Abstract
Objective Cytokines play a central role in both health and disease, modulating immune responses and acting as diagnostic markers and therapeutic targets. This work takes a systems-level approach for integration and examination of immune patterns, such as cytokine gene expression with information from biomedical literature, and applies it in the context of disease, with the objective of identifying potentially useful relationships and areas for future research.
Results We present herein the integration and analysis of immune-related knowledge, namely, information derived from biomedical literature and gene expression arrays. Cytokine-disease associations were captured from over 2.4 million PubMed records, in the form of Medical Subject Headings descriptor co-occurrences, as well as from gene expression arrays. Clustering of cytokine-disease co-occurrences from biomedical literature is shown to reflect current medical knowledge as well as potentially novel relationships between diseases. A correlation analysis of cytokine gene expression in a variety of diseases revealed compelling relationships. Finally, a novel analysis comparing cytokine gene expression in different diseases to parallel associations captured from the biomedical literature was used to examine which associations are interesting for further investigation.
Discussion We demonstrate the usefulness of capturing Medical Subject Headings descriptor co-occurrences from biomedical publications in the generation of valid and potentially useful hypotheses. Furthermore, integrating and comparing descriptor co-occurrences with gene expression data was shown to be useful in detecting new, potentially fruitful, and unaddressed areas of research.
Conclusion Using integrated large-scale data captured from the scientific literature and experimental data, a better understanding of the immune mechanisms underlying disease can be achieved and applied to research.
Keywords: cytokines, disease, expression, MeSH, data integration
BACKGROUND AND SIGNIFICANCE
Cytokines and immune cells play a central role in both health and disease.1–4 Cytokines are small proteins that are secreted from a variety of immune cell types, are involved in many biological processes, and act as key regulators of the immune system.5 The availability of different types of immune-related data is increasing dramatically, providing an attractive source of data for systematic analysis, which could help identify useful immune-related relationships and areas for future research. However, utilizing these data efficiently increasingly requires the integration of different data sources. Researchers must be able to integrate their data with other existing resources and compare the results to prior knowledge. For example, by integrating large numbers of microarray studies, it is possible to compare the results from different studies6 and carry out meta-analyses. Furthermore, different kinds of experimental data (eg, genomics, transcriptomics, and proteomics) can be integrated to gain a better view of an investigated system or disease and to help identify patterns that would otherwise be missed.
One rich, readily available source of disease-related knowledge is the corpus of published scientific research. Many disease-related phenotypic trends are captured in biomedical literature and can be extracted from freely available PubMed7 records. Numerous text mining tools and techniques have been specifically developed for mining and extracting information from PubMed abstracts and full articles.8,9 One such tool is SemRep,10 which specializes in extracting semantic relations from biomedical free text in MEDLINE citations. However, many of these tools and approaches are complex, multi-stage, and task- or domain-specific. Other established tools include MedLEE (Medical Language Extraction and Encoding),11,12 a system based on natural language techniques that has been repeatedly demonstrated to be applicable to many clinical fields, from chest radiographs to pathology reports.11,13,14 However, MedLEE specializes in information extraction from clinical textual records rather than other biomedical text sources, such as PubMed abstracts.
In contrast, an easily accessible source of information is the co-occurrences of entities or concepts in Medical Subject Headings (MeSH) terms associated with PubMed records. MeSH is the National Library of Medicine’s controlled vocabulary thesaurus; it consists of sets of terms naming descriptors in a hierarchical structure and is used for indexing MEDLINE PubMed publications. MeSH descriptors associated with each MEDLINE citation are manually assigned and provide a straightforward, yet potentially very useful, knowledge resource. Numerous works using concept co-occurrences in biomedical texts or in associated MeSH terms have been previously described.15–22 Examples of the usefulness of this data source range from using MeSH descriptor co-occurrence frequencies for the automated annotation of articles19 to constructing and analyzing a network of co-occurring terms.18
Disease-related information can also be gathered from a wide range of experimental data repositories. For example, large-scale expression studies are made freely available by the Gene Expression Omnibus, which currently contains nearly 1.5 million microarrays from almost 60 000 studies.23 Data from such sources can be synthesized, integrated, and compared, to facilitate the investigation of the system or diseases in focus.
To date, integrated immune-related data, such as text-extracted concepts and cytokine level measurements, have not, to our knowledge, been systematically and methodically examined in the context of disease. One database, ImmuneXpresso,24 focuses on the relationships between immune cells and cytokines extracted from the literature. In addition, cell-specific gene expression data was used to cross-validate cytokine-mediated cell-cell interactions and suggest novel interactions. However, data forms other than text-derived data (such as experimental data and clinical data) are not incorporated into ImmuneXpresso, a limitation that is addressed in the work presented here. We present herein a novel integration and analysis of MeSH descriptor-derived cytokine-disease associations and cytokine-specific gene expression data. By integrating these data, we were able to examine relationships between cytokines and diseases that would otherwise be missed. We provide several compelling use cases and examples of the utility of this approach and the knowledge gained by using it.
OBJECTIVES
This work aims to examine immune-related patterns in the context of diseases by suggesting links between different diseases and pinpointing unaddressed areas of research. By intelligently integrating knowledge from different sources, specifically information from the biomedical literature and cytokine gene expression data, we are able to identify and investigate patterns and associations that would otherwise be missed.
