Abstract
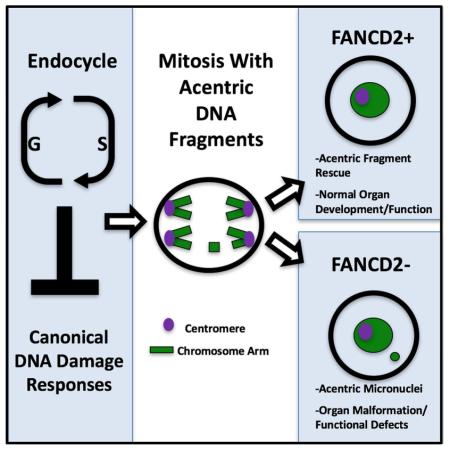
Conserved DNA damage responses (DDRs) sense genome damage and prevent mitosis of broken chromosomes. How cells lacking DDRs cope with broken chromosomes during mitosis is poorly understood. DDRs are frequently inactivated in cells with extra genomes (polyploidy), suggesting study of polyploidy can reveal how cells with impaired DDRs/genome damage continue dividing. Here, we show continued division and normal organ development occurs in polyploid, DDR-impaired Drosophila papillar cells. As papillar cells become polyploid, they naturally accumulate broken acentric chromosomes, but do not apoptose/arrest the cell cycle. To survive mitosis with acentric chromosomes, papillar cells require Fanconi Anemia proteins FANCD2 and FANCI, and Blm helicase, but not canonical DDR signaling. FANCD2 acts independently of previous S-phases to promote alignment and segregation of acentric DNA produced by double-strand breaks, thus avoiding micronuclei and organ malformation. As polyploidy and impaired DDRs can promote cancer, our findings provide insight into disease-relevant DNA damage tolerance mechanisms.
Keywords: Endocycle, Polyploidy, FANCD2
INTRODUCTION
Conserved DNA damage responses (DDRs) prevent mitosis of cells with DNA damage by promoting cell cycle arrest, DNA repair, and apoptosis. Attenuating these responses leads to mitosis with damaged/unstable genomes, which can ultimately enable tumor progression (Hanahan and Weinberg 2011; Halazonetis et al. 2008). Thus, understanding mechanisms enabling expansion of DDR-attenuated, genomically unstable cells is of great interest.
An increasingly recognized cause of DDR attenuation is the endocycle. This modified cell cycle lacks M-phase, and repeated endocycles thus generate polyploid cells (Edgar et al. 2014; Fox and Duronio 2013). In mammalian cancer cells, endocycles can promote resistance to irradiation and cisplatin, common cancer therapies that induce high levels of DNA damage (Shen et al. 2008; 2013b). Further, endocycled mouse embryonic fibroblasts generated by transient telomere DNA breakage are tumorigenic (Davoli et al. 2010; Davoli and de Lange 2012). Thus, continued division of endocycled cells associates with DNA damage therapy resistance and tumor growth.
Developmentally programmed endocycles also induce DNA damage and DDR inactivation. DNA damage can result from under-replication in the endocycled genome (Beliaeva et al. 1998; Gall et al. 1971; Nordman et al. 2011; Yarosh and Spradling 2014; Hannibal et al. 2014). Such under-replicated DNA is prone to deletions and inversions, (Yarosh and Spradling 2014) which may result from DSBs.
Indeed Drosophila endocycled cells accumulate the ATM/ATR phosphorylation mark γ-H2AV (Mehrotra et al. 2008), a readout of double-strand DNA breaks (DSBs). In Drosophila, such DSB accumulation is likely due to low p53 (a core DNA damage-responsive transcription factor) levels and chromatin silencing at p53 pro-apoptotic target genes (Mehrotra et al. 2008; Hassel et al. 2014; Zhang et al. 2014). Similarly, in mice, differentiation of endocycling trophoblast giant cells involves decreased p53 (Soloveva and Linzer 2004), and suppression of the DDR kinase Chk1 (Ullah et al. 2008;2011). Thus, in both developmental and cancerous settings, endocycles promote impaired DDRs and tolerated DNA DSBs.
However, many developmentally endocycled cells do not resume mitosis, and thus these systems cannot be used to identify responses enabling continued mitosis of genome-damaged cells. We recently developed study of Drosophila papillar cells as a developmentally and genetically tractable model of polyploid mitosis after endocycles. Here, using our model, we uncover mechanisms permitting these polyploid cells to undergo viable division with DNA damage. Similar to previous studies, we find endocycled papillar cells lack p53-mediated apoptosis. Further, we find papillar cells lack S-phase checkpoints and enter mitosis without undergoing high fidelity DNA repair. Despite lacking these normally crucial DDRs, both papillar mitosis and organ development are highly resistant to DNA damage by DSBs. By live imaging pupal development, we show an important part of the papillar DDR involves alignment and segregation of broken, acentric chromosome fragments. This response does not depend on p53, or core DNA damage kinases.
Instead, the Fanconi Anemia protein FANCD2, its frequent partner FANCI, and the Bloom helicase (Blm) are a crucial part of this non-canonical DDR. We show FANCD2 acts independently of S-phases prior to mitosis entry, and does not require its core complex partner FANCM to promote segregation of acentric fragments produced by DNA DSBs. This response ensures normal organ development by preventing acentric micronuclei. Our results pinpoint a mechanism enabling viable mitosis despite an impaired DDR.
RESULTS
Lack of apoptosis and S-phase checkpoints during pre-mitotic endocycles
Previous study of endocycle-induced DDR inactivity focused on post-mitotic tissues. To understand the impact of endocycles on subsequent divisions, we turned to an accessible in vivo model: Drosophila rectal papillar cells (hereafter: papillar cells or papillar precursors).
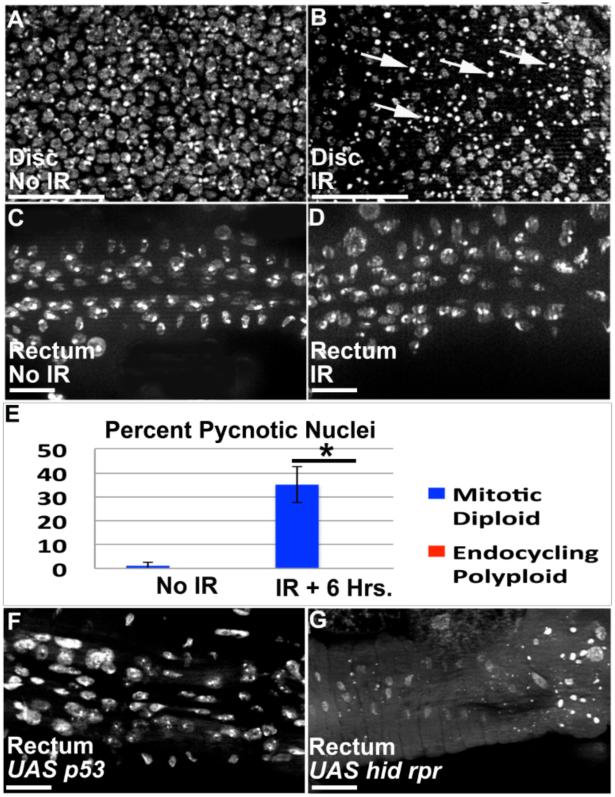
During 2nd larval instar, papillar precursors endocycle, producing octoploid nuclei (Fox et al. 2010; Schoenfelder et al. 2014). Unlike previously studied examples of endocycled cells with an inactive DDR, papillar cells then undergo polyploid divisions. We thus asked if these mitotic endocycled cells also lack an apoptotic response to damaged DNA. It is well established that exposure to Ionizing Radiation (IR) causes DNA damage and apoptotic cell death in diploid cells. Accordingly, we find induction of pycnotic nuclei and TUNEL labeling in diploid wing imaginal tissue after 20 Gy of X-ray induced IR (Fig1A,B,E, FigS1A,B, Methods). In contrast, IR does not induce pycnotic nuclei or TUNEL in endocycling 2nd instar papillar precursors (Fig1C-E, FigS1C,D). The lack of apoptosis in papillar precursors is not due to lack of IR-induced DNA breakage, as IR causes robust γ-H2AV accumulation in endocycling papillar precursors one hour after IR (FigS1E,F).
Figure1. Lack of p53-dependent apoptosis in papillar precursors.
A,B.3rd instar larval wing imaginal discs. A.Nuclei in undamaged disc. B.Pycnotic nuclei (arrows) in irradiated disc, 6 hrs. post IR. C,D.2nd instar rectum (during papillar precursor endocycling). C.Nuclei in unirradiated rectum. D.No pycnotic nuclei in rectum, 6 hrs. post IR. E.% pycnotic nuclei from A-D (wing data-blue, rectum data-red). Numbers indicate mean % pycnotic nuclei/animal (N=minimum 10 animals/condition, multiple replicates). *= significant to p<.001, two-tailed T-test. Bars=standard deviation. F.UAS-p53 expression in rectum (using byn-Gal4) does not induce pycnotic nuclei. G.UAS hid, UAS rpr co-expression in rectum (Methods) does induce pycnotic nuclei. DAPI=DNA in all images. Scale bar=10μm.
