Abstract
miR-17 family microRNAs (miRNAs) are crucial for embryo development, however, their role in muscle development is still unclear. miR-20a-5p and miR-20b-5p belong to the miR-17 family and are transcribed from the miR-17~92 and miR-106a~363 clusters respectively. In this study, we found that miR-20a-5p and miR-20b-5p promoted myoblast differentiation and repressed myoblast proliferation by directly binding the 3′ UTR of E2F transcription factor 1 (E2F1) mRNA. E2F1 is an important transcriptional factor for organism’s normal development. Overexpression of E2F1 in myoblasts promoted myoblast proliferation and inhibited myoblast differentiation. Conversely, E2F1 inhibition induced myoblast differentiation and repressed myoblast proliferation. Moreover, E2F1 can bind directly to promoters of the miR-17~92 and miR-106a~363 clusters and activate their transcription, and E2F1 protein expression is correlated with the expression of pri-miR-17~92 and pri-miR-106a~363 during myoblast differentiation. These results suggested an auto-regulatory feedback loop between E2F1 and miR-20a-5p/20b-5p, and indicated that miR-20a-5p, miR-20b-5p and E2F1 are involved in myoblast proliferation and differentiation through the auto-regulation between E2F1 and miR-20a-5p/20b-5p. These findings provide new insight into the mechanism of muscle differentiation, and further shed light on the understanding of muscle development and muscle diseases.
MicroRNAs (miRNAs) are endogenous noncoding single-stranded RNAs that can play important regulatory roles in animals and plants1. Approximately one-third of all mammalian genes are thought to be targeted by miRNAs2. Many physiological and biochemical processes, such as tumorigenesis, development, cell proliferation, differentiation and apoptosis, have been found to be regulated by miRNAs3,4,5,6,7. During muscle development, many miRNAs have been found to regulate diverse aspects of developmental processes. Some of these miRNAs exhibit muscle-specific expression pattern and play a critical role in muscle cell proliferation and differentiation. These muscle-specific miRNAs are called myogenic miRNAs (MyomiRs), which include miR-1, miR-206 and miR-1338,9. They can interact with myogenic regulatory factors (MRFs) and regulate many muscle development-related genes, therefore play important roles in muscle development4,10. Additionally, many non-MyomiRs also play roles in muscle development. By directly inhibiting MyoD, MEF2C, Pax3, YY1, IGF-2, Cdc25A and the other genes that are related to myoblast proliferation or differentiation, these non-MyomiRs are all involved in the regulation of muscle development4,11,12,13,14,15.
Three paralogous miRNA gene clusters, which include miR-17~92, miR-106a~363 and miR-106b~25, are implicated in a wide variety of cellular processes16. Mice deficient for miR-17~92 die shortly after birth with defects in lung, heart and other organs, while ablation of miR-106a~363 or miR-106b~25 has no obvious phenotypic consequences17. Among these parologous miRNAs, the miR-17 family, composed of miR-17, miR-20a/b, miR-106a/b and miR-93, has been demonstrated to involve in critical pathways that regulate embryo development and stem cell differentiation18. Members of the miR-17 family have same seed sequence, therefore these paralogous miRNAs would sharing some overlapping functions. In human tumours, the expression of miR-17 family miRNAs is correlated with the cell cycle genes expression, and this correlation is resulted from the inhibitory effect of these miRNAs to their common target genes, which can negatively regulate cell cycle progression19. miR-17-5p and miR-20a, which are induced by c-Myc, can regulate E2F1 translational yield by binding to the 3′ UTR of E2F1 mRNA and then regulate cell proliferation20. Additionally, the developmental processes of lung, heart and B cell are all related to the roles of the miR-17 family miRNAs16, demonstrating the critical combined effects of these miRNAs during embryo development. However, as an essential part of embryo development regulation, the roles of miR-17 family during muscle development are still not clear.
Members of the E2F transcription factor family play an essential role in the regulation of cellular proliferation and cell cycle progression during organism’s normal development21,22. Mice lacking E2F1 show testicular atrophy and exocrine gland dysplasia23. During muscle development, dyregulated E2F1 expression can induce the inhibition of myogenic differentiation24. E2F1-mediated transcription plays an essential role in myogenesis25. The transcription activity of myogenic bHLH proteins MyoD and myogenin, which are critical regulators for myogenic differentiation26,27, can be inhibited by E2F125. During C2C12 myocyte differentiation, the expression of E2F1 is down-regulated24. However, the regulation of E2F1 during this process still remains to be demonstrated.
Avian is an ideal model organism for muscle development research. In our previous study, we found that miR-20a-5p and miR-20b-5p have a significantly higher expression level in skeletal muscle at day 14 embryo (E14) than at 7 week (7W) chick28. Considering that the proliferation and differentiation of muscle cells are very different between E14 and 7W, we assume that miR-20a-5p and miR-20b-5p may be involved in these two processes. In this study, we confirmed that miR-20a-5p and miR-20b-5p can regulate myoblast proliferation and differentiation in quail muscle clone 7 (QM-7) cells, which closely resemble their mammalian counterparts in most respects29,30. miR-20a-5p and miR-20b-5p overexpression inhibited myoblast proliferation but promoted myoblast differentiation, whereas inhibition of miR-20a-5p and miR-20b-5p could result in increased proliferation and decreased differentiation of myoblasts. Both miR-20a-5p and miR-20b-5p can directly bind to the 3′ UTR of E2F1 mRNA and inhibit E2F1 expression. Moreover, E2F1 binds directly to the promoters of the miR-17~92 and miR-106a~363 clusters and activates their transcription. The protein level of E2F1 is correlated with the expression of pri-miR-17~92 and pri-miR-106a~363 during myoblast differentiation. Altogether, these results suggest an auto-regulatory feedback loop between E2F1 and miR-20a-5p/20b-5p, and this auto-regulatory loop may play important roles in myoblast proliferation and differentiation.