MATERIALS AND METHODS
Extracting PubMed Records-Associated MeSH Descriptors
A list of disease names and name synonyms was extracted from the Human Disease Ontology25 (downloaded from the Open Biomedical Ontologies Foundry26 in February 2014) and used for capturing PubMed record-associated MeSH disease terms.27 The Human Disease Ontology was selected for this project, for its comprehensive coverage of human disease concepts and its ontological properties, which could also be used to conduct complex queries. A list of cytokine MeSH descriptors (such as “interferon gamma,” “transforming growth factor beta,” and “chemokine CCL3”) was manually compiled by a domain expert, who browsed MeSH’s sub-trees and selected those descriptors that were deemed relevant to this work. Similarly, a list of immune-related cell type MeSH descriptors (such as “lymphocytes,” “Th1 cells,” and “basophils”) was also compiled (see Supplementary File 1). The MeSH Supplementary Concept Records, which include concepts that are supplementary to MeSH descriptors, were not used to capture cytokine concepts, because the majority of these concepts represent species-specific variations and the PubMed records used for this work were limited to those linked to the keyword “human” (see below).
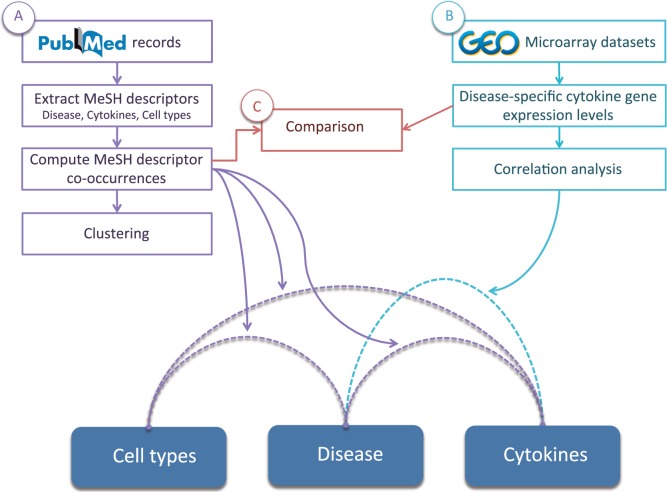
For proof of concept, PubMed records (n = 2 405 255) were exported (in February 2014) from the National Center for Biotechnology Information in the MEDLINE format, using the key word “human” in a text-word search of PubMed. Using a script implemented in Perl, the list of associated MeSH descriptors was recorded for each PubMed record. The lists of cytokine names, cell types, and diseases (compiled as described above) were searched for exact matches with the MeSH descriptors associated with each record (Figure 1A).
Figure 1:
Analysis workflow components. Dashed lines indicate the linking of types of entities via different types of data. Solid lines indicate workflows and analyses that are presented in this article.
Therapeutics- and diagnostics-related MeSH descriptors (from a list compiled by a domain expert, which included terms such as “diagnosis,” “prognosis,” and “therapy”; see Supplementary File 1) were also identified. These MeSH descriptors were used to assign each record to one of the two categories (therapeutics or diagnostics), both categories, or neither category.
Evaluation of MeSH Descriptor Co-occurrences
To evaluate whether co-occurrences of MeSH descriptors (computed as described below) within the same PubMed record represent a true (meaningful and feasible) relationship between the terms, 100 random abstracts were selected from our dataset and evaluated by a domain expert with a significant background in immunology. For each abstract, each pair of MeSH descriptors (disease-cell, disease-cytokine, or cell-cytokine) was evaluated to determine whether it represented a true relationship, an indirect relationship, or an incorrect relationship. The complete evaluation results are available as Supplementary File 2. A true relationship is one that is clearly and explicitly stated in the text. For example, the association between non-small cell lung carcinoma and dendritic cells would be considered a true relationship, based on the text “we investigated the changes in DC phenotype and expression of B7-H molecules induced by non-small cell lung cancer.”28 Indirect relationships are ones that can be understood or inferred from the text but are not explicit. For example, the association between “CD4 positive T-lymphocytes” and “breast neoplasms” would be considered indirect, because, in the text “we review here evidence for the importance of specific CD4+ Th activation in cancer immunotherapy”29 breast neoplasms are not explicitly mentioned (nor are they explicitly mentioned anywhere else in the abstract), although cancer, in general, is. An incorrect relationship between two concepts is one in which, for example, one of the concepts is not mentioned at all in the text or, based on the text, there is no demonstrated relationship between the two concepts (directly or indirectly).
Clustering Analysis of MeSH Descriptor Co-occurrence Data
In order to examine disease similarities based on cytokine co-occurrences in the literature, we set out to cluster these patterns of co-occurrences. To do so, a quantitative cytokine-disease matrix was generated by obtaining, for each disease and cytokine in the database, a count of the number of records mapped to that disease and that cytokine. Similar matrices for cytokine-cells and cells-disease co-occurrences were also generated (see Supplementary File 3).
Using these matrices as input, hierarchical clustering was performed using (1-correlation) as the distance measure, taking into consideration only co-occurrences that were supported by 10 or more records (Figure 1A). To identify statistically significant clusters, the pvclust package in R30 was used, with the following parameters: method – “average,” alpha variable – 0.95 (P < .05), and the number of bootstrapping – 1000. The complete clustering results are provided as Supplementary File 4.
