Using an integrated surveillance platform, we incorporated genetic, antigenic, and epidemiologic indicators to evaluate agent–host factors that contributed to low vaccine effectiveness during the 2014–2015 influenza season, including variation in the viral genome and negative effects of serial vaccination.
Keywords: influenza vaccines, vaccine effectiveness, genomics, antigenic drift, sentinel surveillance
Abstract
Background. The 2014–2015 influenza season was distinguished by an epidemic of antigenically-drifted A(H3N2) viruses and vaccine components identical to 2013–2014. We report 2014–2015 vaccine effectiveness (VE) from Canada and explore contributing agent–host factors.
Methods. VE against laboratory-confirmed influenza was derived using a test-negative design among outpatients with influenza-like illness. Sequencing identified amino acid mutations at key antigenic sites of the viral hemagglutinin protein.
Results. Overall, 815/1930 (42%) patients tested influenza-positive: 590 (72%) influenza A and 226 (28%) influenza B. Most influenza A viruses with known subtype were A(H3N2) (570/577; 99%); 409/460 (89%) sequenced viruses belonged to genetic clade 3C.2a and 39/460 (8%) to clade 3C.3b. Dominant clade 3C.2a viruses bore the pivotal mutations F159Y (a cluster-transition position) and K160T (a predicted gain of glycosylation) compared to the mismatched clade 3C.1 vaccine. VE against A(H3N2) was −17% (95% confidence interval [CI], −50% to 9%) overall with clade-specific VE of −13% (95% CI, −51% to 15%) for clade 3C.2a but 52% (95% CI, −17% to 80%) for clade 3C.3b. VE against A(H3N2) was 53% (95% CI, 10% to 75%) for patients vaccinated in 2014-2015 only, significantly lower at −32% (95% CI, −75% to 0%) if also vaccinated in 2013–2014 and −54% (95% CI, −108% to −14%) if vaccinated each year since 2012–2013. VE against clade-mismatched B(Yamagata) viruses was 42% (95% CI, 10% to 62%) with less-pronounced reduction from prior vaccination compared to A(H3N2).
Conclusions. Variation in the viral genome and negative effects of serial vaccination likely contributed to poor influenza vaccine performance in 2014–2015.
The 2014–2015 influenza season in the northern hemisphere had several distinguishing features including an early and intense epidemic due to the A(H3N2) subtype; dominant circulating strains that were mismatched to vaccine; and vaccine that was unchanged from 2013–2014 [1–4]. Compared with other influenza types/subtypes, predominant A(H3N2) epidemics are typically associated with a greater population burden of serious outcomes, notably involving the elderly [5, 6]. Surveillance indicators from Canada and elsewhere show that the 2014–2015 season was among the worst in recent years with respect to serious outcome statistics [1, 2, 7, 8].
The test-negative design (TND) is an efficient form of case-control study first piloted for influenza vaccine effectiveness (VE) monitoring in 2004–2005 by Canada's Sentinel Practitioner Surveillance Network (SPSN) [9]. Since then, the TND has been used for influenza VE estimation annually in Canada [10–12] and has been adopted for this purpose by multiple countries globally [9], including for mid-season assessment. In 2014–2015, several countries used the TND to assess VE mid-season, reporting negligible protection against the A(H3N2) epidemic strain [13–17].
Over the past decade, the Canadian SPSN has also linked patient clinical data to detailed genomic and antigenic characterization of viruses collected from the same TND study participants to gain insight into the impact of virologic changes on VE [10, 11, 13]. Viral evasion of vaccine-induced antibody protection largely occurs through evolution in the surface hemagglutinin (HA) protein [18, 19]. Historically, vaccine match has been assessed antigenically by hemagglutination inhibition (HI) assay [18], but sequencing of the HA gene can provide more direct and nuanced insight into virus diversity and vaccine relatedness [10, 11]. Genomic analyses are interpreted in the context of anticipated antigenic/immunogenic effects; however, the correlation between molecular-level changes and VE remains uncertain, requiring further integrated analyses [10, 11, 19–24].
In addition to virologic considerations, host factors may also influence VE. In particular, several studies have recently shown that prior vaccination may be associated with reduced immunogenicity and VE [10, 11, 13, 25–29]. Previous modeling simulations have suggested that negative interference from prior influenza vaccination may be pronounced in the context of antigenically drifted virus and successive seasons of homologous (ie, identical) vaccine, as was the scenario in 2014–2015 [30].
We used the integrated platform of the Canadian SPSN to explore these agent–host factors and their impact on VE during the 2014–2015 season, including variation in the viral genome and the effects of serial vaccination.
METHODS
Canadian SPSN
The community-based SPSN includes general practitioners at designated outpatient clinics in British Columbia, Alberta, Ontario, and Quebec. Patients who presented to a sentinel site within 7 days of influenza-like illness (ILI) onset were eligible to participate in the VE study. ILI was defined as acute respiratory illness with fever and cough and at least 1 of the following: sore throat, arthralgia, myalgia, or prostration. Fever was not required for elderly patients (aged ≥65 years). Epidemiologic data, including current and up to 2 previous seasons' vaccine receipt, were collected from consenting patients/guardians using a standard questionnaire at specimen collection. Ethics review boards in each participating province approved the study.
Influenza Vaccines
Influenza vaccines delivered in Canada for the 2014–2015 publicly funded campaign beginning in October were mostly nonadjuvanted, inactivated, split (68%) or subunit (22%) trivalent influenza vaccines. An adjuvanted-subunit vaccine was also publicly funded for community-based elderly adults in British Columbia. Live attenuated influenza vaccine for individuals aged 2–59 years, including trivalent and quadrivalent formulations, was publicly funded and preferentially recommended for children in British Columbia (2–8 years old), Alberta (2–17 years old), and Quebec (2–17 years old).
For the 2014–2015 northern hemisphere trivalent vaccine, the World Health Organization (WHO) recommended the following components, which were the same as those used in 2013–2014 [3, 10, 11]: an A/Texas/50/2012(H3N2)-like (clade 3C.1) virus that is also antigenically related to the A/Victoria/361/2011(H3N2)-like (clade 3C) prototype used in 2012–2013; a B/Massachusetts/02/2012(Yamagata lineage) (clade 2) virus with variable antigenic relatedness to the 2012–2013 B/Wisconsin/1/2010(Yamagata lineage) (clade 3) vaccine; and an A/California/07/2009(H1N1)pdm09-like virus unchanged since 2009.
An egg-adapted, high-growth reassortant (HGR) version of the WHO-recommended cell-passaged prototype is provided to manufacturers for egg-based vaccine production; however, egg-adaptation can introduce mutations in the HGR that may also influence antigenicity/immunogenicity [10]. Egg-adapted HGRs for 2014–2015 were identical to those for 2013–2014 and are called X-223A and BX-51B for the A(H3N2) and influenza B components, respectively [11]. Both are changed from the corresponding HGRs for 2012–2013 called IVR-165 and BX-39, respectively [10].
Epidemiologic Analyses
Patients aged ≥1 year old at specimen collection (ie, age eligible for vaccine throughout the season) and meeting inclusion/exclusion criteria shown in Figure 1 contributed to VE analysis. Patients who self-reported receiving at least one 2014–2015 influenza vaccine dose ≥2 weeks before ILI onset were considered vaccinated. VE against medically attended, laboratory-confirmed influenza was estimated by logistic regression as (1 – odds ratio) × 100%, comparing self-reported vaccination status between influenza test-positive cases and influenza test-negative controls and adjusting for relevant confounders (age group, sex, comorbidity, province, specimen collection interval, and calendar time). Calendar time was based on week of specimen collection and modeled using cubic B-spline functions with 3 equally spaced knots. Stratified VE estimates were derived using logistic regression models with an interaction term for vaccination status and the stratification variable (eg, age group). Serial/repeat vaccination effects were assessed through indicator-variable analyses based on self/guardian report of vaccine receipt in 2013–2014 among participants aged ≥2 years and/or vaccine receipt in 2012–2013 among participants aged ≥3 years.
