Abstract
Background
Salinity stress is a major constrain in the global rice production and hence serious efforts are being undertaken towards deciphering its remedial strategies. The comparative analysis of differential response of salt sensitive and salt tolerant lines is a judicious approach to obtain essential clues towards understanding the acquisition of salinity tolerance in rice plants. However, adaptation to salt stress is a fairly complex process and operates through different mechanisms. Among various mechanisms involved, the reactive oxygen species mediated salinity tolerance is believed to be critical as it evokes cascade of responses related to stress tolerance. In this background, the present paper for the first time evaluates the ROS generating and the scavenging system in tandem in both salt sensitive and salt tolerant cultivars of rice for getting better insight into salinity stress adaptation.
Results
Comparative analysis of ROS indicates the higher level of hydrogen peroxide (H2O2) and lower level of superoxide ions (O2-) in the salt tolerant as compared to salt sensitive cultivars. Specific activity of ROS generating enzyme, NADPH oxidase was also found to be more in the tolerant cultivars. Further, activities of various enzymes involved in enzymatic and non enzymatic antioxidant defence system were mostly higher in tolerant cultivars. The transcript level analysis of antioxidant enzymes were in alignment with the enzymatic activity. Other stress markers like proline were observed to be higher in tolerant varieties whereas, the level of malondialdehyde (MDA) equivalents and chlorophyll content were estimated to be more in sensitive.
Conclusion
The present study showed significant differences in the level of ROS production and antioxidant enzymes activities among sensitive and tolerant cultivars, suggesting their possible role in providing natural salt tolerance to selected cultivars of rice. Our study demonstrates that the cellular machinery for ROS production and scavenging system works in an interdependent manner to offer better salt stress adaptation in rice. The present work further highlights that the elevated level of H2O2 which is considered as a key determinant for conferring salt stress tolerance to rice might have originated through an alternative route of photocatalytic activity of chlorophyll.
Keywords: Rice, Salinity, Reactive oxygen species (ROS), NADPH oxidase, Hydrogen peroxide (H2O2), Antioxidant enzymes
Background
Rice is an important cereal crop that has the potential to provide food security to the increasing world population. Out of the total global agricultural land, 150 million hectares is estimated to be under rice cultivation that leads to an annual production averaging to 500 million metric tons [1]. But a large amount of the rice biomass is not harvested under field conditions due to sensitivity of the crop to various abiotic stresses like drought, salinity, low temperature, heat shock [2, 3]. Among these stresses, salinity is a major constrain to rice production worldwide which adversely affect its growth and development at the molecular, biochemical and physiological level [4, 5]. It affects more than 20 % of total cultivated land worldwide that results in US$12 billion loss in global agricultural production and this loss is further increasing each year [5, 6]. As in the present scenario of global climate change, the level of land salinization is expected to increase and hence there is an immediate world-wide concern for development of better salt tolerant cultivars for future food security. For achieving this objective, a thorough comparative analysis of salt sensitive and tolerant cultivars coupled with increased understanding of the underlying mechanism involved in salt stress adaptation is much warranted.
Reactive oxygen species (ROS) generation, signalling and detoxification are vital components of salt stress adaptation mechanisms and a priority area of research worldwide for the accomplishment of better growth and yield of a crop under salt affected areas [7]. ROS such as superoxide radical (O2-), hydroxyl radical (OH-) and hydrogen peroxide (H2O2) are produced in a low concentration as a result of normal cellular metabolism in plants and plays important role in growth, development and in adaptation to stress [8]. But, when a plant is subjected to stress, the perturbation of ROS homeostasis takes place which triggers multiple signalling responses involved in critical functions of plants [9]. Hence, a fine tuned balance between ROS production and scavenging is crucial for the plant survival under unfavourable conditions.
NADPH oxidases are one of the major enzyme systems involved in production of ROS in the apoplastic space under stress [10]. They catalyze the transfer of electons from NADPH to molecular oxygen (O2) for the generation of free radical O2- [11]. NADPH oxidases hold a distinct position among the various ROS generating enzyme systems in plants because of their role in different signalling pathways involved in plant growth, development and stress tolerance [12]. Superoxide ions produced as a result of enzymatic activity of NADPH oxidase is converted into H2O2 via superoxide dismutase (SOD) and it diffuses to adjacent cellular components where it acts as a signalling molecule that activates various stress responsive pathways. This indicates that the coordination of ROS generating and scavenging system might play a critical role in salt stress adaptation mechanism. However, the exact role of NADPH oxidase in conferring salt stress adaptation is not yet properly understood.
Earlier studies conducted on salt tolerant cultivar Pokali and sensitive cultivar IR64 under the effect of salt treatment has indicated a definitive link between scavenging system and salinity tolerance [13]. However, to best of our knowledge no work has been initiated to compare the ROS generating and scavenging system together in salt sensitive and tolerant cultivars. Therefore, in the present study, a comparative study of ROS dynamics was conducted in salt tolerant (Luna Sankhi and Luna Suvarna) and salt sensitive (IR64 and Pusa Basmati-1) cultivars of rice. The information gained on real time situation on this critical stress alleviating ROS pathway will facilitate in developing cultivars with better salt stress adaptation. Further, this work adds to the existing knowledge of our understanding on the complex physiological, biochemical and genetic mechanisms involved in salt stress adaptation.
Results
Histochemical analysis of ROS
The level of different types of ROS among salt sensitive and tolerant cultivars was monitored histochemically using three different dyes (Fig. 1). Nitrozolium blue (NBT) assay showed higher accumulation of dark blue coloured pigment that estimates the higher level of O2- in the rice cultivars IR64 and Pusa Basmati-1. On the contrary to this, 3, 3-diaminobenzidine (DAB) showed higher level of H2O2 that was assessed through the intensity of dark brown coloured spots in tolerant cultivars (Luna Sankhi and Luna Suvarna). Similarly, staining using 2', 7’-dichlorodihydrofluorescein diacetate (H2DCFDA) which binds to ROS (predominantly H2O2) yielded a higher intensity of green fluorescence in tolerant cultivars as compared to the sensitive cultivars.
Fig. 1.