Results
miR-20a-5p and miR-20b-5p repress myoblast proliferation
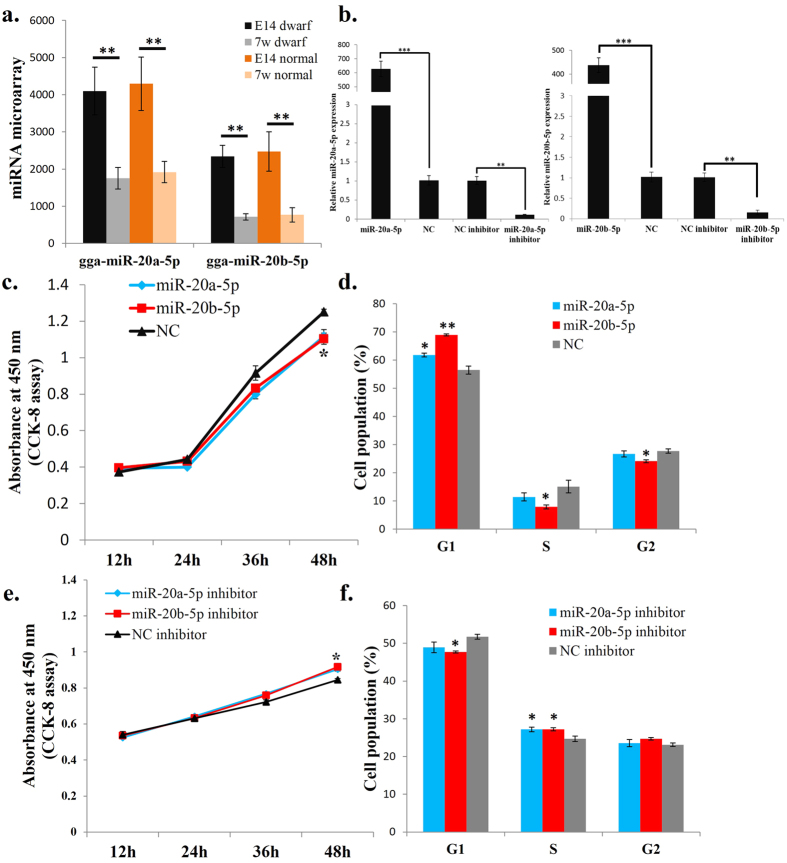
Our previous microarray data (GSE37360, GSE37367 and GSE37368) showed that the expression levels of miR-20a-5p and miR-20b-5p are significantly higher in the leg muscle of E14 normal and dwarf chickens than those in 7w chickens (Fig. 1a). To investigate the effects of miR-20a-5p and miR-20b-5p on avian muscle cell, we used the QM-7 cell lines as a model system to identify the functional characteristics of miR-20a-5p and miR-20b-5p in skeletal myogenesis. Firstly we introduced miR-20a-5p (5′-UAAAGUGCUUAUAGUGCAGGUAG-3′) and miR-20b-5p (5′-CAAAGUGCUCAUAGUGCAGGUAG-3′) mimics into the QM-7 cells (Fig. 1b) and then monitored the proliferation status of cells using the Cell Counting Kit-8. The results showed that overexpression of either miR-20a-5p or miR-20b-5p repressed cell proliferation significantly (Fig. 1c). Next, we used flow cytometry analysis of the cell cycle to test the effects of miR-20a-5p and miR-20b-5p on cell cycle regulation. The results showed that QM-7 transfected with the miR-20a-5p and miR-20b-5p mimic could significantly increase the number of cells in the G0/G1 stage, but decrease the number of cells in the S and G2 stages (Fig. 1d and Supplementary File 1). Additionally, we transfected QM-7 cells with the inhibitor of miR-20a-5p and miR-20b-5p (Fig. 1b). CCK-8 assay showed that either miR-20a-5p or miR-20b-5p knockdown can promote cell proliferation (Fig. 1e), and the results of cell cycle analysis showed that both miR-20a-5p inhibitor and miR-20b-5p inhibitor can increase the number of cells in the S stage (Fig. 1f and Supplementary File 1). Therefore, these data suggest that both miR-20a-5p and miR-20b-5p can repress myoblast proliferation and affect cell cycle progression of avian myoblasts.
Figure 1. miR-20a-5p and miR-20b-5p repress myoblast proliferation.
(a) The miRNA microarray hybridization signals of miR-20a-5p and miR-20b-5p in E14 and 7w of dwarf and normal chickens’ leg muscle. (b) The expression of miR-20a-5p and miR-20b-5p after transfection of the indicated miRNA mimic or miRNA inhibitor. (c) CCK-8 assay was performed to access the effect of miR-20a-5p and miR-20b-5p overexpression on myoblast proliferation. (d) Cell cycle analysis of myoblasts at 48 h after transfection of miR-20a-5p and miR-20b-5p mimics or NC mimic. (e) CCK-8 assay was performed to access the effect of miR-20a-5p and miR-20b-5p loss-of-function on myoblast proliferation. (f) Cell cycle analysis of myoblasts at 48 h after transfection of miR-20a-5p inhibitor, miR-20b-5p inhibitor or NC inhibitor. Results are shown as the mean ± sem of three independent experiments. One sample t test was used to analysis the statistical differences between groups. *p < 0.05; **p < 0.01; ***p < 0.001.
miR-20a-5p and miR-20b-5p promote myoblast differentiation
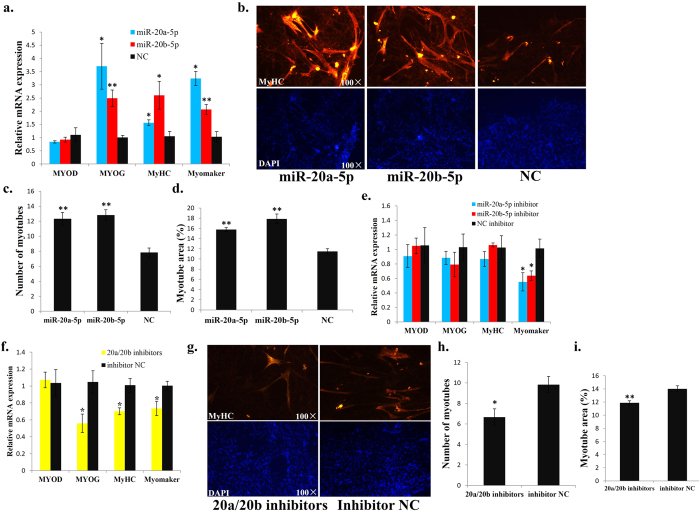
As cell cycle arrest is important for myoblast differentiation, and both miR-20a-5p and miR-20b-5p are implicated in cell cycle arrest, we next studied the potential functions of miR-20a-5p and miR-20b-5p in myoblast differentiation. QM-7 cells cultured in growth medium (GM) were transfected with the miR-20a-5p mimic and miR-20b-5p mimic. After transfection, the cells were induced to differentiation by changing the GM to differentiation medium (DM). Four major myoblast differentiation marker genes, MYOD (named mf1 gene in quail, accession: L16686.1), MYOG (named mf2 gene in quail, accession: L15473.1), MyHC (named slow myosin heavy chain gene in quail, accession: U53862.1) and Myomaker (referred to chicken TMEM8C gene, accession: KP230536.1), were detected at 72 h after differentiation according to qPCR analysis. The expressions of MYOG, MyHC and Myomaker were all significantly up-regulated in the miR-20a-5p- and miR-20b-5p-transfected cells compared with those in the control cells (Fig. 2a), whereas the MYOD expression did not alter between the two groups of cells (Fig. 2a). Moreover, overexpression of miR-20a-5p and miR-20b-5p markedly increased the number of MyHC-positive cells (Fig. 2b,c) and the total areas of myobube (Fig. 2d).
Figure 2. miR-20a-5p and miR-20b-5p promote myoblast differentiation.
(a) The relative mRNA expression of MYOD, MYOG, MyHC and Myomaker after transfection of miR-20a-5p, miR-20b-5p or NC mimic. (b) MyHC staining of myoblasts at 72 h after transfection of miR-20a-5p, miR-20b-5p or NC mimic. (c) Number of myotubes at 72 h after transfection of miR-20a-5p, miR-20b-5p or NC mimic. (d) Myotube area (%) at 72 h after transfection of miR-20a-5p, miR-20b-5p or NC mimic. (e) The relative mRNA expression of MYOD, MYOG, MyHC and Myomaker after transfection of miR-20a-5p inhibitor, miR-20b-5p inhibitor or NC inhibitor. (f) The relative mRNA expression of MYOD, MYOG, MyHC and Myomaker after transfection of miR-20a-5p/miR-20b-5p mixed inhibitors or NC inhibitor. (g) MyHC staining of myoblasts at 72 h after transfection of miR-20a-5p/miR-20b-5p mixed inhibitors or NC inhibitor. (h) Number of myotubes at 72 h after transfection of miR-20a-5p/miR-20b-5p mixed inhibitors or NC inhibitor. (i) Myotube area (%) at 72 h after transfection of miR-20a-5p/miR-20b-5p mixed inhibitor or NC inhibitor. Results are shown as the mean ± sem of three independent experiments. One sample t test was used to analysis the statistical differences between groups. *p < 0.05; **p < 0.01.
