Abstract
Objective The objective of openFDA is to facilitate access and use of big important Food and Drug Administration public datasets by developers, researchers, and the public through harmonization of data across disparate FDA datasets provided via application programming interfaces (APIs).
Materials and Methods Using cutting-edge technologies deployed on FDA’s new public cloud computing infrastructure, openFDA provides open data for easier, faster (over 300 requests per second per process), and better access to FDA datasets; open source code and documentation shared on GitHub for open community contributions of examples, apps and ideas; and infrastructure that can be adopted for other public health big data challenges.
Results Since its launch on June 2, 2014, openFDA has developed four APIs for drug and device adverse events, recall information for all FDA-regulated products, and drug labeling. There have been more than 20 million API calls (more than half from outside the United States), 6000 registered users, 20,000 connected Internet Protocol addresses, and dozens of new software (mobile or web) apps developed. A case study demonstrates a use of openFDA data to understand an apparent association of a drug with an adverse event.
Conclusion With easier and faster access to these datasets, consumers worldwide can learn more about FDA-regulated products.
Keywords: openFDA, drug safety, adverse event, API, application programming interface, open data, open source
BACKGROUND AND SIGNIFICANCE
In the United States, Food and Drug Administration (FDA)-regulated products account for about 25 cents of every dollar spent by American consumers each year—products that touch the lives of every American every day.1 Americans rely on the FDA to keep their food, medical products, and other FDA-regulated products safe, and where applicable, effective. FDA’s major activities regarding marketed products are to:
Require firms that manufacture or distribute FDA-regulated products to register with FDA, list the products, and provide the labeling on those products.
Review reports of adverse events, which may include patient or user harm, use error, and/or a quality problem with the product, to monitor marketed products. FDA accepts all voluntary reports and has issued reporting requirements that vary with the particular type of product.
Inspect firms and/or products, either routinely or as part of an investigation prompted by an adverse event report or other concern.
Monitor product recalls, which most often are manufacturer initiated.
FDA has been making non-confidential portions of these data available on www.fda.gov in 3 modes:
Web-based search tools that return structured and/or unstructured data. These are excellent for occasional use with simple queries.
The entire database downloadable in comma-separated value (CSV) or standard generalized markup language (SGML) format. This mode is only practical for users with broadband internet access, large storage space, statistical software, and the technical skill to properly process the relational database files and any unstructured fields.
Individual files based on popular Freedom of Information Act (FOIA) requests. These files may not be relevant to the user’s questions. Members of the public may also file custom FOIA requests for specific records.
To address these difficulties FDA launched the openFDA project in March 2013.
The first priority were databases with a combination of high general consumer interest, low accessibility on www.fda.gov, and high interest on the part of the FDA organization that stewards the database.
By making the data available to the public in a new harmonized big data format, FDA encourages scientists, clinicians, informaticists, software developers, and other technically focused individuals in both the private and public sectors to explore the data, develop applications that automatically access the data, and offer their own enhancements to the data or the software. The more recent Presidential Executive Order on Open Data2 and the Department of Health and Human Services Health Data Initiative3 require FDA to make its publicly available data more easily accessible in a structured, computer readable format.
On June 2, 2014, openFDA was launched in Beta mode at https://open.fda.gov.4 This project uses cutting edge technologies, and is a pilot for how FDA can develop and deploy novel applications in the public cloud securely and efficiently in the future. In this article, we describe the system and demonstrate how to obtain openFDA data through application programming interface (API) calls. A case study illustrates the use of openFDA in investigating an apparent association between a drug and an adverse event.
METHODS
Data Sources
Four main data sources are currently available in openFDA:
FAERS (FDA Adverse Event Reporting System) for drugs and selected biological products.5–7
SPL (Structured Product Labeling) for drugs and selected biological products.8,9
RES (Recall Enterprise System), primarily recall notices, and also market withdrawals and safety alerts, for drugs, selected biological products, devices, and foods.10–12
MAUDE (Manufacturer and User Device Experience), adverse event reports for medical devices.13
Details for each data source can be found at https://open.fda.gov/updates/. To address issues related to differences in the structure of the three drug databases (adverse event reports, recalls, and labeling), openFDA features harmonization on drug identifiers (generic name, brand name, etc.), to make it easier to both search for and understand the drug products returned by API queries (details can be found at https://open.fda.gov/updates/). When users query a drug database API, they can search either fields original to the database (never deleted) or the harmonized openFDA fields.