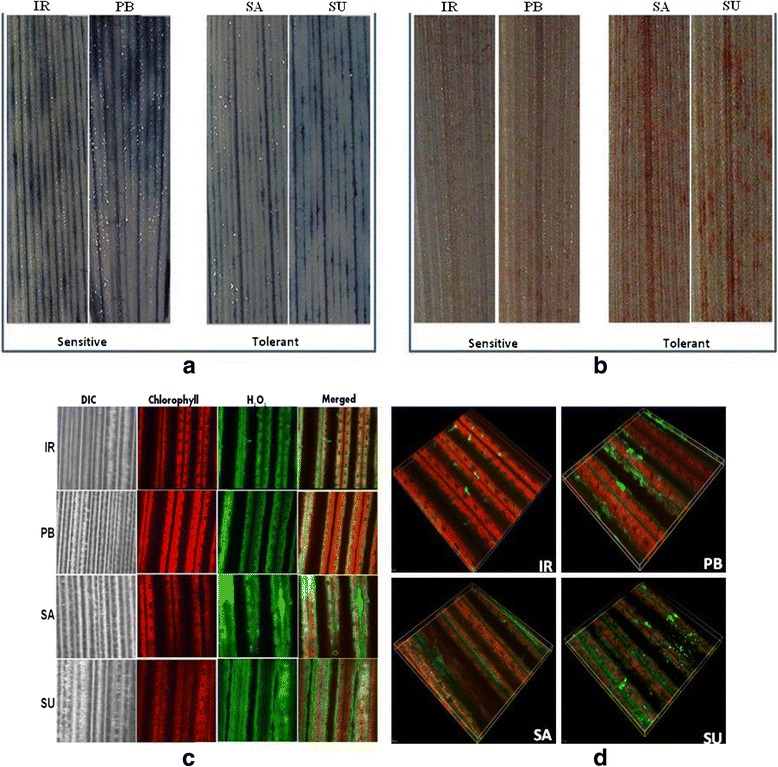
O2- and H2O2 accumulation in leaves of different cultivars of rice. Leaves were excised at the base of stems and were supplied with different stains. (a) NBT staining (b) DAB staining (c) Confocal images of H2DCFDA staining (d) 3D-view of confocal H2DCFDA staining images (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna)
NADPH oxidase activity assay
The quantitative analysis of ROS generating NADPH oxidase enzyme activity showed significant difference among four cultivars of rice (Fig. 2). The enzymatic activity (nmoles min-1 mg-1) was found to be significantly high in Luna Sankhi (0.424 ± 0.009) followed by Luna Suvarna 0.415 ± 0.010 than the sensitive cultivar IR64 (0.182 ± 0.011) and Pusa Basmati-1 (0.177 ± 0.028).
Fig. 2.
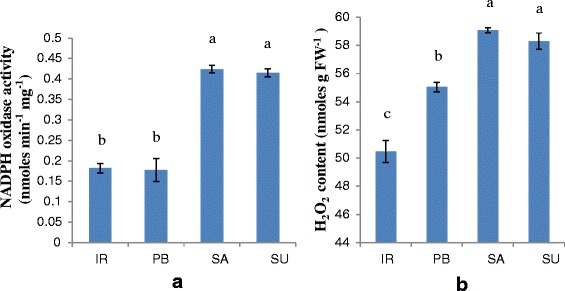
Comparative analysis of (a) NADPH oxidase activity and (b) H2O2 content of 14 days old seedlings of different rice cultivars. (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna). Bars represent mean ± SE (n = 3). Different letters (a, b, c) within cultivars are significantly different (Tukey LSD, p ≤ 0.05)
H2O2 content estimation
The estimation of aqueous H2O2 content in four varieties showed considerable difference as expected on the basis of histochemical assays (Fig. 2). The content of aqueous H2O2 (units nmoles g-1 FW) was found to be significantly more in tolerant cultivars Luna Sankhi (59.076 ± 0.187) and Luna Suvarna (58.296 ± 0.585) as compared to sensitive varieties IR64 (50.467 ± 0.76) and Pusa Basmati-1 (55.042 ± 0.340) which were at par with each other.
Antioxidant enzyme activity
Significant differences in the specific activity of various antioxidant enzymes were observed in different cultivars (Table 1). The overall specific activity of all the antioxidant enzymes, except catalase (CAT) was found to be higher in tolerant cultivars. SOD that converts superoxide ions into hydrogen peroxide showed significantly higher specific activity (units mg protein-1) in Luna Sankhi (0.0661 ± 0.0033) and Luna Suvarna (0.0866 ± 0.0008) with respect to IR64 (0.0243 ± 0.0023) and Pusa Basmati-1 (0.0387 ± 0.0021). The specific activity of ascorbate peroxidase (APX) was augmented to a significantly higher in Luna Sankhi (26.21 ± 0.301) and Luna Suvarna (26.13 ± 0.230) as compared to IR64 (19.85 ± 0.563) and Pusa Basmati-1 (18.55 ± 0.518) but on the contrary the specific activity of CAT was more in sensitive cultivars IR64 (0.486 ± 0.012) and Pusa Basmati-1 (0.581 ± 0.003) in comparison to Luna Sankhi (0.277 ± 0.190) and Luna Suvarna (0.397 ± 0.013) (Table 1). The enzymes involved in the well known glutathione-ascorbate pathway also showed considerable difference in their specific activity among different cultivars. The specific activity of glutathione reductase (GR) was found to be higher in Luna Sankhi (5.98 ± 0.276) and Luna Suvarna (5.91 ± 0.072) than IR64 (3.58 ± 0.045) and Pusa Basmati-1 (4.12 ± 0.112) (Table 1). Similarly, guaiacol peroxidase enzyme (GPX) also showed similar type of trend with higher activity in Luna Sankhi (68.41 ± 3.45) and Luna Suvarna (75.51 ± 1.68) followed by IR64 (51.16 ± 1.65) and Pusa Basmati-1 (51.80 ± 0.40). Dehydroascorbate reductase (DHAR) and monodehydroascorbate reductase (MDHAR) activities were also found to be high in tolerant cultivars (Luna Sankhi and Luna Suvarna) in comparison to the sensitive ones (IR64 and Pusa Basmati-1) (Table 1).
Table 1.