To better understand the physiological relevance of miR-20a-5p and miR-20b-5p during myoblast differentiation, we transfected the QM-7 cells with miR-20a-5p inhibitor and miR-20b-5p inhibitor and then induced the cells to differentiation. After 72 h differentiation, we detected the relative expression of the four genes. The results showed that only Myomaker was significantly down-regulated in the miR-20a-5p inhibitor- and miR-20b-5p inhibitor-transfected cells (Fig. 2e). However, as we co-transfected the QM-7 cells with miR-20a-5p and miR-20b-5p mixed inhibitors and then induced the cells to differentiation, the expressions of MYOG, MyHC and Myomaker were all reduced when compared with those in the control cells, although the MYOD expression also has no differences between the two groups of cells (Fig. 2f). The number of MyHC-positive cells and the total areas of myotubes were also significantly reduced when miR-20a-5p and miR-20b-5p mix inhibitors were transfected during QM-7 differentiation (Fig. 2g–i). Therefore, we argue that both miR-20a-5p and miR-20b-5p promote myoblast differentiation.
E2F1 is a direct target gene of miR-20a-5p and miR-20b-5p
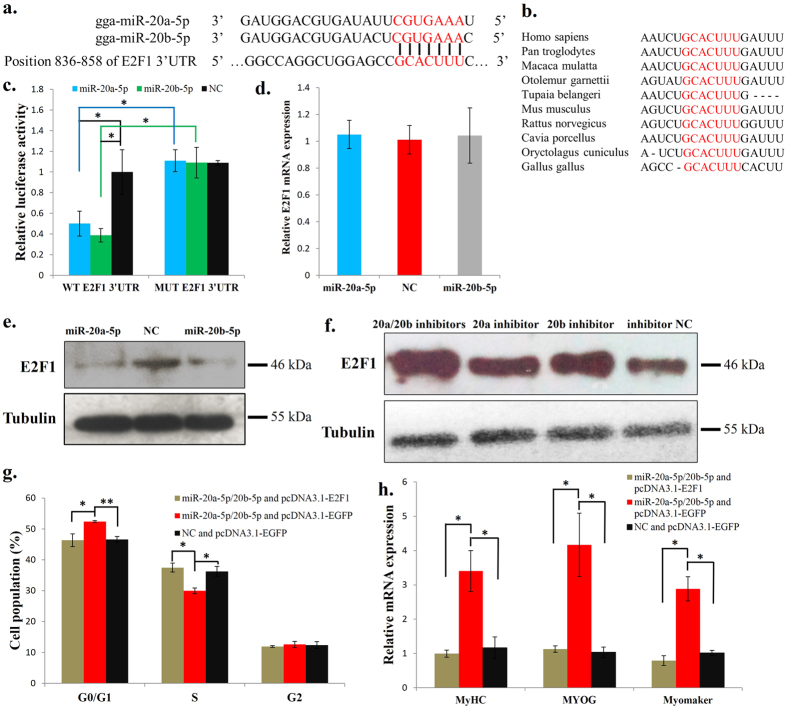
Previous study has demonstrated that E2F1 is a direct target of miR-20a in a mammal prostate cancer cell line31, and the E2F1 transcription factor is an important regulator of embryo development23. Therefore, we examined whether E2F1 is a target gene of miR-20a-5p and miR-20b-5p during avian myoblast differentiation. By searching for the 3′ UTR of chicken E2F1 mRNA (NM_205219.1), we found that the seed sequence of miR-20a-5p and miR-20b-5p can perfectly match the position 851-857 of chicken E2F1 mRNA 3′ UTR (Fig. 3a), and the sequence of this position are highly conserved among vertebrates (Fig. 3b). To further determine whether miR-20a-5p and miR-20b-5p could bind to the 3′ UTR of E2F1 mRNA directly, we constructed two dual-luciferase reporter vectors with the wild-type or mutant 3′ UTR of E2F1 inserted at the 3′ end of the firefly luciferase gene. Then we tested the effects of miR-20a-5p and miR-20b-5p on these reporters in DF-1 cells. The results showed that miR-20a-5p and miR-20b-5p transfection significantly inhibited the reporter activity, whereas they had no effect on the mutant reporter activity (Fig. 3c), suggesting that the predicted binding site in the E2F1 3′ UTR is a bona fide target of miR-20a-5p and miR-20b-5p. The mRNA and protein levels of E2F1 were also detected in miR-20a-5p and miR-20b-5p overexpressing myoblasts, respectively. Myoblasts transfected with either miR-20a-5p or miR-20b-5p would reduce E2F1 protein expression, whereas they had no significant effect on the mRNA level of E2F1 (Fig. 3d,e). Additionally, E2F1 protein level can be increased with the transfection of either miR-20a-5p inhibitor or miR-20b-5p inhibitor (Fig. 3f), and the protein level can be increased more when miR-20a-5p and miR-20b-5p inhibitors were co-transfected into the cells (Fig. 3f).
Figure 3. E2F1 is a direct target gene of miR-20a-5p and miR-20b-5p.
(a) The potential binding site (red) of miR-20a-5p and miR-20b-5p in the E2F1 mRNA 3′ UTR. Sequence in blue indicate the mutation of the binding site. (b) The potential binding site (red) of miR-20a-5p and miR-20b-5p in the E2F1 mRNA 3′ UTR is highly conserved among vertebrates. (c) Dual-luciferase reporter assay indicated that miR-20a-5p and miR-20b-5p can bind to the predicted binding site in the E2F1 mRNA 3′ UTR. (d) E2F1 mRNA expression after transfection of miR-20a-5p and miR-20b-5p mimic. (e) E2F1 protein expression after transfection of miR-20a-5p and miR-20b-5p mimic. (f) E2F1 protein expression after transfection of miR-20a-5p inhibitor, miR-20b-5p inhibitor, miR-20a-5p/miR-20b-5p mixed inhibitors or NC inhibitor. (g) Myoblasts were co-transfected with miR-20a-5p/20b-5p and pcDNA3.1-E2F1, miR-20a-5p/20b-5p and pcDNA3.1-EGFP, or NC and pcDNA3.1-EGFP as control, followed by 48 h culture, and the cell cycle phase was then analysed. (h) Myoblasts were co-transfected with miR-20a-5p/20b-5p and pcDNA3.1-E2F1, miR-20a-5p/20b-5p and pcDNA3.1-EGFP, or NC and pcDNA3.1-EGFP as control, followed by 72 h differentiation, and the expression of MYOG, MyHC and Myomaker was then quantified by q-PCR. Results are shown as the mean ± sem of three independent experiments. One sample t test was used to analysis the statistical differences between groups. *p < 0.05; **p < 0.01.
To further evaluate the principal roles of E2F1 in the functional effects induced by miR-20a-5p and miR-20b-5p, we overexpressed E2F1 in myoblasts with transfection of miR-20a-5p and miR-20b-5p mixed mimics. The results showed that E2F1 overexpression can restore cell cycle arrest in miR-20a-5p/20b-5p transfected cells (Fig. 3g), and the presence of miR-20a-5p/20b-5p with control plasmid (pcDNA3.1-EGFP) in myoblasts led to increased cell cycle arrest in the G0/G1 stage. Furthermore, E2F1 overexpression can also overcome the differentiation effects of miR-20a-5p/20b-5p in myoblasts. The increased expressions of MYOG, MyHC and Myomaker as a consequence of miR-20a-5/20b-5p transfection were attenuated by the co-expression of E2F1 (Fig. 3h). Therefore, E2F1 is a direct target gene of miR-20a-5p and miR-20b-5p, and its overexpression could overcome the cell cycle and differentiation effects of miR-20a-5p and miR-20b-5p in myoblasts.