Logical Architecture
The architecture and technology were chosen to make openFDA scalable; quickly responsive; transferable to new technologies as they mature; easily accessible by application developers, researchers, and the general public; and transparent. The data are on the cloud that has been approved for federal use (Amazon Web Services East).14 Details can be found at https://open.fda.gov/updates/.
Design of open.fda.gov
The design of the open.fda.gov website draws on best practices in agile development, intuitive user experience, and data visualization, aiming to provide one unified, simple presentation to users. The site is organized around broad types of data, rather than FDA’s internal structure. The site is characterized by a combination of interactive programmer-friendly queries, visualizations and examples that help explain the nature of the data and how to use JSON URL query command syntax. Plain language is used throughout.
Design for Engaged, Open Community
Users are encouraged to use GitHub15 to see all the open source code and post their own additions or modifications. StackExchange16 is encouraged for discussion, questions, and answers. In addition, FDA publishes update announcements on open.FDA.gov and an openFDA Twitter account, and maintains an openFDA email account is to widen the options for user participation.
RESULTS
Use of openFDA in Applications
Since the launch of openFDA, a growing community on StackExchange has been engaged in discovering novel ways to use, integrate, and analyze the openFDA data.17 By mid-July 2015, there have been more than 20 million API calls (more than half of which were generated outside the United States), more than 6000 registered API users, and more than 1800 Twitter followers. Twitter has been used to broadcast openFDA news and followers’ feedback and announcements. For example, some followers have shared their experiences of using openFDA to study gender differences in reported drug side effects18 and to cross reference openFDA data with other databases.19
As a result of the US General Services Administration’s Request for Quotes20 on June 17th21 that requires responders to build working prototype apps that use openFDA and post them in GitHub,22 GitLab,23 or BitBucket,24 dozens of prototypes have been posted on GitHub.
Apps have been developed that allow a consumer who experiences an adverse event to determine whether there is a report of anyone else having a similar experience after taking the same drug.25–27 An interactive dashboard display of drug reports was published as a “hobby.”28 More advanced statistics are available on a family of public sites29–33 used in the following case study.
Case Study: The Causal Relationship between Aspirin and Flushing
This case illustrates some of the professional pharmacovigilance processes for assessing an apparent association between a drug and a particular adverse event in a collection of drug adverse event reports summarized in a public FDA guidance document.34 In the past, triggers of increased adverse event reporting to FDA have included news reports, public FDA alerts, and the introduction of new products.35–38 Among the many other issues with drawing causality conclusions from the reports are limited knowledge of the relative extent of use of the drugs involved, and various other alternative explanations for the apparent association. Aspirin is one of the most common drugs listed in openFDA drug adverse event reports (4.2% of all reports). A natural question is what types of adverse events have been reported for aspirin? Or, this could be rephrased as, what types of adverse events were more often reported in the same reports as aspirin, compared to reports that do not mention aspirin? A proportional reporting rate (PRR), a commonly used statistic for this question, of 2 indicates that the proportion of reports for the drug-event combination is twice the proportion of the event in the overall database.34,38,39 Using an interactive program designed to look at openFDA drug reports data,29 we can quickly look at the most common events for aspirin (Table 1). Further details of the steps used in this case can be found at https://open.fda.gov/updates/.
Table 1:
Events reported for generic drug name “aspirin” with PRR > 2.0, ranked by PRR.
| Event rank | Event | No. of reports for both aspirin and event | No. of reports for event | PRR |
|---|---|---|---|---|
| (any) | 169,838 | |||
| 1 | Flushing | 10,071 | 42,843 | 7.6 |
Notes: The query URL for all aspirin reports was http://go.usa.gov/cvRTJ.Finished data and PRR output was from https://open.fda.gov/static/docs/openFDA-analysis-example.pdf.29
PRR is the Proportional Reporting Ratio.
