist lead optimization15.
An alternative approach to hit identification is target-based, which relies on the availability of purified protein against which a HTS can be performed10 and/or knowledge of the target, such as the crystal structure, to guide structure-based drug design. Drawbacks of this approach include the difficulty in balancing target activity with the physicochemical properties needed to enter whole cells and evade efflux. This approach also requires extensive validation of the target. A recent review summarized the results of target-based and phenotypic screens conducted at the Novartis Institute for Tropical Diseases. After failing with target-based screens, phenotypic screens led to the identification of 5 chemical series in 7 years. Of these, 3 series were terminated due to glycerol-dependent activity which is irrelevant in Mtb, lack of in vivo activity, or limited maximum exposure10. Target-based screening has also been reviewed to identify the key properties of promising targets such as essentiality for growth, vulnerability, druggability, reduced propensity for resistance, and target localization as well as amenability to chemotherapy16.
For a known target, computational approaches such as docking the molecules (into the protein structure), quantitative structure-activity relationship (QSAR), pharmacophore or machine learning models can be developed to screen chemical libraries12. We have previously used 3D pharmacophore models, alone or in combination with Bayesian models to identify compounds with antitubercular whole-cell activity17,18, as a bridge between phenotypic screening and rational structure-based drug design.
The current study focuses on naphthoquinone (NQ) compounds which have widely reported biological activities including anti-cancer and anti-malarial activities. For instance, atovaquone (2-(trans-4-(P-chlorophenyl)cyclohexyl)-3-hydroxy-1,4-naphthoquinone), a well-known 2-OH-1,4-NQ, targets the respiratory electron transfer chain, and is clinically used in anti-pneumocystis, anti-toxoplasmosis and anti-malarial treatments. NQs also have anti-microbial activity against different bacterial pathogens, including Mtb19,20,21,22,23,24,25. Recently, we showed that NQ-based compounds inhibit the activities of PBCV-1 and Helicobacter pylori thymidylate synthase ThyX26,27 as well as Mtb DNA gyrase28. These observations led us to investigate inhibition of Mtb ThyX by NQs and develop pharmacophore models for these two essential enzymes that are both required for DNA replication29.
ThyX is an essential thymidylate synthase (TS) that is both mechanistically and structurally unrelated to the analogous human enzyme30,31. These enzymes catalyze the methylation of 2′-deoxyuridine-5′-monophosphate (dUMP) to synthesize 2′-deoxythymidine-5′-monophosphate (dTMP), an essential DNA precursor. In this reaction, 5,10-methylenetetrahydrofolate (CH2H4folate) and nicotinamide adenine dinucleotide phosphate (NADPH) are used as carbon and hydride donors, respectively. In the case of Paramecium bursaria chlorella virus-1 ThyX, structural data have revealed stacking of NQ against the flavin adenine dinucleotide (FAD) co-factor, partially overlapping with the dUMP-binding pocket27. As dUMP acts in the ThyX reaction both as the activator and the substrate32, NQ binding at the ThyX active site results in potent inhibition of ThyX activity. Importantly, unlike human TS, ThyX produces tetrahydrofolate (H4folate) as a byproduct explaining why many thyX-carrying organisms do not require dihydrofolate reductase (FolA)33. Strikingly, although mycobacteria possess two distinct families of thymidylate synthases, the canonical ThyA as well as the non-canonical ThyX, only ThyX is essential in these organisms34. ThyX thus represents a promising target against Mtb34,35, with a crystal structure available [PDB:2AF636]. Several laboratories have developed dUMP analogs to target Mtb ThyX, although most of the hits to date are non-selective and also inhibit ThyA37,38. More recently, conditional depletion of ThyX was shown to result in modest hypersensitivity of Mtb to the thymidylate synthase inhibitor and anticancer drug, 5-fluorouracil (5-FU)39, suggesting that inhibition of ThyX through metabolic conversion of 5-FU to 5-FdUMP comprises one element of the complex mechanism of anti-tubercular action of this drug.
NQs have also been shown to be active against DNA gyrase28 and appear to bind at the N-terminal domain of GyrB26 at a novel site that is distinct from the ATPase active site and the well-established binding site for aminocoumarin antibiotics40. This enzyme is a topoisomerase present in bacteria and plants but not animals, and is a validated target for antibacterials that include the fluoroquinolones, which are important second-line drugs for TB. It consists of two subunits, GyrA and GyrB, which form an A2B2 complex in the active enzyme. DNA gyrase catalyzes supercoiling of DNA in an ATP-dependent reaction; the ATPase site resides in the GyrB subunit41.
The observed overlap of NQs binding and inhibiting both ThyX and GyrB from Mtb motivated the current study to identify new inhibitors suggested using computational approaches.
Results
Identification of NQs as inhibitors of ThyX and gyrase
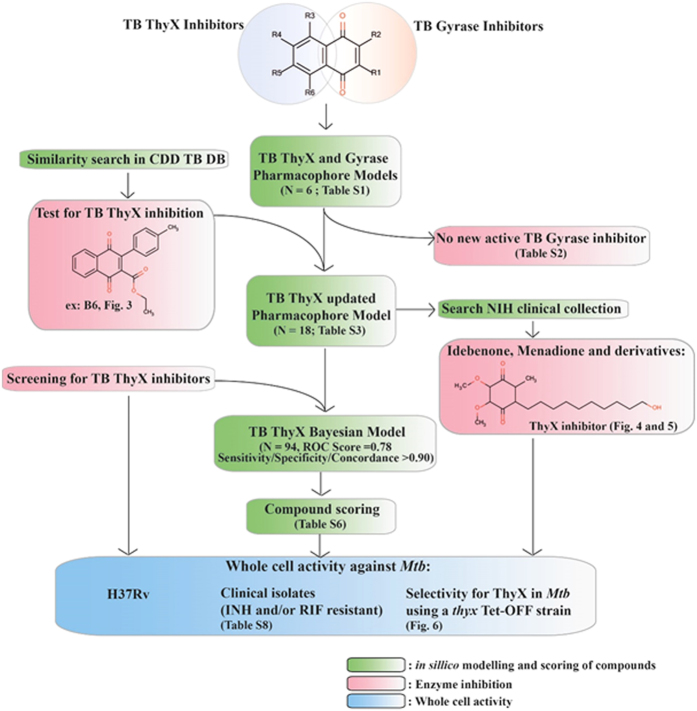
In this study, we utilized a combined computational and experimental workflow (Fig. 1) to obtain new insight into Mtb ThyX and DNA gyrase inhibition, and identify new inhibitors in the case of ThyX. A starting point for the study was the identification of NQs as inhibitors of Mtb ThyX and DNA gyrase (Supplementary Table 1). The compounds 2EO4 and C8-C1, originally identified as the inhibitors of the PBCV-1 ThyX enzyme, were found to also inhibit Mtb ThyX, but were inactive against Mtb gyrase. Diospyrin inhibits only Mtb gyrase whereas other tested molecules showed comparable activity against both enzymes (Supplementary Table 1). These results revealed that selective or dual inhibition of these enzymes is feasible and prompted further computational analyses to identify additional inhibitors.