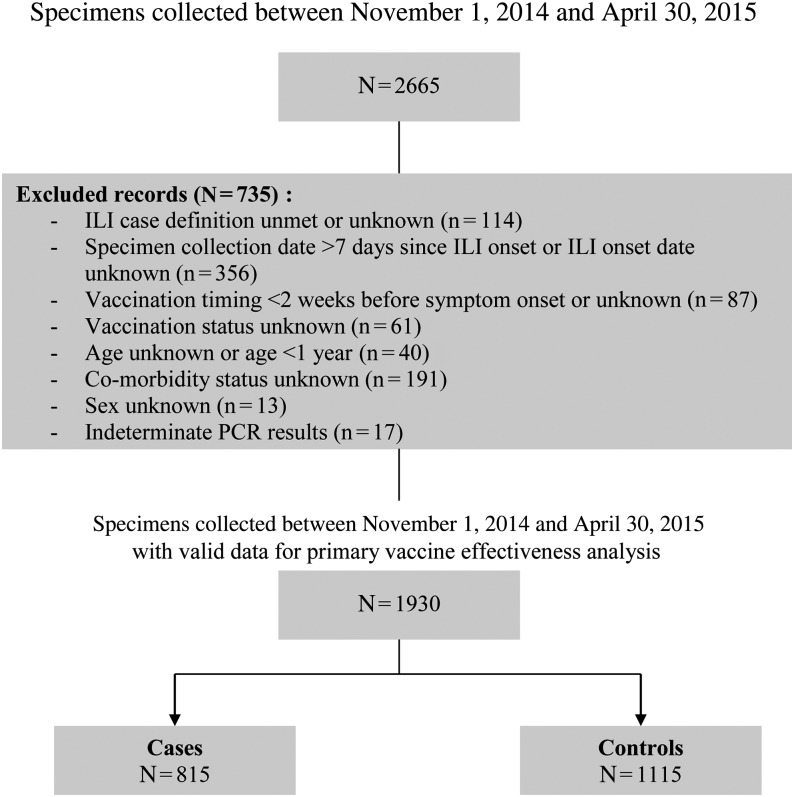
Figure 1.
Vaccine effectiveness study inclusion and exclusion criteria for the 2014–2015 season, Canadian Sentinel Practitioner Surveillance Network. aExclusions are not mutually exclusive; specimens may have >1 exclusion criterion that applies. Counts for each criterion will sum to more than the total number of specimens excluded. Abbreviations: ILI, influenza-like illness; PCR, polymerase chain reaction.
Virologic Characterization
Nasal/nasopharyngeal swabs were tested for influenza viruses by reverse-transcription polymerase chain reaction at provincial reference laboratories. Viruses contributing to VE analysis were further characterized genetically and antigenically.
Genetic Characterization
Sanger sequencing of the viral HA gene was attempted directly on all original patient specimens testing influenza-positive to establish clade designation and to detect amino acid differences between circulating viruses and the vaccine HGR at established antigenic sites, labelled A–E for A(H3N2) [11, 20]. Amino acid substitutions close to the receptor binding site (RBS) and involving antigenic site A and immunodominant antigenic site B of A(H3N2) viruses are considered most relevant to antigenicity/immunogenicity [20, 21]. Mutations at position 145 of antigenic site A and positions 155, 156, 158, 159, 189, and 193 of antigenic site B are emphasized because they have been associated with all major A(H3N2) antigenic cluster-transition events since 1968 [22]. Substitutions associated with potential gain/loss of glycosylation are also emphasized for their potential effects in masking/uncovering antibody epitopes [23, 24]. Phylogenetic analysis used the approximate likelihood method to determine clade distribution. Deduced amino acids of HA1 were aligned in FastTree [31] and visualized in FigTree [32]. GenBank accession numbers are as follows: KP701523-KP701743; KU729277-KU729659.
Antigenic Characterization
Viruses isolated in established mammalian cell lines (MDCK, MDCK-SIAT1) or primary rhesus monkey kidney cells at provincial reference laboratories were submitted to Canada's National Microbiology Laboratory (NML) for antigenic characterization by HI assay as described elsewhere [33]. Virus isolation was attempted on all influenza-positive specimens in all SPSN provinces except Ontario where a shortage of reagents limited that capacity. HI characterization was conducted on virus isolates with sufficient hemagglutination titer using guinea pig erythrocytes for A(H3N2) and turkey erythrocytes for A(H1N1)pdm09 and B(Yamagata) based on post-infection ferret antisera raised against cell- and/or egg-passaged vaccine reference viruses [18]. To address potential neuraminidase-mediated binding to erythrocytes by A(H3N2) viruses, HI assays were conducted in the presence of 20 nM oseltamivir-carboxylate following single passage in MDCK-SIAT1 cells to improve viral titers where indicated [33]. Antigenic relatedness of a sentinel isolate was defined by ≤4-fold reduction in HI antibody titer compared with the titer of the homologous reference strain [18].
RESULTS
Participant Characteristics
From November 1, 2014 to April 30, 2015, 1930 specimens met inclusion/exclusion criteria (Figure 1). As with previous seasons [10, 11], the largest proportion (63%) were collected from nonelderly adults (aged 20–64 years; Table 1). However, compared to the 2013–2014 season of A(H1N1)pdm09 dominance [11], a greater proportion of patients in 2014–2015 were elderly adults (13% vs 9%; P < .01), more notable among cases (14% vs 6%; P < .01) than controls (12% vs 10%; P = .13).
Table 1.
Profile of Participants Included in 2014–2015 Influenza Vaccine Effectiveness Evaluation, Canadian Sentinel Practitioner Surveillance Network
| Characteristic | Overall, n (%) | Distribution by Case Status, n (%) |
Vaccination Coverage Within Strata, n (%) |
|||
|---|---|---|---|---|---|---|
| Cases | Controls | P Valuea | Vaccinated | P Valuea | ||
| N (%) | 1930 | 815 (42) | 1115 (58) | 684 (35) | ||
| Age group (y) | .12 | <.01 | ||||
| 1–8 | 224 (12) | 88 (11) | 136 (12) | 49 (22) | ||
| 9–19 | 242 (13) | 111 (14) | 131 (12) | 41 (17) | ||
| 20–49 | 774 (40) | 304 (37) | 470 (42) | 216 (28) | ||
| 50–64 | 438 (23) | 199 (24) | 239 (21) | 185 (42) | ||
| ≥65 | 252 (13) | 113 (14) | 139 (12) | 193 (77) | ||
| Median (range) | 39 (1–103) | 40 (1–103) | 38 (1–94) | .07 | … | |
| Sex | .01 | <.01 | ||||
| Female | 1182 (61) | 472 (58) | 710 (64) | 449 (38) | ||
| Male | 748 (39) | 343 (42) | 405 (36) | 235 (31) | ||
| Comorbidityb | .43 | <.01 | ||||
| No | 1489 (77) | 636 (78) | 853 (77) | 436 (29) | ||
| Yes | 441 (23) | 179 (22) | 262 (24) | 248 (56) | ||
| Province | <.01 | <.01 | ||||
| Alberta | 560 (29) | 220 (27) | 340 (30) | 232 (41) | ||
| British Columbia | 292 (15) | 99 (12) | 193 (17) | 91 (31) | ||
| Ontario | 657 (34) | 273 (34) | 384 (34) | 270 (41) | ||
| Quebec | 421 (22) | 223 (27) | 198 (18) | 91 (22) | ||
| Collection interval (d) | <.01 | .25 | ||||
| ≤4 | 1432 (74) | 654 (80) | 778 (70) | 497 (35) | ||
| 5–7 | 498 (26) | 161 (20) | 337 (30) | 187 (38) | ||
| Median (range) | 3 (0–7) | 3 (0–7) | 3 (0–7) | <.01 | … | |
| Calendar timec | <.01 | <.01 | ||||
| November | 107 (6) | 22 (3) | 85 (8) | 23 (22) | ||
| December | 473 (25) | 262 (32) | 211 (19) | 171 (36) | ||
| January | 589 (31) | 252 (31) | 337 (30) | 218 (37) | ||
| February | 351 (18) | 117 (14) | 234 (21) | 143 (41) | ||
| March | 269 (14) | 115 (14) | 154 (14) | 89 (33) | ||
| April | 141 (7) | 47 (6) | 94 (8) | 40 (28) | ||
| Received 2014–2015 influenza vaccine | ||||||
| Any vaccinationd | 734/1980 (37) | 300/836 (36) | 434/1144 (38) | .35 | … | |
| ≥2 wk before onset | 684 (35) | 279 (34) | 405 (36) | .34 | … | |
| LAIV overalle | 33/391 (8) | 14/162 (9) | 19/229 (8) | .90 | … | |
| LAIV childrenf | 31/64 (48) | 13/34 (38) | 18/30 (60) | .08 | ||
| Adjuvantedg | 37/108 (34) | 14/45 (31) | 23/63 (37) | .56 | … | |
| Prior vaccination history | ||||||
| 2013–2014 vaccineh | 758/1801 (42) | 330/779 (42) | 428/1022 (42) | .84 | 564/758 (74) | <.01 |
| 2012–2013 vaccinei | 733/1719 (43) | 333/757 (44) | 400/962 (42) | .32 | 530/733 (72) | <.01 |
Abbreviation: LAIV, live attenuated influenza vaccine.