E2F1 promotes myoblast proliferation and represses myogenic differentiation
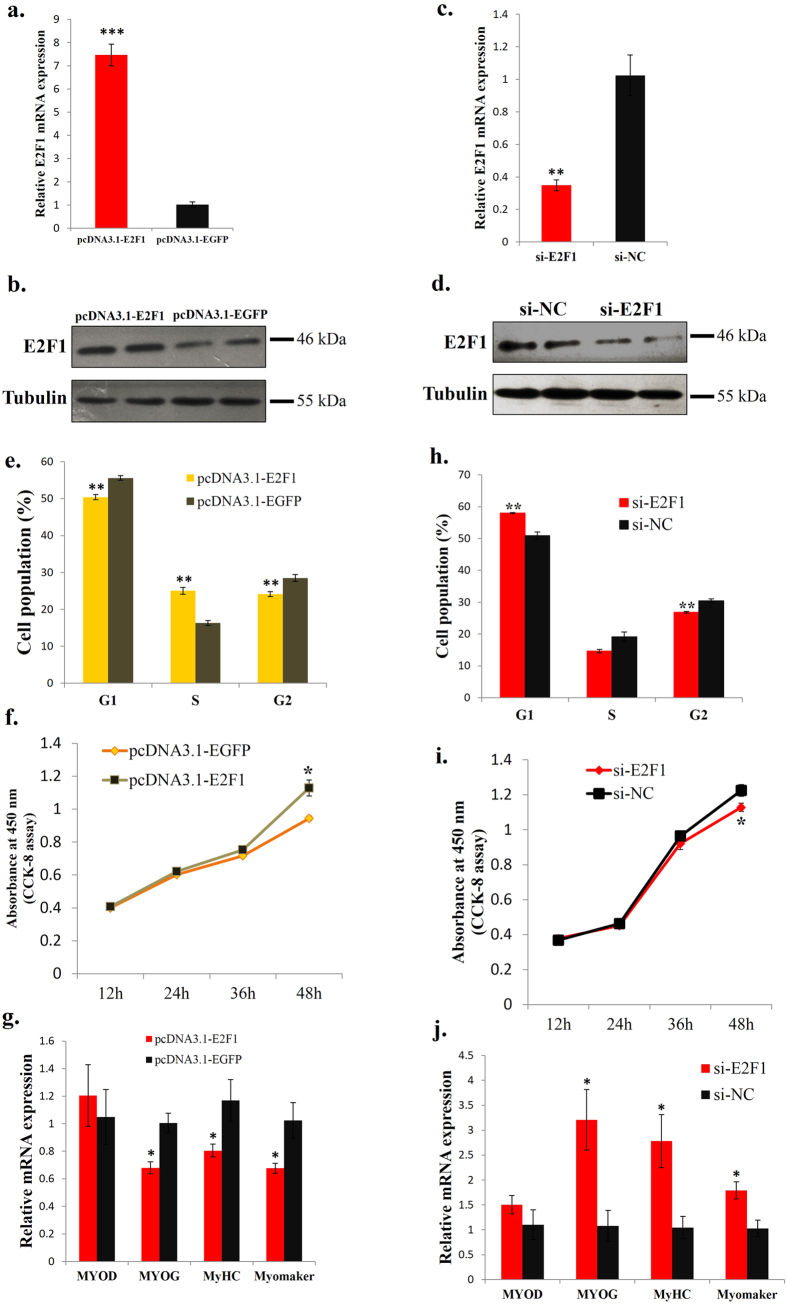
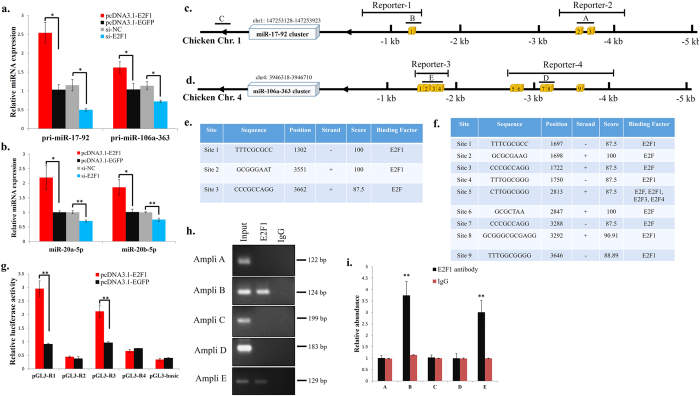
In this study, we performed E2F1 overexpression and knockdown assays in QM-7 cells to verify its functions in avian myogenic proliferation and differentiation. As shown in Fig. 4, transfecting pcDNA3.1-E2F1 into myoblasts increased E2F1 mRNA and protein expression (Fig. 4a,b), whereas transfecting si-E2F1 into myoblasts decreased E2F1 mRNA and protein expression (Fig. 4c,d). E2F1 overexpression significantly promoted myoblast proliferation, reduced the number of cells that progressed to G0/G1 phase and increased the number of cells that progressed to S phase (Fig. 4e,f and Supplementary File 1). Additionally, E2F1 overexpression significantly reduced MYOG, MyHC and Myomaker expression (Fig. 4g), suggesting an inhibitory role of E2F1 in myogenic differentiation. On the other hand, knockdown of the E2F1 expression in QM-7 cells significantly increased the number of cells in the G0/G1 stage and reduced the number of cells in the S and G2 stages (Fig. 4h and Supplementary File 1). The CCK-8 assay also showed that E2F1 knockdown significantly reduced cell proliferation (Fig. 4i). Moreover, we transfected QM-7 cells with anti-E2F1 siRNA in GM and then induced the cell to differentiation. The results showed that E2F1 knockdown significantly increased MYOG, MyHC and Myomaker expression, whereas the expression of MYOD had not been affected (Fig. 4j). Therefore we conclude that E2F1 can regulate avian myoblast proliferation and differentiation.
Figure 4. E2F1 promotes myoblast proliferation and represses myogenic differentiation.
(a) E2F1 mRNA expression after pcDNA3.1-E2F1 and pcDNA3.1-EGFP transfection. (b) E2F1 protein expression after pcDNA3.1-E2F1 and pcDNA3.1-EGFP transfection. (c) E2F1 mRNA expression after si-E2F1 and si-NC transfection. (d) E2F1 protein expression after si-E2F1 and si-NC transfection. (e) Cell cycle analysis of myoblasts at 48 h after pcDNA3.1-E2F1 and pcDNA3.1-EGFP transfection. (f) CCK-8 assay was performed to access the effects of E2F1 overexpression on myoblast proliferation. (g) The relative mRNA expression of MYOD, MYOG, MyHC and Myomaker after pcDNA3.1-E2F1 and pcDNA3.1-EGFP transfection. (h) Cell cycle analysis of myoblasts at 48 h after si-E2F1 and si-NC transfection. (i) CCK-8 assay was performed to access the effects of E2F1 loss-of-function on myoblast proliferation. (j) The relative mRNA expression of MYOD, MYOG, MyHC and Myomaker after si-E2F1 and si-NC transfection. Results are shown as the mean ± sem of three independent experiments. One sample t test was used to analysis the statistical differences between groups. *p < 0.05; **p < 0.01; ***p < 0.001.