To examine molecular regulation of this apoptotic inactivity, we examined the consequences of expressing p53 and its pro-apoptotic targets. Using a UAS-p53 construct used previously in salivary glands (Mehrotra et al. 2008), we find p53 expression during papillar endocycles is insufficient to induce apoptosis (Fig1F), whereas the same construct expressed with the same promoter (Methods) causes robust cell loss in the diploid hindgut pylorus (FigS1G,H). In contrast to p53 expression, co-expression of p53 pro-apoptotic targets hid and reaper during endocycles induces robust papillar precursor death (Fig1G). We conclude that as in non-mitotic endocycling cells, mitosis-capable papillar precursor cells attenuate a DNA damage-responsive, p53-dependent apoptotic response during endocycles.
In addition to apoptosis, cell cycle arrest is an important DDR. We thus tested if, during endocycles, papillar precursors lack DNA damage checkpoints for S-phase entry or progression. One hour after IR, there is no change in the number of papillar precursor cells in endocycle S-phases (using EdU, FigS1I-K). Further, by examining late S-phase patterns of EdU incorporation, we find the proportion of endocycling cells that progress to late S-phase does not decrease after IR (FigS1L). Thus, during pre-mitotic endocycles of papillar cells, DNA breakage fails to activate either p53-dependent apoptosis or S-phase entry/progression checkpoints.
Acentric chromosomes accumulate but segregate during papillar mitosis
Given the inactive responses to broken DNA in endocycling papillar cells, we next examined whether such inactivity leads to unrepaired/aberrant chromosomes after endocycles. During metamorphosis (4-5 days after endocycles), papillar cells undergo 1-3 complete mitotic cycles as octoploid cells. Even in the absence of exogenous DNA damage, these divisions are frequently error-prone. While some errors are due to extra centrosomes (Schoenfelder et al. 2014), we also observed chromosome structure aberrations, which could also contribute to mitotic errors (Fox et al. 2010).
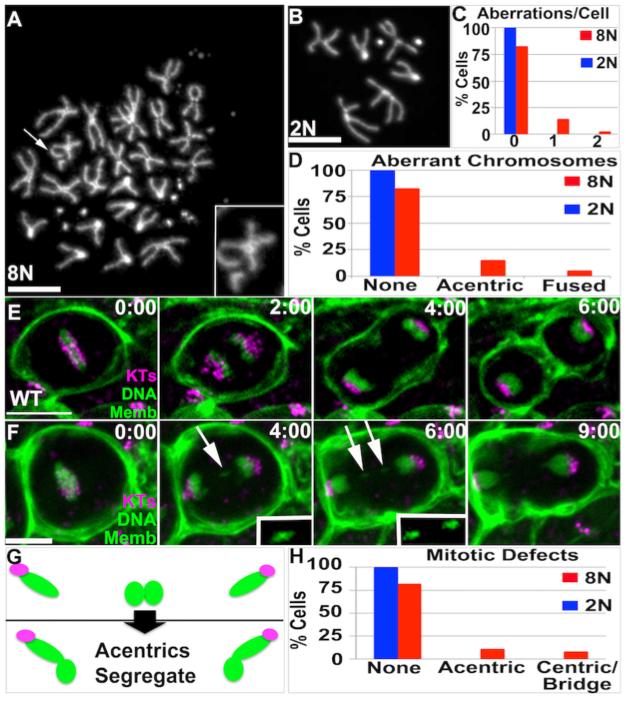
We thus examined the nature of papillar chromosome aberrations in detail, and asked if aberrations are more common in these polyploid cells relative to diploid cells. Indeed, in the absence of exogenous DNA damage, papillar cells naturally accumulate two recurring chromosome aberrations: acentric fragments (Fig2A) and chromosome fusions (dicentric chromosomes, FigS2A). We detected at least one of these aberrations in 19.8% of papillar cells, with acentric chromosomes being most common (Fig2C,D). In contrast, no such aberrations are found in diploid imaginal disc cells (Fig2B-D). Therefore, endocycled polyploid papillar cells naturally accumulate chromosomal aberrations that persist into mitosis.
Figure2. Acentric chromosomes accumulate and segregate during Wild Type papillar development.
A.Example papillar cell with acentric chromosome (arrow, 2X magnified inset). B.Example diploid imaginal disc cell with normal karyotype. DAPI=DNA in A,B. C.Number aberrations/cell for cells examined in A,B. D.Distribution of acentric/fused chromosomes for cells examined in A,B. A-D: data from N=96 8N and N=93 2N cells respectively, from 8 replicates. The increased incidence of aberrant karyotypes in 8N vs. 2N is significant when accounting for increased chromosomes in 8N cells (Chi square, p<.05). E,F.Time-lapses of papillar mitosis. CenpC-Tomato=kinetochores (KTs, purple), histone H2AV=DNA (green, nuclear), and Moesin-GFP=cell membranes (Memb, green). Time is in minutes relative to anaphase onset. E.Example of normal mitotic segregation. F. Example of acentric chromatids that segregate into daughter nuclei. White arrow and 2X magnified, contrast-enhanced insets highlight segregating acentric DNA. G.Diagram of fate of acentric and fused papillar chromosomes. Green=DNA, Purple=Centromeres. H.Frequency of mitotic errors in 8N papillar and 2N imaginal disc tissue. Data from N=92 (papillar) and N=89 (imaginal disc) movies (numerous replicates). Scale bar=5μm.
To gain insight into the fate of chromosome aberrations in polyploid mitotic papillar cells, we used live imaging. To distinguish between errant chromosomes containing or lacking centromeres, we used the markers histone-GFP (to mark DNA) and CenpC-Tomato (to mark kinetochores/centromeres), as well as a fragment of Moesin-GFP (to mark cell membranes and cytokinesis). In WT, we observed normal mitotic chromosome segregation in 82% of cells (Fig2E,H, Movie S1), in general agreement with previous measurements (Fox et al. 2010; Schoenfelder et al. 2014). Our approach also enabled us to detect cells with mitotic defects, in which DNA lags in anaphase. In these aberrant mitoses, CenpC enabled us to distinguish lagging DNA lacking (Fig2F, MovieS2) or containing centromeres (FigS2B). Lagging DNA with centromeres frequently localizes in an anaphase bridge, which persists until cytokinesis (FigS2B). Such bridges could represent separation of under-replicated DNA (Unhavaithaya and Orr-Weaver 2012). Alternatively, as part of the bridge-breakage-fusion cycle (Titen and Golic 2008), bridges are expected for cells with fused (dicentric) chromosomes, which we detect in our metaphase chromosome squashes (FigS2A, C).
Much less expected was the fate of acentric DNA fragments. In 11/92 WT papillar divisions we could clearly detect acentric fragments, in general agreement with our chromosome squash data (12.0%, Fig2F,H vs. 15.6%, Fig2D). Despite lacking detectable centromeres, these acentric fragments successfully incorporate into daughter nuclei before cytokinesis (Fig2F,G, MovieS2). By comparison, in 89 WT time-lapse movies of diploid imaginal disc tissue, we did not detect acentric DNA (Fig2H). Taken together, our data show chromosome structural aberrations arise in normal development of apoptosis-deficient papillar cells. The most common aberration is acentric DNA, which remarkably segregates into daughter cells in mitosis (Fig2G).
Papillar development is highly tolerant of DNA breakage
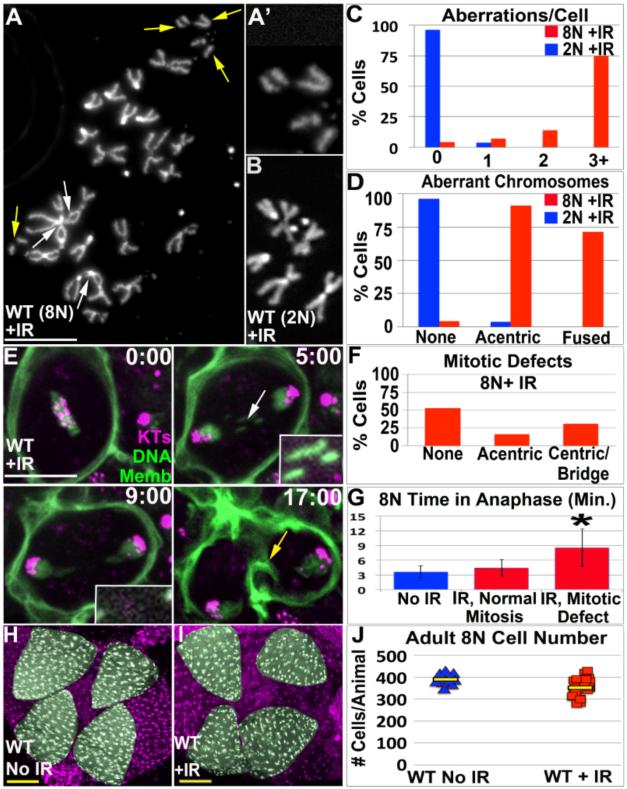
The purpose of canonical DDRs is to prevent mitotic expansion of genome-damaged cells. However, endocycled papillar cells lack canonical DDRs and enter mitosis with chromosome aberrations. It remained possible that the fraction of cells with such aberrations are eliminated by a non-apoptotic mechanism. We thus tested if increasing the incidence and number of papillar chromosome aberrations would cause mitotic or developmental defects. We allowed animals exposed to IR during papillar endocycles to progress through the 4-5-day period between endocycle completion and mitosis initiation. In these animals, chromosome aberrations induced during endocycles persist several days later and remain when papillar cells re-enter mitosis. This result mirrors the persistence of mitotic chromosome aberrations in papillar cells without exogenous DNA breakage. In mitotic papillar cells, IR most noticeably causes an increase in acentric (Fig3A,C,D vs. Fig2A,C,D) and fused chromosomes (FigS2D and Fig3A,C,D, vs. FigS2A and Fig2C,D). Thus, IR primarily amplifies the two major classes of naturally occurring DNA damage.