Figure 1. Workflow for combined computational and experimental approaches.
In silico modelling and scoring of compounds is boxed in green. Enzyme assays are boxed in pink. Whole cell activity measurements are boxed in blue.
Substructure searching and common features pharmacophores used for virtual screening with ThyX
Using the experimental data described in Supplementary Table 1, we were able to build common features pharmacophores for Mtb ThyX and gyrase that consisted of excluded volumes, two hydrogen bond acceptors and one hydrophobic feature (Fig. 2). The Mtb GyrB pharmacophore used 6 NQs (Fig. 2A) and resulted in the same features as for the Mtb ThyX pharmacophore (Fig. 2B), albeit in a different arrangement. Isodiospyrin which inhibits GyrB was predicted to have a poor fit score against Mtb ThyX, as shown in Fig. 2C. After similarity searching previously identified whole-cell active compounds in the CDD TBDB42,43, using the napthoquinone substructure we identified a Mtb ThyX inhibitor, ethyl 3-(4-methylphenyl)-1,4-dioxonaphtalene-2-carboxylate (molecule B6, Fig. 3A), with a Ki of 4.5 μM (Fig. 3B). This molecule as well as others screened in this process were added into the models to update them. All 19 compounds that we selected for GyrB at this stage were inactive (Supplementary Table 2); however, our approach led to additional compounds that showed substantial activity against ThyX. For example, one pharmacophore with 18 molecules (N18 Pharmacophore, Supplementary Table 3) was used to search the NIH clinical collection of over 700 compounds, and one quinone compound, idebenone (Fig. 4A), was selected for testing based on its fit to the model (Fig. 4B). This molecule was found to be an uncompetitive inhibitor of Mtb ThyX with respect to dUMP (Ki = 3.3 μM, Fig. 5), suggesting that it binds preferentially to the ThyX-dUMP complex. It also has weak whole-cell activity against Mtb, (MIC90 = 125 μM; Supplementary Table 4). This table also shows the whole-cell activity of several other compounds against replicating Mtb, including B6, D4, D5, E1, E10, F1 and F2 which were identified by screening against the Mtb ThyX (Fig. 1). Interestingly, menadione inhibited growth of Mtb while some additional analogs were less active or had solubility issues (Supplementary Table 5).
Figure 2. Initial common feature pharmacophores for ThyX and GyrB.
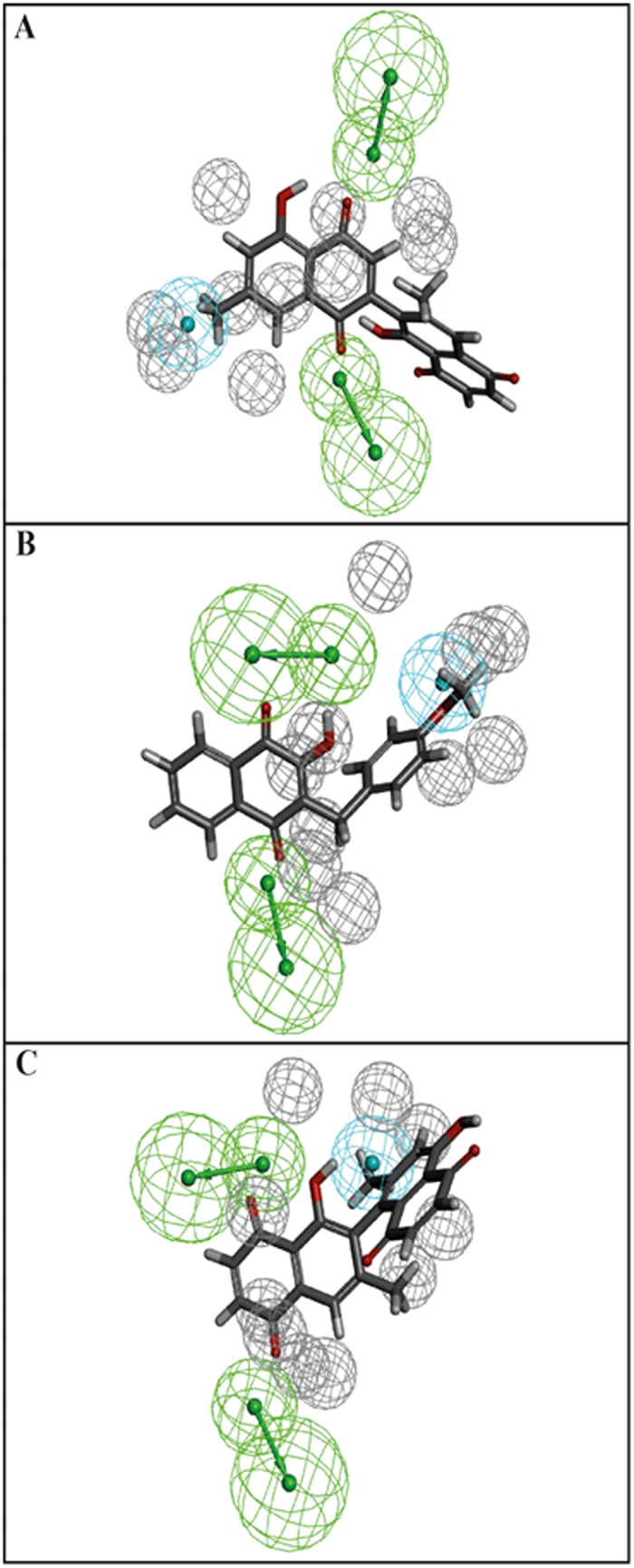
(A) Diospyrin mapped to the GyrB model, (B) C8-C1 mapped to ThyX model. (C) ThyX model was used to score isodiospyrin (the fit was poor 0.004). Blue = hydrophobe, green = hydrogen bond acceptor, grey = excluded volume.