E2F1 regulates miR-17~92 and miR-106a~363 expression directly
When we transfected QM-7 cells with pcDNA3.1-E2F1 or anti-E2F1 siRNA, we found that the expression of both miR-17~92 and miR-106a~363 pri-precursors was significantly changed (Fig. 5a). Additionally, the mature miR-20a-5p and miR-20b-5p expression also significantly changed (Fig. 5b). Considering that E2F1 is a transcriptional factor, we explored the possibility that E2F1 may directly regulate the transcription of the miR-17~92 and miR-106a~363 clusters. By using MATCH online software, a tool for predicting transcription factor binding sites in DNA sequence32, we found that there are three and nine putative E2F-binding sites within the first 5 kb of the promoters of the chicken miR-17~92 cluster and miR-106a-363 cluster, respectively (Fig. 5c–f). To confirm that E2F1 can regulate the expression of miR-17~92 and miR-106a~363 clusters directly, we cloned four reporters containing the potential E2F-binding sites of these promoters into the pGL3-basic luciferase reporter plasmid. The luciferase activity of both reporter-1 and reporter-3 was significantly increased when the two reporters were co-transfected with pcDNA3.1-E2F1 plasmid, respectively (Fig. 5g). However, reporter-2 and reporter-4 did not alter their luciferase activities when the pcDNA3.1-E2F1 plasmid were co-transfected, respectively (Fig. 5g). These results suggested that E2F1 can directly bind the regions of reporter-1 and reporter-3 and activate their transcriptions.
Figure 5. E2F1 regulates miR-17~92 and miR-106a~363 expression directly.
(a) The relative expression of pri-miR-17~92 and pri-miR-106a~363 after transfection with indicated plasmid or siRNA. (b) The relative expression of miR-20a-5p and miR-20b-5p after transfection with the indicated plasmid or siRNA. (c) Location of the putative E2F binding sites in the miR-17~92 cluster promoter. Yellow boxes represent the putative E2F binding sites. Horizontal bars represent the amplicons A to C used in the ChIP assay. Reporter-1 and Reporter-2 represent the promoter regions used in luciferase reporter assay. (d) Location of the putative E2F binding sites in the miR-106a~363 cluster promoter. Yellow boxes represent the putative E2F binding sites. Horizontal bars represent the amplicons D and E used in the ChIP assay. Reporter-3 and Reporter-4 represent the promoter regions used in luciferase reporter assay. (e) Basic information of the predicted E2F binding sites in the miR-17~92 cluster promoter. (f) Basic information of the predicted E2F binding sites in the miR-106a~363 cluster promoter. (g) The relative luciferase activity of pGL3-R1, pGL3-R2, pGL3-R3, pGL3-R4 and pGL3-Basic after pcDNA3.1-E2F1 or pcDNA3.1-EGFP transfection. (h) ChIP and PCR amplification of regions from the miR-17~92 and miR-106a~363 cluster promoter. (i) Quantitation of the ChIP assay results by real-time PCR. Results are shown as the mean ± sem of three independent experiments. One sample t test was used to analysis the statistical differences between groups. *p < 0.05; **p < 0.01.
To further confirm the binding of endogenous E2F1 to the promoters, we performed chromatin immunoprecipitation (ChIP) experiments to detect the association of E2F1 with the miR-17~92 and miR-106a-363 promoters in chicken primary myoblasts. In the miR-17~92 promoter, we generated amplicon A which overlaps site 2 and 3, amplicon B which overlaps site 1, and amplicon C which was used as a negative control. In the miR-106a~363 promoter, we generated amplicon D which overlaps site 7 and 8, and amplicon E which overlaps site 1, 2, 3 and 4 (Fig. 5c,d). The ChIP-PCR showed that only amplicon B and amplicon E were amplified after ChIP assay with anti-E2F1 antibody (Fig. 5h), and the relative abundance of the PCR product was more than two-fold compared to the control group (Fig. 5i). On the other hand, amplicon A, amplicon C and amplicon D were all negative. Therefore, these results suggest that the fragments of the amplicon B and amplicon E contain E2F1 binding site. Altogether, the above results present an auto-regulatory feedback loop, where translation of the E2F1 mRNA is controlled by miR-20a-5p and miR-20b-5p, which pri-precursors are regulated by E2F1 at the transcriptional level.
Correlation between pri-miR-17~92, pri-miR-106a~363 and E2F1 protein levels in QM-7 and chicken primary myoblasts undergoing differentiation in vitro
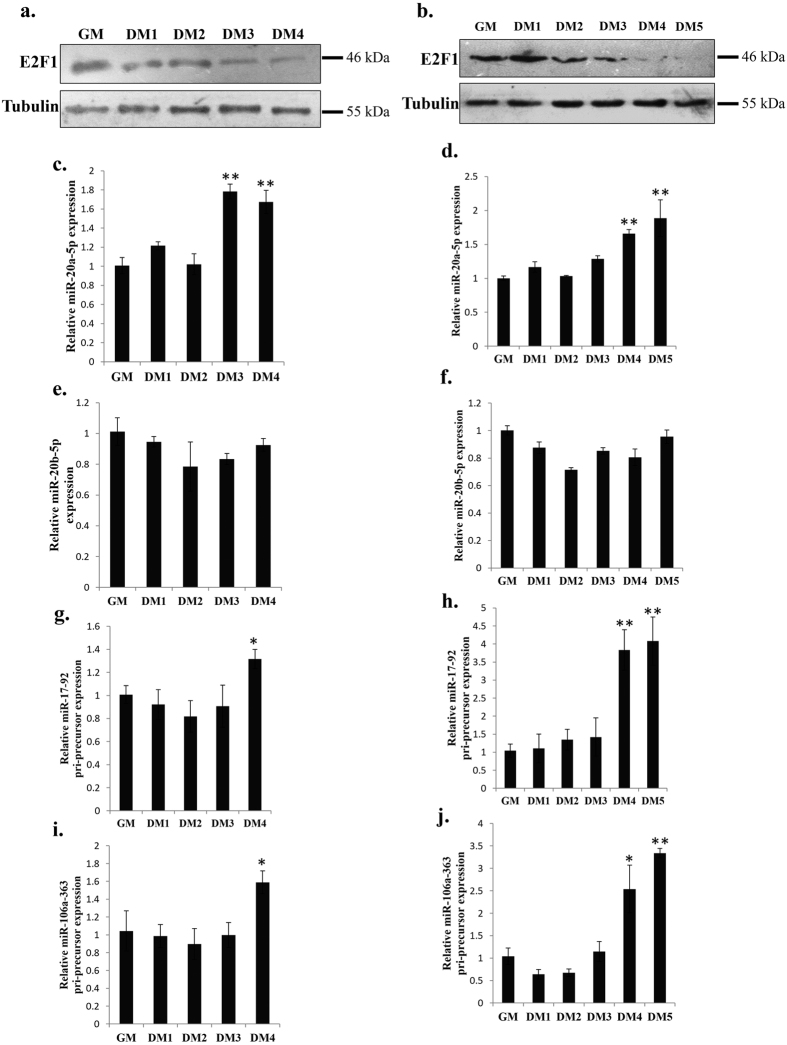
To better understand the potential physiologic relevance of miR-20a-5p, miR-20b-5p and E2F1 interaction during myogenesis, we next examined the expression of miR-20a-5p, miR-20b-5p and E2F1 during QM-7 and chicken primary myoblasts undergoing differentiation in vitro. Western blot assays showed that the E2F1 protein level was down-regulated gradually from GM to DM during both QM-7 and primary myoblast differentiation (Fig. 6a,b). On the other hand, miR-20a-5p expression was significantly up-regulated during myoblast differentiation (Fig. 6c,d), and there is no significant change in miR-20b-5p expression during the differentiation process (Fig. 6e,f). For the expression of miRNA pri-precursors, both pri-miR-17~92 and pri-miR-106a~363 were significantly up-regulated during myoblast differentiation (Fig. 6g–j). Therefore, the results of up-regulated pri-miR-17~92 and pri-miR-106a~363 expression and down-regulated E2F1 expression suggest a correlation relationship between these regulators during myoblast differentiation.