Gene Expression Analysis
Processed gene expression data for a variety of disease phenotypes were obtained from the Gene Expression Omnibus,23 with selection limited to only those studies that used the Affymetrix Human Genome U133, U133A, or U133 Plus 2.0 arrays (Figure 1B). Only studies that examined a disease state in comparison to a healthy (or equivalent) control were selected (resulting in a total of 22 datasets, representing 28 disease phenotypes, as described in a previous article31). These studies are listed in Supplementary File 5. The diseases included in this dataset are: Huntington’s disease, melanoma, polycystic ovary syndrome, bipolar disorder, vulvar intraepithelial neoplasia, acne, psoriasis, Parkinson’s disease, teratozoospermia, endometriosis, adenoma, Down’s syndrome, multiple sclerosis, heart failure, squamous cell cervical cancer, sickle cell anemia, obesity, renal clear cell carcinoma, and type 1 and type 2 diabetes.
To identify diseases with similar cytokine expression patterns, fold changes between the disease and the control states were calculated for probes representing cytokine genes, which were manually selected (n = 51). A median of the fold changes was calculated for cytokine genes represented by more than one probe. Fold changes were then used to calculate Pearson correlations between diseases (Supplementary File 6). Because we used a within-study measure (i.e. the fold change between disease and control states), to compare patterns between different studies, it was not necessary to conduct uniform processing, such as normalization, for the datasets that were examined.
Comparing MeSH Descriptor-Derived Co-occurrences and Cytokine Gene Expression
To examine similarities and differences between MeSH descriptor-derived co-occurrences and cytokine gene expression, data for matching diseases and cytokines from both datasets were collected. For each of the diseases for which data in our expression dataset was available, the matching MeSH descriptor-derived cytokine-disease co-occurrence counts were collected into a matrix and transformed into z-scores. Cytokine gene expression fold changes were also collected into a matrix and transformed into z-scores. The transformation of values from both datasets to z-scores, a method for data normalization regularly used in expression microarray analysis, was necessary, in order to standardize the fold changes and co-occurrence counts (which represent very different types of values) and make them comparable. Twenty-six disease phenotypes were matched in the MeSH descriptor dataset (excluding vulvar intraepithelial neoplasia and teratozoospermia, which were unmatched). A differential matrix of 26 matching disease phenotypes over 51 matching cytokines was generated by subtracting the cytokine gene expression fold changes z-score (absolute values) matrix from the MeSH descriptor co-occurrences z-score matrix (Figure 1C). The resulting differential matrix was plotted using the color2D.matplot function in the plotrix R package.
The dataset is freely available in MySQL,30 as Supplementary File 7.
RESULTS
Extracting Cytokine, Cell Type, and Disease Co-occurrences from Biomedical Literature
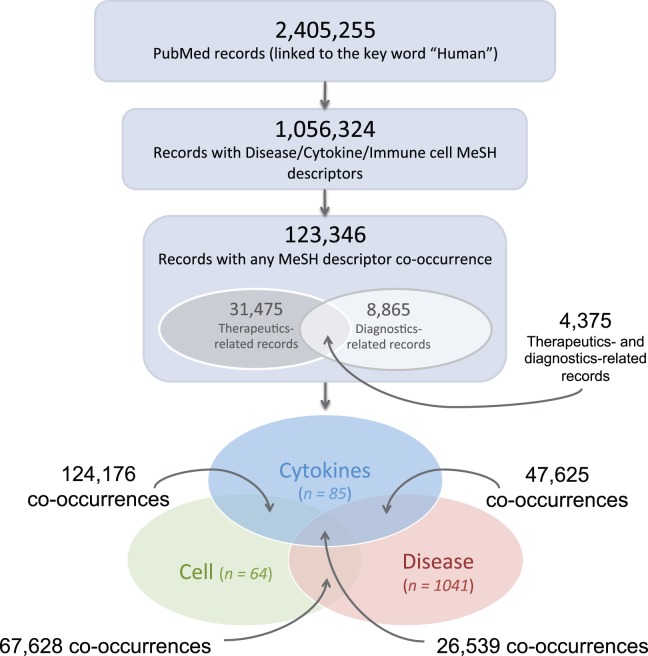
MeSH descriptors associated with over 2.4 million PubMed records were studied for occurrences of cytokines, disease, and cell type terms (Figure 2), and the co-occurrences of these different types of entities within the biomedical literature were captured. This dataset includes 265 968 co-occurrences between 85 different cytokines, 1041 diseases, and 64 cell types. MeSH descriptors were also used to categorize records into those associated with therapeutics or diagnostics (see Materials and Methods section).
Figure 2:
Extracting PubMed record-associated MeSH descriptors. The lower Venn diagram represents the occurrences of the different types of MeSH descriptors (disease, cell, or cytokine), with overlapping areas representing the co-occurrences between the different term types. The numbers of co-occurrences illustrated here correspond to the full dataset of records linked to the key word “Human.”
We next set out to evaluate the nature and capability of MeSH descriptor co-occurrences to capture true relationships and dependencies between concepts, as they are described in the biomedical literature. From 100 randomly selected records, 253 pairs of MeSH descriptors of different types (cytokine, disease, or cell type) were captured by our computational analysis. Of these co-occurrences, 61.7% were found to represent real relationships or dependencies in the abstracts associated with the records, and an additional 10.7% were found to represent indirect relationships between the terms. The remaining co-occurrences (27.7%) were found to represent incorrect relationships. These findings are a strong indication that the co-occurrence of MeSH descriptors linked to any given PubMed record are a good source for mining dependencies between different types of biomedical entities.