Comparative analysis of different antioxidant enzymes and ascorbate content in 14 days old seedlings of different rice cultivars
| Cultivar | SOD (units mg protein-1) | APX (μmol min-1 mg protein-1) | CAT (μmol min-1 mg protein-1) | GPX (μmol min-1 mg protein-1) | GR (μmol min-1 mg protein-1) | DHAR (μmol min-1 mg protein-1) | MDHAR (μmol min-1 mg protein-1) | Reduced ascorbate/Oxidised ascorbate ratio |
|---|---|---|---|---|---|---|---|---|
| IR64 | 0.0243 ± 0.0023d | 19.85 ± 0.563b | 0.486 ± 0.012a | 51.16 ± 1.65b | 3.58 ± 0.045b | 1.02 ± 0.016b | 1.76 ± 0.068b | 0.739 ± 0 .027b |
| PB | 0.0387 ± 0.0021c | 18.55 ± 0.518b | 0.581 ± 0.003a | 51.80 ± 0.40b | 4.12 ± 0.112b | 1.05 ± 0.014b | 1.60 ± 0.063b | 0.698 ± 0.011b |
| Luna Sankhi | 0.0661 ± 0.0033b | 26.21 ± 0.301a | 0.277 ± 0.190b | 68.41 ± 3.45a | 5.98 ± 0.276a | 1.43 ± 0.05a | 2.24 ± 0.060a | 0.884 ± 0.017a |
| Luna Suvarna | 0.0866 ± 0.0008a | 26.13 ± 0.230a | 0.397 ± 0.013b | 75.51 ± 1.68a | 5.91 ± 0.072a | 1.49 ± 0.05a | 2.28 ± 0.075a | 0.905 ± 0.033a |
(SOD Superoxide Dismutase, APX Ascorbate Peroxidase, CAT Catalase, GPX Guaicol Peroxidase, GR Glutathione Reductase, DHAR Dehydroascorbate Reductase, MDHAR Monodehydroascorbate Reductase). Values represent mean ± SE (n = 3)
Different letters (a, b, c, d) within cultivars are significantly different (Tukey LSD, p ≤ 0.05)
Expression analysis of antioxidant enzymes
The semi-RT PCR analysis of various antioxidant genes also showed higher level of key antioxidant enzymes in tolerant cultivars except CAT and Mn-SOD at the transcript level (Fig. 3). Among the different isoforms of SOD, the transcript level of Fe-SOD and Cu/Zn-SOD showed much higher accumulation in Luna Sankhi and Luna Suvarna. These results were further confirmed at the protein level with in–gel SOD assay (Fig. 4). On the contrary, Mn-SOD showed lesser expression in tolerant cultivars when compared against the sensitive cultivars. The transcript level of CAT was also found to be more in the sensitive cultivars (IR64 and Pusa Basmati-1). However, APX and GR enzymes were found in the same type of trend at the transcript level as followed in their respective enzymatic activities. Their gene expression was also higher in Luna Sankhi and Luna Suvarna as compared to the sensitive ones (IR64 and Pusa Basmati-1).
Fig. 3.
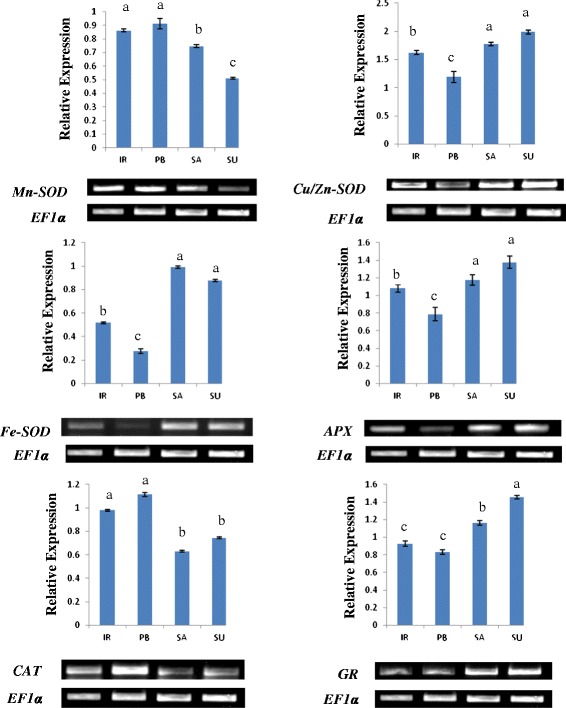
An ethidium bromide stained agarose gel harbouring products from reverse transcriptase-PCR of different key antioxidant genes of 14 days old seedlings of different rice cultivars. Gel shows the PCR products after 35 cycles of PCR and the results were first normalized to the housekeeping gene EF1α. Bars represent mean ± SE (n = 3). Different letters (a, b, c, d) within cultivars are significantly different (Tukey LSD, p ≤ 0.05). (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna)
Fig. 4.
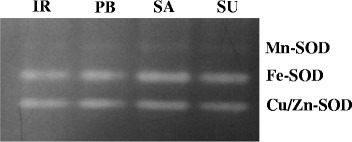
In-gel analysis of the pattern of SOD isoenzymes in two salt sensitive and two salt tolerant cultivars of rice (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna)
Ascorbate content
A notable difference in the reduced to oxidised ascorbate ratio was found in 4 cultivars of rice (Table 1). Tolerant cultivars Luna Suvarna and Luna Sankhi showed significantly higher ratio of (0.905 ± 0.033) and (0.884 ± 0.017), respectively in contrast to the sensitive cultivars IR64 (0.739 ± 0.027) and Pusa Basmati-1 (0.698 ± 0.011).
Total protein, free proline and MDA content analysis
In this study, we estimated the total protein content in the four cultivars. The protein content (mg FW -1) was found to be significantly high in tolerant cultivars Luna Sankhi (14.73 ± 0.275) and Luna Suvarna (15.22 ± 0.403) with respect to the sensitive cultivars IR64 (12.75 ± 0.275) and Pusa Basmati-1(12.85 ± 0.352) (Table 2). The significantly enhanced content of proline (μmoles g FW -1) was found in Luna Sankhi (6.90 ± 0.0513) and Luna Suvarna (6.78 ± 0.2450) against the sensitive cultivar IR64 (4.29 ± 0.0177) and Pusa Basmati-1 (4.58 ± 0.0199) (Table 2). Further, the level of lipid peroxidation (μmoles g FW -1) was determined through the estimation of MDA equivalents content in plants. The level of MDA was found to be more in sensitive cultivars IR64 (33.831 ± 0.058) and Pusa Basmati-1 (33.945 ± 0.047) than the tolerant cultivars Luna Sankhi (31.502 ± 0.206) and Luna Suvarna (29.414 ± 0.222) (Table 2).
Table 2.
Comparative analysis of total protein, proline and MDA of 14 days old seedlings of different rice cultivars
| Cultivar | Protein content (mg FW-1) | Proline content (μmoles g FW-1) | MDA content (μmoles g FW-1) |
|---|---|---|---|
| IR64 | 12.75 ± 0.275b | 4.29 ± 0.0177c | 33.831 ± 0.058a |
| PB | 12.85 ± 0.352b | 4.58 ± 0.0199b | 33.945 ± 0.047a |
| Luna Sankhi | 14.73 ± 0.275a | 6.90 ± 0.0513a | 31.502 ± 0.206b |
| Luna Suvarna | 15.22 ± 0.403a | 6.78 ± 0.2450a | 29.414 ± 0.222c |
Values represent mean ± SE (n = 3)
Different letters (a, b, c) within cultivars are significantly different (Tukey LSD, p ≤ 0.05)
Chlorophyll content analysis
A significant difference in the content of chlorophyll a, chlorophyll b and total chlorophyll was observed in sensitive and tolerant cultivars of rice (Fig. 5). The maximum content of chlorophyll a (μg/ml) was found in IR64 (30.598 ± 0.076) followed by Pusa Basmati-1(28.418 ± 0.995) that was significantly higher from the Luna Sankhi (26.334 ± 0.128) and Luna Suvarna (23.569 ± 0.195). However, the highest content of chlorophyll b (41.537 ± 0.868) and total chlorophyll (69.931 ± 0.772) was found in Pusa Basmati-1 whereas the minimum content of these pigments was found in Luna Suvarana.
Fig. 5.
