Abstract
Heterozygous SHANK3 mutations are associated with idiopathic autism and Phelan-McDermid syndrome. SHANK3 is a ubiquitously expressed scaffolding protein that is enriched in postsynaptic excitatory synapses. Here we used engineered conditional mutations in human neurons to show that heterozygous and homozygous SHANK3 mutations severely and specifically impair Ih-channels. SHANK3 mutations caused alterations in neuronal morphology and synaptic connectivity; chronic pharmacological blockage of Ih-channels reproduced these phenotypes, suggesting they may be secondary to Ih-channel impairment. Moreover, mouse Shank3-deficient neurons also exhibited severe decreases in Ih-currents. SHANK3 protein interacted with hyperpolarization-activated cyclic nucleotide-gated channel proteins (HCN proteins) forming Ih-channels, indicating that SHANK3 functions to organize HCN-channels. Our data suggest SHANK3 mutations predispose to autism, at least partially, by inducing an Ih-channelopathy that may be amenable to pharmacological intervention.
Haploinsufficiency of SHANK3 constitutes one of the more frequent single-gene mutations in autism spectrum disorders (ASDs; 1-5). Moreover, SHANK3 is among several genes deleted in Phelan-McDermid syndrome (a.k.a. 22q13.3 deletion syndrome), and SHANK3 haploinsufficiency may be the most important contributing factor to Phelan-McDermid syndrome pathology (1-5). In addition, emerging evidence links SHANK3 mutations to schizophrenia (6,7), and SHANK3 overexpression has also been implicated in neuropsychiatric disorders (8). Analysis of human neurons differentiated from induced pluripotent stem cells (iPSC) from patients with Phelan-McDermid syndrome have revealed two apparently disparate phenotypes, an impairment in synaptic transmission and an increase in input resistance (9). The synaptic impairments of Phelan-McDermid neurons are rescued with SHANK3 (9), consistent with the presence of SHANK3 in postsynaptic specializations where SHANK3 is thought to function as a scaffolding protein that organizes receptor signaling (10-12). Thus, Phelan-McDermid syndrome may result from a synaptic impairment caused by SHANK3 haploinsufficiency. The dramatically increased input resistance of Phelan-McDermid neurons, however, remains an enigma (9). Because human neurons with sole SHANK3 mutations have not been analyzed, the role of SHANK3 in the phenotypes of Phelan-McDermid neurons remains incompletely understood, as does the functional effect of a pure SHANK3 haploinsufficiency on human neurons. In mice, heterozygous and homozygous Shank3 mutations cause a behavioral phenotype resembling autism and/or obsessive-compulsive disorders, and produce changes in synaptic transmission (13-21). However, the underlying molecular mechanisms and possible contributory non-synaptic effects are unclear, as are the cellular effects of murine Shank3 mutations.
To address these issues that are crucial for progress towards understanding ASDs and Phelan-McDermid syndrome, we generated human neurons with conditional heterozygous and homozygous SHANK3 loss-of-function mutations. Unexpectedly, we found that besides causing synaptic impairments, loss of SHANK3 function selectively and severely impaired Ih-currents. We observed that SHANK3 protein directly bound to hyperpolarization-activated cyclic nucleotide-gated channel (HCN) proteins mediating Ih-currents, and that at least some of the synaptic impairments in SHANK3-mutant human neurons are an indirect result of the Ih-channelopathy produced by the SHANK3 mutations during neuronal development. Finally, we observed a similar phenotype in Shank3-deficient mouse neurons, suggesting a general function for SHANK3 in scaffolding HCN-channels.