Discovering Biomedical Relationships Through MeSH Descriptor Co-occurrences
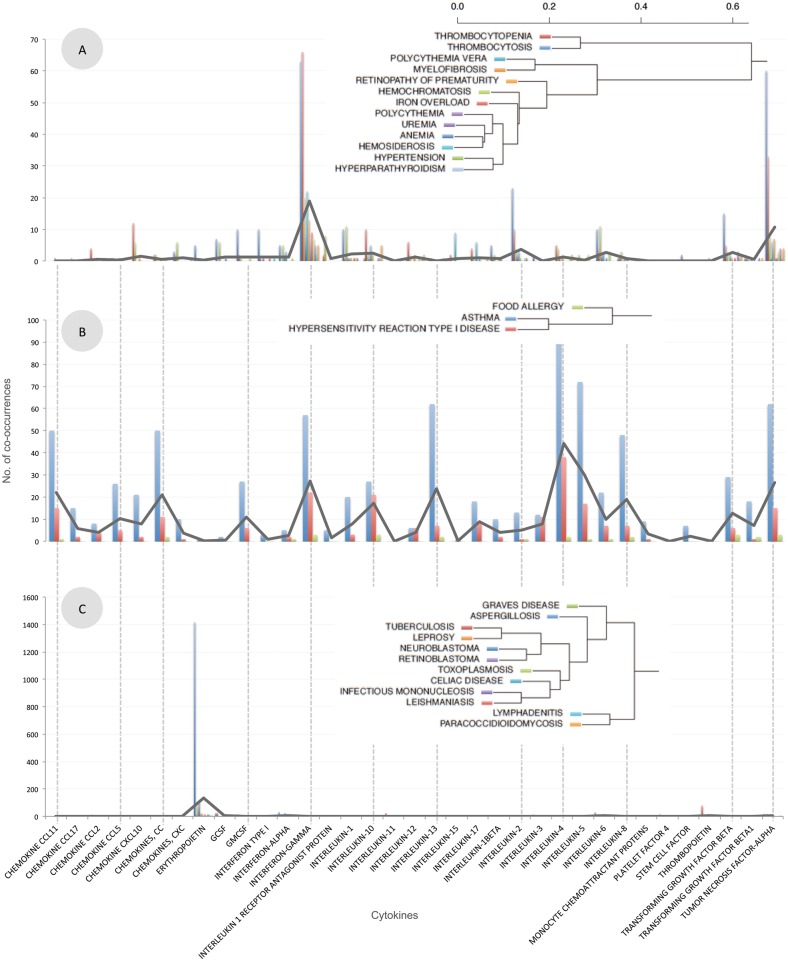
Using the MeSH descriptor co-occurrence counts, 8693 diseases were clustered based on their co-occurrence with cytokines, generating 45 significant clusters (P < .05).
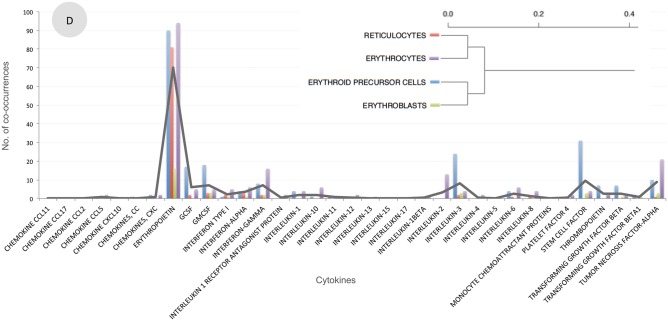
The resulting disease clusters reflect, at least in some cases, current medical knowledge. In the example provided in Figure 3A, many blood-related diseases (such as anemia, uremia, hypertension, polycythemia vera hemochromatosis, thrombocytopenia, and iron overload) are grouped together and characterized by a high association with erythropoietin, a hormone that controls red blood cell production. In another example (Figure 3B), allergy-related diseases (food allergy, asthma, and hypersensitivity reaction type I disease) were grouped together based on a strong association between these diseases and a variety of different cytokines, such as interleukin 4 (IL-4), IL-5, IL-13, and interferon gamma (IFNγ). These conditions all involve some immune response to allergens, and the cytokines associated with this cluster have all been shown to be major contributors to conditions such as asthma and allergies.31 Clustering of cell types, using MeSH descriptor co-occurrence counts, also captures common medical knowledge. In the example illustrated in Figure 3D, different forms of red blood cells were clustered together. Taken together, these clusters of diseases or cell types based on their association with cytokines, as captured by MeSH descriptor associations in the literature, represent current medical views, thus validating our dataset.
Figure 3:
Selected disease and cell type clusters. Diseases (shown in A, B, and C) or cell types (shown in D) were clustered based on their pattern of co-occurrence with cytokines. For each cluster, the hierarchical dendrogram is shown, along with a plot illustrating the number of co-occurrences for each disease/cell and each cytokine. Black lines illustrate the average of the co-occurrences in each cluster.