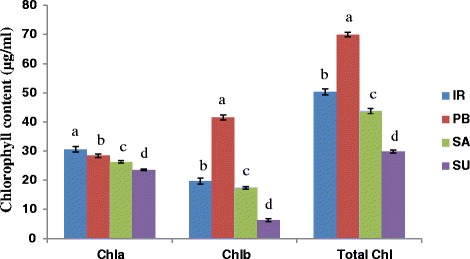
Comparative analysis of chlorophyll a, chlorophyll b and total chlorophyll in 14 days old seedlings of different rice cultivars. Bars represent mean ± SE (n = 3). Different letters (a, b, c, d) within cultivars are significantly different (Tukey LSD, p ≤ 0.05). (Chl: Chlorophyll, IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna)
Discussion
In the present study, it was observed that the tolerant cultivars (Luna Sankhi and Luna Suvarna) maintain a higher level of H2O2 and lower level of O2- as compared to the sensitive cultivars (IR64 and Pusa Basmati-1). H2O2 acts as a secondary messenger that activates the various stress adaptive signalling pathways at both genetic and physiological levels [14]. It is known to activate various genes involved in the acclimation and tolerance to salt stress [15]. Further, enhanced H2O2 production in halophytes is essential for their “salt stress preparedness” [16]. Our study indicates that salt tolerant cultivars Luna Sankhi and Luna Suvarna are capable of constitutive activation of plant defence pathways due to the higher basal level of H2O2 that in turn keeps the tolerant cultivars ready for adaptation to salt stress conditions. Hence, H2O2 ‘signatures’ may be operating in salt stress signalling pathways in rice that keeps the defence pathway networks activated in tolerant cultivars [17]. Furthermore, the elevated basal level of H2O2 found in the tolerant cultivars could be attributed to the higher activity of NADPH oxidase and SOD enzyme in Luna Sankhi and Luna Suvarna that was further confirmed through the estimation of specific enzymatic activity of SOD. These findings suggest that the higher generation of O2- in a NADPH dependent manner and its rapid conversion into signalling molecule H2O2 could be considered as a critical clue in understanding the model depicting better adaptation of tolerant cultivars towards salinity.
Once the signalling cascade has been initiated, plants are defended to the toxic effects of H2O2 with the help of APX, CAT and other peroxidases in a way similar to the Ca2+ efflux system that operates in well-known cytosolic calcium ‘signatures’ [6]. Similarly, in the present work, the specific activity of peroxidases APX and GPX was found to be high in tolerant varieties which could be scavenging the excess of H2O2 to prevent the plant from its ill effects, whereas the specific activity of CAT was found to be low. The less expression of CAT in tolerant cultivars could be due to the common substrate of APX and CAT and because of the higher affinity of APX for hydrogen peroxide than CAT [18]. Moreover, APX regulates the role of H2O2 as a signalling molecule unlike CAT which is more involved in detoxification [15].
Till now, the majority of research on the relation between ROS and salinity tolerance has been focussed on the enzymatic antioxidants although non-enzymatic antioxidants also play crucial role in scavenging excess of ROS and conferring tolerance [6]. Ascorbate is a known non enzymatic antioxidant that plays a central role in the ascorbate–glutathione cycle [19]. APX uses ascorbate as a reductant for the scavenging of H2O2 and converts it into dehydroascorbate. But for the maintenance of antioxidative capacity of ascorbate, its rapid regeneration into reduced form is necessary. APX functions in combination with GR, DHAR and MDHAR in glutathione-ascorbate pathway for the regeneration of ascorbate. In our studies, the specific activities of GR, DHAR and MDHAR enzymes were also found to be high in tolerant cultivars. Further the ratio of reduced to oxidised ascorbate was also observed to be high in Luna Sankhi and Luna Suvarna. This could be implied to the better operation of non-enzymatic glutathione-ascorbate pathway in tolerant cultivars which may be one of the many reasons for their superior salt tolerance ability [19]. Furthermore, the higher activity of various antioxidant enzymes (SOD, CAT, GPX, APX, and GR) in a coordinated manner in salt tolerant cultivars suggests that they are the major determinants in the model for depicting salt tolerance.
To further understand the dynamic role of antioxidant enzymes in conferring salt tolerance to halophytic varieties of rice, their transcript level expression study was conducted. Among the different isoforms of SOD, the expression of Fe-SOD and Cu/Zn-SOD was found to be high in tolerant cultivars whereas, the expression of Mn-SOD was more in sensitive cultivars. In an earlier study from our laboratory, we observed a higher expression of Fe-SOD and Cu/Zn-SOD in response to salt treatment in Pusa Basmati-1 [20]. Therefore, the higher basal expression level of Fe-SOD and Cu/Zn-SOD in tolerant cultivars could be one of the many reasons for their better adaptation in salt stress.
Furthermore, to get a better insight into the mechanism conferring elevated adaptation to salinity stress in Luna Sankhi and Luna Suvarna, some of the biochemical stress markers were studied. In the present study, enhanced level of total protein in Luna Sankhi and Luna Suvarna was observed which could be attributed to the synthesis of new or elevated level of proteins linked to stress tolerance in these cultivars. This result is further strengthened by the report that enhanced synthesis level of proteins is one of the major determinants of stress adaptation in halophytes [21]. Proline is a known osmoprotectant that also plays multiple antioxidant roles. Hyperaccumulation of free proline in plants is known in conferring salinity tolerance [22, 23]. The higher accumulation of free proline in Luna Sankhi and Luna Suvarna may be playing an important role in quenching of excess of ROS and stabilization of ROS-scavenging enzymes along with its well known role in osmoticum maintainence [24]. Further, with regard to reduced MDA content in tolerant cultivars, our results are in agreement to the previous studies where it has been shown that higher antioxidant enzyme activities are responsible for low MDA content which in turn inhibits the membrane damage by ROS and hence confers tolerance [25, 26]. It was interesting to note that the content of chlorophyll a, chlorophyll b and total chlorophyll was less in the tolerant cultivars in comparison to sensitive cultivars. Under salt stress conditions, plants are often observed to have reduced chlorophyll content [20, 27]. However, analysis of chlorophyll content in salt tolerant and sensitive cultivars will provide the physiological explanation to adaptation of plant to salinity stress and hence it is a subject of worth investigation. In our investigation the less chlorophyll contents in tolerant cultivars may be either due to the ROS mediated degradation of cholorophyll or it could be photocatalytic activity of cholorophyll itself for the production of H2O2 [28, 29]. In our case, we presume that in tolerant cultivars the salt tolerance could be attributed to the enhanced production of H2O2, which may be due to the photocatalytic activity of chlorophyll [30]. Moreover, the role of light in chlorophyll degradation has also been suggested [31].