Figure 6. Correlation between pri-miR-17~92, pri-miR-106a~363 and E2F1 protein levels in QM-7 and chicken primary myoblast undergoing differentiation in vitro.
(a) E2F1 protein expression during QM-7 myoblast differentiation. (b) E2F1 protein expression during chicken primary myoblast differentiation. (c) The relative miR-20a-5p expression during QM-7 myoblast differentiation. (d) The relative miR-20a-5p expression during chicken primary myoblast differentiation. (e) The relative miR-20b-5p expression during QM-7 myoblast differentiation. (f) The relative miR-20b-5p expression during chicken primary myoblast differentiation. (g) The relative pri-miR-17~92 expression during QM-7 myoblast differentiation. (h) The relative pri-miR-17~92 expression during chicken primary myoblast differentiation. (i) The relative pri-miR-106a~363 expression during QM-7 myoblast differentiation. (j) The relative pri-miR-106a~363 expression during chicken primary myoblast differentiation. Results are shown as the mean ± sem of three independent experiments. One sample t test was used to analysis the statistical differences between groups. *p < 0.05; **p < 0.01.
Discussion
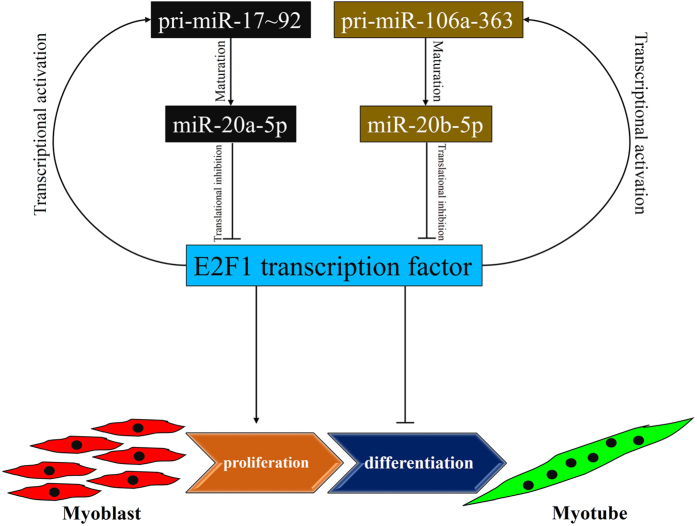
In this study, we found that the miR-17~92 cluster and miR-106a~363 cluster are directly regulated by the transcription factor E2F1. As miR-20a-5p and miR-20b-5p, which belong to the two clusters respectively, can inhibit E2F1 protein expression, an auto-regulatory feedback loop can be presented between E2F1 and the two clusters (Fig. 7). The auto-regulatory feedback loop is an interesting model for the regulation of gene expression, especially with the participation of miRNAs in this loop. miRNA regulation in this loop not only act as a simple linear regulator, but also contain check-and-balance function33. An example of this auto-regulatory loop is miR-15a and its target gene c-myb. miR-15a directly binds and regulates c-myb expression, while c-Myb protein binding site is occupied in miR-15a promoter and is required for miR-15a expression34. The presence of miR-15a in this auto-regulatory loop is to regulate c-myb expression and to play an importance role in human hematopoiesis34. Another example is a muscle-specific miRNA miR-1 and its target gene Yin Yang 1 (YY1), which is a repressor of muscle genes13. This auto-regulatory feedback loop not only maintain the balance of the expression level of transcription factors and miRNAs, but also is involved in the regulation of myogenesis13. In this study, we found another auto-regulatory feedback loop, E2F1-miR-20a-5p/20b-5p, which might also be involved in myogenesis (Fig. 7). Previous study has demonstrated that E2F1 down-regulation is required for myoblast differentiation24. Our results have indicated that miR-20a-5p and miR-20b-5p can directly inhibit E2F1 expression in myoblasts, and miR-20a-5p is up-regulated during myoblast differentiation. Therefore, the auto-regulatory feedback loop of E2F1-miR-20a-5p/20b-5p might play roles in the regulation of E2F1 during myoblast differentiation, specifically in repressing E2F1 expression to relieve the inhibitory role of E2F1 in this process.
Figure 7. Model for the proposed E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop in the regulation of myoblast proliferation and differentiation.
Skeletal muscle differentiation is a complex process with the regulation of diverse genetic and environmental factors. These factors can interact with each other and construct multiple signaling pathways to regulate the cell differentiation process. The network between transcription factors and miRNAs is an interesting global feature and has been extensive focused on35,36. An auto-regulatory feedback loop with transcription factor and miRNA can make the regulatory network more abundant and of highly organization. However, few of these kinds of feedback loops have been found in myogenesis. Additionally, as an important regulator of embryo development, miR-17 family has been rarely studied in myogenesis. Our results not only showed that both miR-20a-5p and miR-20b-5p can regulate the proliferation and differentiation of myoblasts, but also proposed an E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop that might be involved in myogenesis. These findings provide new information into the regulation of skeletal muscle differentiation.
The E2F-miR-20a auto-regulatory feedback loop not only exists in avian myoblast cell, but also exists in human cell. In a prostate cancer cell line, miR-20a directly inhibits the translation of the E2F1, E2F2 and E2F3 mRNA, and the endogenous E2F1, E2F2 and E2F3 bind to the promoter of the miR-17~92 cluster and activate its transcription31. In HEK-293 cells, miR-17~92 cluster and E2Fs can also construct an auto-regulatory feedback loop37. This auto-regulatory feedback loop is important for balancing endogenous E2Fs expression level and may be involved in the regulation of cellular proliferation, transformation and apoptosis31,37. In this study, we not only found the auto-regulatory feedback loop of E2F1-miR-20a-5p, but also found the auto-regulatory feedback loop of E2F1-miR-20b-5p. Both these feedback loops might be involved in avian myoblast proliferation and differentiation. miR-20a-5p and miR-20b-5p inhibit the protein level of E2F1 directly, and the endogenous E2F1 binds to the promoters of the miR-17~92 cluster and miR-106a~363 cluster and activates their transcription. However, there are still some questions existing in myoblast differentiation process. Endogenous E2F1 can bind and activate pri-miR-17~92 and pri-miR-106a~363 expression. The protein level of E2F1 is down-regulated during myoblast differentiation, whereas the expression of pri-miR-17~92 and pri-miR-106a~363 are up-regulated, suggesting that there be another transcriptional factor that can promote pri-miR-17~92 and pri-miR-106a~363 transcription during the differentiation process. Previous studies have demonstrated that c-Myc directly activate transcription of the miR-17~92 and miR-106a~363 clusters20,38. In addition, p53, STAT5 and MYCN can also regulate the transcription of the miR-17~92 or miR-106a~363 clusters directly39,40,41,42. Therefore, E2F1 is not the only factor that can regulate the transcription of these two clusters. Other transcriptional factors may also be involved in the regulation of miR-17~92 and miR-106a~363 clusters transcription during myoblast differentiation.
The protein level of E2F1 is down-regulated during avian myoblast differentiation. Similarly, E2F1 mRNA level in C2 myocytes is also reduced during differentiation process24. E2F1 is important for cell proliferation and development, but the regulation of this protein during muscle differentiation still remains unclear. Our data showed that miR-20a-5p and miR-20b-5p directly regulated E2F1 protein level by binding to the 3′ UTR of E2F1 mRNA. However, miR-20b-5p expression level was relatively consistent during myoblast differentiation, and the expression of miR-20a-5p was modestly up-regulated. These expression patterns may not fully explain the down-regulated expression of E2F1 protein level. There may be other factors involved in the inhibition of E2F1 protein expression. The members of miR-17 family have same seed sequences, suggesting that these miRNAs might sharing common target genes. Previous studies have demonstrated that miR-17-5p, miR-106a-5p, miR-106b-5p and miR-93-5p can also inhibit E2F1 expression directly43,44,45. Therefore, it is possible that all members of the miR-17 family can directly inhibit E2F1 expression during myoblast differentiation. Considering that pri-miR-17~92 and pri-miR-106a~363 expression are all significantly up-regulated during myoblast differentiation, the down-regulation of E2F1 protein level might be resulted from the up-regulated expression of all of the miR-17 family members.