“Flushing” is the most common event, with 6% (10 071/169 838) of all reports mentioning aspirin also mentioning flushing. Furthermore, the PRR indicates that a report containing aspirin is more than seven times as likely to include flushing as a report that does not contain aspirin. Labeling for aspirin does not include flushing in the list of adverse events. Before concluding that aspirin causes flushing, one must rule out noncausal explanations of the association, including, but not limited to: 1) the association was a chance occurrence, 2) an extraneous event resulted in the apparent association, 3) the event was related to the underlying condition that prompted the medication use, and 4) other medications are responsible for the relationship.
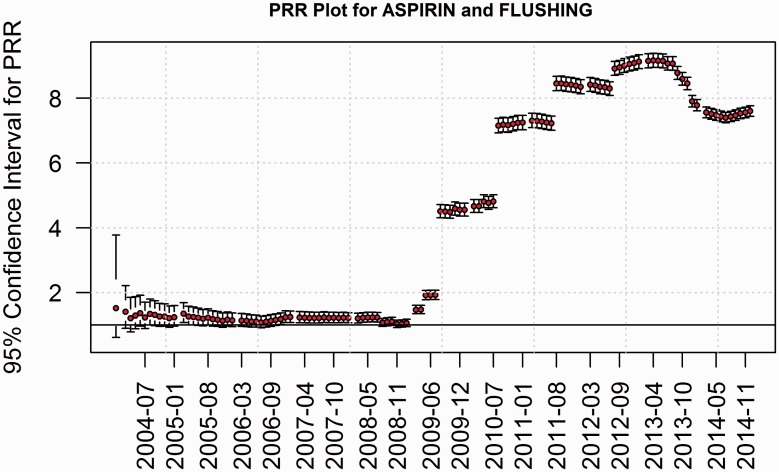
Explanations 1) and 2) can be investigated using a dynamic PRR graph (Figure 1).
Figure 1.
Dynamic proportional reporting ratios (PRR) for reports with aspirin and flushing. At each month, the accumulated reports were used to calculate the PRR and its 95% confidence interval.26
In Figure 1 we see that before 2009 there was little or no statistical association between aspirin and flushing, with the PRR values only slightly above 1, and the 95% confidence intervals often including 1. After 2008 we see the PRR rapidly increase to 4, and then increase further to between 7 and 9. The confidence intervals for post 2008 data all exclude 1, so these are unlikely to be a chance association. The dramatic increase after 2008 suggests an explanatory event in 2008.
In addition, other medication(s) could explain the relationship. Each report allows mentions of multiple drugs and multiple events. Table 2 shows the most common drugs mentioned in reports that also mention aspirin and flushing.
Table 2.
Drugs most frequently mentioned in reports with “flushing” events, restricted to those with PRR >2.0.
| Drug rank | Drug | No. of reports for both flushing and drug | No. of reports for drug | PRR | Drug labeling lists flushing in the field “information_for_patients” |
|---|---|---|---|---|---|
| (any drug) | 42,841 | ||||
| 1 | Niacin | 15,303 | 36,434 | 66 | Yes |
| 2 | Niacin and simvastatin | 4975 | 10,446 | 55 | Yes |
| 3 | Dimethyl fumarate | 6234 | 24,771 | 30 | Yes |
| 4 | Aspirin | 10,071 | 169,838 | 7.6 | No |
| 5 | Lisinopril | 2822 | 90,470 | 3.3 | Yes |
Using openFDA drug labeling, we found that the drugs in Table 2 that list “flushing” are “niacin,” “niacin and simvastatin,” and “lisinopril.” The combination of niacin with simvastatin was first approved as Simcor, February 19, 2008, just before the rise in reports noted in Figure 1. Niacin was reported to reduce the risk of myocardial infarction and stroke in 1975, and to reduce atherosclerosis beginning in 1987.40 Consensus guidelines for niacin therapy were published in 2012 and 2013.41,42 Lisinopril was approved in 1988.43 We then tested whether these three drugs explain all of the apparent association between aspirin and flushing. Looking at aspirin reports that do not mention niacin or lisinopril results in the complete absence of flushing from the list of associated adverse events.