a Differences between cases and controls and vaccinated and unvaccinated participants were compared using the χ2 test or Wilcoxon rank sum test.
b Chronic comorbidities that place individuals at higher risk of serious complications from influenza, as defined by Canada's National Advisory Committee on Immunization, include heart, pulmonary, renal, metabolic, blood, cancer, and immune comprising conditions or those that compromise management of respiratory secretions, or morbid obesity. Questionnaire answered “yes,” “no,” or “unknown” without specifying comorbidity.
c Based on month of specimen collection. Missing collection dates were imputed as the laboratory accession date minus 2 days, which is the average time period between collection date and laboratory accession date for records with complete data for both fields.
d Participants who received seasonal 2014–2015 influenza vaccine <2 weeks before influenza-like illness (ILI) onset or for whom vaccination timing was unknown were excluded from the primary analysis. They were included for assessing “any” immunization, regardless of timing, for comparison with other sources of vaccination coverage.
e Among participants aged 2–59 years who received 2014–2015 influenza vaccine ≥2 weeks before ILI onset and had complete data for type of vaccine (includes 2 adult recipients).
f Among participants aged 2–17 years who received 2014–2015 influenza vaccine ≥2 weeks before ILI onset and had complete data for type of vaccine.
g Among participants aged ≥65 years who received 2014–2015 influenza vaccine ≥2 weeks before ILI onset and had complete data for adjuvanted vaccine receipt.
h Children aged <2 years in 2014–2015 were excluded from 2013–2014 vaccine uptake analysis as they may not have been eligible for vaccination during the fall 2013 immunization campaign.
i Children aged <3 years in 2014–2015 were excluded from 2012–2013 vaccine uptake analysis as they may not have been eligible for vaccination during the fall 2012 immunization campaign.
Overall, 35% of patients were considered vaccinated, without significant difference between cases (34%) and controls (36%; P = .34; Table 1). Among patients who received the 2014–2015 influenza vaccine and were old enough to be vaccinated in previous seasons, 87% (564/651) of those aged ≥2 years had also received vaccine in 2013–2014 and 82% (506/616) of those aged ≥3 years had received vaccine each year since 2012–2013.
Influenza Detection
Influenza virus was detected in 815 (42%) specimens, including 590 (72%) influenza A and 226 (28%) influenza B, peaking at >60% test positivity in late December (Table 2; Supplementary Figure 1).
Table 2.
Influenza Virus Characterization by Type and Subtype, 2014–2015 Influenza Vaccine Effectiveness Evaluation, Canadian Sentinel Practitioner Surveillance Network
| Specimen | Alberta, n (%) | British Columbia, n (%) | Ontario, n (%) | Quebec, n (%) | Overall, n (%) |
|---|---|---|---|---|---|
| N | 560 | 292 | 657 | 421 | 1930 |
| Influenza negative | 340 (61) | 193 (66) | 384 (58) | 198 (47) | 1115 (58) |
| Influenza positive | 220 (39) | 99 (34) | 273 (42) | 223 (53) | 815 (42) |
| Influenza Aa | 165 (75) | 84 (85) | 215 (79) | 126 (57) | 590 (72) |
| A(H3N2) | 161 (98) | 81 (96) | 206 (96) | 122 (97) | 570 (97) |
| A(H1N1)pdm09 | 2 (1) | 1 (1) | 3 (1) | 1 (1) | 7 (1) |
| Subtype unknown | 2 (1) | 2 (2) | 6 (3) | 3 (2) | 13 (2) |
| Influenza Ba | 55 (25) | 15 (15) | 58 (21) | 98 (44) | 226 (28) |
| Yamagata lineage | 43 (78) | 13 (87) | 50 (86) | 87 (89) | 193 (85) |
| Victoria lineage | 1 (2) | 1 (7) | 2 (3) | 2 (2) | 6 (3) |
| Lineage unknown | 11 (20) | 1 (7) | 6 (10) | 9 (9) | 27 (12) |
| Antigenic characterizationb | |||||
| Influenza A(H3N2)c,d | 105/161 (65) | 39/81 (48) | 25/206 (12) | 100/122 (82) | 269/570 (47) |
| A/Switzerland/9715293/2013-likee,f,g | 24 (23) | 18 (46) | 7 (28) | 1 (1) | 50 (19) |
| Insufficient titer to run HI assayh | 81 (77) | 21 (54) | 18 (72) | 99 (99) | 219 (81) |
| Influenza A(H1N1)pdm09 | 0/2 (0) | 1/1 (100) | 3/3 (100) | 1/1 (100) | 5/7 (71) |
| A/California/7/2009-likei,j | 0 | 1 (100) | 3 (100) | 1 (100) | 5 (100) |
| Influenza Bk | 26/55 (47) | 12/15 (80) | 20/58 (34) | 83/98 (85) | 141/226 (62) |
| B/Massachusetts/2/2012-likel | 25 (96) | 12 (100) | 19 (95) | 78 (94) | 134 (95) |
| B/Brisbane/60/2008-likem | 1 (4) | 0 | 1 (5) | 2 (2) | 4 (3) |
| Insufficient titer to run HI assay | 0 | 0 | 0 | 2 (2) | 2 (1) |
| Genetic characterizationn | |||||
| Influenza A(H3N2) | 161 (100) | 81 (100) | 206 (100) | 122 (100) | 570 (100) |
| Clade 3C.2a | 139 (86) | 43 (53) | 123 (60) | 104 (85) | 409 (72) |
| Clade 3C.3 | 1 (1) | 1 (1) | 6 (3) | 1 (1) | 9 (2) |
| Clade 3C.3a | 0 | 1 (1) | 2 (1) | 0 | 3 (1) |
| Clade 3C.3b | 6 (4) | 26 (32) | 7 (3) | 0 | 39 (7) |
| Sequencing attempted but failed | 15 (9) | 10 (12) | 68 (33) | 17 (14) | 110 (19) |
| Influenza B(Yamagata) lineage | 43 (100) | 13 (100) | 50 (100) | 87 (100) | 193 (100) |
| Clade 3 (B/Wisconsin/1/2010-like) | 39 (91) | 13 (100) | 37 (74) | 76 (87) | 165 (85) |
| Sequencing attempted but failed | 4 (9) | 0 | 13 (26) | 11 (13) | 28 (15) |
Abbreviation: HI, hemagglutination inhibition.
a One participant coinfected with influenza A(H3N2) and influenza B has been included in totals for both influenza A and B.
b Antigenic characterization of viruses with sufficient hemagglutination titer was by HI assay.
c Culture isolation was attempted on all viruses detected in British Columbia, Alberta, and Quebec where 244/364 (67%) A(H3N2) detections could be successfully cultivated. In Ontario, 25/206 (12%) A(H3N2) detections were cultivated owing to a shortage of reagents and virus growth issues.
d Includes 234 clade 3C.2a, 25 clade 3C.3b, 4 clade 3C.3, and 2 clade 3C.3a viruses; 4 with unknown clade.
e Ferret antisera to cell-passaged reference viruses provided by the US Centers for Disease Control and Prevention (CDC).
f Of the 50 A(H3N2) viruses with sufficient hemagglutination titer, all 50 (100%) had ≤4-fold reduction to cell-passaged A/Switzerland/9715293/2013 virus. Of the 49/50 A/Switzerland/9715293/2013-like viruses with sequencing results, 31 (63%) were clade 3C.2a, 15 (31%) were clade 3C.3b, 2 (4%) were clade 3C.3a, and 1 (2%) was clade 3C.3; 1 could not be sequenced.