Figure3. Papillar development is refractory to frequent chromosome structural aberrations.
A.Example of chromosome structure in 8N mitotic papillar cell 5 days after IR induced during endocycles. Acentric fragments (yellow arrows, 2X magnified inset in A’) and chromosome fusions (white arrows) are indicated. B.Diploid genital imaginal disc karyotype, from animal irradiated at the same time as cells in A. C.Number aberrations/cell from irradiated wild-type octoploid (8N WT) papillar and diploid WT (2N WT) imaginal disc cells. D.Distribution of acentric/fused chromosomes from same animals as in C. Compare C,D to unirradiated data in Fig2C,D. A-D from N=70 and N=54 polyploid and diploid cells respectively. The increased incidence of aberrant karyotypes in 8N vs. 2N is significant when accounting for increased chromosomes in 8N cells (Chi square, p<.0001). E.Time-lapse of papillar mitosis after IR induced during endocycles. CenpC-Tomato=kinetochores (KTs, purple), histone H2AV=DNA (green, nuclear), and Moesin-GFP=cell membranes (Memb, green). Time is in minutes relative to anaphase onset. Example of lagging acentric DNA (white arrow, 2x magnified and contrast-enhanced insets) that segregates into daughter nuclei. Yellow arrow at 17:00 highlights contractile ring. F.Distribution of mitotic defects in 8N papillar cells after IR, scored from time-lapse data. Note- the percentage of cells with acentric DNA is likely an under-estimate, due to the co-existence of centromere-containing DNA that overlaps acentric DNA in some cases. G.Avg. time from anaphase onset to cytokinesis furrow initiation. IR divisions were binned into those with/without detectable errors. *= significant change from No IR or IR with normal mitosis (Two-tailed T-test, p<.001). Error bars= standard deviation. Data in E-G from N=113 IR divisions, from numerous replicates. H.WT adult rectum. DNA-purple, papillae-pseudo-colored green. I.WT adult rectum in animals irradiated during endocycles. J.Adult papillar cell number in WT, from control and IR (induced during endocycles). N= 13-23 animals counted/condition from multiple replicates. Yellow bars=mean. DAPI=DNA in all images. White scale bar=5 μm, Yellow scale bar=50 μm.
The increased incidence of persistent damage days after IR suggests papillar cells lack high-fidelity DNA repair (however, the presence of chromosome fusions suggests papillar cells likely engage in limited repair). To compare the lack of high fidelity repair in papillar cells to diploid cells irradiated at the same stage, we examined genital imaginal disc cells at the same time point (5 days after IR), when they divide while enveloping forming papillae (Fox et al. 2010). Chromosome aberrations were only present in a small number of these diploid cells during metamorphosis (Fig3B-D) suggesting repair of damage occurs prior to re-entering mitosis or that damaged cells are cleared from the tissue. Thus, unlike diploid cells, endocycling papillar cells not only lack apoptosis and S-phase checkpoints, but also lack high-fidelity DNA repair.
Given the persistence of chromosome aberrations in mitotic papillar cells, we could next examine if elevating the level of DNA breaks alters the fate of papillar chromosome aberrations in mitosis. In animals irradiated during 2nd larval instar papillar endocycles, the number of error-prone divisions increases significantly (Fig3F vs. Fig2H-8N). Many of these defects are present during the first papillar division, confirming DNA damage left either un-repaired or aberrantly repaired after IR causes mitotic errors. Mitotic errors associate with a longer anaphase (Fig3G). As in un-irradiated animals, centromere-containing DNA that lags forms a bridge that is resolved in cytokinesis (FigS2E, MovieS3). Due to co-existence of centric and acentric aberrations in some irradiated papillar cells, we could only score/follow the existence and fate of acentric DNA in a subset of irradiated papillar cells. As in un-irradiated animals, the acentric DNA that we could clearly observe segregated into daughter cells prior to cytokinesis (Fig3E, MovieS4). These results reinforce the model that papillar cells possess a robust response to segregate acentric DNA in mitosis.
Given that papillar cells undergo mitosis with chromosome aberrations, we next asked if increasing such aberrations affects papillar development. During development papillar cells undergo 1-3 divisions while forming four cone shaped structures of 100 cells each (Fig3H, Fox et al. 2010; Schoenfelder et al. 2014). Papillar development also leads to patterned gene expression, reflected in activation of enhancers such as AICR210H05-Gal4 at each papillar base (FigS2F). We examined AICR210H05-Gal4 expression in animals exposed to IR during endocycles. Additionally, to precisely quantify the effect of IR on papillar development, we hand-counted adult papillar cells (390+/−24 per animal without IR in WT, Fig3J). Strikingly, papillar development proceeds normally despite increased DNA breaks induced by IR (Fig3H-J, FigS2G). We detect only a minor reduction in mean adult papillar cell number after IR (352+/−43 cells/animal after IR, Fig3J). This suggests that even when containing a high number of chromosomal aberrations, papillar cells execute multiple rounds of division while undergoing proper morphogenesis and organ patterning. Thus, polyploid mitotic papillar cells are remarkably tolerant of acentric and fused chromosome structure aberrations.
FANCD2 plays a key role in acentric chromosome segregation and papillar cell survival
To begin to understand the robust DNA breakage tolerance of papillar cells, which lack canonical DDRs, we took a candidate approach. Given that: 1)acentric chromosomes are the most prominent chromosome aberration in both naturally occurring and irradiated papillar cells, and 2)the remarkable ability of such acentric DNA to segregate, we hypothesized that genes important for segregation of acentric DNA are critical to papillar DNA damage tolerance. We thus looked for genes involved in segregating acentric DNA.
The Spindle Assembly Checkpoint gene BubR1 was recently shown to function as part of a “tether” connecting acentric DNA and its centromere when breaks are induced during Drosophila neuroblast mitosis (Royou et al. 2010; Karg et al. 2015; Derive et al. 2015). Using the I-Cre1 system, an established method of generating an acentric chromosome (Rong et al. 2002; Royou et al. 2010), we generated acentric X-chromosomes in diploid neuroblasts. We readily detected BubR1-positive tethers in neuroblasts (FigS3A). In contrast, while we clearly detected BubR1 on metaphase papillar kinetochores, no BubR1 tethers were detected in papillar anaphase cells following I-Cre1 induction (FigS3B) or after IR (data not shown). Further, bubR1(KEN) mutants, which are defective in tether formation and cause diploid cell lethality following I-Cre induction (Royou et al. 2010), have normal papillar structure/cell number after IR (FigS3C). Thus, BubR1 tethers do not appear to be essential for acentric DNA segregation in papillar cells.
As an alternative mechanism of acentric DNA segregation, we considered a role for the Fanconi Anemia protein FANCD2. Together with FANCI and Blm helicase, FANCD2 was previously implicated in resolving “ultra-fine DNA bridges” that persist into mitosis (Chan et al. 2007; Chan and Hickson 2009; Naim and Rosselli 2009). We thus investigated if FANCD2 plays a role in the papillar DDR. To do so, we expressed either one of two fancd2 RNAi constructs (Methods, hereafter fancd2) specifically in papillar cells and the associated hindgut throughout development. In the absence of any exogenous DNA damage, fancd2 loss primarily increases acentric DNA in mitotic papillar cells (by roughly 2-fold, Fig4A,B). We thus used IR to amplify the two main naturally occurring forms of chromosome aberrations (acentric and fused chromosomes, FigS4A,B) and assayed the effect of fancd2 depletion. IR causes papillae of reduced size and significantly decreased cell number in fancd2 relative to WT (Methods, Fig4C-F, FigS5A vs. Fig3H-J). This phenotype is not due to additional chromosome aberrations in fancd2 after IR, as papillar chromosomes of irradiated fancd2 animals do not display novel/increased chromosome defects after IR (FigS4A-C, Methods). Additionally, the mitotic index is similar between fancd2 and WT after IR (FigS4D), suggesting papillar cells continue dividing without fancd2, but exhibit reduced survival.