Conclusion
The tolerant cultivars of rice maintain a high threshold level of H2O2 due to more enzymatic activity of NADPH oxidase and enough SOD “in stock” that may constitutively activates the defence pathways resulting in higher intrinsic activity of various antioxidant enzymes. This provides certain adaptive advantage to halophytes over glycophytes because of which salt stress does not lead to oxidative stress in tolerant cultivars. Further, the first time comparative analysis of ROS generating system NADPH oxidase in halophytes and glycophytes provide a critical clue on understanding the model depicting salt tolerance in rice. We propose that these two systems work together in synchronization with each other for achieving salt tolerance. It will be interesting to further explore the crosstalk that exists between ROS generating and scavenging system for achieving salinity tolerance and whether this crosstalk operates in feedforward or feedback manner. Furthermore, the observation of degradation of chlorophyll for producing higher level of H2O2 and in turn offering tolerance points out towards an uncharted mechanism in salt stress adaptation biology.
Methods
Plant material
Four rice cultivars differing in salt stress tolerance level were selected for this work. Seeds of two salt sensitive cultivars viz. IR64 and Pusa Basmati-1 were procured from IARI (Indian Agricultural Research Institute, New Delhi) and the seeds of two salt tolerant cultivars Luna Sankhi and Luna Suvarna were collected from CRRI (Central Rice Research Institute Cuttack, Odisha, India). Luna Sankhi (CR Dhan 405) and Luna Suvarna (CR Dhan 403) are salt tolerant varieties of Indian origin that can tolerate salinity stress in the range of 6-8 dS m-1 [31].
Surface sterilization and inoculation
Surface sterilization of the seeds was carried out according to Sharma’s method [26]. Healthy and mature seeds were selected and were sterilized using sodium hypochlorite and mercuric chloride. Sterilized seeds were then inoculated in autoclaved sand moistened with autoclaved double distilled water in plastic boxes and were kept at 25 ± 2 °C temperature with 14 h photoperiod under light intensity of 2000 lux for 14 days.
Histochemical ROS detection
The histochemical level of different types of ROS was detected using three different dyes. For the detection of O2-, the second leaf of 14 d seedlings was cut into small pieces and these pieces were vacuum infiltered in 6 mM NBT prepared in 10 mM of sodium citrate buffer (pH = 6) for 10 min. They were incubated for 2 h at room temperature and the chlorophyll content of these pieces was then removed by boiling them in absolute ethanol till the complete removal of chlorophyll. After the removal of chlorophyll, pieces were observed under the stereo-microscope [32].
For the detection of H2O2, two different stains were used, DAB and H2DCFDA. For the DAB assay, small pieces of second leaf of 14 d seedlings were vacuum infiltered in DAB (1 mg/ml in water) for 10 min and were then incubated for 2 h under the dark conditions. Chlorophyll content was removed and they were observed under the stereo-microscope [32]. For H2DCFDA assay, pieces of second leaf were incubated in 10 μM H2DCFDA and were vacuum infiltered for 5 min. The leaves were then washed with double distilled water and were observed under the confocal microscope using laser beam of excitation 488 nm [33, 34].
NADPH oxidase assay
Isolation of plasma membrane proteins
Plasma membrane proteins were extracted from the 0.5 g of fresh tissue [13]. The plantlets were homogenized in protein extraction buffer containing 0.25 M sucrose, 50 mM HEPES (pH 7.2), 3 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 3.6 mM L-cysteine, 0.1 mM MgCl2, 0.6 % PVP and PIC (Protease Inhibitory Cocktail). The homogenate was filtered through two layers of muslin cloth and was centrifuged at 10,000 g for 45 min at 4 °C. Supernatant was ultracentrifuged at 1, 80, 000 g for 60 min at 40 C. The pellet was resuspended in 10 mM chilled Tris-HCl. The protein content was determined according to method described by Bradford [35] using Bovine Serum Albumin (BSA) as a standard.
Spectrophotometric assay of NADPH oxidase activity
The O2- generating activity of NADPH oxidase was measured by following the Kaundal’s method [13]. The assay mixture was prepared with 50 mM Tris-HCl buffer (pH 7.5), 1 mM XTT (sodium, 3-[1-[phenylamino-carbonyl]-3, 4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate), 1 mM NADPH, and 20 μg of membrane protein. The rate of reduction of XTT by O2- was determined spectrophotometrically at 492 nm using an extinction coefficient of 2.16 x 104 M-1 cm-1.
Estimation of aqueous H2O2 content
The content of aqueous H2O2 was estimated using the modified ferrous oxidation-xylenol orange (FOX) assay [13]. 0.5 g of fresh seedlings of 14 d plants was homogenised in activated charcoal (0.1 g) prepared in 5 ml of 5 % trichloroacetic acid. Homogenate was filtered through Whatmann filter No.1 and was centrifuged at 8000 rpm for 10 min. The supernatant was used for the estimation of aqueous H2O2. FOX reagent was prepared using 1 ml of reagent ‘a’ (25 mM ammonium ferrous sulfate prepared in 2.5 M sulfuric acid), 50 μl of reagent ‘b’ (0.25 M xylenol orange prepared in HPLC-grade methanol), and 90 ml of reagent ‘c’ (9.69 mg of butylated hydroxytoluene prepared in 90 ml of HPLC- grade methanol). The total volume was raised to 100 ml with distilled water. In a test tube, 0.2 ml of plant extract was taken and 1 ml FOX reagent was added to it. The reaction mix was mixed properly and was incubated at room temperature for 15 min. All solutions were prepared fresh and used within 2 h. The content of aqueous H2O2 was estimated by recording the absorbance at 560 nm. Concentration was calculated using a standard curve prepared by taking varying concentrations of H2O2.
Extraction of protein
For the estimation of antioxidant enzyme activities, 1 g of fresh 14 d seedlings were homogenised in 3 ml of chilled buffer containing 50 mM phosphate buffer (pH = 7.8), 2 mM EDTA, 1 mM DTT, 1 mM PMSF (Phenylmethylsulfonyl Fluoride), 0.5 %(v/v) Triton X-100 and 10 % (w/v) PVP-40 (Polyvinylpyrrolidone). The homogenate was centrifuged for 20 min at 12,000 rpm and the supernatant was collected. This supernatant was then used for various enzymatic assays [26]. Protein concentration was determined using Bradford assay with BSA as a standard [35].
Estimation of antioxidant enzyme activities
The enzymatic activities of different antioxidant enzymes were determined spectrophotometrically. The EC (Enzyme Commission) number mentioned against each enzyme is a unique number in the numerical classification scheme for enzymes and represent the chemical reactions they catalyze.
SOD assay (EC 1.15.1.1)
The enzymatic activity of SOD was determined using the method given by Beauchamp and Fridovich [36]. 1 ml of reaction mixture was prepared in 50 mM potassium phosphate buffer using 2 μM riboflavin, 75 μM Nitrotetrazolium blue (NBT), 100 μM EDTA, 13 mM DL-methionine and 50 μl of enzyme extract and the absorbance was taken at 560 nm. The reaction mix was illuminated for 20 min at 25 °C for initiation. 1 unit of enzyme activity is expressed as the amount of enzymes required for 50 % inhibition of NBT reduction at 25 °C.