Figure 3. Structure and dose response curve for B6 against ThyX.
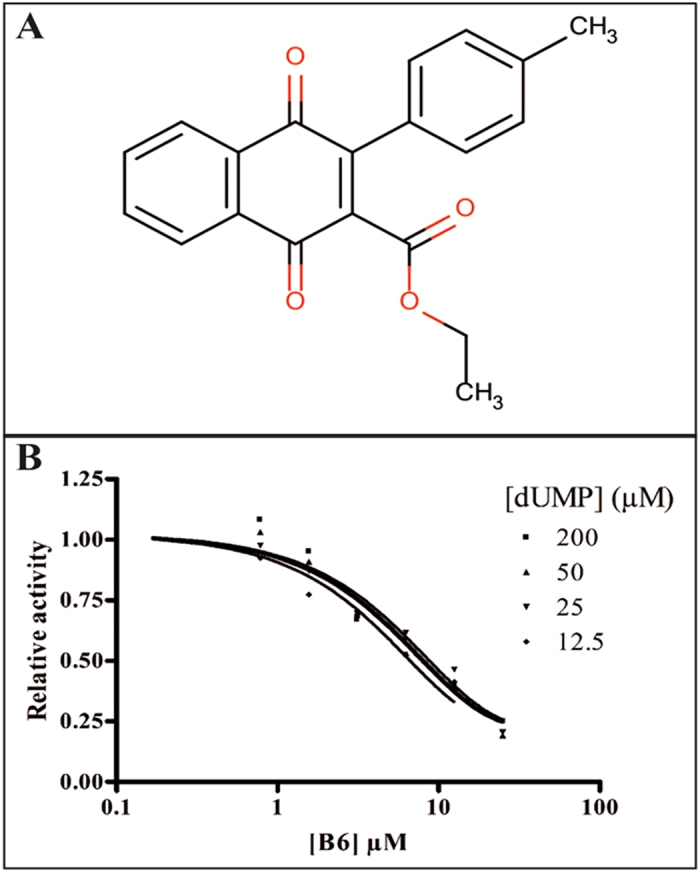
(A) Structure of ethyl 3-(4-methylphenyl)-1,4-dioxonaphtalene-2-carboxylate (B6), a NQ identified by similarity searching in the CDD Public database of previously tested compounds against Mtb with known whole cell activity. (B) B6 dose response curves versus ThyX.
Figure 4. Idebenone mapped to the ThyX N = 18 pharmacophore model.
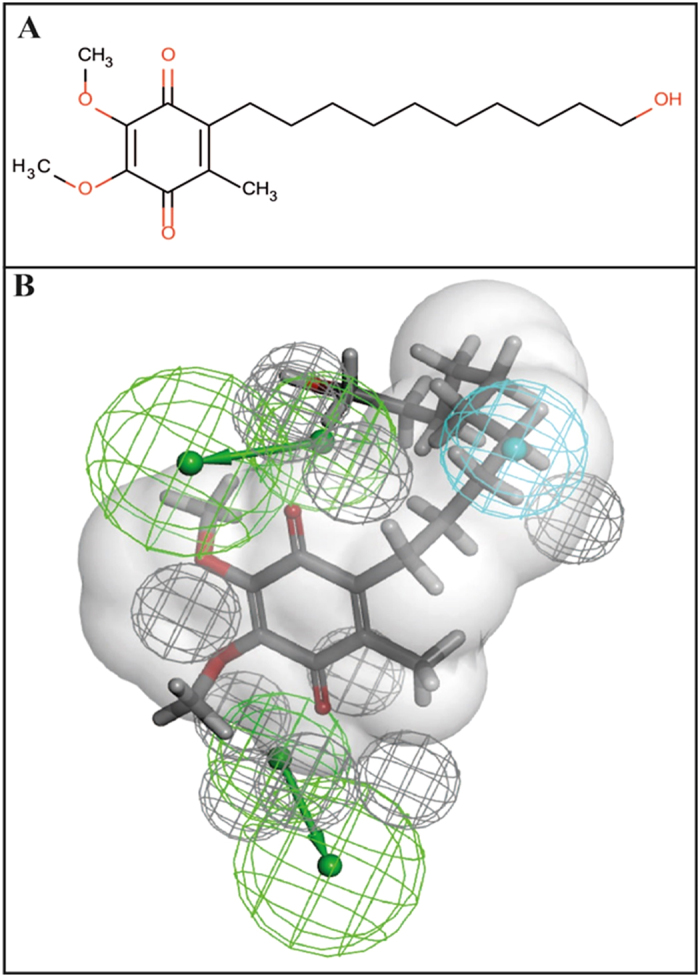
(A) idebenone 2D structure. (B) Pharmacophore showing van der Waals surface based on C8-C1, Blue = hydrophobe, green = hydrogen bond acceptor, grey = excluded volume.
Figure 5. Idebenone dose response against ThyX and effect of dUMP.