Generation of heterozygous SHANK3 conditional knockout (cKO) mutations
To investigate the role of SHANK3 mutations in human neurons, we constructed conditional mutations in the SHANK3 gene in human H1 embryonic stem cells (ES cells) using homologous recombination (Fig. 1A, 1B, S1; 22). We chose this approach because it enables generation of matching control and mutant neurons from the same ES cell clones, thus eliminating subclone-to-subclone variability. The SHANK3 locus expresses multiple transcripts that encode different SHANK3 isoforms with distinct protein interaction domains (Fig. 1A, S2A). To conditionally delete major SHANK3 isoforms, we targeted exon 13 of the SHANK3 gene whose deletion causes a frame-shift in all major SHANK3 mRNAs.
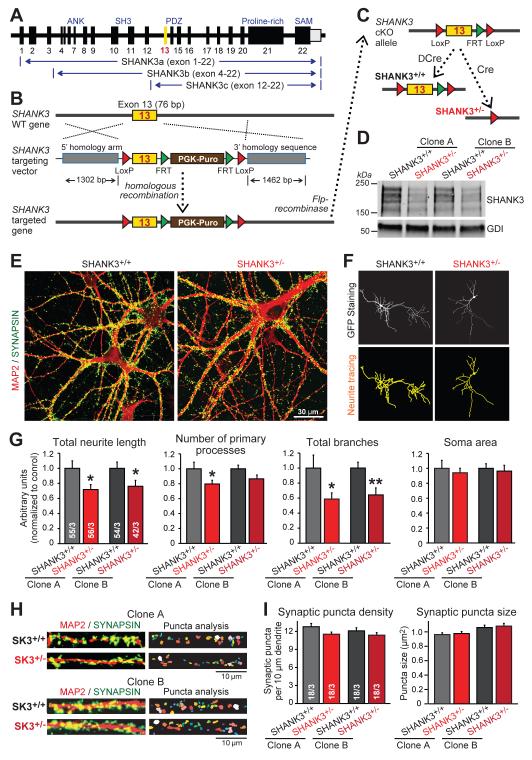
Figure 1. SHANK3 haploinsufficiency impairs dendritic development of human neurons.
A) Diagram of the SHANK3 gene and of the three major SHANK3 transcripts that are blocked by conditional deletion of exon 13 (yellow).
B & C) SHANK3 targeting strategy in human ES cells. Homologous recombination was mediated using recombinant adeno-associated virus (AAV) (B) and confirmed by PCR (Fig. S1B). The PGK-puromycin resistance cassette (brown box) was excised by Flp-recombinase to generate the conditional KO (cKO) allele (SHANK3+/cKO; panel C). SHANK3+/cKO ES cells were converted into human SHANK3+/+ or SHANK3+/− neurons by co-expression of Ngn2 with either mutant inactive Cre-recombinase (ΔCre) or active Cre-recombinase (Cre).
D) Reduction of SHANK3 protein levels in human SHANK3+/− neurons derived from two independent SHANK3+/cKO ES cell clones.
E) Representative images of SHANK3+/+ and SHANK3+/− neurons (day 21) labeled by double immunofluorescence for MAP2 (red) and synapsin (green).
F) Representative dendritic arborization analyses by MetaMorph software of SHANK3+/+ and SHANK3+/− neurons (sparsely transfected with EGFP).
G) SHANK3 haploinsufficiency impairs dendritic arborization (summary graphs of indicated parameters measured in matching SHANK3+/+ and SHANK3+/− neurons derived from independent SHANK3+/cKO ES cell clones; normalized to SHANK3+/+ controls).
H) Representative images of SHANK3+/+ and SHANK3+/− dendrites stained for MAP2 (red) and synapsin (green) for analysis of synaptic puncta by MetaMorph software.
I) Summary graphs of dendritic synaptic puncta density and size in SHANK3+/+ and SHANK3+/− neurons derived from two independent SHANK3+/cKO ES cell clones.
Data in G and I are means ± SEM. Numbers of cells/independent cultures analyzed are shown in the bars. Statistical significance was evaluated by Student’s t-test, (*, p<0.05; **, p<0.01). For additional data, see Figs. S1, S2.