-
b)
CAT assay (EC 1.11.1.6)
CAT specific activity was estimated at 25 °C using the method given by Aebi [37]. Decrease in absorbance of H2O2 was measured in 1 ml of reaction mixture having 10 mM H2O2 and 20 μl of enzyme extract in 50 mM of potassium phosphate buffer (pH = 7). Specific enzyme activity was expressed as l mole of H2O2 decomposed mg protein-1 min-1.
-
c)
APX assay (EC 1.11.1.11)
The specific activity of APX was assayed as the rate of oxidation of ascorbate in the presence of H2O2 [26]. The reaction mixture of 1 ml comprised of 0.5 mM ascorbate, 0.1 mM H2O2 and 0.1 mM EDTA, potassium phosphate buffer(pH = 7) and 10 μl of enzyme extract. Decrease in absorption was observed spectrophotometerically at 290 nm at 25 °C. One unit of enzyme activity was expressed as the amount of enzyme required to oxidise 1 μM of ascorbate/minute/g tissue.
-
d)
GR assay (EC 1.6.4.2)
The specific activity of GR is determined by analysing the decrease in absorbance at 340 nm [26]. 1 ml reaction mixture used for the assay had 50 mM potassium phosphate buffer (pH = 7.8), 1 mM EDTA, 1 mM oxidized glutathione (GSSG) and 25 μl enzyme extract. The reaction was started by adding 0.1 mM NADPH at last. Enzyme activity was expressed as μmol of NADPH oxidised min-1 mg protein-1.
-
e)
GPX assay (EC 1.11.1.7)
The activity of GPX is observed by measuring the increase in absorbance at 436 nm [26]. The reaction mixture was prepared in 50 mM potassium phosphate buffer (pH = 7) having 9 mM guaiacol, 10 mM H2O2 and 33 μl of enzyme extract. The enzymatic activity of GPX is expressed as the amount of enzyme required to produce 1 μmol guaiacol dehydrogenation product min-1 mg protein-1.
-
f)
DHAR assay (EC 1.8.5.1)
The specific activity of DHAR was estimated by measuring the increase in absorbance at 25 °C due to the formation of ascorbate from dehydroascorbate at 265 nm [26]. The reaction mixture of 1 ml consisted of 50 mM of potassium phosphate buffer (pH = 7), 0.1 mM EDTA, 0.5 mM dehydroascorbate, 2.5 mM GSH and 25 μl of enzyme extract. One unit of enzyme activity was expressed as amount of enzyme required to produce 1 μmol of ascorbate min-1 mg protein-1.
-
g)
MDHAR assay (EC 1.6.5.4)
The specific activity of MDHAR was determined at 25 °C by measuring the increase in absorbance at 340 nm [26]. The 1 ml reaction mixture used for this estimation consisted of 50 mM Tris-HCl buffer (pH = 7.6), 0.15 units of ascorbate oxidase enzyme, 2.5 mM ascorbic acid and 0.2 mM NADPH/NADH. One unit of enzyme activity was given as the amount of enzyme required to oxidase 1 μmol of NADPH min-1 mg protein-1.
Expression analysis of antioxidant enzymes
For the gene expression studies of various antioxidant genes, semi quantitative primers were designed using the IDT software (www.idtdna.com) (Table 3). Total RNA from the 14 d plants was extracted using the trizol method as per the manufacturer’s instructions. A total of 3 μg of RNA was used for the cDNA preparation with commercial cDNA synthesis kit. Gene expression was studied in 50 μl of polymerase chain reaction (PCR) using 50 ng of cDNA template. The PCR parameters used were: predenaturation at 94 °C for 4 min, followed by 35 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min, with a final extension step of 72 °C for 7 min. Rice elongation factor ef1α was used as internal control. All the PCR’s were performed at least with three independent samples and intensity of the products was confirmed with 1 % agarose gel with ethidium bromide. The relative level of transcript in each PCR reaction was determined using the integerated density value (IDV), with Alpha 2000TM Image Analyzer software.
Table 3.
List of primers for RT-PCR
| S. No. | Name of the gene | Database | Accession number | Primer sequence |
|---|---|---|---|---|
| 1 | EF1α | GenBank | D63580.1 | F: 50-GTACAAGATCGGTGGTATT-30 |
| R: 50-GGGTACTCAGAGAAGGTCT-30 | ||||
| 2 | Cu/Zn-SOD | GenBank | L19435.1 | F: 50-CCTCAAGCCTGGTCTCCAT-30 |
| R:50-CAGCCTTGAAGTCCGATGAT-30 | ||||
| 3 | Fe-SOD | GenBank | AY770495.1 | F: 50-CTTGATGCCCTGGAACCTTA-30 |
| R: 50-GCCAGACCCCAAAAGTGATA-30 | ||||
| 4 | Mn-SOD | GenBank | L19436.1 | F: 50-GCCATTGATGAGGATTTTGG-30 |
| R: 50-CAAGCAGTCGCATTTTCGTA-30 | ||||
| 5 | CAT | GenBank | D26484.1 | F: 50-GTTCGGTTCTCCACAGTCGT-30 |
| R: 50-CCCTCCATGTGCCTGTAGTT-30 | ||||
| 6 | APX | GenBank | D45423.1 | F: 50-CCAAGGGTTCTGACCACCTA-30 |
| R: 50-CAGTTCGGAGAGCTTGAGGT-30 | ||||
| 7 | GR | GenBank | AB009592.1 | F: 50-AACAGCCGATGGCATAAAAG-30 |
| R:50-CAACCACCAGTTTCATGACG-30 |
In-gel SOD assay
For in-gel SOD assay, 100 mg of fresh seedling tissue of each cultivar was homogenized with 50 mM Tris HCl buffer (pH = 7.5). The homogenate was centrifuged at 12000 rpm for 15 min at 4 °C [38]. The protein content was quantified in the supernatant using Bradford assay with BSA as a standard. Native-PAGE analysis was carried out with 50 μg protein of each cultivar at 80 V stacking and 100 V resolving. The gel was then stained according to Rucinska’s method using 2.45 mM NBT prepared in 50 mM potassium phosphate buffer for 20 min in dark [39]. The gel was again washed with autoclaved water followed by immersion in potassium phosphate (50 mM, pH 7.8) containing TEMED (28 mM) and riboflavin (3 μM) for 15 min. The gel was then kept on dry white illumination tray till the gel became uniformly blue except at positions containing SOD and maximum contrast between the achromatic zones and general blue colour was achieved.