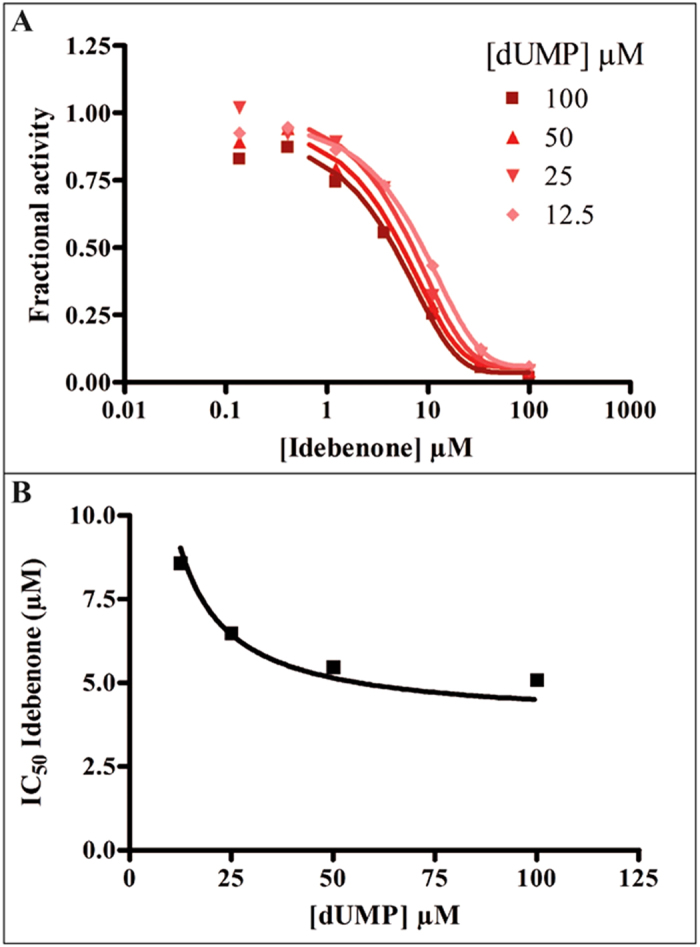
(A) Idebenone dose response curves versus ThyX. (B) Effect of the dUMP concentration on the IC50 value of Idebenone.
Predicting new ThyX inhibitors using Bayesian machine learning models
In prior work, we have used Bayesian machine learning to build models of whole-cell screens of small molecule compounds active against Mtb14, which led to the identification of new active compounds13,14,44. After testing 94 molecules against Mtb ThyX at 100 μM (the training set, Supplemental Table 6), we generated a Bayesian model using molecules with >70% inhibition as actives. This resulted in a promising model with a ROC score of 0.78 after 5-fold cross validation, alongside sensitivity, specificity and concordance values greater than 0.90 (Supplementary Fig. 1A). The ‘good’ features were predominantly quinones and NQs (Supplementary Fig. 1B), while ‘bad’ features included amines and sulfonamides (Supplementary Fig. 1C). A model with similar statistics was generated in CDD models using FCFP6 descriptors alone, with a 3-fold cross validation ROC score of 0.80 (Supplementary Fig. 2).
Experimental testing of Bayesian model for NQ inhibitors of ThyX
The Bayesian model generated with Discovery Studio was used to predict activity of 14 compounds (007B-010K, Supplementary Table 7) which were not included in the training set. The closest distance calculation uses the calculated Euclidean distance between each molecule and those in the training set and suggests they are different (a zero distance corresponds to identical molecules). Two of these molecules were predicted as “inactive” (010-I and 010-C, Supplementary Table 7). However, six of the remaining 12 compounds (50%) were predicted as actives and exhibited over 70% inhibition (the selected cut-off) of Mtb ThyX activity in vitro at 100 μM. These experimental results illustrate the robustness of the models.
Whole-cell activity against Mtb
We next investigated whole-cell activity of the NQs against laboratory and clinical isolates of Mtb with different drug resistance profiles to standard anti-tuberculars (Supplementary Table 8) under replicating conditions. MIC90 values of the molecules in the test set (Supplementary Table 7) ranged from 20 to 200 μM (Supplementary Table 9).
Additional compounds that either acted as MtbThyX inhibitors in vitro or were selected by the N18 pharmacophore model were also evaluated to identify those with growth inhibitory activity against replicating Mtb H37Rv (Supplementary Table 4). Molecules with whole-cell activity were then tested against a conditional knockdown mutant of Mtb H37Rv, thyX Tet-OFF, in which thyX is expressed under the control of a Tet-regulated promoter39. In this target-based whole-cell assay, compounds with ThyX-selective activity in Mtb can be identified on the basis of whether ThyX depletion confers hypersensitivity to the compound39,45. As expected, the positive control, 5-FU, showed a progressive, ATc-dependent shift in MIC90 from 3.1 μM in the absence of ATc to 0.3 μM at an ATc concentration of 6.2 ng/ml. Among the other compounds tested in this assay, two showed ≥ 4-fold increase in potency upon ThyX depletion, namely, E1 (2-chloro-3-(4-methanesulfonylpiperazin-1-yl)-1,4-dihydronaphthalene-1,4-dione)) and idebenone (Fig. 6). These compounds demonstrated the same activity against thyX Tet-OFF in the absence of ATc compared to the H37Rv control, with MIC90 values of 62.5 and 125 μM, respectively. Addition of ATc resulted in a progressive increase in susceptibility to E1, with the MIC90 value decreasing to 15.6 μM at an ATc concentration of 6.2 ng/ml. ThyX depletion also sensitized Mtb to idebenone with the MIC90 shifting ~5-fold, from 125 μM to 22 μM. However, for reasons that are unclear, idebenone displayed atypical behavior in this checkerboard assay compared to 5-FU39 and E1, showing ~90% growth inhibition over an unusually large concentration range (62.5–7.8 μM) when added to thyX Tet-OFF in the presence of ATc at 6.2 ng/ml (Fig. 6B). Of the other compounds tested, F2 showed a slight (~2-fold) MIC90 shift (62.5 to 31.2 μM) in the presence of ATc at 6.2 ng/ml, whereas no shift in MIC90 was observed for B6, D4, D5, E10 and F1. Together, these results implicate ThyX as a potential target for E1 and idebenone in Mtb.
Figure 6. Depletion of ThyX in Mtb confers modest hypersensitivity to E1 and idebenone.