g A subset of A(H3N2) viruses with sufficient hemagglutination titer were additionally compared against egg-passaged reference virus based on ferret antisera provided by the CDC. Of the 36 A(H3N2) viruses with sufficient titer, 35 (97%) had ≤4-fold reduction to egg-passaged A/Switzerland/9715293/2013 virus and 1 (3%; clade 3C.3b) had 8-fold reduction. Of the 35/36 A/Switzerland/9715293/2013-like viruses with sequencing results, 25 (71%) were clade 3C.2a, 7 (20%) were clade 3C.3b, 2 (6%) were clade 3C.3a, and 1 (3%) was clade 3C.3; 1 could not be sequenced.
h Of the 216/219 viruses with insufficient titer to run HI assay and with sequencing results, 203 were clade 3C.2a, 10 were clade 3C.3b, and 3 were clade 3C.3.
i Ferret antisera produced to the egg-passaged reference virus at Canada's reference laboratory (the National Microbiology Laboratory [NML]).
j Of the 5 A(H1N1)pdm09 viruses characterized by HI assay, all had reductions ≤4-fold to a cell-passaged A/California/07/2009 virus.
k Culture isolation was attempted on all viruses detected in British Columbia, Alberta, and Quebec where 121/168 (72%) influenza B detections could be successfully cultivated. In Ontario, 20/58 (34%) influenza B detections were cultivated owing to a shortage of reagents and virus growth issues.
l Yamagata lineage, clade 2. Ferret antisera to cell-passaged reference virus provided by the CDC. Of the 135 B(Yamagata) lineage viruses characterized, 134 (99%) had ≤4-fold reduction and 1 (1%; not displayed) had 8-fold reduction to cell-passaged B/Massachusetts/02/2012 (clade 2) virus.
m Victoria lineage. Ferret antisera to the egg-passaged reference virus provided by the CDC.
n Genetic clade-level characterization based on sequencing of original patient specimen (provincial reference labs) or cultured isolate (NML).
Of the 577 influenza A specimens with known subtype, virtually all (570; 99%) were A(H3N2). Of 460/570 (81%) A(H3N2) viruses sequenced, most (409; 89%) belonged to clade 3C.2a or clade 3C.3b (39; 8%). None clustered with the 2014–2015 clade 3C.1 vaccine strain (A/Texas/50/2012) [3] and very few (n = 3; <1%) clustered with the clade 3C.3a strain (A/Switzerland/9715293/2013) recommended for the updated 2015–2016 northern hemisphere vaccine [4] (Table 2; Supplementary Figure 2A).
Of the 199 influenza B specimens with known lineage, most (193; 97%) were B(Yamagata). Of 165/193 (85%) B(Yamagata) viruses sequenced, none clustered with the B(Yamagata) clade 2 vaccine strain for 2014–2015 (B/Massachusetts/2/2012) [3]. Instead, all clustered with the B(Yamagata) clade 3 vaccine strain used in 2012–2013 (B/Wisconsin/1/2010) [10] (Table 2; Supplementary Figure 2B).
VE Estimates
Overall and Clade-Specific VE
Overall adjusted VE against medically attended laboratory-confirmed influenza was 9% (95% confidence interval [CI], −14% to 27%; Table 3). Adjusted VE against the predominant A(H3N2) subtype was −17% (95% CI, −50% to 9%). Adjusted VE against the clade 3C.2a epidemic strain was comparable at −13% (95% CI, −51% to 15%), while adjusted VE against clade 3C.3b viruses was higher but not statistically significant at 52% (95% CI, −17% to 80%). Adjusted VE against influenza B(Yamagata) was statistically significant at 42% (95% CI, 10% to 62%), virtually identical with restriction to clade 3 viruses (Table 3).
Table 3.
Vaccine Effectiveness Estimates and 95% Confidence Intervals by Influenza Type, Subtype/Lineage, and Clade, 2014–2015 Season, Canadian Sentinel Practitioner Surveillance Network
| Model | Any Influenza | Any Influenza A | Any A(H3N2) | A(H3N2) Clade 3C.2a |
A(H3N2) Clade 3C.3b |
Any Influenza B | Any B(Yamagata) | B(Yamagata) Clade 3 |
|---|---|---|---|---|---|---|---|---|
| N | 1930 | 1705 | 1685 | 1524 | 1154 | 1341 | 1308 | 1280 |
| n case (% vac) | 815 (34) | 590 (39) | 570 (39) | 409 (40) | 39 (18) | 226 (23) | 193 (24) | 165 (23) |
| n control (% vac) | 1115 (36) | 1115 (36) | 1115 (36) | 1115 (36) | 1115 (36) | 1115 (36) | 1115 (36) | 1115 (36) |
| Primary analysis | ||||||||
| Unadjusted | 9 (−10–24) | −10 (−36–10) | −13 (−40–8) | −15 (−45–9) | 62 (12–83) | 49 (29–63) | 45 (22–61) | 48 (23–64) |
| Age groupa | 15 (−5–30) | −5 (−31–16) | −8 (−35–14) | −7 (−38–17) | 57 (−3–82) | 54 (33–68) | 50 (28–66) | 50 (25–67) |
| Sex (female/male) | 7 (−12–23) | −13 (−39–8) | −16 (−43–6) | −18 (−49–7) | 60 (8–83) | 49 (29–64) | 45 (22–61) | 47 (23–64) |
| Comorbidity (no/yes) | 7 (−12–24) | −10 (−36–11) | −13 (−40–8) | −13 (−44–11) | 61 (9–83) | 46 (24–62) | 43 (18–60) | 45 (18–63) |
| Province (Alberta, British Columbia, Ontario, Quebec) | 3 (−17–20) | −12 (−38–9) | −15 (−42–6) | −20 (−52–5) | NRb | 39 (14–57) | 33 (3–54) | 36 (5–57) |
| Collection interval (≤4/5–7 d) | 8 (−12–24) | −12 (−38–9) | −15 (−42–6) | −17 (−48–8) | 60 (10–83) | 48 (28–63) | 45 (21–61) | 47 (22–64) |
| Calendar timec | 12 (−6–28) | −10 (−37–11) | −14 (−43–8) | −15 (−47–11) | 62 (12–83) | 53 (32–67) | 50 (27–65) | 51 (27–67) |
| Age, sex, comorbidity, province, interval, timec | 9 (−14–27) | −13 (−45–12) | −17 (−50–9) | −13 (−51–15) | 52 (−17–80)d | 45 (18–64) | 42 (10–62) | 42 (8–63) |
| Stratified analysis, by age group | ||||||||
| 1–19 y | ||||||||
| Main effects and interaction onlye | 2 (−55–39) | −31 (−111–19) | −34 (−117–17) | −31 (−123–23) | NRb | 82 (25–96) | 78 (7–95) | 73 (−15–94) |
| Adjustedf | −5 (−71–35) | −57 (−167–7) | −64 (−180–3) | −56 (−182–14) | NRb | 84 (29–96) | 80 (11–96) | 76 (−10–95) |
| 20–64 y | ||||||||
| Main effects and interaction onlye | 14 (−10–32) | −4 (−36–20) | −8 (−42–18) | −7 (−46–22) | NRb | 47 (20–65) | 45 (15–64) | 46 (15–66) |
| Adjustedf | 7 (−20–28) | −8 (−45–20) | −13 (−52–17) | −10 (−57–22) | NRb | 33 (−6–57) | 29 (−16–56) | 30 (−17–58) |
| ≥65 y | ||||||||
| Main effects and interaction onlye | 20 (−43–56) | 17 (−54–56) | 19 (−52–56) | 20 (−54–59) | NRb | 31 (−93–75) | 23 (−131–74) | 31 (−135–80) |
| Adjustedf | 20 (−47–57) | 15 (−67–57) | 17 (−64–58) | 24 (−59–64) | NRb | 19 (−160–75) | 17 (−187–76) | 25 (−194–81) |
| Stratified analysis, by comorbidity | ||||||||
| Participants without comorbidity | ||||||||
| Main effects and interaction onlyg | 8 (−15–27) | −15 (−47–10) | −18 (−51–8) | −15 (−53–13) | NRb | 52 (28–68) | 50 (23–68) | 50 (21–69) |
| Adjustedh | 6 (−20–27) | −24 (−64–6) | −28 (−69–4) | −23 (−70–11) | NRb | 48 (17–67) | 45 (11–67) | 44 (6–67) |
| Participants with comorbidity | ||||||||
| Main effects and interaction onlyg | 6 (−38–36) | 2 (−48–35) | −2 (−54–33) | −8 (−71–32) | NRb | 24 (−50–62) | 15 (−75–58) | 24 (−65–65) |
| Adjustedh | 16 (−28–44) | 14 (−36–46) | 11 (−43–44) | 11 (−50–48) | NRb | 37 (−37–71) | 29 (−61–69) | 32 (−63–72) |
Abbreviations: % vac, percentage vaccinated; NR, not reported.
a Age group categories: 1–8, 9–19, 20–49, 50–64, ≥65 years.
b Model did not converge and/or sample sizes do not support reliable estimation.
c Calendar time was modeled by week of specimen collection using cubic B-spline functions with 3 equally spaced knots.
d Adjusted model for influenza A(H3N2) clade 3C.3b outcome not adjusted for province due to small number of cases within strata.
e Adjusted for age group (1–19, 20–64, ≥65 years) and vaccine × age group interaction.
f Adjusted for age group (1–19, 20–64, ≥65 years), sex, comorbidity, province, collection interval, calendar time (spline), and vaccine × age group interaction.
g Adjusted for comorbidity and vaccine × comorbidity interaction.
h Adjusted for age group, sex, comorbidity, province, collection interval, calendar time (spline), and vaccine × comorbidity interaction.