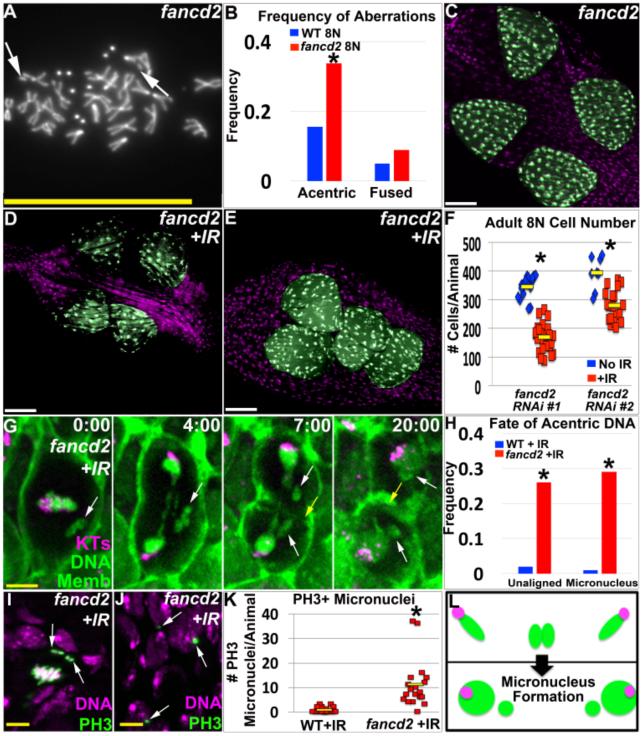
Figure4. FANCD2 promotes papillar cell viability and prevents micronucleus formation after IR.
A.Representative image of papillar chromosomes in fancd2 RNAi#1 animal. Arrows indicate acentric chromosomes. DAPI indicates DNA. B.Frequency of acentric and fused chromosomes in WT vs. fancd2 RNAi#1 papillar cells +/− IR. C.Adult fancd2RNAi#1 rectum. D.Adult fancd2RNAi#1 rectum from animal irradiated during papillar endocycles. E.Adult fancd2RNAi#2 rectum from animal irradiated during papillar endocycles. Papillae false-colored in green, DNA in purple in C-E. For comparable controls see Fig3H,I. F.Adult papillar cell number in fancd2 +/−IR. N=8-30 animals/condition. Yellow bars=mean. *=significant change between NoIR vs. +IR compared to WT (Methods). G.Time-lapse of fancd2RNAi#1 papillar cell after IR. CenpC-Tomato=kinetochores (KTs, purple), histone H2AV=DNA (green, nuclear), and Moesin-GFP=cell membranes (Memb, green). Time is in min. relative to anaphase onset. White arrows=unaligned DNA that form micronuclei. Yellow arrow=cytokinetic furrow. H.Frequency unaligned DNA and micronuclei +/−IR in WT vs. fancd2RNAi#1. Data from 84-113 divisions/condition, from multiple replicates. *=significant change from no IR by Chi-squared (p<.00001, Methods). I.Unaligned PH3+ chromosomes (green labeling/arrows, DNA in purple) in fancd2RNAi#1 after IR. J.Persistent PH3+ micronuclei (green labeling/arrows, DNA in purple) in fancd2RNAi#1 after IR. K.Frequency PH3+ micronuclei +/−IR in WT vs. fancd2RNAi#1. From N=18-27 animals/condition from multiple replicates. Yellow bars=mean. *=significant change from no IR (Two-tailed T-test, p<.01). L.Diagram of acentric chromosome fate in fancd2 after IR. Green=DNA, Purple=Centromeres. Instead of segregating completely into daughter nuclei as in Fig3E, acentric DNA accumulates in micronuclei (indicated by small green circles without centromeres). DAPI=DNA in all images. White scale bar=50μm, yellow scale bar=5μm.
To understand why fancd2 decreases cell survival after IR, we used live imaging. Similar to WT, IR causes lagging DNA with centromeres in fancd2, which localize in an anaphase bridge until cytokinesis (FigS4E). Therefore, it is unlikely that FANCD2 resolves centromere-containing anaphase bridges in papillar cells. In contrast, we noted two significantly enhanced mitotic abnormalities related to acentric DNA in fancd2 after IR. First, prior to anaphase, acentric DNA frequently fails to align on the metaphase plate during fancd2 divisions (Fig4G,H). Second, at anaphase, acentric DNA lags substantially and often fails to incorporate into daughter nuclei, resulting in acentric micronuclei (Fig4G,H, MovieS5). We detect unaligned acentric DNA both during the first and subsequent fancd2 divisions (FigS4F). We also find unaligned metaphase DNA in fancd2 mitotic cells accumulates the mitotic marker phospho-histone H3 (PH3, Fig4I). Interestingly, we see a robust increase in micronuclei in fancd2 animals after IR, which retain PH3 even after cells exit mitosis (when the rest of the nucleus has lost the marker, Fig4J,K). This latter observation is consistent with aberrant cell cycle activity in micronuclei (Crasta et al. 2012). Taken together, lack of FANCD2 leads to unaligned acentric DNA, acentric micronuclei, and papillar cell number loss following IR (Fig4L).
FANCD2 responds specifically to DSBs and acts independently of previous S-phases
FANCD2 prevents genome instability in response to a host of stimuli, including inter-strand cross-links, recombination intermediates, replication stress/late replication, and lesions created by IR (Grompe 2002; Walden and Deans 2014; Moldovan and D'Andrea 2009; Pontel et al. 2015). In papillar cells, we find acentric DNA fragments are the most common aberration, and FANCD2 loss leads to inaccurate segregation of these fragments. These data suggest in genome-damaged papillar cells, FANCD2 may play an important role by responding specifically to double-strand break (DSB)-induced acentric DNA.
To test this idea, we employed the I-Cre system to generate papillar cells with acentric X-chromatids (Fig5A-C). We used I-Cre because IR causes a host of chromosome aberrations, and it was unclear which causes cell loss in fancd2. We optimized conditions so the number of acentric fragments/cell was similar to following IR (Fig5B-Meta vs. Fig3D-acentric, Methods). As described previously for diploid Drosophila neuroblasts, and as we observe after IR, acentric papillar DNA generated by I-Cre lags in anaphase but eventually segregates into daughter nuclei (Fig5C, MovieS6). As seen after IR, I-Cre induction during papillar endocycles causes essentially no change in adult papillar structure/cell number (Fig5D,F). In contrast, I-Cre causes significant cell number reduction in multiple fancd2 RNAi lines (Fig5E,F, FigS5B). As for IR, we detect PH3+ unaligned DNA (Fig5G) and an increase in persistent PH3+ micronuclei in multiple fancd2 RNAi lines (Fig5H,I). Thus, I-Cre largely phenocopies IR induction in fancd2. We also note that, for multiple fancd2 RNAi lines, the number of PH3+ micronuclei significantly increases without exogenous DNA breakage (Fig5I), underscoring the importance of fancd2 in papillar cells not only following exogenous DNA damage but also under physiological conditions.
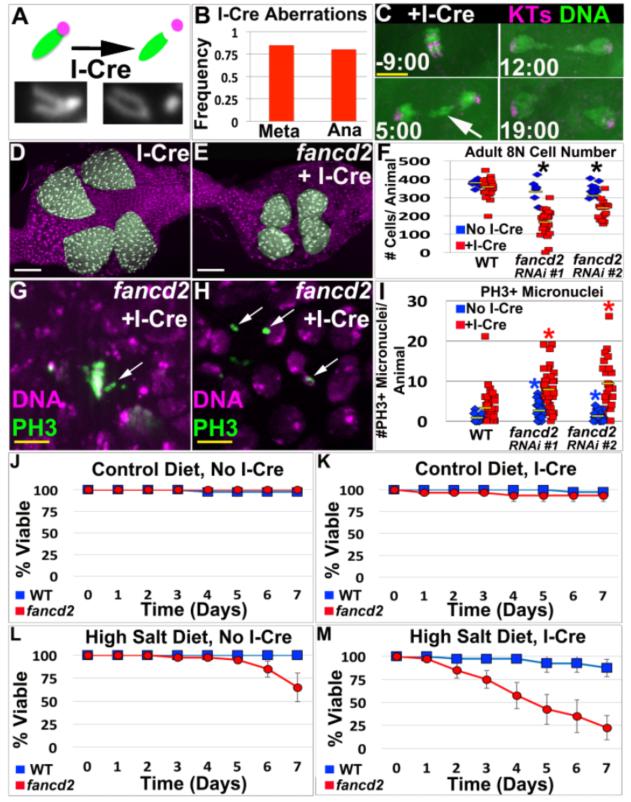
Figure5. FANCD2 promotes papillar cell viability and prevents micronucleus formation specifically in response to acentric chromosomes.