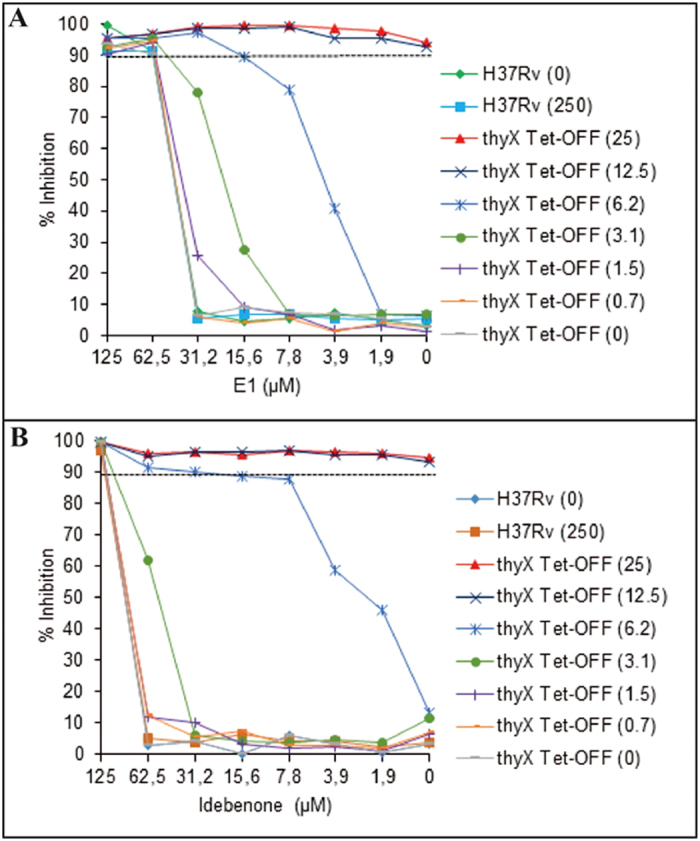
The effect of ThyX depletion on the susceptibility to E1 and idebenone was assessed using the thyX Tet-OFF mutant in a checkerboard assay. The H37Rv strain was used as a control. Bacterial viability was assessed by the Alamar Blue assay, measuring fluorescence at 544/590 nm. The numbers in parentheses denote the concentration of anhydrotetracycline (ATc, ng/ml), which modulates the expression of thyX in thyX Tet-OFF39. Ninety percent growth inhibition (MIC90) is represented by the dashed horizontal line. The results shown are representative of one of the three biological replicates.
Discussion
We have used a combination of cheminformatics and experimental strategies (Fig. 1) to identify new inhibitors of Mtb ThyX. Many previous studies have shown that NQs possess activity against Mtb19,20,21,22,23,24,25,28 and other bacteria27. It has long been suggested that such quinones are reactive and generally non-specific46, yet these natural products are already in therapeutic use, demonstrating that selective toxicity can be attained47,48. Starting with a set of NQs active against Mtb ThyX and/or GyrB (Supplementary Table 1), and employing a common feature pharmacophore approach, we showed that while a small set of compounds had identical pharmacophore features in each model, their arrangement was unique (Fig. 2). As we updated these models we noted little apparent change in the Mtb ThyX model and retrieved several compounds of interest for testing. However, the Mtb GyrB pharmacophore was unable to retrieve any additional active molecules containing NQ or other features (Supplementary Table 2). One of the compounds identified as active against Mtb ThyX using a pharmacophore based on NQs was idebenone (Figs 4 and 5), which demonstrate that the models can also retrieve non-NQs as ThyX inhibitors. Unlike the competitive mode of inhibition of NQs against PBCV-1 ThyX, idebenone exhibited uncompetitive inhibition of Mtb ThyX with respect to dUMP binding suggesting that it binds to, in addition to the free enzyme, to the Mtb ThyX-dUMP complex27. Idebenone has recently completed a phase III clinical trial for Duchene Muscular Dystrophy49. It is a potent antioxidant50 and can donate electrons to complex III of the electron transport chain. This drug has also found utility in studies on mitochondrial diseases including Friedreich’s Ataxia51 and is approved in Europe for Leber’s Hereditary Optic Neuropathy52. It is therefore possible that idebenone, while exhibiting only weak activity on Mtb whole cells, could have potential for repurposing against additional diseases as well. We also tested menadione and analogs that had improved whole cell activity (Supplementary Table 5) compared to idebenone. Menadione was inactive against Mtb gyrase but 7-methyljuglone, an analogue of menadione, was active against both Mtb ThyX and gyrase, indicating that modification of a single hydroxyl in methyljuglone to a methyl group in menadione, participates in dual inhibition of Mtb ThyX and gyrase (Supplementary Table 1).
As we collected and tested a relatively large number of molecules (N = 94) against Mtb ThyX, we were able to use a machine learning approach. To our knowledge this represents the first target-based machine learning approach towards the identification of enzyme inhibitors in Mtb. Clearly, quinone and NQ substructures were identified as important for activity and molecules with amines and sulfonamides were less desirable. These machine learning results could help us focus on compounds to test in the future. The Bayesian model has the added advantage of being able to quickly score compounds so could narrow down potential compounds for testing. We evaluated the Bayesian model with additional NQs (Supplementary Table 7). Our model predicted potent new Mtb ThyX inhibitors that had some whole-cell activity against Mtb, thus attesting to the utility of our approach. In addition we have used a second software tool (CDD Models) which produces a similar leave-out-cross-validation ROC value, with the advantage that this Bayesian model can be shared openly with others so that they can benefit from these modeling efforts53.
In this study we had greater success identifying additional inhibitors of Mtb ThyX than GyrB. While there is some inhibition overlap revealed by the NQs, this could suggest that the chemical property/feature permissiveness of Mtb ThyX is greater than GyrB due to differences in the binding site interactions. Substructure searching for compounds previously identified in whole-cell screens against Mtb identified a NQ (B6) with activity against Mtb ThyX (Fig. 3) and moderate antibacterial activity on Mtb (Supplementary Table 4). Idebenone, while not particularly active in whole Mtb cells may target ThyX as part of its mechanism of action in Mtb. Therefore, idebenone could be a starting point for structure-based design or by creating new derivatives using our previous Bayesian models based on whole cell activity data14. The progress of idebenone in clinical trials and as an approved drug suggests that it is likely very acceptable from the point of view of physicochemical properties, formulation and safety.