Prior Vaccination Effects
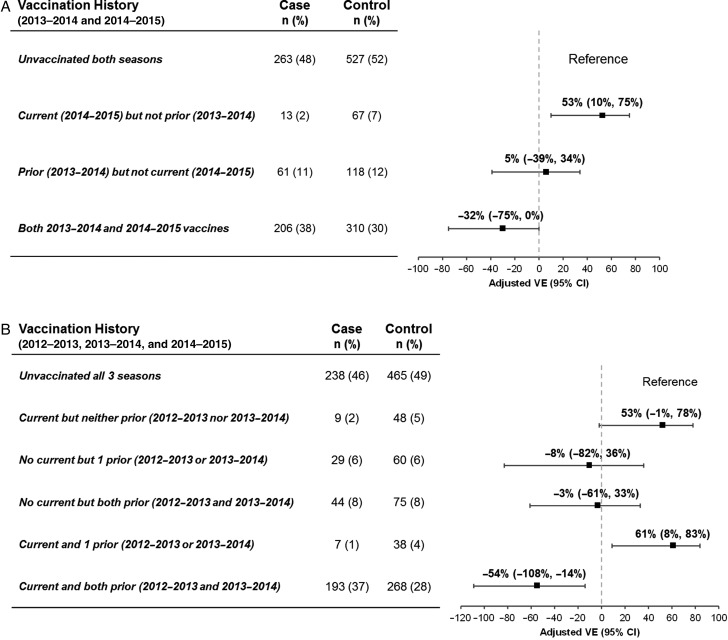
Compared with participants who were unvaccinated both seasons, adjusted VE against A(H3N2) for those vaccinated in 2014–2015 but not 2013–2014 was 53% (95% CI, 10% to 75%), significantly lower at −32% (95% CI, −75% to 0%) for participants also vaccinated in 2013–2014 (Figure 2A; Supplementary Table 1A). In a separate model that also considered vaccination in 2012–2013, VE was −54% (95% CI, −108% to −14%) for participants serially vaccinated each year since 2012–2013 (Figure 2B; Supplementary Table 1A). A similar pattern was observed with restriction to nonelderly adults (Supplementary Table 1B).
Figure 2.
Effect of prior 2012–2013 and/or 2013–2014 season influenza vaccine receipt on current 2014–2015 influenza vaccine effectiveness for influenza A(H3N2). Analyses are based on the same exclusion criteria as primary analysis, adjusted for age group (<9, 9–19, 20–49, 50–64, ≥65 years), sex, comorbidity, province, collection interval, and calendar time (spline). Calendar time was modeled by week of specimen collection using cubic B-spline functions with 3 equally spaced knots. A, The effect of prior 2013–2014 vaccine receipt in participants aged ≥2 years in 2014–2015 and with complete data for 2013–2014 and 2014–2015 influenza vaccine receipt. B, The effect of prior 2013–2014 and/or 2012–2013 vaccine receipt in participants aged ≥3 years in 2014–2015 and those with complete data for 2012–2013, 2013–2014, and 2014–2015 influenza vaccine receipt. Abbreviations: CI, confidence interval; VE, vaccine effectiveness.
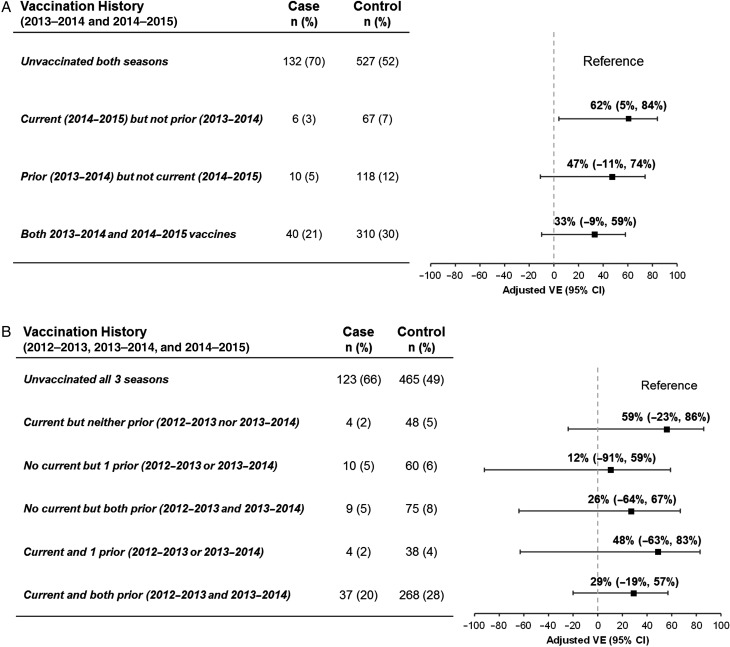
For influenza B(Yamagata), VE was 62% (95% CI, 5% to 84%) among participants vaccinated in 2014–2015 but not 2013–2014, with lower VE (but with overlapping CIs) of 33% (95% CI, −9% to 59%) for participants also vaccinated in 2013–2014 (Figure 3A; Supplementary Table 1A). When vaccination in 2012–2013 was also considered, VE was comparable at 29% (95% CI, −19% to 57%) for participants serially vaccinated each year since 2012–2013 (Figure 3B; Supplementary Table 1A).
Figure 3.
Effect of prior 2012–2013 and/or 2013–2014 season influenza vaccine receipt on current 2014–2015 influenza vaccine effectiveness for influenza B(Yamagata). Analyses are based on the same exclusion criteria as primary analysis, adjusted for age group (<9, 9–19, 20–49, 50–64, ≥65 years), sex, comorbidity, province, collection interval, and calendar time (spline). Calendar time was modeled by week of specimen collection using cubic B-spline functions with 3 equally spaced knots. A, The effect of prior 2013–2014 vaccine receipt in participants aged ≥2 years in 2014–2015 and with complete data for 2013–2014 and 2014–2015 influenza vaccine receipt. B, The effect of prior 2013–2014 and/or 2012–2013 vaccine receipt in participants aged ≥3 years in 2014–2015 and those with complete data for 2012–2013, 2013–2014, and 2014–2015 influenza vaccine receipt. Abbreviations: CI, confidence interval; VE, vaccine effectiveness.
Virologic Characterization
Influenza A(H3N2)
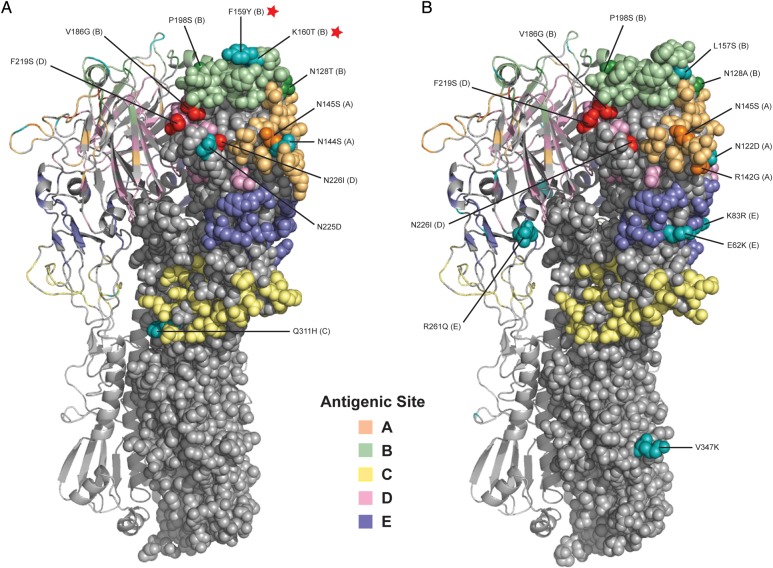
Both clade 3C.2a and clade 3C.3b viruses bore multiple (10–12) antigenic site amino acid mutations relative to the 2014–2015 A(H3N2) HGR vaccine strain, including the following 6 shared substitutions: N145S (site A; cluster transition), N128T/A (site B; N128T is a potential gain of glycosylation in clade 3C.2a viruses), and P198S (site B) as well as V186G (site B), F219S (site D), and N226I (site D) present in the egg-adapted HGR rather than circulating viruses (Supplement Table 2A).