In summary, we have demonstrated that a drug-event association is unlikely to be causal. Research beyond the reporting data is usually essential to fully understand the relationship between drug-event pairs. For example, Cefali et al.44 found that aspirin is a good way to treat flushing. Our case may be a drug (niacin) causing the event (flushing), and an event (flushing) leading to use of the drug (aspirin).
DISCUSSION
The openFDA initiative makes it possible for technology specialists to effectively, automatically, and quickly search, query, or pull massive amounts of public information directly from FDA datasets via URL queries to APIs. As the case study illustrated, openFDA allows users to quickly conduct a variety of analyses to explore the nature of posted data.
As we focus on making existing public datasets accessible in new ways, it is important to note that only data that has already been cleared for public use is considered for openFDA. This is the practical reason that narrative descriptions of drug adverse event reports are not in the public datasets. The absence of the narratives from the drug reports is enough to prevent drawing any valid conclusions from solely the openFDA drug report data.
For the first time, the recalls data are more readily searchable and the drug labeling data are searchable on any of the standard labeling fields.
CONCLUSION
OpenFDA brings a new model of big data search and analytics across disparate and complex sources by simplifying dataset structures. With easier and better access, users can learn more about FDA-regulated products, as shown in the case study of aspirin and flushing. A new open community shares code, documentation, examples, apps, and ideas related to openFDA. As the president’s executive order stated, “openness in government…promotes the delivery of efficient and effective services to the public.”2 We invite use of openFDA for entrepreneurship, innovation, and scientific discovery.
FUNDING
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
COMPETING INTERESTS
The authors have no competing interests to declare.
Acknowledgments
The authors wish to acknowledge the ongoing support of the core openFDA team of FDA employees (Steven Hubbard and Kelemework Yimam) and the wider past and current openFDA team (Sharon Campbell, Sean Herron, and Joe Neubauer). The openFDA team has relied heavily on participation by data owners in the FDA programs (Sandra Abbott, Steven Anderson, Yulia Borodina, Erin Brandt, Eric Brodsky, Margarita Brown, Isaac Chang, Lisa Creason, Suranjan De, Ann Ferriter, David Gartner, Marni Hall, David Heller, Randy Levin, Canida Lyle, Elias Mallis, Stanley Milstein, Jonathan Montgomery, Franklin Ohaegbu, Glenn Peterson, John Quinn, Greg Parkover, Lilliam Rosario, Nancy Sager, Jeff Shuren, Ella Smith, Lonnie Smith, Daniel St Laurent, Kevin Starry, Katherine Vierk, and Linda Walter-Grimm); they provided crucial help to prioritize which datasets to make available, get the right data, ensure data quality, and properly document the data. The authors also appreciate support from Ricky Cokely, Mildred Cooper, Andrea Fischer, Janet Gentry, Amber Griffin, Margaret Hamburg, Janet Woodcock, Walter Harris, Joshua Lehman, Angelique Hebert, Milan Kubic, Alan McClelland, Rosie Owens, Jeff Ventura, Bradford Wintermute, Sean Wybenga, and Carolyn Yancey.
CONTRIBUTORS
Dr T.A.K.-H. spearheaded the conception and design of the openFDA project. He made significant contributions to the conceptualization of the article and review and synthesis of the literature. Dr Z.X. wrote the initial draft of the manuscript and conducted data analysis. M.M., H.N., A.B., and Russell Power developed openFDA API. Dr J.L. and E.J. conducted case study analysis. Dr R.A.B. contributed content, critical review, and revision of the manuscript.
REFERENCES
- 1.Executive Summary: Strategic Plan for Regulatory Science. Food and Drug Administration (FDA). Updated: January 16, 2013. http://www.fda.gov/ScienceResearch/SpecialTopics/RegulatoryScience/ucm268095.htm. Accessed December 10, 2014.
- 2.Executive Order – Making Open and Machine Readable the New Default for Government Information. Office of the Press Secretary, The White House. May 9, 2013. http://www.whitehouse.gov/the-press-office/2013/05/09/executive-order-making-open-and-machine-readable-new-default-government-. Accessed May 7, 2014.
- 3.HHS Open Government Plan. US Department of Health and Human Services. Last reviewed January 2, 2015. http://www.hhs.gov/open/plan/opengovernmentplan/initiatives/initiative.html. Accessed March 1, 2014.