In future work, new compounds could be evaluated with our previously published Bayesian models for whole cell activity14 alongside the current Mtb ThyX Bayesian model in order to prioritize molecules for testing. This would enable the optimization of both target and whole cell activity in parallel. We propose that a combination of pharmacophore modeling, target-based whole-cell assay and resultant machine learning molecules using this data could result in the identification of novel scaffolds for pursuit.
Methods
Compounds
Tested compounds were purchased from ChemBridge, Vitas-M Laboratory Ltd., InterBioScreen or Sigma and were >90% pure (see Supplementary Table 6 for more details).
ThyX assays
Mtb ThyX activity was measured using the tritium release assay, as previously described for Helicobacter pylori ThyX26. Reaction mixture included 10 mM MgCl2, 300 mM NaCl, 500 μM FAD, bovine serum albumin (200 μg/ml), 250 μM CH2H4folate (Eprova, Merck), 1 mM NADPH, 100 μM dUMP, [5H3]dUMP (Moravek Biochemicals, CA, USA) and 1 μM Mtb ThyX in 50 mM HEPES pH 8. DMSO concentration was maintained constant at 1%. Reactions were initiated by addition of NADPH (1 mM) at 37 °C and were stopped after 7 mins. For idebenone, IC50 value decreased as a function of increasing dUMP concentration indicating an un-competitive mode of inhibition. Therefore, the formula IC50 = Ki * (1 + Km/[S]) + [E]/2 where Km is the Michaelis constant for dUMP, [S] is the dUMP concentration, and [E] is the enzyme concentration in the assay, was used to convert a measured IC50 value to the corresponding Ki.
Gyrase assays
Mtb gyrase supercoiling assays were carried out as described previously54.
Drug susceptibility testing against Mtb
A resazurin (Alamar Blue) assay was used to assess activity against strains of Mtb55. The antimicrobial susceptibility test was performed in a clear-bottomed, round well, 96-well microtiter plate. Compounds were tested at 8 concentrations ranging between 40 and 0.31 μg/mL with a final DMSO concentration of 1.25% in each well. After a growth medium containing ~104 bacteria was added to each well, the different dilutions of compounds were added. Controls included wells containing concentrations of rifampin and isoniazid ranging from 0.00039 to 8.0 μg/mL to control for assay performance; wells with bacteria, growth medium, and vehicle (1.25% DMSO); and sterility control wells with medium. Plates were incubated at 37 °C for 6 d in an ambient incubator at which time 5 μL of 1% resazurin dye was added to each well. After 2 days of incubation, visual inspection of color (pink, periwinkle or blue) was recorded for each well along with measurements of fluorescence in a microtiter plate fluorimeter with excitation at 530 nm and emission at 590 nm. The lowest drug concentration that inhibited growth of ≥90% of Mtb bacilli in the broth was considered the MIC90 value56. Rifampicin and isoniazid were used as positive controls and were consistently in the acceptable range. Values were converted from μg/ml to μM throughout the paper, for consistency.
Target-based whole-cell screening
The susceptibility of Mtb H37Rv and a conditional knockdown of ThyX in Mtb, thyX Tet-OFF, to a subset of test compounds was also determined by the broth microdilution method, as described previously39. Briefly, bacteria were grown in Middlebrook 7H9 broth (BD) supplemented with OADC (BD), 0.2% glycerol and 0.05% Tween-80 to mid-exponential phase. The culture was diluted and ~105 CFU/ml was added to a 96-well microtiter plate containing 2-fold serial dilutions of drug which was then incubated at 37 °C. For the pairwise combination (anhydrotetracycline (ATc) vs. test compound) assay using the thyX Tet-OFF strain, a two-dimensional array of serial dilutions of ATc and the test compound was prepared in a 96-well plate. Control wells consisting of bacteria only or medium only were treated with the same concentration of DMSO as used in drug-containing wells. At day 7, 10 μl of Alamar Blue solution (Invitrogen) was added and plates were reincubated at 37 °C. After 24 h, fluorescence (excitation 544 nm; emission 590 nm) was measured in a FLUOstar OPTIMA plate reader (BMG LABTECH, Offenberg, Germany). Data were normalised to the minimum and maximum inhibition controls to generate a dose response curve (% inhibition) from which the MIC90 was determined.
Substructure searching
The CDD database (Collaborative Drug Discovery Inc. Burlingame, CA) has been described previously and applied for TB research14. The literature data on Mtb drug discovery has been curated and 26 Mtb specific datasets (including published data from high throughput screens performed by NIAID/Southern Research Institute have been compiled in the CDD Public database) are hosted, representing over 350,000 compounds derived from patents, literature and HTS data, and we have termed this the CDD TB DB. The NQ substructure was used to query these NIAID/Southern Research Institute data. Molecules which showed previously published good whole-cell activity (MIC90) were selected for testing. The data generated in this study were collected and uploaded in the CDD private Vault (http://www.collaborativedrug.com/register) from sdf files and mapped to custom protocols.
Common features pharmacophores
A set of NQs (Supplemental Table 1) was used to build common features pharmacophores for Mtb ThyX and GyrB with Discovery Studio 4.1 (Biovia, San Diego, CA) from 3D conformations of the molecules generated with the CAESAR algorithm. This identified key features for each protein. The pharmacophores were then used to search various databases (for which up to 100 molecule conformations with the FAST conformer generation method with the maximum energy threshold of 20 kcal/mol, were created) such as the NIH clinical drugs set containing over 700 molecules. The pharmacophore models were updated as additional data were generated.
Bayesian models
We have previously described the generation and validation of the Laplacian-corrected Bayesian classifier models for Mtb14 using Discovery Studio 3.5 (San Diego, CA). The models were all generated using the following molecular descriptors: molecular function class fingerprints of maximum diameter 6 (FCFP_6), AlogP, molecular weight, number of rotatable bonds, number of rings, number of aromatic rings, number of hydrogen bond acceptors, number of hydrogen bond donors, and molecular fractional polar surface area which were all calculated from input sdf files. This approach was applied to the ThyX data generated in this study using the cut off of 70% inhibition at 100 μM to define actives. The resulting models were also validated using leave-one-out cross-validation; 5-fold validation to generate the receiver operator curve area under the curve (ROC AUC); concordance; specificity, and selectivity, as described previously. In the current study, as well as using the datasets individually, we also combined the datasets. Bayesian models were also generated in the CDD Vault using CDD Models, as described previously53. The current implementation used the FCFP6 fingerprints alone, and by default 3-fold cross-validation is performed. The model can also be exported from CDD Vault for use in other open source software and mobile apps53.