A.Top-Diagram of I-Cre system. One acrocentric X chromatid (green) shown before/after I-Cre induced DSB severs the connection with the centromere (purple). Bottom-Example of uncut and cut X-chromosomes in papillar cells from I-Cre expressing flies. B.Frequency of mitotic papillar cells after I-Cre (expressed during endocycles) with at least 1 aberrant X chromosome at metaphase (Meta, from chromosome preparation data) or with lagging DNA at anaphase (Ana, from live imaging data). From N=58-82 cells/condition, from multiple replicates. C.Representative time-lapse papillar DNA segregation following I-Cre (expressed during endocycles). CenpC-Tomato=kinetochores (KTs, purple), histone H2AV=DNA (green). Time in min. relative to anaphase onset. Arrow indicates lagging DNA, which segregates. D.WT adult rectum after I-Cre (expressed during endocycles). E.fancd2RNAi#1 adult rectum after I-Cre (expressed during endocycles). Papillae false-colored green, DNA in purple in D and E. F.Avg. adult papillar cell number/animal for WT and fancd2RNAi#1 +/− I-Cre. From N=8-23 animals/condition, multiple replicates. *=significant change between +/− I-Cre compared to WT (Methods). Yellow bars= mean. G.Unaligned PH3+ chromosomes (green labeling/white arrows, DNA in purple) in fancd2RNAi#1 after I-Cre. H.Persistent PH3+ micronuclei (green labeling/white arrows, DNA in purple) in fancd2RNAi#1 after I-Cre. I. Number PH3+ micronuclei +/− I-Cre in WT vs. fancd2RNAi#1 and fancd2RNAi#2. Yellow bars= mean. From N=17-23 animals/condition, multiple replicates. *=significant difference from WT (Two-tailed T-test, p<.005). DAPI=DNA in all images. J-M.Survival of adults of WT (blue) and fancd2 RNAi#1 (red) animals over time for the indicated I-Cre and diet conditions. Bars= standard error. Each genotype/condition represents 3 replicates with 10 animals/replicate. White scale bar=50μm, yellow scale bar=5μm.
As a further test of the physiological importance of FANCD2 in papillar cells, we employed a test of papillar function. We previously established that animals with decreased rectal papillar cells cannot survive high levels of dietary salt, due to the role of papillar structures in salt absorption (Schoenfelder et al. 2014). To test if fancd2 loss impacts the physiological function of adult papillae following DSB induction, we fed WT and fancd2 animals either control or a high-salt diet in the presence or absence of I-Cre expression. Indeed, fancd2 loss in developing papillae renders adult flies sensitive to high-salt, a result that is enhanced by I-Cre expression (Fig5J-M). Together, these data show FANCD2’s response to DSB-induced acentric DNA is a key aspect of the resistance of mitotic papillar cells to DNA damage.
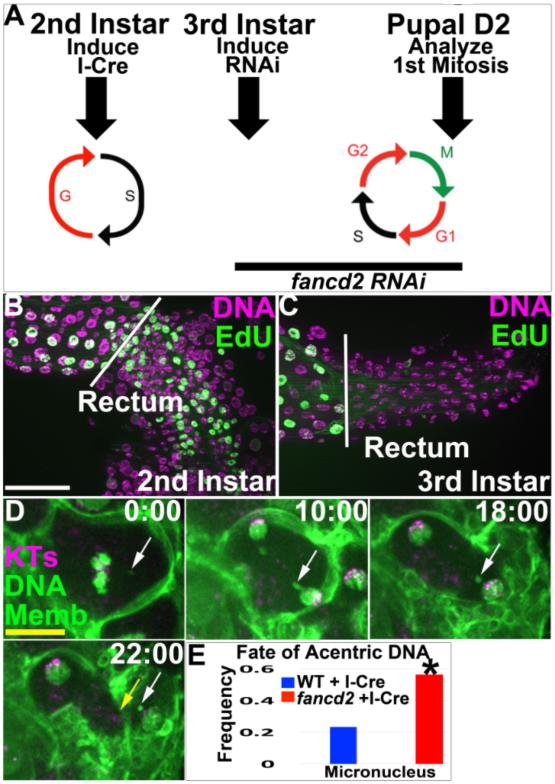
Our data point to a role for fancd2 in promoting acentric DNA segregation. As loss of fancd2 can promote replicative stress (Lossaint et al. 2013; Howlett et al. 2005) it was possible that our observed role for FANCD2 was due to secondary consequences of depleting fancd2 during endocycle DNA replication, which occurs prior to mitosis of acentric DNA. We thus tested if fancd2’s role in preventing acentric DNA-related defects arises during endocycle DNA replication. If so, depleting fancd2 after endocycle S-phases (but before mitosis) should not affect the papillar response to acentric DNA. To test this idea, we exploited the changing cell cycle biology of papillar cells. A large window of time (several days) separates the last endocycle S-phase (2nd larval instar stage) from the next S-phase, the latter of which occurs after the first papillar mitosis (Pupal Day 2). We thus induced I-Cre as before during endocycles, but then used temporal control of RNAi (Fig6A, Methods) to only deplete fancd2 after completion of the last endo-S-phase (as confirmed by EdU labeling, Fig6A-C). We then live imaged the first mitosis following this temporally restricted fancd2 depletion. Remarkably, fancd2 depletion in the absence of endocycle S-phase still leads to unaligned metaphase chromosomes and acentric micronuclei (Fig6D,E, MovieS7). From these data, we conclude fancd2’s role in preventing micronuclei in response to acentric DNA occurs independently of prior S-phases.
Figure6. Depletion of fancd2 in the absence of endocycle S-phases does not alter the acentric DNA phenotype.
A.Experimental procedure. I-Cre was induced during 2nd larval instar papillar endocycles. Following endocycles (3rd larval instar), fancd2 RNAi was induced. On pupal Day 2 (D2), the first papillar mitosis was examined. B.EdU labeling (green, DNA in purple), showing S-phase in the 2nd instar larval rectum (area to right of white line). C.EdU/DNA labeling as in B, now showing endocycles are complete in the rectum by 3rd instar stage (note-endocycles continue in the ileum, left of the white line). D.Time-lapse of I-Cre/fancd2 RNAi#1 animal cultured under conditions shown in A. Example unaligned acentric DNA fragment (white arrow) that ends up in a micronucleus following cytokinesis (yellow arrow). Purple-kinetochores (KTs, CenpC Tomato) green-DNA (DNA, his GFP) and cell membranes (Memb, Moesin GFP). Time in minutes relative to anaphase onset. E.Graph of incidence of unaligned metaphase acentric DNA and micronuclei after anaphase in WT or fancd2 RNAi#1 following I-Cre expression, as in panel A. From N= 30-32 cells/condition. *=significant change by Chi-squared, p<.001. White scale bar=25 μm, Yellow scale bar=5μm.
FANCI and Bloom also facilitate papillar DSB survival
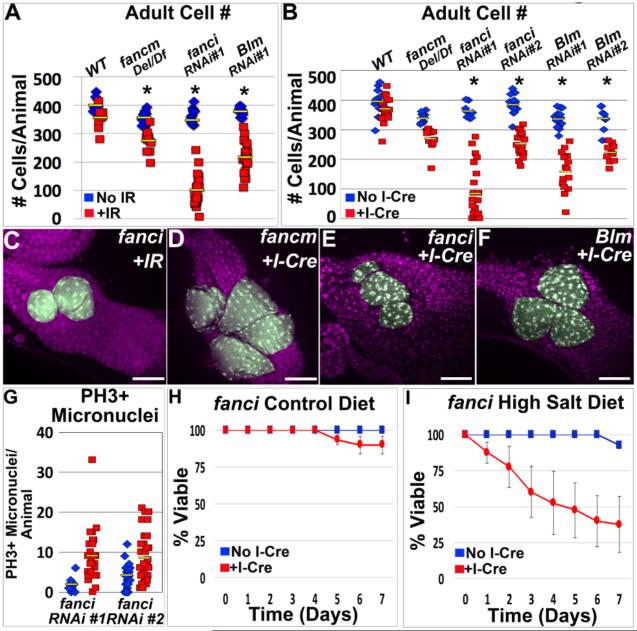
Having found a role for FANCD2 in response to acentric DNA, we next asked if other Drosophila Fanconi Anemia proteins play similar roles. We first examined FANCM, a conserved helicase and member of the Fanconi core complex that recognizes DNA damage and recruits additional FANC proteins during cross-link repair (Kuo et al. 2014). As for FANCD2, we examined papillar survival in fancm mutants (Methods) following IR and I-Cre. fancm mutants are mildly sensitive to IR during papillar development (Fig7A, FigS5C). In contrast, fancm mutants do not exhibit a significantly increased sensitivity to DNA damage specifically caused by I-Cre-induced acentric DNA (Fig7B,D). Thus, FANCM responds to IR in papillar cells, but unlike FANCD2 is dispensable for the papillar response to acentric DNA. This result is consistent with previous reports of FANCD2 functions independent of the FANCM-containing core complex (Yeo et al 2014, Raghunandan et al 2015).
Figure7. FANCI and Blm also facilitate papillar DSB survival.