- 4.Kass-Hout T. OpenFDA: Innovative Initiative Opens Door to Wealth of FDA’s Publicly Available Data. Food and Drug Administration. June 2, 2014. FDA. http://blogs.fda.gov/fdavoice/index.php/2014/06/openfda-innovative-initiative-opens-door-to-wealth-of-fdas-publicly-available-data. Accessed March 5, 2015. [Google Scholar]
- 5.Reports Received and Reports Entered into FAERS by Year. Food and Drug Administration. August 6, 2014. http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm070434.htm. Accessed July 18, 2015.
- 6.The Adverse Event Reporting System (AERS): Older Quarterly Data Files. Food and Drug Administration. August 15, 2013. http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm083765.htm. Accessed July 18, 2015.
- 7.FDA Adverse Event Reporting System (FAERS): Latest Quarterly Data Files. Food and Drug Administration. June 16, 2015. http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm082193.htm. Accessed July 18, 2015.
- 8.Kass-Hout T. Providing easy public access to prescription drug, over-the-counter drug, and biological product labeling. Food and Drug Administration. August 18, 2014. https://open.fda.gov/update/drug-product-labeling/. Accessed December 10, 2014. [Google Scholar]
- 9.DailyMed. National Library of Medicine, US National Institutes of Health. http://dailymed.nlm.nih.gov/dailymed/. Accessed July 18, 2015.
- 10.Enforcement Reports. Food and Drug Administration. July 15, 2015. http://www.fda.gov/Safety/Recalls/EnforcementReports/default.htm. Accessed July 18, 2015.
- 11.FDA 101: Product Recalls – From First Alert to Effectiveness Checks. Food and Drug Administration. Updated April 29, 2015. http://www.fda.gov/ForConsumers/ConsumerUpdates/ucm049070.htm. Accessed July 18, 2015.
- 12.Kass-Hout T. OpenFDA provides ready access to recall data. Food and Drug Administration. August 8, 2014. https://open.fda.gov/update/openfda-provides-ready-access-to-recall-data. Accessed July 2015. [Google Scholar]
- 13.Manufacturer and User Facility Device Experience Database – (MAUDE). Food and Drug Administration. May 7, 2015. http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/PostmarketRequirements/ReportingAdverseEvents/ucm127891.htm. Accessed July 18, 2015.
- 14.FedRAMP Compliant Systems. FedRAMP, US General Services Administration. https://www.fedramp.gov/marketplace/compliant-systems/. Accessed July 18, 2015.
- 15.FDA/openFDA. GitHub, Inc. 2015. https://github.com/FDA/openfda. Accessed July 18, 2015.
- 16.StackExchange. StackExchange. http://stackexchange.com/search?q=openFDA. Accessed July 18, 2015.
- 17.OpenFDA. Stack Exchange. https://opendata.stackexchange.com/search?q=openFDA. Accessed July 18, 2015.
- 18.James G. (@gupta_james). “see how men & women get different drug side effects genderedreactions.com – a @GenderMedKI and @TheThinkTrain project using @openFDA data.” September 1, 2014, 2:23pm. Tweet.
- 19.Glenn V. (@TusculumGolfer). “do any of your drug databases have both the @US_FDA @FDA_Drug_Info NDC and @WHO ATC codes listed for each drug? To cross reference.” December 10, 2014, 7:07pm. Tweet.
- 20.Request for Quotation (RFQ) 4QTFHS150004 for the Establishment of Multiple Award Blanket Purchase Agreements (BPAs for Agile Delivery Services (ADS I). US General Services Administration. https://pages.18f.gov/ads-bpa/assets/ADS_RFQ_Final.pdf. Accessed July 18, 2015.
- 21.The Agile Delivery BPA is (almost) here! US General Services Administration. June 15, 2015. https://18f.gsa.gov/2015/06/15/agile-bpa-is-here/. Accessed July 18, 2015.
- 22.GitHub. GitHub, Inc. https://github.com. Accessed July 18, 2015.
- 23.Create, review and deploy code together. GitLab. https://about.gitlab.com/. Accessed July 18, 2015.
- 24.Your team, your code, connected. Atlassian, Inc. https://bitbucket.org. Accessed July 18, 2015.