Ascorbate content
The content of ascorbic acid was estimated using the Dutilleul’s method with minor modifications [40]. 1 g of fresh leaf tissues of four cultivars was homogenized with 3 % metaphosphoric acid (3 %) containing EDTA (1 mM). The homogenate was centrifuged at 8000 × g for 15 min. 0.1 ml of supernatant was taken and was mixed with 1 ml citrate phosphate buffer (0.1 M, pH 2.1). The absorbance was taken at 265 nm and 1 unit of APX was added. The solution was incubated for 5 min at room temperature and absorbance was again noted at 265 nm. The difference in two readings of absorbance was calculated that indicated the reduced ascorbate content utilized specifically by APX. For the estimation of total ascorbate content, 0.1 ml of DTT (0.1 M) was added in another set and absorbance was read at 265 nm after 5 min. 1 unit of APX was added and absorbance was taken after 5 min. For the determination of oxidized ascorbate content, reduced ascorbate content was subtracted from the total ascorbate content. Calibration curve was prepared using graded concentration of L-ascorbic acid in 3 % HPO3. The ratio of reduced ascorbate and oxidized ascorbate was also determined.
Proline content
Proline content was determined using the method given by Bates [41]. 0.5 g of total seedlings were crushed in liquid nitrogen and homogenised in 3 % aqueous sulfosalicyclic acid. The samples were then centrifuged at 12000 rpm for 15 min and 2 ml of the supernatant was taken in a test tube. Equal amount of acid ninhydin and glacial acetic acid was added to the supernatant and the reaction mix was kept in boiling water for 1 h. The reaction was stopped by keeping the tubes on ice. Added 4 ml of toluene to the reaction mixture and toluene layer separated from the aqueous phase was collected. The proline content was determined by measuring the absorbance of the toluene at 520 nm. The amount of proline was estimated using the proline standard curve and was expressed as μmoles gFW-1.
Lipid peroxidation
The level of lipid peroxidation was determined using the TBARS assay [42]. 1 g of tissue sample was homogenised in 3 ml of 0.1 % of chilled trichloroacetic acid (TCA) and 3 ml of solution containing 0.5 % TBA (Thiobarbituric acid) in 20 % TCA was added to it. The reaction mixture was incubated at 95 °C for 30 min and the reaction was stopped by keeping the tube on ice. Centrifuged the mixture at 10,000 rpm for 15 min. The supernatant was collected and its absorbance was measured at 532 nm, with a reading at 600 nm subtracted from it to account for nonspecific turbidity. 155 mM-1cm-1 extinction coefficient was used for the quantification of MDA-TBA complex and was expressed as μmoles gFW-1.
Chlorophyll content
Chlorophyll content was estimated using the method given by Arnon with some modifications [43]. 100 mg of fresh leaves of each cultivar were taken and were homogenised in liquid nitrogen. To the homogenised tissue sample, 1.5 ml of 80 % acetone was added and the reaction was incubated in dark for 1 h. Samples were centrifuged at 15,000 rpm for 3 min. The supernatant was collected and the absorbance was measured spectrophotometrically at 645 and 663 nm against 80 % acetone as blank. The chlorophyll content was determined as follows:
Statistical analysis
Data from different experiments was analysed statistically with one-way analysis of variance (ANOVA). The value of different parameters is expressed as mean ± SE of three independent replicates. The Tukey LSD test was applied for multiple comparisons using Sigma stat version 3.5, and significance of difference between the cultivars were set as p ≤ 0.05.
Abbreviations
APX, ascorbate peroxidase; CAT, catalase; DAB, 3, 3-diaminobenzidine; DHAR, dehydroascorbate reductase; GPX, guaicol peroxidase; GR, glutathione reductase; H2DCFDA, 2',7'-dichlorodihydrofluorescein diacetate; H2O2, hydrogen peroxide; MDA, malondialdehyde; MDHAR, monodehydroascorbate reductase; NADPH, nicotinamide adenine dinucleotide phosphate hydrogen; NBT, nitrozolium blue; O2-, superoxide ions; PCR, polymerase chain reaction; PIC, protease inhibitory cocktail; PMSF, phenylmethylsulfonyl fluoride; ROS, reactive oxygen species; semi-RT, semi quantitative reverse transcriptase; SOD, superoxide dismutase; XTT, sodium3-[1-[phenylamino-carbonyl]-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate
Acknowledgements
Navdeep Kaur is thankful to the Department of Biotechnology (Government. of India) for Senior Research Fellowship. We thank the Department of Genetics IARI, New Delhi, India for providing the seeds of IR64 and Pusa Basmati-1 and Director, CRRI, Odisha, India for providing the seeds of Luna Sankhi and Luna Suvarna. Authors acknowledge the Department of Biotechnology, Government of India for financial support (Project No. BT/PR13965/BRB/10/883/2010).
Availability of data and materials
All the data supporting the findings is contained within the manuscript.
Authors’ contributions
NK, PKP and IS conceived and designed the experiments. NK and MD performed the experiments. PKP and NK analysed and interpreted the data. NK carried out statistical analysis. PKP, NK and IS wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
References
- 1.Onyango AO. Exploring options for improving rice production to reduce hunger and poverty in Kenya. World Environ. 2014;4:172–179. [Google Scholar]
- 2.Thakur P, Kumar S, Malik JA, Berger JD, Nayyar H. Cold stress effects on reproductive development in grain crops: an overview. Environ Exp Bot. 2010;67:429–443. doi: 10.1016/j.envexpbot.2009.09.004. [DOI] [Google Scholar]
- 3.Mantri N, Patade V, Penna S, Ford R, Pang E. Abiotic stress responses in plants: present and future. In: Ahmad P, Prasad MNV, editors. Abiotic stress responses in plants: metabolism, productivity and sustainability. New York: Springer; 2012. pp. 1–19. [Google Scholar]
- 4.Todaka D, Nakashima K, Shinozaki K, Yamaguchi SK. Towards understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice. 2012;5:6. doi: 10.1186/1939-8433-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta B and Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular Characterization. Int J Genomics. 2014. doi:org/10.1155/2014/701596. [DOI] [PMC free article] [PubMed]
- 6.Bose J, Rodrigo MA and Shabala S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J Exp Bot. 2013. doi:10.1093/jxb/ert430. [DOI] [PubMed]
- 7.Choudhury S, Panda P, Sahoo L, Panda SK. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal Behav. 2013. doi:10.4161/psb.23681. [DOI] [PMC free article] [PubMed]
- 8.Jajic I, Sarna T, Strzalka K. Senescence, stress, and reactive oxygen species. Plants. 2015;4:393–411. doi: 10.3390/plants4030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baxter A, Mittler R, Suzuki N. ROS as key players in plant stress signalling. J Exp Bot. 2013. doi:10.1093/jxb/ert375. [DOI] [PubMed]
- 10.Srivastava AK, Srivastava S, Lokhande HV, Souza SFD, Suprasanna P. Salt stress reveals differential antioxidant and energetic responses in glycophyte (Brassica juncea L.) and halophyte (Sesuvium portulacastrum L.). Front Environ Sci. 2015. doi:10.3389/fenvs.2015.00019.