Sentinel clade 3C.2a strains additionally bore the following substitutions: N144S (site A; potential loss of glycosylation), F159Y (site B; cluster transition), K160T (site B; potential gain of glycosylation), Q311H (site C), and N225D (nonantigenic site, but within the RBS; Figure 4A; Supplementary Table 2A) [34, 35]. Sentinel clade 3C.3b strains instead bore R142G (site A), N122D (site A; potential loss of glycosylation), L157S (site B), and the following site E substitutions: E62K, K83R, and R261Q (Figure 4B; Supplementary Table 2A) [34, 35].
Figure 4.
Crystal structure of hemagglutinin (HA) of circulating A(H3N2) clade 3C.2a and clade 3C.3b viruses relative to the 2014–2015 egg-adapted high-growth reassortant (HGR) vaccine strain. Three-dimensional structural model shows antigenic site substitutions in the HA1 of representative sentinel influenza A(H3N2) viruses compared with the 2014–2015 egg-adapted A/Texas/50/2012-like (clade 3C.1) HGR vaccine strain (X-223A) for (A) clade 3C.2a and (B) clade 3C.3b. The homology models of HA were generated using the SWISS-MODEL web-based automated modeling server [34]. The final images of the HA structures were generated using Pymol (Schrödinger, LLC) [35]. The trimeric HA protein of A(H3N2) was constructed using the A/Victoria/361/2011 human A(H3N2) virus (PDB accession number 4WE8.1) with 97% identity to circulating strains. Antigenic sites (A–E) are shown in pastel colors. Substitutions in antigenic sites are labelled, and those identifying clade designations are shown in cyan, those arising from egg passage and/or in the HGR are shown in red, and other substitutions in sentinel viruses are shown in darker shading of the corresponding antigenic site color. Mutations at pivotal antigenic site B positions 159 (a cluster-transition position) and 160 (associated with a potential gain of glycosylation) are indicated with a red star, present in clade 3C.2a viruses but absent from clade 3C.3b viruses relative to X-223A. (Panel A republished with modifications on permission of Oxford University Press from Skowronski DM et al [11], J Infect Dis 2015; 212(5).)
Culture isolates from 269/570 (47%) A(H3N2) viruses detected by provincial laboratories were submitted to the NML for antigenic characterization by HI assay, of which 234/269 (87%) belonged to clade 3C.2a (Table 2). Viruses collected early in the season were antigenically distinct from the 2014–2015 cell-passaged A/Texas/50/2012 (clade 3C.1) vaccine prototype [13]. After December 18, 2014, antisera to the A/Switzerland/9715293/2013 (clade 3C.3a) prototype for the 2015–2016 vaccine became available [4], and culture isolates were subsequently characterized only in relation to that. However, as we described in detail in Skowronski et al [33], only 50/269 (19%) A(H3N2) virus isolates overall and only 31/234 (13%) belonging to clade 3C.2a could be successfully characterized by HI. All were considered antigenically related to the cell-passaged A/Switzerland/9715293/2013 prototype as were 35 (97%) of a further subset of 36 sentinel A(H3N2) viruses also characterized in relation to the egg-passaged prototype (Table 2) [33].
Influenza B(Yamagata)
Circulating clade 3 influenza B(Yamagata) viruses bore 6–7 antigenic site amino acid differences from the 2014–2015 B(Yamagata) clade 2 vaccine (Supplementary Table 2B). Culture isolates from 141/226 (62%) influenza B viruses detected by provincial laboratories were submitted to the NML for HI characterization, including 135 B(Yamagata), 4 B(Victoria), and 2 of unknown lineage. Of the 135 influenza B(Yamagata) clade 3 viruses successfully characterized, 134 (99%) were considered antigenically related to the cell-passaged 2014–2015 clade 2 B/Massachusetts/2/2012 vaccine strain; 1 showed >4-fold reduction in HI titer and was considered antigenically distinct.
DISCUSSION
In this analysis for the 2014–2015 influenza season in Canada, overall VE was <10%, which is the lowest recorded in more than a decade of annual monitoring by the Canadian SPSN [12]. Suboptimal VE estimates have been highlighted previously, notably during the 2010–2011 and 2012–2013 seasons of predominant A(H3N2) activity in Canada and the United States [10, 12, 26, 29, 36, 37] and in a recent meta-analysis of TND studies globally reporting pooled average VE < 40% for A(H3N2) viruses [38]. In the current 2014–2015 analysis, however, we found no vaccine protection against the dominant A(H3N2) epidemic strain in Canada. Such historically low VE requires in-depth examination of agent–host factors that may have contributed.
Clade 3C.2a viruses that dominated in 2014–2015 were first detected by the Canadian SPSN in January 2014 and comprised about one-quarter of the few A(H3N2) viruses identified by the network during the 2013–2014 season [11]. By autumn 2014, clade 3C.2a viruses had become the predominant variant in Canada, ultimately comprising about 90% of the 2014–2015 A(H3N2) epidemic, with mostly clade 3C.3b viruses comprising the remaining 10% [13]. In clade-specific analyses, VE estimates differed substantially between these 2 genetic subgroups. Whereas the 2014–2015 vaccine provided no protection against the clade 3C.2a epidemic strain, vaccination reduced the risk of medically attended clade 3C.3b illness by about half. Although not statistically significant, these differences in clade-specific VE are consistent with the clade variation in VE also observed in the United States [39].
Genomic analysis revealed several key differences between sentinel clade 3C.2a and clade 3C.3b viruses, potentially informing variation in VE findings. Both clades exhibited multiple amino acid mutations at antigenic sites of the HA protein and were considered antigenically distinct from the 2014–2015 A/Texas/50/2012 (clade 3C.1) vaccine strain [3]. However, clade 3C.2a viruses were distinguished by additional F159Y mutation affecting a major cluster-transition position at the highly exposed tip of immuno-dominant antigenic site B [21, 22]. Serologic analyses have also highlighted mutation at position 159 as likely responsible for the 2014–2015 antigenic drift [40]. In addition, clade 3C.2a viruses possess an adjacent K160T mutation, conferring a potential glycosylation motif at residues 158–160 of the HA [23, 24, 33]. Such site-specific glycosylation can hinder antibody access to viral epitopes, a particular concern when affecting pivotal antigenic site B [23, 33, 40].
In our analysis, as elsewhere, only a small proportion of A(H3N2) virus isolates, particularly those belonging to clade 3C.2a, could be successfully characterized by HI assay [1, 2, 33]. For the majority of A(H3N2) viruses that could not be characterized, laboratories globally have imputed antigenic relatedness on the basis of genetic sequencing [1, 2, 41]. However, as shown in a recent analysis by the Canadian SPSN comparing viral sequences before and after growth in cell culture, the minority (<15%) of clade 3C.2a virus isolates that could be HI characterized may not be representative of viruses circulating in nature [33]. Cell culture isolation introduced mutations resulting in the full or partial loss of the potential glycosylation motif at residues 158–160, a signature feature of clade 3C.2a viruses potentially relevant to antibody binding [33]. Variability and uncertainty in laboratory findings underscore the need to more fully investigate genomic and antigenic indicators of vaccine–virus relatedness and their correlation with actual epidemiologic measures of vaccine protection.