The inhibitory effect of E2F1 in myogenic differentiation is through the induction of genes which repress the transcription activity of MYOG and MYOD25. E2F1 overexpression cannot inhibit MYOD expression, but the expression of MYOG and MyHC, which can be transcribed by MYOD46,47, would be reduced in E2F1 overexpressed myoblasts24. This regulatory effect also exists in our results, which were performed in avian myoblasts. Additionally, either miR-20a-5p or miR-20b-5p is able to regulate MYOG and MyHC expressions by inhibiting E2F1 expression directly. However, MYOG and MyHC expressions were downregulated only when both miR-20a-5p and miR-20b-5p were inhibited. This result might be due to the insufficient release of E2F1 expression from miR-20a-5p and miR-20b-5p inhibition by only one of the inhibitors transfection. The E2F1 protein level would be reduced more when the inhibitors of miR-20a-5p and miR-20b-5p were co-transfected in myoblasts. Additionally, the potential complementary effect between miR-20a-5p and miR-20b-5p can also influence the effect of only one of the inhibitors transfection. On the other hand, Myomaker is an important transmembrane protein in myoblast fusion48,49,50. Its expression during myoblast differentiation depends on the transcription activity of MYOD and MYOG49,51. In this study, inhibition of miR-20a-5p and miR-20b-5p cannot reduce myogenic differentiation, but the myoblast fusion might be reduced based on the down-regulation of Myomaker. As E2F1 expression is reduced in the transfection of one of the miRNA inhibitors, the transcription activity of both MYOD and MYOG would be inhibited. Therefore, Myomaker expression can also be down-regulated in the case that only one of the inhibitors was transfected.
In conclusion, both miR-20a-5p and miR-20b-5p inhibit myoblast proliferation and promote myoblast differentiation by directly binding and inhibiting E2F1 mRNA expression. E2F1 binds to the promoters of the miR-17~92 and miR-106a~363 clusters and induces their transcriptional expression. Our findings present an E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop and suggest that this loop is important for myoblast proliferation and differentiation. Additionally, this auto-regulatory loop might also be involved in the regulation of E2F1 expression during myoblast differentiation. These findings offer new insight into the mechanisms and regulatory networks of muscle differentiation, and further shed light on the understanding of muscle development and muscle diseases.
Methods
Cell culture
Chicken primary myoblasts were isolated and cultured as previously described15. QM-7 cells were cultured in Medium 199 (Gibco) with 10% (v/v) fetal bovine serum (Gibco), 10% tryptose phosphate broth (Sigma Life Science) and 0.2% penicillin/streptomycin (Invitrogen). When switched to medium without serum, the QM-7 cells cease dividing and fuse to form large multinucleated myotubes. DF-1 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Gibco) with 10% (v/v) fetal bovine serum (Gibco) and 0.2% penicillin/streptomycin (Invitrogen). All experimental protocols were approved by the South China Agricultural University Institutional Animal Care and Use Committee (approval ID: SCAU#0014). And the methods were carried out in accordance with the regulations and guidelines established by this committee.
RNA isolation, RT-PCR and quantitative real-time PCR
RNAiso reagent (Takara) was used for isolating the total RNA from cells. cDNA synthesis was performed using PrimeScriptTM RT reagent Kit (Perfect Real Time) (Takara). qPCR was carried out using KAPA SYBR® FAST qPCR Kit (KAPA Biosystems) by Bio-rad CFX96 Real-Time Detection system (Bio-Rad). For miRNA quantitation, miRNA reverse transcription was performed using Bulge-loop miRNA qRT-PCR Primer Sets (RiboBio) and First-Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer’s instructions. The qPCR primers specific for gga-miR-20a-5p, gga-miR-20b-5p and U6 were designed by RiboBio, and the qPCR was also performed using KAPA SYBR FAST qPCR Kit (KAPA). The quantification of qPCR results was done as previously described15.
Western blot and immunofluorescence
Western blot and immunofluorescence assays were performed as previously described15. The following antibodies were used for Western blot: E2F1 (Santa Cruz Biotechnology) and Tubulin (Bioworld). The following antibody was used for immunoblotting: MyHC (DSHB). The total myotube area was calculated as the percentage of the total image area covered by myotubes, and the measurement was performed using ImageJ software (National Institutes of Health) on cells labelled with anti-MyHC.
ChIP assays
ChIP assay was performed using Pierce Agarose ChIP Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Briefly, 106 cells of the chicken primary myoblasts were fixed in 1% formaldehyde to cross-link DNA and proteins. After incubation of 10 min at 37 °C, 125 nM glycine was added to neutralization. Then, the cells were lysed and chromatin was sheared to 200–1000 bp DNA fragments using micronuclease digestion. Input DNA was stored for following assay. Remaining sheared DNA was incubated with anti-E2F1 antibody (Santa Cruz Biotechnology). Normal Rabbit IgG was used as negative control. The samples were then reversed and purified using a DNA clean-up column. ChIP products were finally subjected to normal PCR and quantitative PCR. Samples were done in triplicate. The primer sequences for PCR analysis are listed in Supplementary File 2.
RNA oligonucleotides and transfection
The miR-20a-5p mimic, miR-20b-5p mimic, mimic control duplexes, miR-20a-5p inhibitor, miR-20b-5p inhibitor and inhibitor NC were purchased from RiboBio. siRNA against E2F1 was purchased from GenePharma. Lipofectamine 3000 reagent (Invitrogen) was used for transfection according to the manufacturer’s instructions. 50 nM miRNA mimics, 100 nM miRNA inhibitor (as well as the mixed inhibitors) or 100 nM siRNA was used in the transfection assay.
Plasmid construction
For E2F1 overexpression vector, the E2F1 coding sequences were amplified and inserted into the pcDNA-3.1 vector (Invitrogen) using the XhoI and EcoRI restriction sites. For pmirGLO dual-luciferase miRNA target reporter vector, the E2F1 3′ UTRs were amplified and inserted into the pmirGLO dual-luciferase reporter vector (Promega) using the DraI and SalI restriction sites. E2F1-3′ UTR mutant plasmid was generated by changing the binding site of miR-20a-5p and miR-20b-5p from CACTTT to AGAGCG. PCR amplification was performed to do the mutagenesis and DpnI digestion to remove the parental DNA. For miR-17~92 and miR-106a~363 promoter reporter plasmids, four fragments of the miR-17~92 and miR-106a~363 promoters were amplified using the primers listed in Supplementary File 2. Then the PCR products of Reporter-1 and Reporter-2 were digested with KpnI and SmaI, the PCR products of Reporter-3 was digested with SacI and SmaI, and the PCR products of Reporter-4 was digested with KpnI and XhoI. After digestion, the products were inserted into the pGL3-basic reporter vector (Promega) to create the reporters of pGL3-R1, pGL3-R2, pGL3-R3 and pGL3-R4.