- 11.Kaundal A, Rojas CM, Mysore KS. Measurement of NADPH oxidase activity in plants. Bio-Protocol. 2012. http://www.bio-protocol.org/e278.
- 12.Kaur G, Sharma A, Guruprasad K, Pati PK. Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnol Adv. 2014;32:551–563. doi: 10.1016/j.biotechadv.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Shabrawi HE, Kumar B, Kaul T, Reddy MK, Singla P, Sopory SK. Redox homeostasis, antioxidant defense, and methylglyoxal detoxification as markers for salt tolerance in Pokkali rice. Protoplasma. 2010;245:85–96. doi: 10.1007/s00709-010-0144-6. [DOI] [PubMed] [Google Scholar]
- 14.Salwa AO, Mona GD, Salman SR. Comparative study between the physiological role of hydrogen peroxide and salicylic acid in alleviating the harmful effect of low temperature on tomato plants grown under sand-ponic culture. Sci Agric. 2015;9:49–59. [Google Scholar]
- 15.Sofo A, Scopa A, Nuzzaci M, Vitti A. Ascorbate peroxidase and CAT activities and their genetic regulation in plants subjected to drought and salinity stresses. Int J Mol Sci. 2015;16:13561–13578. doi: 10.3390/ijms160613561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shabala S, Wua H, Bose J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015;241:109–119. doi: 10.1016/j.plantsci.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- 18.Cuypers A, Smeets K, Ruytinx J, Opdenakker K, Keunen E, Remans T, et al. The language of calcium signaling. Annu Rev Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- 19.Ozgur R, Uzilday B, Sekmen AH, Turkan I. Reactive oxygen species regulation and antioxidant defence in halophytes. Funct Plant Biol. 2013;40:832–847. doi: 10.1071/FP12389. [DOI] [PubMed] [Google Scholar]
- 20.Sharma I, Bhardwaj R, Pati PK. Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J Plant Growth Regul. 2015. doi:10.1007/s00344-015.
- 21.Kosová K, Prášil IT, Vítámvás P. Protein contribution to plant salinity response and tolerance acquisition. Int J Mol Sci. 2013;14:6757–6789. doi: 10.3390/ijms14046757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radyukina NL, Kartashov AV, Ivanov YV, Shevyakova NI, Kuznetsov VV. Functioning of defense systems in halophytes and glycophytes under progressing salinity. Russ J Plant Physiol. 2007;54:806–815. doi: 10.1134/S1021443707060131. [DOI] [Google Scholar]
- 23.Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Signorelli S, Coitiño EL, Borsani O, Monza J. Molecular mechanisms for the reaction between •OH radicals and proline: insights on the role as reactive oxygen species scavenger in plant stress. J Phys Chem. 2013;118:37–47. doi: 10.1021/jp407773u. [DOI] [PubMed] [Google Scholar]
- 25.André DAN1, José TP, Joaquim EF, Carlos EBdA, Enéas GF. Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ Exp Bot. 2006;56:87–94. doi: 10.1016/j.envexpbot.2005.01.008. [DOI] [Google Scholar]
- 26.Sharma I, Ching E, Saini S, Bhardwaj R, Pati PK. Exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety Pusa Basmati-1. Plant Physiol Biochem. 2013;69:17–26. doi: 10.1016/j.plaphy.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Hu L, Zhang L, Pan X, Hu X. Exogenous spermidine is enhancing tomato tolerance to salinity–alkalinity stress by regulating chloroplast antioxidant system and chlorophyll metabolism. BMC Plant Biol. 2015. doi:10.1186/s12870-015-0699-7. [DOI] [PMC free article] [PubMed]
- 28.Chen XB, Zhao XH, Zhu Y, Gong YD, Li LB, Zhang JP, et al. Hydrogen peroxide-induced chlorophyll a bleaching in the cytochrome b6f complex: a simple and effective assay for stability of the complex in detergent solutions. Photosynth Res. 2006;90:205–14. doi: 10.1007/s11120-006-9118-0. [DOI] [PubMed] [Google Scholar]
- 29.Lobanov AV, Rubtsova NA, Vedeneeva YA, Komissarov GG. Photocatalytic activity of chlorophyll in hydrogen peroxide generation in water. Dokl Chem. 2008;421:190–193. doi: 10.1134/S0012500808080065. [DOI] [Google Scholar]
- 30.Merzlyak MN, Gitelson AA, Pogosyan SI, Likhimena L, Chivkunova OB. Light- induced pigment degradation in leaves and ripening fruits studied in sity with reflectance spectroscopy. Physiol Plant. 1998;104:661–667.
- 31.Gregorio GB, Islam MR, Vergara GV, Thirumeni S. Recent advances in rice science to design salinity and other abiotic stress tolerant rice varieties. SABRAO J Breed Genet. 2013;45:31–41. [Google Scholar]
- 32.Wu GL, Cui J, Tao L, Yang H. Fluroxypyr triggers oxidative damage by producing superoxide and hydrogen peroxide in rice (Oryza sativa) Ecotoxicology. 2010;19:124–132. doi: 10.1007/s10646-009-0396-0. [DOI] [PubMed] [Google Scholar]
- 33.Kim A, Poul EJ, Ian MM, Alexander S. Monitoring reactive oxygen species formation and localisation in living cells by use of the fluorescent probe CM-H2DCFDA and confocal laser microscopy. Physiol Plant. 2009;136:369–383. doi: 10.1111/j.1399-3054.2009.01243.x. [DOI] [PubMed] [Google Scholar]
- 34.Sharma A, Vats SK, Pati PK. Post-infectional dynamics of leaf spot disease in Withania somnifera. Ann Appl Biol. 2013;165:429–440. doi: 10.1111/aab.12148. [DOI] [Google Scholar]
- 35.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Beauchamp CU, Fridovich I. Improved assays for superoxide dismutase and an assay applicable to polyacrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 37.Aebi H. CAT in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 38.Kuo WY, Huang CH, Liu AC, Cheng CP, Li SH, Chang WC, et al. Chaperonin 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol. 2013;197:99–110. doi: 10.1111/j.1469-8137.2012.04369.x. [DOI] [PubMed] [Google Scholar]
- 39.Rucinska R, Waplak S, Gwozdz E. Free radical formation and activity of antioxidant enzymes in lupin roots exposed to lead. Plant Physiol Biochem. 1999;37:187–194. doi: 10.1016/S0981-9428(99)80033-3. [DOI] [Google Scholar]
- 40.Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, et al. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;15:1212–1226. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bates LS, Waldeen RP, Teare ID. Rapid determination of free proline for water stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 42.Hodges MD, DeLong JM, Forney CF, Prange RK. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta. 1999;207:604–611. doi: 10.1007/s004250050524. [DOI] [PubMed] [Google Scholar]
- 43.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data supporting the findings is contained within the manuscript.