A.Avg. adult papillar cell number/animal for WT and indicated FANCD2 network mutants +/−IR. From N=7-25 animals/condition, multiple replicates. *=significant change +/−IR compared to WT (Methods). Yellow bars=mean. B.Avg. adult papillar cell number/animal for WT and indicated mutants +/−I-Cre. From N=9-11 animals/condition, multiple replicates. *= significant change +/−I-Cre compared to WT (Methods). Yellow bars=mean. C.fanci RNAi#1 adult rectum after IR. D.fancm adult rectum after I-Cre. E.fanci RNAi#1 adult rectum after I-Cre. F.blm RNAi#1 adult rectum after I-Cre. Papillae false-colored green, DNA in purple in C-F. G.Number PH3+ micronuclei +/− I-Cre in WT vs. fanciRNAi#1 and fanciRNAi#2. Yellow bars= mean. From N=9-27 animals/condition, multiple replicates. H,I.Survival of fanci RNAi#1 adults without (blue) or with (red) I-Cre expression, plotted over time for the indicated diet conditions. Bars= standard error. Each genotype/condition represents 3 replicates with 10 animals/replicate. DAPI=DNA in all images. White scale bar=50μm, yellow scale bar=5μm.
We next examined FANCI, a DSB-responsive nuclease known to function in a heterodimer with FANCD2 (Kondo and Perrimon 2011; Moldovan and D'Andrea 2009). fanci loss decreases adult papillar cell number not only after IR, but also after I-Cre (Fig7A,B,C,E, FigS5D-F). Further, following I-Cre induction, PH3+ micronuclei accumulate in fanci animals at increased frequencies relative to WT (Fig7G), and fanci loss leads to salt-stress sensitivity (Fig7H,I). We also note that, as for fancd2, the level of PH3+ micronuclei without exogenous DNA damage is significantly higher in fanci than in WT (Fig7G vs. Fig5I). These data suggest that like FANCD2, FANCI is required for the papillar acentric DNA response. We then asked if Blm helicase is active in papillar cells following DSB induction. We examined Blm because of its role in FANCD2-mediated segregation of ultra-find DNA bridges (Chan et al. 2007; Naim and Rosselli 2009). Much like FANCD2 and FANCI, Blm loss causes cell loss not only after IR, but also after I-Cre (Fig7A,B,F, FigS5G-J). We were unable to examine micronuclei in Blm-deficient animals due to additional, I-Cre-independent mitotic defects, which have been described previously (FigS5K, McVey et al. 2007). Taken together, our data implicate FANCD2, FANCI, and Blm as regulators of an acentric DNA response that ensures survival of DSB-resistant endocycled cells.
Core DDR components are not required for papillar DSB survival
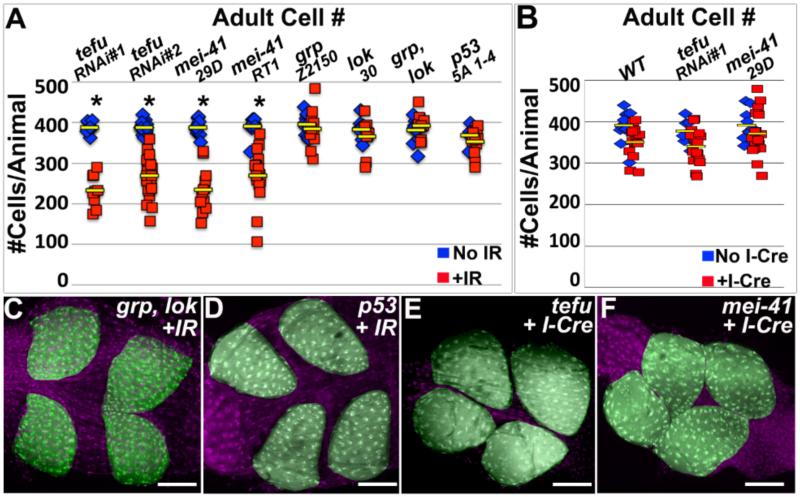
Finally, we asked if our identified FANCD2-dependent response to acentric DNA requires core DDR signaling. The well-known DDR kinases ATM and ATR have both been implicated in FANCD2-dependent repair in specific contexts (Andreassen et al. 2004; Sobeck et al. 2006; Taniguchi et al. 2002). We first induced IR during endocycles in ATM (Drosophila tefu, hereafter ATM) or ATR (Drosophila mei-41, hereafter ATR) mutants (Methods). Both ATM and ATR animals have mild IR-specific decreases in adult papillar cell number (Fig8A, FigS5L,M), implicating these upstream kinases in a papillar response to IR. In contrast, null mutations in one or both of the canonical ATM/ATR downstream kinases chk1 (Drosophila grp) or chk2 (Drosophila lok) have no effect on papillar cell number after IR (Fig8A,C). Similarly, p53 null animals have no impact on post-IR papillar cell survival (Fig8A,D). Thus, papillar cells retain an IR-responsive role for ATM and ATR, but not their common downstream effectors Chk1, Chk2, or p53.
Figure8. Canonical DNA damage regulators are not required for papillar cell survival in response to acentric chromosomes.
A-B.Avg. adult papillar cell number/animal for WT and indicated mutants +/− IR (A) or I-Cre (B). From N=7-17 animals/condition, multiple replicates. *=significant change +/− IR or I-Cre compared to WT (Methods). Yellow bars=mean. C.grp, lok adult rectum after IR. D.p53 adult rectum after IR. E.tefuRNAi#1 adult rectum after I-Cre. F.mei-41 adult rectum after I-Cre. DNA (labeled with DAPI) in purple and papillae pseudo-colored in green in all images. Scale bar=50μm.
Given our demonstrated role for ATM and ATR after IR in papillar cells, we examined if, as for FANCD2, FANCI, and Blm, these kinases are required for papillar cell viability in response to DSB-induced acentric DNA. Both ATM and ATR are dispensable for cell survival following I-Cre mediated induction of acentric chromosomes (Fig8B,E,F). These results differ from the clear requirement for FANCD2, FANCI, and Blm after I-Cre and suggest ATM/ATR play an IR-responsive role independent from responding to acentric DNA in papillar cells. Taken together, our data suggest that successful mitosis with DSB-induced acentric DNA does not require the core DDR regulators ATM, ATR, Chk1, Chk2, or p53.
DISCUSSION
Through activation of apoptosis, cell cycle checkpoints, and high fidelity DNA repair, canonical DDRs prevent cells from entering mitosis with broken chromosomes. In this study, we find naturally endocycling Drosophila papillar cells fail to apoptose, arrest the endocycle, or accomplish accurate repair, leading to mitosis with broken chromosomes. Despite this, papillar development is not hindered by chromosome breakage. In these damage-resistant polyploid cells, DNA fragments that appear to be completely separate from DNA containing centromeres align and segregate during mitosis. Our data suggest that this segregation is mediated by FANCD2, FANCI, and Blm, and serves to prevent micronucleus formation and promote cell viability and proper organ function. As endocycling cells can also acquire resistance to cancer therapies that induce DNA breaks, further study in this area is likely to shed light on cancer-relevant biology.
The endocycle is a recurrent source of DNA damage response attenuation
Previous work in post-mitotic Drosophila and mouse cells identified the endocycle as a source of DDR inactivation. Similar to previous work in non-mitotic endocycling cells and in mouse trophoblasts (Soloveva and Linzer 2004; Mehrotra et al. 2008; Zhang et al. 2014), we find papillar cells do not rely on/activate p53 in response to DNA damage. We recently showed papillar cells undergo a distinct endocycle: the pre-mitotic endocycle (Schoenfelder et al. 2014). Thus, a lack of a requirement for p53 activity appears common in diverse endocycling cells.
In mice, another DDR regulator- Chk1- is inactive in endocycling trophoblast cells (Ullah et al. 2011; 2008). As a potential indicator that this inactivity may be conserved, we find Chk1 (and also Chk2) are not required for the papillar acentric DNA response. In the Drosophila embryo, a Chk1-dependent checkpoint eliminates cells with replication errors (Fogarty et al. 1997; Sibon et al. 1997; 2000; Takada et al. 2007). Given that endocycles frequently cause replication-induced DSBs (Yarosh and Spradling 2014; Mehrotra et al. 2008; Nordman et al. 2011; Hannibal et al. 2014), we speculate a conserved property of endocycles is inactivity of this Chk1 replication checkpoint. This inactivity would explain how cells progress through what are often error-prone endo-S phases and accumulate DNA breaks. Such Chk1-independency may be reversible, as endocycled Drosophila follicle cells require Chk1 for replication fork elongation during gene amplification (Alexander et al. 2015).
Beyond endocycles, polyploidy also inactivates the DDR in other ways. In mammalian cells, cytokinesis failure arrests the cell cycle. Continued division of resulting polyploid cells requires p53 inactivity (Ganem et al. 2014; Wong and Stearns 2005; Fujiwara et al. 2005). Polyploid cancer cells also re-wire canonical DDRs (Zheng et al. 2012). These examples and others (Schoenfelder and Fox 2015) highlight numerous connections between polyploidy and DDR inactivation. However, to this point little was known about how polyploid cells proliferate without functional DDR components.