Participants who received the 2014–2015 vaccine without vaccination the year before had significant protection against A(H3N2) illness, whereas VE was significantly diminished in those who had also received the identical 2013–2014 vaccine. Reduced VE associated with prior vaccination has been reported previously [10, 11, 26–29], including for 2014–2015 in mid-season analysis against A(H3N2) by the Canadian SPSN [13] and end-of-season analysis in a European multicenter case-control study [42], neither of which showed statistically significant effects. Our findings also align with earlier modeling simulations that predict negative interference from prior vaccination when the antigenic distance between vaccine and circulating strains is large but between consecutive vaccine components is small [30]. Such effects are anticipated to be pronounced during epidemics of antigenically drifted virus and successive seasons of identical (but mismatched) vaccine antigen [30], as were the particular conditions for A(H3N2) in 2014–2015.
We observed a greater negative dose-response pattern for A(H3N2) in those who had additionally received the 2012–2013 vaccine that was also antigenically related to the 2013–2014 and 2014–2015 A(H3N2) vaccine components (Figure 2) [3, 10, 11]. Negative VE for A(H3N2), with 95% CIs less than zero, suggests that participants who were vaccinated every year since 2012–2013 were at significant 1.54 times (54%) increased risk of A(H3N2) illness compared with those consistently unvaccinated. Others have also reported negative, but nonsignificant, point estimates of VE against A(H3N2) during the 2014–2015 season, interpreted as consistent with a true null effect, but without further exploration based on prior serial vaccination over 2 seasons, as undertaken here [14, 15]. Statistically significant increased risk from prior seasonal vaccination was reported during the 2009 pandemic context of a novel virus subtype substantially mismatched to vaccine, but has not previously been recognized in relation to seasonal influenza drift variants [43–48]. Few other studies have examined the cumulative effects of habitual influenza vaccination across several seasons [27], and the precise virologic, immunologic, or epidemiologic conditions required for such negative effects, if real, are unknown. Pending additional investigation, our findings should be interpreted cautiously. Among our participants vaccinated in 2014–2015, >80% were repeatedly vaccinated each year. We adjusted for potential confounders but cannot rule out underlying differences between the repeatedly vaccinated and the much smaller subset of infrequently vaccinated participants. VE estimates in the latter subgroup may also be less stable owing to limited sample size.
Despite antigenic relatedness based on HI assay, the 2014–2015 influenza B(Yamagata) clade 2 vaccine also provided suboptimal protection against the clade-mismatched circulating B(Yamagata) clade 3 viruses (VE < 45%); similarly low VE was reported in the United States [39]. This 2014–2015 VE estimate is lower than the clade-specific VE of 60%–70% reported from Canada during the 2013–2014 season using the same B(Yamagata) clade 2 vaccine against virtually identical circulating clade 3 viruses [11]. However, as with the A(H3N2) outcome, variation by prior vaccination history was observed. VE against B(Yamagata) strains was higher among recipients of 2014–2015 vaccine alone (62%), comparable to the prior season's estimate [11] but reduced (with overlapping CIs) among repeat recipients of the same clade 2 vaccine in 2013–2014 and 2014–2015 (33%). However, in contrast to A(H3N2), further attenuation of VE was not observed among those who additionally received the heterologous but clade 3-matched 2012–2013 vaccine (Figure 3).
There are limitations to this analysis, including those related to statistical power. As VE estimates on either side of the null approach zero, sample size requirements to demonstrate statistically significant effects increase substantially. Ideally, clade-specific VE estimates would have been further stratified by prior vaccination and other subgroup status to better understand their separate effects; however, sample size did not support that exploration. Vaccine status is based on a combination of self-report and sentinel practitioner documentation that may be subject to information bias, notably in recalling prior seasons' vaccination. Current and prior seasons' vaccine coverage among our test-negative controls was comparable to previous reports by the SPSN [10, 11] and to other Canadian population survey estimates [49]. Although we identified no obvious flags for concern in our participant profiles, as for any observational design we cannot rule out random variation, residual bias, or confounding to explain findings.
In summary, integrated genetic, antigenic, and epidemiologic analysis from Canada suggests that a combination of agent–host factors, including viral genomic variation and repeat vaccination effects, likely contributed to the historically low VE observed during the 2014–2015 influenza season. Further investigation linking virologic and epidemiologic analyses is needed to advance our understanding of these critical agent–host interactions and to improve annual influenza vaccine reformulation and program recommendations.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We gratefully acknowledge the contribution of those at the sentinel sites whose regular submission of specimens and data provided the basis of our analyses. We acknowledge the coordination and technical support provided by epidemiologic and laboratory staff in all participating provinces. We thank the following for network coordination and data entry activities in each province: Elaine Douglas and Kinza Rizvi for TARRANT in Alberta; Romy Olsha for Public Health Ontario; and Sophie Auger for the Institut national de santé publique du Québec. We thank those who provided laboratory support at the British Columbia Public Health Microbiology and Reference Laboratory, the Alberta Provincial Laboratory for Public Health (ProvLab), the Public Health Ontario Laboratory, and the Laboratoire de santé publique du Québec. We further acknowledge the virus detection and gene sequencing support provided by Kanti Pabbaraju, Sallene Wong, and Danielle Zarra of the Alberta Provincial Laboratory; Aimin Li and Stephen Perusini of Public Health Ontario; and Joel Ménard and Lyne Désautels of the Québec Provincial Laboratory. Finally, we acknowledge the authors, originating and submitting laboratories of the reference virus sequences from Global Initiative on Sharing All Influenza Data's EpiFlu Database (www.gisaid.org).
Financial support. Funding was provided by the Canadian Institutes of Health Research (grant number TPA-90193), the British Columbia Centre for Disease Control, Alberta Health and Wellness, Public Health Ontario, Ministère de la santé et des services sociaux du Québec, l'Institut national de santé publique du Québec, and the Public Health Agency of Canada.
Potential conflicts of interest. Within 36 months of manuscript submission, G. D. S. received research grants and compensation for travel costs to attend an ad hoc advisory board meeting for GlaxoSmithKline, a research grant from Pfizer for unrelated studies, and separate compensation for participation as an expert witness in a legal challenge of enforced healthcare worker influenza vaccination. J. B. G. has received a research grant from Pfizer. M.K. has received research grants from Roche, Merck, Hologic, Boerhinger Ingelheim, and Siemens. S. S. and T. L. K. were funded by the Canadian Institutes of Health Research (grant number TPA-90193). All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Centers for Disease Control and Prevention. Influenza (flu). Past weekly surveillance reports. Available at: http://www.cdc.gov/flu/weekly/pastreports.htm Accessed 4 April 2016.
- 2.Public Health Agency of Canada. FluWatch. Weekly reports 2014–2015 season. Available at: http://www.phac-aspc.gc.ca/fluwatch/14-15/index-eng.php Accessed 4 April 2016.
- 3.World Health Organization. Recommended composition of influenza virus vaccines for use in the 2014–2015 northern hemisphere influenza season. Geneva: WHO, 2014. Available at: http://www.who.int/influenza/vaccines/virus/recommendations/2014_15_north/en/ Accessed 4 April 2016. [Google Scholar]
- 4.World Health Organization. Recommended composition of influenza virus vaccines for use in the 2015–2016 northern hemisphere influenza season. Geneva: WHO, 2015. Available at: http://www.who.int/influenza/vaccines/virus/recommendations/2015_16_north/en/ Accessed 4 April 2016. [Google Scholar]
- 5.Schanzer DL, Sevenhuysen C, Winchester B, Mersereau T. Estimating influenza deaths in Canada, 1992–2009. PLoS One 2013; 8:e80481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson WW, Shay DK, Weintraub E et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003; 289:179–86. [DOI] [PubMed] [Google Scholar]
- 7.Public Health Agency of Canada. Reported influenza hospitalizations and deaths in Canada: 2011–12 to 2015–16 (data to March 26, 2016). Available at: http://www.phac-aspc.gc.ca/influenza/flu-stat-eng.php Accessed 4 April 2016.
- 8.United Kingdom Office for National Statistics. Statistical Bulletin. Excess winter mortality in England and Wales 2014/15 (provisional) and 2013/14 (final). Available at: http://www.ons.gov.uk/ons/dcp171778_425192.pdf Accessed 4 April 2016.