Other clusters can be used to generate interesting hypotheses regarding links between diseases, based on their association patterns with cytokines. In one example, Figure 3C shows a cluster of diseases characterized by their high association with IFNγ and tumor necrosis factor alpha (TNFα). This cluster contains an unusual grouping of several different types of diseases: different parasitic infections (such as leishmaniasis, leprosy, and toxoplasmosis), different types of cancer (such as retinoblastoma and neuroblastoma) and other diseases (such as celiac disease and Graves’ disease). Clusters of this kind could be used to generate hypotheses regarding the link between these different types of diseases and a possible shared immunological mechanism. In this case, it could be hypothesized that, although these diseases do not share an obvious common mechanism, because they are all highly associated with IFNγ and TNFα in our MeSH descriptor co-occurrence data, they may share some common immunological basis. One possible theory is that the commonality between these diseases is the formation of granulomas, a form of localized nodular inflammation found in tissues and comprising mostly mononuclear cells.34 Tuberculosis, aspergillosis, leishmaniasis, infectious mononucleosis, and toxoplasmosis are all granulomatous infections. On the other hand, although Graves’ disease, celiac disease, retinoblastoma, and neuroblastoma are not granulomatous by definition, there have reportedly been cases of each of the diseases in which granulomas were observed.35–38 Although this could be a possible explanation for the grouping of these diseases seen in our analysis, this theory is only one of many possibilities and must be further investigated and validated before any conclusions can be drawn.
Cytokine Gene Expression
In order to further validate the MeSH descriptor-derived co-occurrences dataset, a correlation analysis was performed on cytokine gene expression levels in a variety of disease phenotypes (Figure 4).
Figure 4:
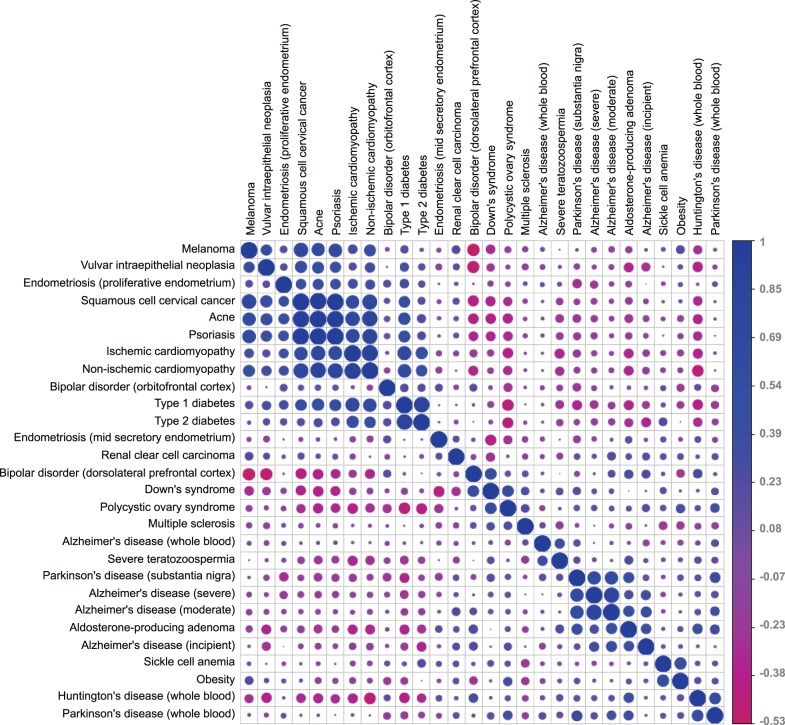
Cytokine gene expression correlations between all pairs of diseases. Pearson correlations were calculated for the cytokine gene fold change patterns between all possible pairs of diseases. The circles in the intersection of each disease pair illustrate the size and direction of the correlation (the larger the circle is, the stronger the correlation; the closer the color to blue, the more positive the correlation; and the closer the color to pink, the more negative the correlation).
Several disease pairs were found to be highly correlated based on cytokine gene expression. In one example, type 1 and type 2 diabetes were highly correlated (r = 0.85). Both types of diabetes share several similar etiology aspects, including immune system involvement. Death of pancreatic β-cells occurs in both types of diabetes, and, in both diseases, it seems that inflammatory processes and cytokines are involved.37 We observed that, in peripheral blood mononuclear cells samples collected from children with type 1 or type 2 diabetes, there was a significant over-expression of chemokine (C-C motif) ligand 2 (CCL2) in the disease states compared with the control samples (fold changes of 4.3 and 4.6, respectively). Additionally, chemokine (C-X-C motif) ligand 1 (CXCL1), IL-1b, and IL-8 were significantly over-expressed in both diseases and IL-18, IL-5, and platelet factor 4 (PF4) were significantly under-expressed in both forms of diabetes. Thus, our results further demonstrate the immunological link between the two diseases.
A second pair of diseases correlated on cytokine expression are cervical cancer and psoriasis (r = 0.96). In addition to a similar pattern of cytokine expression, the single nucleotide polymorphism (SNP) rs6887695, located in the IL-12b gene, was found to be associated with an increased risk for both cervical cancer and psoriasis,40,41 indicating that there is a genetic commonality between these two diseases in addition to the transcriptional similarities. Patterns of co-occurrence of cervical cancer and psoriasis with cytokines in biomedical texts were also found to correlate with one another (r = 0.75, P < .05), thus validating that disease similarities found by gene expression and genetic markers can also be captured by MeSH descriptor co-occurrences.