FANCD2 as an emergency DNA damage regulator
Emerging evidence suggests in the absence of canonical DDRs, emergency DDRs make mitosis compatible with genome damage. BubR1, Polo, and Aurora B respond to broken chromosomes in Drosophila neuroblasts (Royou et al. 2010; Karg et al. 2015; Derive et al. 2015), and Aurora B and INCENP form heterochromatic DNA threads that segregate achiasmate chromosomes during Drosophila meiosis (Hughes et al. 2009). Here, we propose that FANCD2 performs a similar function.
We argue a key role of emergency DDRs is prevention of micronuclei. We previously showed papillar cells are highly tolerant of whole chromosome aneuploidy (Schoenfelder et al. 2014). In contrast to whole chromosome aneuploidy, we find papillar cells are intolerant of mis-segregated acentric DNA fragments and micronuclei. Recent work implicated micronuclei as catalysts in chromothripsis, a genome-shattering event linked to cell death and cancer (Hatch and Hetzer 2015; Zhang et al. 2015). Interestingly, polyploid cancer cells were recently shown to be more likely to undergo chromothripsis (Mardin et al. 2015). Further, Fanconi Anemia and Bloom syndrome patients are cancer-prone, and cells from these patients accumulate micronuclei (Naim and Rosselli 2009). Our data suggest polyploid papillar cells are a useful model to identify other factors like FANCD2 that act in mitotic cells lacking canonical DDRs.
Our data suggest FANCD2, Blm, and FANCI can recognize and respond to DSB-induced acentric DNA, and may act to tether acentric DNA to its centromere-containing fragment. However, it remains possible that instead the chromosomes that we score as acentric contain ultra-fine DAPI-negative DNA bridges, which are technically challenging to visualize. In mitotic mammalian cells, FANCD2, FANCI, and Blm have been shown previously to localize to a specific class of these ultra-fine bridges (UFBs) between daughter nuclei (Chan et al. 2009; Naim and Rosselli 2009). The exact nature and various classes of these bridges is still being determined, although at least some of these structures contain DNA that fails to replicate until cells enter mitosis (Minocherhomji et al 2015).
Thus, it is possible that while I-Cre initially severs the DNA completely, limited DNA repair may join the centromere with its acentric fragment, creating an ultra-fine bridge structure. Similarly, during papillar development, under-replication may naturally occur, as it does during many endocycles that fail to initiate late replication (Lily and Spradling 1996; Nordman et al. 2011; Yarosh and Spradling 2014). If so, such un-replicated DNA may bridge apparent centric and acentric chromosomes, and may be recognized by FANCD2 early in the first mitosis. However, we previously showed that papillar cells are distinct from other endocycled cells in that they do initiate late replication and achieve (at least very close to) full genome duplications (Fox et al. 2010). Future experiments will therefore have to determine exactly what FANCD2 recognizes following endocycles/DSB induction. Regardless, our data suggest that FANCD2 specifically responds to endocycle-induced DNA breakage and is required for segregation of such damaged chromosomes.
Another possible interpretation of our work is that loss of fancd2 increases lesions such as inter-strand cross-links or lesions similar to those caused by IR, which sensitizes papillar cells with apparent acentric DNA. However, our data argue against this model. First, FANCM, a component of the Fanconi core complex, is not required for the response to acentric DNA. A previously established function of this FANCM core complex is to ubiquitinate FANCD2, which is important for specific repair events such as repair of DNA cross-links (Alpi et al. 2008; Marek and Bale 2006). Second, ATM is also not required for the FANCD2 acentric DNA response, suggesting that ATM-mediated FANCD2 phosphorylation, which is important for FANCD2’s previously established response to IR, is also not involved (Sobeck et al. 2006; Taniguchi et al. 2002). Because of these results, we consider it unlikely that excess DNA cross-links and IR-related DNA breaks caused by absence of fancd2 somehow synergize with I-Cre-induced DNA breakage to cause the fancd2 phenotypes, because if this were the case we would see the same result with fancm and ATM mutants following I-Cre (although it remains possible that fancd2 elevates some other class of DNA damage that we did not analyze in this study). Third, FANCD2 acts independently of DNA replication prior to mitotic entry to prevent acentric micronuclei, suggesting that excessive replication stress from endocycles is not the cause of fancd2 micronuclei. Thus, our data suggest that papillar cells are a useful model of the in vivo consequences of FANCD2’s response to DSBs and apparent acentric DNA.
Finally, in a clinical setting, combining current cancer therapies with FANCD2 deficiency has recently emerged as a promising strategy in treating human tumors (Shen et al. 2013a; Burdak-Rothkamm et al. 2015; Liu et al. 2015). Our work may argue that such therapies would be effective in radio-resistant cancers exhibiting either polyploidy or lack of canonical DDR functions.
EXPERIMENTAL PROCEDURES
Drosophila stocks and genetics
Unless indicated, WT was brachyenteron (byn) Gal4, UAS Moesin-GFP. byn-Gal4 expressed all UAS transgenes except in FigS2F,G. All experiments were at 22°C except Fig1G, where animals were kept at 18°C before shifting to 29°C just prior to 2nd larval instar, to inactivate Tub-Gal80(ts), and Fig6, where animals were similarly kept at 18°C until the indicated time period. Salt stress was performed as in Schoenfelder et al. (2014). flybase.org lists the full genotypes for all stocks used. Alleles and transgenes used in this study: byn-Gal4, Tub-Gal80(ts), UAS-p53 (P Gus p53 2.1), UAS-hid, UAS-rpr, CenpC-Tomato, UAS-Moesin-GFP, hisH2AV-GFP, hisH2AV-RFP, hs-I-Cre1 (1A), hs-I-Cre1 (2A), UAS tefu RNAi (v108074, referred to as tefu RNAi #1), UAS tefu RNAi (JF0142,2 referred to as tefu RNAi #2), mei-41 (29D), mei-41 (RT1), grp (Z2150), lok (30), p53 (5A 1-4), UAS fancd2 dsRNA (referred to as fancd2 RNAi #1), UAS fancd2 RNAi (v45433, referred to as fancd2 RNAi #2 in figure 5 and figure 6), UAS fancd2 RNAi (HMC03558, referred to as fancd2 RNAi #2 in figure 4),UAS fanci RNAi (HMS00769, referred to as fanci RNAi#1), UAS fanci RNAi (v24655, referred to as fanci RNAi #2), fancm (Del), Df 3R (ED6058)-a fancm-spanning Deficiency, UAS Blm RNAi (v13309, referred to as Blm RNAi #1), UAS Blm RNAi (v13310, referred to as Blm RNAi #2), AICR-Gal4 (10H05), UAS-GFP, UAS-RFP, BubR1-GFP, UAS Moesin-mCherry, and BubR1 (KEN).
DNA damage
IR was performed in 2nd instar larvae aged to the stage of papillar precursor endocycles. Animals in 60 or 100mm petri dishes with a thin layer of standard Drosophila food were placed in an X-RAD 160 PXI precision X-ray Irradiator (calibrated by a Dosimetrist) at 20 Gy. To compare the number of aberrant chromosomes between WT and fancd2 under the same IR conditions, GFP-marked WT larvae were mixed with RFP-marked fancd2 larvae in the same dish, which was then irradiated. For I-Cre, animals were heat shocked in a 37°C water bath in vials for 90 min. All experiments represent at least 2 separate IR/I-Cre treatments.
Tissue Preparation/Microscopy
Fixation, chromosome preparations, and live imaging were as in Schoenfelder et al. 2014. Antibodies: Mouse Phospho-Histone H3 Ser10, (1:1000, Cell Signaling), Mouse Drosophila Gamma H2AV, (1:2500, Lake et al. 2013), Rabbit GFP (1:1000, Life Technologies). Nuclear labeling in all fixed images: DAPI. For EdU (Invitrogen) labeling, tissue was pulsed with EdU for 15 min., and detection was according to manufacturer’s instructions. TUNEL labeling was as in Schoenfelder et al 2014. Fixed images were acquired using a Zeiss AxioImager M.2 with Apotome processing (20X, 40X or 63X). Live imaging used an Andor XD Spinning Disk Confocal Microscope (60X silicon or 100X oil).
Image analysis
Z-projections were assembled using ImageJ. Movies were assembled using MetaMorph (Molecular Devices). Adobe Photoshop was used to adjust brightness/contrast. ImageJ’s “cell counter” was used to count cell number.
Statistical analyses of adult cell numbers
To determine if papillar cells of a given mutant led to significant cell number decreases after IR/I-Cre relative to WT, the distribution of adult papillar cell number before and after IR/I-Cre was compared for each genotype using a Z-test. Resulting p values determined if any decrease in adult papillar cell number after IR/I-Cre was significantly different than WT. Significant differences (p values ranged from p<.05 to p<1× 10−9) are noted in the figures with *.