Additional Information
How to cite this article: Djaout, K. et al. Predictive modeling targets thymidylate synthase ThyX in Mycobacterium tuberculosis. Sci. Rep. 6, 27792; doi: 10.1038/srep27792 (2016).
Supplementary Material
Acknowledgments
H.M. is supported by the Centre National de la Recherche Scientifique. The work of V.K. in the Epicentre lab was partially supported by The Council for Frontiers of Knowledge. The work was partially supported by a grant from the European Community’s Seventh Framework Program (grant 260872, MM4TB Consortium) to S.E., A.M. and V.M. Work in A.M.’s lab is also supported by grant BB/J004561/1 from BBSRC (UK) and the John Innes Foundation, and work in V.M.’s laboratory by grants from the South African Medical Research Council and the National Research Foundation of South Africa. The CDD TB database was made possible with funding from the Bill and Melinda Gates Foundation (Grant#49852 “Collaborative drug discovery for TB through a novel database of SAR data optimized to promote data archiving and sharing”). The project described was supported by Award Number 2R42AI088893-02 from the NIH/NIAID. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID/NIH. Biovia is kindly acknowledged for providing Discovery Studio to S.E.
Footnotes
S.E. is a consultant for Collaborative Drug Discovery and Collaborations in Chemistry. A.M. is a consultant for Inspiralis Ltd. The funder provided support in the form of salaries for authors [S.E.], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions K.D., V.S. and Y.B. performed in vitro experiments, analyzed results and wrote the paper. V.K., H.B., N.G.B., S.J.H., J.E.P. and P.B. performed in vitro experiments. P.B.M. and A.M. designed in vitro experiments and wrote the paper. V.M. edited the manuscript. H.M. designed the study, analyzed the results and wrote the paper. S.E. designed and performed in silico experiments, analyzed results and wrote the manuscript.
References
- World Health Organization. Global tuberculosis report 2015, http://www.who.int/tb/publications/global_report/en/, data of access 06/05/2016 (2015).
- Lonnroth K. et al. Towards tuberculosis elimination: an action framework for low-incidence countries. Eur Respir J 45, 928–952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab Z., Acosta C. D., Kluge H. H. & Dara M. Consolidated Action Plan to Prevent and Combat Multidrug- and Extensively Drug-resistant Tuberculosis in the WHO European Region 2011–2015: Cost-effectiveness analysis. Tuberculosis (Edinb) 95 Suppl 1, S212–S216 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol 45, 529–564 (2005). [DOI] [PubMed] [Google Scholar]
- Ballel L., Field R. A., Duncan K. & Young R. J. New small-molecule synthetic antimycobacterials. Antimicrob Agents Chemother 49, 2153–2163 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A. I. et al. New antituberculosis drugs, regimens, and adjunct therapies: needs, advances, and future prospects. Lancet Infect Dis 14, 327–340 (2014). [DOI] [PubMed] [Google Scholar]
- Maddry J. A. et al. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 89, 354–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan S. et al. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 89, 334–353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. C. et al. High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 92, 72–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunatha U. H. & Smith P. W. Perspective: Challenges and opportunities in TB drug discovery from phenotypic screening. Bioorg Med Chem 23, 5087–5097 (2015). [DOI] [PubMed] [Google Scholar]
- Andries K. et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227 (2005). [DOI] [PubMed] [Google Scholar]
- Ekins S., Freundlich J. S., Choi I., Sarker M. & Talcott C. Computational Databases, Pathway and Cheminformatics Tools for Tuberculosis Drug Discovery. Trends in Microbiology 19, 65–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S. et al. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models PLOSONE 8, e63240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S. et al. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chem Biol 20, 370–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Freundlich J. S., Hobrath J. V., Lucile White E. & Reynolds R. C. Combining computational methods for hit to lead optimization in Mycobacterium tuberculosis drug discovery. Pharm Res 31, 414–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana B. D., Karakousis P. C., Parish T. & Dick T. Future target-based drug discovery for tuberculosis? Tuberculosis (Edinb) 94, 551–556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker M. et al. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium tuberculosis. Pharm Res 29, 2115–2127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S. et al. Combining Metabolite-Based Pharmacophores with Bayesian Machine Learning Models for Mycobacterium tuberculosis Drug Discovery. PLoS One 10, e0141076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall N. & Meyer J. J. In vitro inhibition of drug-resistant and drug-sensitive strains of Mycobacterium tuberculosis by ethnobotanically selected South African plants. J Ethnopharmacol 66, 347–354 (1999). [DOI] [PubMed] [Google Scholar]
- Lall N. & Meyer J. J. Antibacterial activity of water and acetone extracts of the roots of Euclea natalensis. J Ethnopharmacol 72, 313–316 (2000). [DOI] [PubMed] [Google Scholar]
- Lall N. & Meyer J. J. Inhibition of drug-sensitive and drug-resistant strains of Mycobacterium tuberculosis by diospyrin, isolated from Euclea natalensis. J Ethnopharmacol 78, 213–216 (2001). [DOI] [PubMed] [Google Scholar]
- van der Kooy F., Meyer J. J. & Lall N. Antimycobacterial activity and possible mode of action of newly isolated neodiospyrin and other naphthoquinones from Euclea natalensis. South African Journal of Botany 72, 349–352 (2006). [Google Scholar]
- Mahapatra A. et al. Activity of 7-methyljuglone derivatives against Mycobacterium tuberculosis and as subversive substrates for mycothiol disulfide reductase. Bioorg Med Chem 15, 7638–7646 (2007). [DOI] [PubMed] [Google Scholar]