- 9.Sullivan SG, Feng S, Cowling BJ. Potential of the test-negative design for measuring influenza vaccine effectiveness: a systematic review. Expert Rev Vaccines 2014; 13:1571–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skowronski DM, Janjua NZ, De Serres G et al. Low 2012–13 influenza vaccine effectiveness associated with mutations in the egg-adapted H3N2 vaccine strain, not antigenic drift in circulating viruses. PLoS One 2014; 9:e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skowronski DM, Chambers C, Sabaiduc S et al. Integrated sentinel surveillance linking genetic, antigenic, and epidemiologic monitoring of influenza vaccine-virus relatedness and effectiveness during the 2013–2014 influenza season. J Infect Dis 2015; 212:726–39. [DOI] [PubMed] [Google Scholar]
- 12.Canadian Sentinel Practitioner Surveillance Network (SPSN). Vaccine effectiveness (VE) estimates against laboratory-confirmed, medically-attended influenza, 2004–15 seasons. Available at: http://www.bccdc.ca/NR/rdonlyres/334939C9-3F64-4C22-8683-4067AC95CA9B/0/SPSN_VE_20042015_June_3_2015_dated.pdf Accessed 4 April 2016.
- 13.Skowronski DM, Chambers C, Sabaiduc S et al. Interim estimates of 2014/15 vaccine effectiveness against influenza A(H3N2) from Canada's Sentinel Physician Surveillance Network, January 2015. Euro Surveill 2015; 20:pii:21022. [DOI] [PubMed] [Google Scholar]
- 14.Gilca R, Skowronski DM, Douville-Fradet M et al. Mid-season estimates of influenza vaccine effectiveness against A(H3N2) hospitalization in the elderly in Quebec, Canada, January 2015. PLoS One 2015; 10:e0132195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNeil SA, Andrew MK, Ye L et al. Interim estimates of 2014/15 influenza vaccine effectiveness in preventing laboratory-confirmed influenza-related hospitalisation from the Serious Outcomes Surveillance Network of the Canadian Immunization Research Network, January 2015. Euro Surveill 2015; 20:pii:21024. [DOI] [PubMed] [Google Scholar]
- 16.Flannery B, Clippard J, Zimmerman RK et al. Early estimates of seasonal influenza vaccine effectiveness—United States, January 2015. MMWR Morb Mortal Wkly Rep 2015; 64:10–5. [PMC free article] [PubMed] [Google Scholar]
- 17.Pebody R, Warburton F, Andrews N et al. Effectiveness of seasonal influenza vaccine in preventing laboratory-confirmed influenza in primary care in the United Kingdom: 2014/15 end of season results. Euro Surveill 2015; 20:pii:30013. [PubMed] [Google Scholar]
- 18.Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther 2011; 9:669–83. [DOI] [PubMed] [Google Scholar]
- 19.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science 1999; 286:1921–5. [DOI] [PubMed] [Google Scholar]
- 20.Ndifon W, Wingreen NS, Levin SA. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc Natl Acad Sci U S A 2009; 106:8701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popova L, Smith K, West AH et al. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One 2012; 7:e41895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koel BF, Burke DF, Bestebroer TM et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013; 342:976–9. [DOI] [PubMed] [Google Scholar]
- 23.Tate MD, Job ER, Deng Y-M, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014; 6:1294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An Y, McCullers JA, Alymova I, Parsons LM, Cipollo JF. Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J Proteome Res 2015; 14:3957–69. [DOI] [PubMed] [Google Scholar]
- 25.Thompson MG, Naleway A, Fry AM et al. Effects of repeated annual influenza vaccination among healthcare personnel on serum hemagglutination inhibition antibody response to A/Perth/16/2009 (H3N2)-like virus during 2010-11. Vaccine 2016; 34:981–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohmit SE, Petrie JG, Malosh RE et al. Influenza vaccine effectiveness in the community and the household. Clin Infect Dis 2013; 56:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLean HQ, Thompson MG, Sundaram ME et al. Impact of repeated vaccination on vaccine effectiveness against influenza A(H3N2) and B during 8 seasons. Clin Infect Dis 2014; 59:1375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmit SE, Thompson MG, Petrie JG et al. Influenza vaccine effectiveness in the 2011–2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis 2014; 58:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohmit SE, Petrie JG, Malosh RE, Fry AM, Thompson MG, Monto AS. Influenza vaccine effectiveness in households with children during the 2012–13 season: assessments of prior vaccination and serologic susceptibility. J Infect Dis 2015; 211:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc Natl Acad Sci U S A 1999; 96:14001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rambaut A. FigTree v1.4.0, a graphical viewer of phylogenetic trees. Edinburgh: University of Edinburgh; Available at: http://tree.bio.ed.ac.uk/software/figtree/ Accessed 4 April 2016. [Google Scholar]
- 33.Skowronski DM, Sabaiduc S, Chambers C et al. Mutations acquired during cell culture isolation may affect antigenic characterisation of influenza A(H3N2) clade 3C.2a viruses. Euro Surveill 2016; 21:pii:30112. [DOI] [PubMed] [Google Scholar]
- 34.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 2003; 31:3381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The PyMOL Molecular Graphics System, Version 1.6. Portland: Schrödinger, LLC.
- 36.McLean HQ, Thompson MG, Sundaram ME et al. Influenza vaccine effectiveness in the United States during 2012–2013: variable protection by age and virus type. J Infect Dis 2015; 211:1529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowling BJ, Feng S, Finelli L, Steffens A, Fowlkes A. Assessment of influenza vaccine effectiveness in a sentinel surveillance network 2010–13, United States. Vaccine 2016; 34:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belongia EA, Simpson MD, King JP et al. Variable influenza vaccine effectiveness by subtype: a meta-analysis of test negative design studies. Lancet Infect Dis 2016; doi:10.1016/S1473-3099(16)00129-8. [DOI] [PubMed]
- 39.Flannery B, Clippard J. End-of-season influenza vaccine effectiveness estimates for the 2014–15 season: US influenza vaccine effectiveness (Flu VE) network Advisory Committee on Immunization Practice presentation slides: June 2015. Available at: http://www.cdc.gov/vaccines/acip/meetings/downloads/slides-2015-06/flu-02-flannery.pdf Accessed 4 April 2016. [Google Scholar]
- 40.Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep 2015; 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Appiah GD, Blanton L, D'Mello T et al. Influenza activity – United States, 2014–15 season and composition of the 2015–16 influenza vaccine. MMWR Morb Mortal Wkly Rep 2015; 64:583–90. [PMC free article] [PubMed] [Google Scholar]
- 42.Valenciano M, Kissling E, Reuss A et al. Vaccine effectiveness in preventing laboratory-confirmed influenza in primary care patients in a season of co-circulation of influenza A(H1N1)pdm09, B and drifted A(H3N2), I-MOVE Multicentre Case–Control Study, Europe 2014/15. Euro Surveill 2016; 21:pii:30139. [DOI] [PubMed] [Google Scholar]
- 43.Skowronski DM, De Serres G, Crowcroft NS et al. Association between the 2008–09 seasonal influenza vaccine and pandemic H1N1 illness during spring-summer 2009: four observational studies from Canada. PLoS Med 2010; 7:e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilca R, Deceuninck G, De Serres G et al. Effectiveness of pandemic H1N1 vaccine against influenza-related hospitalization in children. Pediatrics 2011; 128: e1084–91. [DOI] [PubMed] [Google Scholar]
- 45.Crum-Cianflone NF, Blair PJ, Faix D et al. Clinical and epidemiologic characteristics of an outbreak of novel H1N1 (swine origin) influenza A virus among United States military beneficiaries. Clin Infect Dis 2009; 49:1801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuchihashi Y, Sunagawa T, Yahata Y et al. Association between seasonal influenza vaccination in 2008–2009 and pandemic influenza A(H1N1) 2009 infection among school students from Kobe, Japan, April-June 2009. Clin Infect Dis 2012; 54:381–3. [DOI] [PubMed] [Google Scholar]
- 47.Cowling BJ, Ng S, Ma ES et al. Protective efficacy of seasonal influenza vaccination against seasonal and pandemic influenza virus infection during 2009 in Hong Kong. Clin Infect Dis 2010; 51:1370–9. [DOI] [PubMed] [Google Scholar]
- 48.Skowronski DM, Hamelin ME, De Serres G et al. Randomized controlled ferret study to assess the direct impact of 2008–09 trivalent inactivated influenza vaccine on A(H1N1)pdm09 disease risk. PLoS One 2014; 9:e86555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gionet L. Flu vaccination rates in Canada. Health at a Glance. Statistics Canada catalogue no. 82-624-X. Available at: http://www.statcan.gc.ca/pub/82-624-x/2015001/article/14218-eng.htm Accessed 4 April 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.