Identifying Potentially Interesting Cytokine-Disease Associations
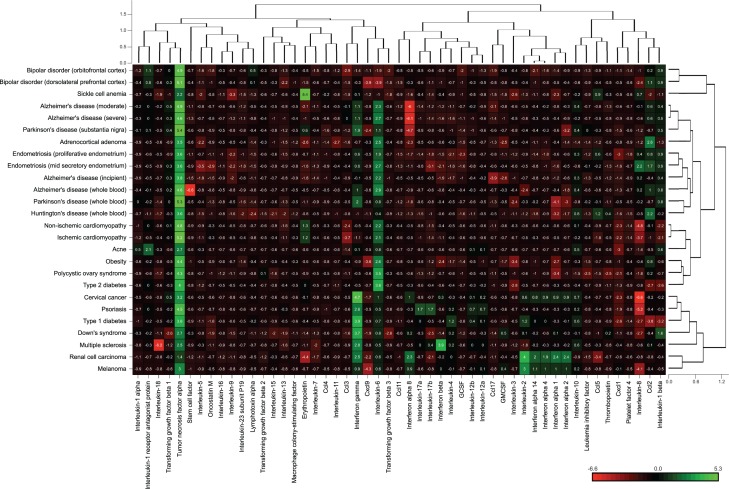
To examine cytokine-disease associations that are of potential interest but have not been examined in the past, cytokine gene expression fold changes in various diseases were compared to cytokine co-occurrence counts from the biomedical literature (Figure 5). A differential matrix of disease phenotypes matched in both datasets was created by calculating the difference between cytokine expression fold change z-scores (absolute value) and MeSH descriptor co-occurrences z-scores. If a given cytokine is highly up- or down-regulated in a given disease and there is also a high cytokine co-occurrence count with that disease based on the data mined from the biomedical literature, the difference between the cytokine expression fold change and the MeSH descriptor co-occurrence z-scores will be around zero, indicating that although such cytokines are interesting in the context of that disease, these associations have already been extensively investigated. In contrast, if a cytokine gene is highly up- or down-regulated in a given disease but has a low MeSH descriptor co-occurrence with that disease, the difference between the cytokine expression fold change and the MeSH descriptor co-occurrence z-scores will be a negative value. A very negative value (represented in red in Figure 5) indicates that that disease-cytokine association is potentially interesting but has not been extensively reported on in the biomedical literature.
Figure 5:
A differential matrix representing the differences between cytokine gene expression and MeSH descriptor co-occurrence counts in various diseases. Red values represent cytokines that are highly up- or down-regulated in a disease with a low count of MeSH descriptor co-occurrences for that disease. Green values represent cytokines that have a high count of MeSH descriptor co-occurrences and a low differential cytokine gene expression fold change value. Black values represent a difference between MeSH descriptor co-occurrence counts z-scores and absolute cytokine gene expression fold change z-scores that are close to zero.
In one example, interferon alpha 8 (IFNα 8) is highly up-regulated in severe and moderate Alzheimer’s disease (over four times higher gene expression levels than in control samples), yet the association between IFNα 8 and Alzheimer’s disease has not been extensively investigated in the biomedical literature. On the other hand, although the expression of TNFα is down-regulated in obesity, the association between the two has been extensively reported on in the biomedical literature and thus is less potentially interesting for future investigations.
DISCUSSION
In this work, we examined the usefulness of exploring and integrating literature-derived data with gene expression data for discovery and hypothesis generation regarding potential links between diseases and immunity. As demonstrated in other fields of research, integrating several sources of information provides a holistic view of the domain that would otherwise not have been achieved.
Integrating Literature-Based and Molecular Data for Research and Discovery
The potential usefulness of MeSH descriptor co-occurrences is demonstrated in several examples. By clustering these co-occurrences, we showed that, although some clusters reflected current medical views (thus validating this dataset), others can be used to generate interesting hypotheses regarding links between diseases based on their cytokine association patterns. To validate MeSH descriptor-derived disease-cytokine patterns and to illustrate the usefulness of incorporating cytokine gene expression data into such analyses, cytokine fold changes in gene expression were compared between several diseases. The examples we present herein clearly demonstrate that disease similarities found by examining gene expression and genetic markers can also be captured by MeSH descriptor co-occurrences. By integrating our MeSH descriptor co-occurrence data with gene expression data, we were able to identify cytokine-disease associations that are potentially interesting but have not been extensively investigated in the past. This type of analysis can be used to assist in detecting new, potentially fruitful, and unaddressed areas of research. Furthermore, such analyses can easily be adopted and applied to other biomedical fields (such as cancer) in which gene expression data or other experimental data, such as proteomic or SNP data, is available.
Evaluation and Limitations of Using MeSH Descriptor Co-occurrences
The approach we took to capturing disease- and immune-related associations from MeSH descriptors has some limitations. One of the assumptions of our method is that co-occurrences of MeSH descriptors within a PubMed record represent a true relationship or dependency. In our evaluation of the extent to which MeSH descriptor co-occurrences represent real relationships, we found that over 70% of co-occurrences of different types of entities (disease, cell type, or cytokine) were found to represent true direct or indirect dependencies. Furthermore, because our evaluation approach was based on abstracts rather than the full text of articles, we have likely underestimated the accuracy of our MeSH descriptor-derived data, missing associations captured by MeSH descriptors that are mentioned in the full texts of the articles but not in the abstracts. One possible way to increase the ability of MeSH descriptor co-occurrences to accurately represent real relationships is to limit the selection of PubMed records to those linked to relevant MeSH descriptors that are designated as the major topics of the article. Another possibility is to leverage MeSH qualifiers. MeSH qualifiers are mainly used to group citations that are concerned with a particular aspect of a MeSH descriptor of interest. Qualifiers such as “diagnosis,” “immunology,” and “blood” can be used to filter MeSH descriptors considered in co-occurrence analyses. However, both of these approaches are likely to greatly reduce the size of the dataset. The ability of our MeSH descriptor-derived data to capture medical knowledge, as demonstrated by our clustering results and by cytokine gene expression patterns, affirms this dataset’s quality and its representation of real immune-disease associations and actual medical views.