- 25.Track, report, or subscribe to side effects of drugs, vaccines and medical devices. RecallGuide. https://www.recallguide.org. Accessed July 18, 2015.
- 26.Drug Side Effects. PatientsVille. 2015. http://www.Patientsville.com. Accessed February 20, 2015.
- 27.Welcome to ResearchAE.com. Social Health Insights LLC. 2014. http://www.researchae.com. Accessed December 10, 2014.
- 28.Ramstein E. openFDA drug adverse events dataviz. YouTube. June 8, 2014. https://www.youtube.com/watch?v=qtSfJUSbMmI&feature=youtu.be. Accessed May 15, 2015. [Google Scholar]
- 29.Levine J. openFDA RR. https://open.fda.gov/static/docs/openFDA-analysis-example.pdf. Accessed July 18, 2015. [Google Scholar]
- 30.Levine J. openFDA RR-Event. https://open.fda.gov/static/docs/openFDA-analysis-example.pdf. Accessed July 18, 2015. [Google Scholar]
- 31.Levine J. openFDA Dynamic PRR. https://open.fda.gov/static/docs/openFDA-analysis-example.pdf. Accessed July 18, 2015. [Google Scholar]
- 32.Levine J. openFDA Change Point Analysis. https://open.fda.gov/static/docs/openFDA-analysis-example.pdf. Accessed July 18, 2015. [Google Scholar]
- 33.Levine J. Report Browser. https://open.fda.gov/static/docs/openFDA-analysis-example.pdf. Accessed July 18, 2015. [Google Scholar]
- 34.Guidance for Industry. Good Pharmacovigilance Practices and Pharmacoepidemiologic Assessment. Food and Drug Administration, US Department of Health and Human Services. March, 2005. http://www.fda.gov/downloads/regulatoryinformation/guidances/ucm126834.pdf. Accessed February 20, 2015.
- 35.Tsong Y. Comparing reporting rates of adverse events between drugs with adjustment for year of marketing and secular trends in total reporting. J Biopharm Stat. 1995;5:95–114. [DOI] [PubMed] [Google Scholar]
- 36.McAdams M, Staffa J, Dal Pan G. Estimating the extent of reporting to FDA: a case study of statin-associated rhabdomyolysis. Pharmacoepidemiol Drug Safety. 2008;17(3):229–239. [DOI] [PubMed] [Google Scholar]
- 37.Dasgupta N, Mandl KD, Brownstein JS. Breaking the news or fueling the epidemic? Temporal association between news media report volume and opioid-related mortality. PLoS One. 2009;4(11):e7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bate A, Evans S. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf 2009;18(6):427–436. [DOI] [PubMed] [Google Scholar]
- 39.Szarfman A, Tonning JM, Doraiswamy PM. Pharmacovigilance in the 21st century: new systematic tools for an old problem . Pharmacotherapy 2004;24(9):1099–1104. [DOI] [PubMed] [Google Scholar]
- 40.Subedi FH, Joshi PH, Jones SR, et al. Current guidelines for high-density lipoprotein cholesterol in therapy and future directions. Vasc Health Risk Manag. 2014;10:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evaluation and treatment of hypertriglyceridemia: an Endocrine Society clinical practice guideline. National Guideline Clearinghouse, Agency for Healthcare Research and Quality. 2012. www.guideline.gov/content.aspx?id=38432&search=niacin. Accessed May 15, 2015.
- 42.Stone NJ, Robinson J, Lichtenstein AH, et al. ACC/AHA Guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults. Circulation. 2013;129(25 Suppl 2):S1–S45. DOI: 10.1161/01.cir.0000437738.63853.7a. [DOI] [PubMed] [Google Scholar]
- 43.Drugs@FDA: FDA Approved Drug Products. Searched for “Zestril”. Food and Drug Administration. http://www.accessdata.fda.gov/scripts/cder/drugsatfda/index.cfm. Accessed May 15, 2015.
- 44.Cefali EA, Simmons PD, Stanek EJ, et al. Aspirin reduces cutaneous flushing after administration of an optimized extended-release niacin formulation . Int J Clin Pharmacol Ther. 2007;45(2):78–88. [DOI] [PubMed] [Google Scholar]