- Dey D., Ray R. & Hazra B. Antitubercular and antibacterial activity of quinonoid natural products against multi-drug resistant clinical isolates. Phytother Res 28, 1014–1021 (2014). [DOI] [PubMed] [Google Scholar]
- Tran T. et al. Quinones as antimycobacterial agents. Bioorg Med Chem 12, 4809–4813 (2004). [DOI] [PubMed] [Google Scholar]
- Skouloubris S. et al. Targeting of Helicobacter pylori thymidylate synthase ThyX by non-mitotoxic hydroxy-naphthoquinones. Open Biol 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta T. et al. Mechanistic and structural basis for inhibition of thymidylate synthase ThyX. Open Biol 2, 120120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkare S. et al. The naphthoquinone diospyrin is an inhibitor of DNA gyrase with a novel mechanism of action. J Biol Chem 288, 5149–5156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner D. F., Evans J. C. & Mizrahi V. Nucleotide Metabolism and DNA Replication. Microbiol Spectr 2 (2014). [DOI] [PubMed] [Google Scholar]
- Myllykallio H. et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297, 105–107 (2002). [DOI] [PubMed] [Google Scholar]
- Koehn E. M. et al. An unusual mechanism of thymidylate biosynthesis in organisms containing the thyX gene. Nature 458, 919–923 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H. F. et al. Substrate interaction dynamics and oxygen control in the active site of thymidylate synthase ThyX. Biochem J 459, 37–45 (2014). [DOI] [PubMed] [Google Scholar]
- Leduc D. et al. Flavin-dependent thymidylate synthase ThyX activity: implications for the folate cycle in bacteria. J Bacteriol 189, 8537–8545 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivian-Hughes A. S., Houghton J. & Davis E. O. Mycobacterium tuberculosis thymidylate synthase gene thyX is essential and potentially bifunctional, while thyA deletion confers resistance to p-aminosalicylic acid. Microbiology 158, 308–318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. H., Gujjar R., Pang C. K. & Rathod P. K. Kinetics and ligand-binding preferences of Mycobacterium tuberculosis thymidylate synthases, ThyA and ThyX. PLoS One 3, e2237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar P. et al. Structure of the Mycobacterium tuberculosis flavin dependent thymidylate synthase (MtbThyX) at 2.0A resolution. J Mol Biol 352, 1091–1104 (2005). [DOI] [PubMed] [Google Scholar]
- Kogler M. et al. Synthesis and evaluation of 6-aza-2′-deoxyuridine monophosphate analogs as inhibitors of thymidylate synthases, and as substrates or inhibitors of thymidine monophosphate kinase in Mycobacterium tuberculosis. Chem Biodivers 9, 536–556 (2012). [DOI] [PubMed] [Google Scholar]
- Kogler M. et al. Synthesis and evaluation of 5-substituted 2′-deoxyuridine monophosphate analogues as inhibitors of flavin-dependent thymidylate synthase in Mycobacterium tuberculosis. J Med Chem 54, 4847–4862 (2011). [DOI] [PubMed] [Google Scholar]
- Singh V. et al. The complex mechanism of antimycobacterial action of 5-fluorouracil. Chem Biol 22, 63–75 (2015). [DOI] [PubMed] [Google Scholar]
- Maxwell A. & Lawson D. M. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr Top Med Chem 3, 283–303 (2003). [DOI] [PubMed] [Google Scholar]
- Bush N. G., Evans-Roberts K. & Maxwell A. DNA Topoisomerases, in InEcoSal—Escherichia coli and Salmonella: cellular and molecular biology. (eds. Böck A. et al.) (ASM Press, Washington DC, 2015). [Google Scholar]
- Ekins S. et al. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis Mol BioSyst 6, 2316–2324 (2010). [DOI] [PubMed] [Google Scholar]
- Ekins S. et al. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems 6, 840–851 (2010). [DOI] [PubMed] [Google Scholar]
- Ekins S., Casey A. C., Roberts D., Parish T. & Bunin B. A. Bayesian models for screening and TB Mobile for target inference with Mycobacterium tuberculosis. Tuberculosis (Edinb) 94, 162–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams G. L. et al. Pathway-selective sensitization of Mycobacterium tuberculosis for target-based whole-cell screening. Chem Biol 19, 844–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y., Shinkai Y., Miura T. & Cho A. K. The chemical biology of naphthoquinones and its environmental implications. Annu Rev Pharmacol Toxicol 52, 221–247 (2012). [DOI] [PubMed] [Google Scholar]
- Schuck D. C. et al. Biological evaluation of hydroxynaphthoquinones as anti-malarials. Malar J 12, 234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. H. et al. Synthesis and Biological Evaluation of Lipophilic 1,4-Naphthoquinone Derivatives against Human Cancer Cell Lines. Molecules 20, 11994–12015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyse G. M. et al. Efficacy of idebenone on respiratory function in patients with Duchenne muscular dystrophy not using glucocorticoids (DELOS): a double-blind randomised placebo-controlled phase 3 trial. Lancet 385, 1748–1757 (2015). [DOI] [PubMed] [Google Scholar]
- Erb M. et al. Features of idebenone and related short-chain quinones that rescue ATP levels under conditions of impaired mitochondrial complex I. PLoS One 7, e36153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpa J. et al. Triple therapy with darbepoetin alfa, idebenone, and riboflavin in Friedreich’s ataxia: an open-label trial. Cerebellum 12, 713–720 (2013). [DOI] [PubMed] [Google Scholar]
- Iyer S. Novel therapeutic approaches for Leber’s hereditary optic neuropathy. Discov Med 15, 141–149 (2013). [PMC free article] [PubMed] [Google Scholar]
- Clark A. M. et al. Open source bayesian models: 1. Application to ADME/Tox and drug discovery datasets. J Chem Inf Model 55, 1231–1245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkare S., Yousafzai F., Mitchenall L. A. & Maxwell A. The role of Ca(2)(+) in the activity of Mycobacterium tuberculosis DNA gyrase. Nucleic Acids Res 40, 9774–9787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino J. C. et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46, 2720–2722 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L. & Franzblau S. G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother 41, 1004–1009 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.