A second shortcoming of using only MeSH descriptor co-occurrences data is that even if these co-occurrences correctly capture an association between two concepts, the type of association (such as “stimulates,” “inhibits,” etc.) the direction of the association or the sentiment of the relationship (positive or negative) are not captured. Future development of this dataset will include attempts to capture specific semantic relations between cytokines, cells, and diseases and could be aided by the use of natural language processing tools such as SemRep.10
Future Directions
The work presented here, specifically, the analyses of the MeSH descriptor-derived co-occurrences data, indicates that the knowledge extracted from this data is sufficient and accurate enough to reflect current medical knowledge. Future work will include the addition of well-established cytokine-cell and disease-cell associations from several resources (such as medical text-books42 and canonical pathways43–47). By incorporating well-established relationships between cytokines, diseases, and cell types, a better model of these relationships can be generated and used to validate the other forms of knowledge (such as that generated from experimental data).
This work also sets out to examine the differences between cytokines and cells as regards their use as diagnostic measures and therapeutic agents (as captured in the biomedical literature). To construct a basis for this, we captured subsets of diseases, cytokines, and cell co-occurrences that were found in PubMed records also linked to diagnostic or therapeutic-related MeSH descriptors (Figure 2). Future work will focus on utilizing this basis to investigate the differences between cytokine associations of diseases in the context of diagnostics and in the context of therapeutics. The strength of such an analysis will come from the “automated” review and summarization of many publications, which could suggest several factors that could be used, possibly together, as diagnostic markers or as therapeutic agents.
Another key future development will focus on immune-related clinical data. This type of data is starting to become available through various platforms, such as ImmPort48 and Clinical Study Data Request.49 However, appropriate methods for these data's capture, representation, and integration need to be identified. Overall, this study has demonstrated that the currently available data has already proved to be very useful for investigating the role that cytokines play in disease.
CONCLUSIONS
Integrating MeSH descriptor-derived association and cytokine gene expression data holds the potential for new discoveries in disease-related immunity. Herein, we demonstrated the usefulness of this approach for capturing immune-related patterns, generating feasible hypotheses about disease and immunological mechanisms, and focusing on potentially useful avenues of future research. We have shown that, using integrated large-scale data, such as that presented here, can help achieve a better understanding of the immune mechanisms underlying disease.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Marina Sirota and Dr Sandra Andorf for helpful discussion and their useful suggestions.
CONTRIBUTORS
N.G., S.B., and A.J.B. conceived the idea, designed the research, and wrote the article. N.G. conducted the analyses. All the authors read and approved the final manuscript.
FUNDING
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grant number: HHSN272201200028C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
COMPETING INTEREST
None.
REFERENCES
- 1.Hamze M, Desmetz C, Guglielmi P. B cell-derived cytokines in disease. Eur Cytokine Network. 2013;24(1):20–26. [DOI] [PubMed] [Google Scholar]
- 2.Cromheecke JL, Nguyen KT, Huston DP. Emerging role of human basophil biology in health and disease. Curr Allergy Asthma Rep. 2014;14(1):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melo RC, Liu L, Xenakis JJ, Spencer LA. Eosinophil-derived cytokines in health and disease: unraveling novel mechanisms of selective secretion. Allergy. 2013;68(3):274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu L, Van Kaer L. Natural killer T cells in health and disease. Front Biosci. 2011;3:236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oppenheim JJ. Cytokines: past, present, and future. Int J Hematol. 2001;74(1):3–8. [DOI] [PubMed] [Google Scholar]
- 6.Burgun A, Bodenreider O. Accessing and integrating data and knowledge for biomedical research. Yearb Med Inform. 2008:91–101. [PMC free article] [PubMed] [Google Scholar]
- 7.PubMed. http://www.ncbi.nlm.nih.gov/pubmed/. Accessed February 2014.