Dual luciferase reporter assay
DF-1 cell, an immortalized cell line of chicken embryo fibroblasts52, was chosen to do the reporter experiment because of its more stable status than the QM-7 cell. The dual luciferase reporter assay was performed as previously described15. Briefly, for the promoter assays of pri-miRNAs, DF-1 cells were co-transfected with reporter plasmid and E2F1 overexpression vector or control vector, and the TK-Renilla reporter was also co-transfected to each sample as an internal control. For miRNA target validation assays, DF-1 cells were co-transfected with wild-type or mutant E2F1 3′UTR dual-luciferase reporter (100 ng) with miRNAs mimic or NC mimic (50 nM) by using Lipofectamine 3000 reagent (Invitrogen) in 96-well plates. The activities of luciferase were measured using Dual-luciferase reporter assay system according to the manufacturer’s instructions (Promega) after 36 h transfection. The luminescent signal was quantified using Synergy 2 Multi-mode Microplate Reader (Biotek) and analysed with Gene5 software (Biotek).
Cell cycle analysis
After 36 h transfection, QM-7 cells cultured in growth medium were collected, fixed in 75% ethanol overnight at −20 °C, and then stained with 50 μg/mL propidium iodide (Sigma) containing 10 μg/mL RNase A (TaKaRa) and 0.2% (v/v) Triton X-100 (Sigma) for 30 min at 4 °C. Subsequently the cell cycle were analysed using a BD Accuri C6 flow cytometer (BD Biosciences), and the data analysis was performed using FlowJo 7.6 software (Verity Software House).
CCK-8 assays
The transfected myoblasts were seeded in 96-well plates (1 × 103 cells/well) and cultured in growth medium. Every 12 h, 10 μL of Cell-Counting Kit-8 (CCK-8) solution (Dojindo Laboratories) was added to each well for 1 h, and absorbance was measured at 450 nm using a Model 680 Microplate Reader (Bio-Rad).
Statistical analysis
Each experiment was repeated three times, and all results are represented as mean ± sem. One-sample t test was used to perform the statistical significance test between groups.
Additional Information
How to cite this article: Luo, W. et al. E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci. Rep. 6, 27904; doi: 10.1038/srep27904 (2016).
Supplementary Material
Acknowledgments
This work was supported by the China Agriculture Research System (CARS-42-G05), and Natural Scientific Foundation of China (31172200), the Program for New Century Excellent Talents in University (NCET-13-0803), and the Foundation for High-level Talents in Higher Education of Guangdong, China.
Footnotes
Author Contributions Study design: W.L. and X.Z. Collecting the samples and performed the experiments: W.L., G.L. and Z.Y. Data interpretation and analysis: W.L. and X.Z. Wrote the manuscript: W.L., Q.N. and X.Z. All authors have read and approved the final manuscript.
References
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–97 (2004). [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005). [DOI] [PubMed] [Google Scholar]
- Kloosterman W. P. & Plasterk R. H. The diverse functions of microRNAs in animal development and disease. Dev Cell 11, 441–50 (2006). [DOI] [PubMed] [Google Scholar]
- Luo W., Nie Q. & Zhang X. MicroRNAs involved in skeletal muscle differentiation. J Genet Genomics 40, 107–16 (2013). [DOI] [PubMed] [Google Scholar]
- Hwang H. W. & Mendell J. T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 96, R40–4 (2007). [PubMed] [Google Scholar]
- Amiel J., de Pontual L. & Henrion-Caude A. miRNA, development and disease. Adv Genet. 80, 1–36 (2012). [DOI] [PubMed] [Google Scholar]
- Jovanovic M. & Hengartner M. O. miRNAs and apoptosis: RNAs to die for. Oncogene 25, 6176–87 (2006). [DOI] [PubMed] [Google Scholar]
- Chen J. F. et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 190, 867–79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. F. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 38, 228–33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D. et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 321, 491–9 (2008). [DOI] [PubMed] [Google Scholar]
- Ge Y., Sun Y. & Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 192, 69–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liang S., Zhao Y. & Han Z. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development 139, 3543–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. et al. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One 7, e27596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist C. G. et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA 106, 13383–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W. et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 5, e1347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A. et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foshay K. M. & Gallicano G. I. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol. 326, 431–43 (2009). [DOI] [PubMed] [Google Scholar]
- Ivanovska I. et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 28, 2167–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V. & Mendell J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–43 (2005). [DOI] [PubMed] [Google Scholar]
- Attwooll C., Lazzerini D. E. & Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 23, 4709–16 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim Biophys Acta. 1602, 131–50 (2002). [DOI] [PubMed] [Google Scholar]
- Yamasaki L. et al. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85, 537–48 (1996). [DOI] [PubMed] [Google Scholar]
- Wang J., Helin K., Jin P. & Nadal-Ginard B. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1. Cell Growth Differ. 6, 1299–306 (1995). [PubMed] [Google Scholar]
- Wang J., Huang Q., Tang W. & Nadal-Ginard B. E2F1 inhibition of transcription activation by myogenic basic helix-loop-helix regulators. J Cell Biochem. 62, 405–10 (1996). [DOI] [PubMed] [Google Scholar]
- Braun T., Buschhausen-Denker G., Bober E., Tannich E. & Arnold H. H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 8, 701–9 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P. et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364, 501–6 (1993). [DOI] [PubMed] [Google Scholar]
- Lin S. et al. Let-7b regulates the expression of the growth hormone receptor gene in deletion-type dwarf chickens. BMC Genomics 13, 306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antin P. B. & Ordahl C. P. Isolation and characterization of an avian myogenic cell line. Dev Biol. 143, 111–21 (1991). [DOI] [PubMed] [Google Scholar]
- Song L., Ke Y. & Zhang Z. Q. High level expression and purification of recombinant PEX protein in cultured skeletal muscle cell expression system. Biochem Biophys Res Commun. 357, 258–63 (2007). [DOI] [PubMed] [Google Scholar]
- Sylvestre Y. et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 282, 2135–43 (2007). [DOI] [PubMed] [Google Scholar]
- Kel A. E. et al. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31, 3576–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci USA 108, 10355–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Kalota A., Jin S. & Gewirtz A. M. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood 113, 505–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R., Lieber D., Oren M. & Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol. 3, e131 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. J. & Walhout A. J. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays 31, 435–45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K., Thomson J. M. & Hammond S. M. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 282, 2130–4 (2007). [DOI] [PubMed] [Google Scholar]
- Kumar P., Luo Y., Tudela C., Alexander J. M. & Mendelson C. R. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol Cell Biol. 33, 1782–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann Y. et al. The miR-17/92 cluster is targeted by STAT5 but dispensable for mammary development. Genesis 50, 665–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brouwer S. et al. Dickkopf-3 is regulated by the MYCN-induced miR-17-92 cluster in neuroblastoma. Int J Cancer 130, 2591–8 (2012). [DOI] [PubMed] [Google Scholar]
- Yan H. L. et al. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 28, 2719–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J. et al. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA 107, 1553–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G. et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J Mol Med (Berl). 89, 1037–50 (2011). [DOI] [PubMed] [Google Scholar]
- Petrocca F. et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13, 272–86 (2008). [DOI] [PubMed] [Google Scholar]
- Pickering M. T., Stadler B. M. & Kowalik T. F. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene 28, 140–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18, 662–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. et al. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25, 502–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millay D. P. et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Li E., Nie Q. & Zhang X. Myomaker, regulated by MYOD, MYOG and miR-140-3p, promotes Chicken Myoblast Fusion. Int J Mol Sci. 16, 26186–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landemaine A., Rescan P. Y. & Gabillard J. C. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun. 451, 480–4 (2014). [DOI] [PubMed] [Google Scholar]
- Millay D. P., Sutherland L. B., Bassel-Duby R. & Olson E. N. Myomaker is essential for muscle regeneration. Genes Dev. 28, 1641–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himly M., Foster D. N., Bottoli I., Iacovoni J. S. & Vogt P. K. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248, 295–304 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.