Supplementary Material
HIGHLIGHTS.
The endocycle inactivates canonical apoptotic and DNA repair responses
Endocycled cells can divide and survive despite high levels of DNA breakage
During division, endocycled cells align and segregate acentric DNA after DSBs
FANCD2 promotes alignment/segregation of acentric DNA
ACKNOWLEDGMENTS
The following kindly provided reagents: Bloomington and Vienna Stock Centers, Allen Bale, Roger Karess, Dan Kiehart, Christian Lehner, Jeff Sekelsky (who provided the fancm deletion stock before publication), Tin-Tin Su (who also provided technical advice on IR), William Sullivan (who also provided technical advice on imaging tethers), and Will Wood. We thank David Kirsch, Daniel Lew, and members of the Fox and MacAlpine laboratories for valuable comments on the manuscript. D.F. is supported by a Pew Scholar Award and NIH grant GM118447. H.B. is supported by NIH grant CA186545.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
DF and HB designed and conducted the experiments, made figures and wrote the manuscript.
REFERENCES
- Alexander JL, Barrasa MI, Orr-Weaver TL. Replication Fork Progression during Re-replication Requires the DNA Damage Checkpoint and Double-Strand Break Repair. Curr Biol. 2015;25:1654–1660. doi: 10.1016/j.cub.2015.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Molecular Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliaeva ES, Alekseenko AA, Moshkin IM, Koriakov DE, Zhimulev IF. [A genetic factor, suppressing DNA underreplication in Drosophila melanogaster polytene chromosomes] Genetika. 1998;34:762–770. [PubMed] [Google Scholar]
- Burdak-Rothkamm S, Rothkamm K, McClelland K, Rashid Al ST, Prise KM. BRCA1, FANCD2 and Chk1 are potential molecular targets for the modulation of a radiation-induced DNA damage response in bystander cells. Cancer Lett. 2015;356:454–461. doi: 10.1016/j.canlet.2014.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Hickson ID. On the origins of ultra-fine anaphase bridges. cc. 2009;8:3065–3066. doi: 10.4161/cc.8.19.9513. [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-Driven Tetraploidization Occurs in Human Cells Undergoing Crisis and Promotes Transformation of Mouse Cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derive N, Landmann C, Montembault E, Claverie MC, Pierre-Elies P, Goutte-Gattat D, Founounou N, McCusker D, Royou A. Bub3-BubR1-dependent sequestration of Cdc20Fizzy at DNA breaks facilitates the correct segregation of broken chromosomes. J. Cell Biol. 2015;211:517–532. doi: 10.1083/jcb.201504059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nature Reviews Molecular Cell. 2014 doi: 10.1038/nrm3756. [DOI] [PubMed] [Google Scholar]
- Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: new insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Gall JG, Spradling AC. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 2010;24:2294–2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nat Cell Biol. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Gall J, Cohen E, Polan M. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33 doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Cornils H, Chiu S-Y, O'Rourke KP, Arnaud J, Yimlamai D, Théry M, Camargo FD, Pellman D. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M. FANCD2: a branch-point in DNA damage response? Nature medicine. 2002 Jun; doi: 10.1038/nm0602-555. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science (New York, NY) 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hannibal RL, Chuong EB, Rivera-Mulia JC, Gilbert DM, Valouev A, Baker JC. Copy number variation is a fundamental aspect of the placental genome. PLoS Genet. 2014;10:e1004290. doi: 10.1371/journal.pgen.1004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel C, Zhang B, Dixon M, Calvi BR. Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Development. 2014;141:112–123. doi: 10.1242/dev.098871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. Linking Micronuclei to Chromosome Fragmentation. Cell. 2015;161:1502–1504. doi: 10.1016/j.cell.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi Anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- Hughes SE, Gilliland WD, Cotitta JL, Takeo S, Collins KA, Hawley RS. Copenhaver GP, editor. Heterochromatic Threads Connect Oscillating Chromosomes during Prometaphase I in Drosophila Oocytes. PLoS Genet. 2009;5:e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg T, Warecki B, Sullivan W. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol Biol Cell. 2015;26:2227–2241. doi: 10.1091/mbc.E15-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Perrimon N. A genome-wide RNAi screen identifies core components of the G2-M DNA damage checkpoint. Sci Signal. 2011;4:rs1. doi: 10.1126/scisignal.2001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HK, McMahan S, Rota CM, Kohl KP, Sekelsky J. Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics. 2014;198:935–945. doi: 10.1534/genetics.114.168096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake CM, Holsclaw JK, Bellendir SP, Sekelsky J, Hawley RS. The development of a monoclonal antibody recognizing the Drosophila melanogaster phosphorylated histone H2A variant (γ-H2AV) G3 (Bethesda) 2013;3:1539–1543. doi: 10.1534/g3.113.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lily MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ghosh P, Magpayo N, Testa M, Tang S, Gheorghiu L, Biggs P, Paganetti H, Efstathiou JA, Lu H-M, et al. Lung cancer cell line screen links fanconi anemia/BRCA pathway defects to increased relative biological effectiveness of proton radiation. Int J Radiat Oncol Biol Phys. 2015;91:1081–1089. doi: 10.1016/j.ijrobp.2014.12.046. [DOI] [PubMed] [Google Scholar]
- Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Decaillet C, Gari K, Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of S phase checkpoint signaling. Mol Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Drainas AP, Waszak SM, Weischenfeldt J, Isokane M, Stütz AM, Raeder B, Efthymiopoulos T, Buccitelli C, Segura-Wang M, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828–828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek LR, Bale AE. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst) 2006;5:1317–1326. doi: 10.1016/j.dnarep.2006.05.044. [DOI] [PubMed] [Google Scholar]
- McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics. 2007;176:1979–1992. doi: 10.1534/genetics.106.070052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- Moldovan G-L, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009 doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL. Developmental control of the DNA replication and transcription programs. Genome Res. 2011;21:175–181. doi: 10.1101/gr.114611.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, et al. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Molecular Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia Core Complex. Cell Cycle. 2015;14:342–353. doi: 10.4161/15384101.2014.987614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A, Gagou ME, Karess R, Sullivan W. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell. 2010;140:235–245. doi: 10.1016/j.cell.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Fox DT. The expanding implications of polyploidy. J Cell Biol. 2015;209:485–491. doi: 10.1083/jcb.201502016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Montague RA, Paramore SV, Lennox AL, Mahowald AP, Fox DT. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development. 2014;141:3551–3560. doi: 10.1242/dev.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Oswald D, Phelps D, Cam H, Pelloski CE, Pang Q, Houghton PJ. Regulation of FANCD2 by the mTOR pathway contributes to the resistance of cancer cells to DNA double-strand breaks. Cancer Research. 2013a;73:3393–3401. doi: 10.1158/0008-5472.CAN-12-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moran DM, Maki CG. Transient nutlin-3a treatment promotes endoreduplication and the generation of therapy-resistant tetraploid cells. Cancer Research. 2008;68:8260–8268. doi: 10.1158/0008-5472.CAN-08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Perez RE, Davaadelger B, Maki CG. Two 4N cell-cycle arrests contribute to cisplatin-resistance. PLoS ONE. 2013b;8:e59848. doi: 10.1371/journal.pone.0059848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, Wallisch M, Akkari Y, Olson S, Wang W, et al. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Molecular and Cellular Biology. 2006;26:425–437. doi: 10.1128/MCB.26.2.425-437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloveva V, Linzer DIH. Differentiation of placental trophoblast giant cells requires downregulation of p53 and Rb. Placenta. 2004;25:29–36. doi: 10.1016/S0143-4004(03)00215-7. [DOI] [PubMed] [Google Scholar]
- Takada S, Kwak S, Koppetsch BS, Theurkauf WE. grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development. 2007;134:1737–1744. doi: 10.1242/dev.02831. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim S-T, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Titen SWA, Golic KG. Telomere Loss Provokes Multiple Pathways to Apoptosis and Produces Genomic Instability in Drosophila melanogaster. Genetics. 2008;180:1821–1832. doi: 10.1534/genetics.108.093625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, de Renty C, DePamphilis ML. Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Molecular and Cellular Biology. 2011;31:4129–4143. doi: 10.1128/MCB.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008;22:3024–3036. doi: 10.1101/gad.1718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26:31–36. doi: 10.1101/gad.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–278. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W, Spradling AC. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 2014;28:1840–1855. doi: 10.1101/gad.245811.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo JE, Lee EH, Hendrickson EA, Sobeck A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 2014;23:3695–3705. doi: 10.1093/hmg/ddu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Mehrotra S, Ng WL, Calvi BR. Low levels of p53 protein and chromatin silencing of p53 target genes repress apoptosis in Drosophila endocycling cells. PLoS Genet. 2014;10:e1004581. doi: 10.1371/journal.pgen.1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-Z, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z, Wu X, Wu J, Wang C, Zhong J, et al. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat Comms. 2012;3:815. doi: 10.1038/ncomms1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.