- 8.de Bruijn B, Martin J. Getting to the (c)ore of knowledge: mining biomedical literature. Int J Med Inform. 2002;67(1-3):7–18. [DOI] [PubMed] [Google Scholar]
- 9.Shatkay H, Feldman R. Mining the biomedical literature in the genomic era: an overview. J Comput Biol. 2003;10(6):821–855. [DOI] [PubMed] [Google Scholar]
- 10.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–477. [DOI] [PubMed] [Google Scholar]
- 11.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. JAMIA. 1994;1(2):161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language. JAMIA. 1999;6(1):76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain NL, Knirsch CA, Friedman C, Hripcsak G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp. 1996:542–546. [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Friedman C. Facilitating research in pathology using natural language processing. AMIA Annual Symposium; 2003: American Medical Informatics Association; 2003: 1057. [PMC free article] [PubMed] [Google Scholar]
- 15.Doerks T, van Noort V, Minguez P, Bork P. Annotation of the M. tuberculosis hypothetical orfeome: adding functional information to more than half of the uncharacterized proteins. PloS one. 2012;7(4):e34302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. Proceedings / AMIA Annual Symposium AMIA Symposium. 2002:722–726. [PMC free article] [PubMed] [Google Scholar]
- 17.Stapley BJ, Benoit G. Biobibliometrics: information retrieval and visualization from co-occurrences of gene names in Medline abstracts. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2000:529–540. [DOI] [PubMed] [Google Scholar]
- 18.Kastrin A, Rindflesch TC, Hristovski D. Large-scale structure of a network of co-occurring MeSH terms: statistical analysis of macroscopic properties. PloS One. 2014;9(7):e102188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kavuluru R, Lu Y. Leveraging output term co-occurrence frequencies and latent associations in predicting medical subject headings. Data Knowl Eng. 2014;94:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowell V, Bodenreider O. Using dependence relations in MeSH as a framework for the analysis of disease information in medline. Proceedings of the Second International Symposium on Semantic Mining in Biomedicine (SMBM-2006); 2006. 2006:83. [Google Scholar]
- 21.Avillach P, Dufour JC, Diallo G, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU-ADR project. JAMIA. 2013;20(3):446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimino JJ, Barnett GO. Automatic knowledge acquisition from MEDLINE. Methods Inform Med. 1993;32(2):120–130. [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen-Orr SS, Goldberger O, Garten Y, et al. Towards a cytokine-cell interaction knowledgebase of the adaptive immune system. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2009:439–450. [PMC free article] [PubMed] [Google Scholar]
- 25.The Human Disease Ontology. 2014. http://do-wiki.nubic.northwestern.edu. Accessed February 2014.
- 26.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medical Subject Headings. http://www.nlm.nih.gov/mesh/
- 28.Schneider T, et al. Non-small cell lung cancer induces an immunosuppressive phenotype of dendritic cells in tumor microenvironment by upregulating B7-H3. Journal of Thoracic Oncology. 2011;6(7):1162–1168. [DOI] [PubMed] [Google Scholar]
- 29.Xu M, Nikoletta LK, Eric von H, et al. CD4+ T-cell activation for immunotherapy of malignancies using Ii-Key/MHC class II epitope hybrid vaccines. Vaccine. 2012;30(18):2805–2810. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–1542. [DOI] [PubMed] [Google Scholar]
- 31.Suthram S, Dudley JT, Chiang AP, Chen R, Hastie TJ, Butte AJ. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol. 2010;6(2):e1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MySQL. http://www.mysql.com/.
- 33.Ngoc PL, Gold DR, Tzianabos AO, Weiss ST, Celedon JC. Cytokines, allergy, and asthma. Curr Opin Allergy Clin Immunol. 2005;5(2):161–166. [DOI] [PubMed] [Google Scholar]
- 34.Zumla A, James DG. Granulomatous infections: etiology and classification. Clin Infect Dis. 1996;23(1):146–158. [DOI] [PubMed] [Google Scholar]
- 35.Dabski K, Winkelmann RK. Generalized granuloma annulare: clinical and laboratory findings in 100 patients. J Am Acad Dermatol. 1989;20(1):39–47. [DOI] [PubMed] [Google Scholar]
- 36.Loche F, Bazex J. [Celiac disease associated with cutaneous sarcoidosic granuloma]. Rev Med Interne. 1997;18(12):975–978. [DOI] [PubMed] [Google Scholar]
- 37.Hojo H, Suzuki S, Kikuta A, Ito M, Abe M. Sarcoid reaction in primary neuroblastoma: case report. Pediatr Dev Pathol. 2000;3(6):584–590. [DOI] [PubMed] [Google Scholar]
- 38.Casson AG, Maroun FB, Arnold AM, Newman CE. Retinoblastoma, eosinophilic granuloma, and malignant melanoma: a case report. Med Pediatr Oncol. 1984;12(5):347–348. [DOI] [PubMed] [Google Scholar]
- 39.Donath MY, Storling J, Maedler K, Mandrup-Poulsen T. Inflammatory mediators and islet beta-cell failure: a link between type 1 and type 2 diabetes. J Mol Med. 2003;81(8):455–470. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Han S, Wang S, et al. Interactions of IL-12A and IL-12B polymorphisms on the risk of cervical cancer in Chinese women. Clin Cancer Res. 2009;15(1):400–405. [DOI] [PubMed] [Google Scholar]
- 41.Nair RP, Ruether A, Stuart PE, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J Invest Dermatol. 2008;128(7):1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallach J. Interpretation of Diagnostic Tests. 6th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 1998. [Google Scholar]
- 43.Tato CM, Cua DJ. SnapShot: cytokines IV. Cell. 2008;132(6):1062 e1–2. [DOI] [PubMed] [Google Scholar]
- 44.Tato CM, Cua DJ. SnapShot: cytokines III. Cell. 2008;132(5):900. [DOI] [PubMed] [Google Scholar]
- 45.Tato CM, Cua DJ. SnapShot: cytokines II. Cell. 2008;132(3):500. [DOI] [PubMed] [Google Scholar]
- 46.Tato CM, Cua DJ. SnapShot: cytokines I. Cell. 2008;132(2):324, e1. [DOI] [PubMed] [Google Scholar]
- 47.Protein Lounge - Cytokine network. 2014. http://www.proteinlounge.com/Pathway/Cytokine Network. Accessed February 2014.
- 48.Bhattacharya S, Andorf S, Gomes L, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2-3):234–239. [DOI] [PubMed] [Google Scholar]
- 49.Clinical Study Data Request. 2014. https://http://www.clinicalstudydatarequest.com