We converted heterozygous conditionally mutant SHANK3+/cKO ES cells into neurons by forced expression of the transcription factor Ngn2, which generates a homogenous population of glutamatergic excitatory neurons with abundant synapse formation (23). We co-expressed active (Cre) or mutant Cre-recombinase (ΔCre) with Ngn2 during neuronal differentiation to produce precisely matching wild-type (ΔCre [SHANK3+/+]) and heterozygous mutant neurons (Cre [SHANK3+/−]; 24,25), using two independently targeted heterozygous clones of SHANK3+/cKO ES cells to control for clonal variation (Fig. 1C). Immunoblotting and polymerase chain reaction (PCR) demonstrated that the heterozygous SHANK3 KO decreased SHANK3 expression but left SHANK1 and SHANK2 expression unchanged (Fig. 1D, S1D, S1E, S2B, S3). No significant changes in other synaptic proteins were observed except for a decrease in PSD95 that is known to bind to SHANKs (Fig. S2C, S2D; 10-12).
SHANK3 haploinsufficiency impairs dendritic arborization, intrinsic electrical properties, and synaptic transmission in human neurons
Human SHANK3+/− neurons exhibited a typical neuronal morphology with abundant synapse formation (Fig. 1E). When we quantified the morphological features of matching SHANK3+/+ and SHANK3+/− neurons derived from independently derived ES cell clones, we observed that SHANK3 haploinsufficiency significantly decreased the length and branching of neurites, but had no effect on the density or size of synapsin-positive synapses (Fig. 1F-1I).
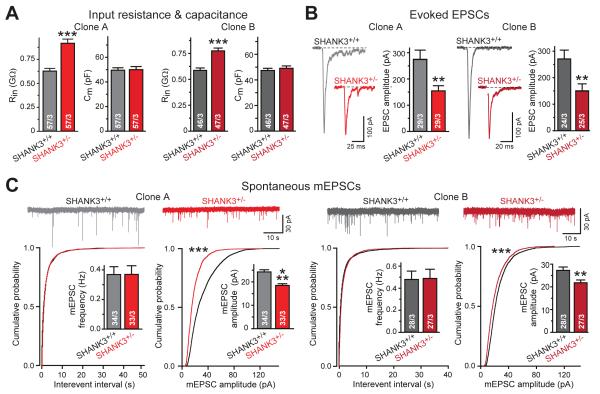
Next, we performed whole-cell patch-clamp recordings from matching SHANK3+/+ and SHANK3+/− neurons. SHANK3 haploinsufficiency caused a large increase (~25-33%) in input resistance without a change in capacitance (Fig. 2A). Moreover, SHANK3+/− neurons exhibited a major decrease in evoked excitatory postsynaptic currents (EPSCs; ~40-50% decrease) and in the amplitude of spontaneous miniature EPSCs (mEPSCs; ~25% decrease) but not in other mEPSC parameters (Fig. 2B, 2C, S2E). Overall, these results resemble those obtained with Phelan-McDermid neurons (9), supporting the notion that despite the large genomic deletion present in Phelan-McDermid neurons, SHANK3 haploinsufficiency may account for most Phelan-McDermid syndrome phenotypes.
Figure 2. SHANK3 haploinsufficiency increases electrical input resistance of human neurons and decreases overall synaptic strength.
A) Input resistance (Rin) but not capacitance (Cm) is increased in SHANK3+/− neurons (summary graphs from matching human SHANK3+/+ and SHANK3+/− neurons derived from two independent SHANK3+/cKO ES cell clones).
B) Evoked synaptic transmission is decreased in SHANK3+/− human neurons (representative EPSC traces (left) and EPSC amplitude summary graphs (right) for independent sets of neurons).
C) Amplitudes but not frequencies of spontaneous mEPSCs (monitored in 1 μM tetrodotoxin) are impaired in SHANK3+/− neurons (top, representative traces; bottom, cumulative plots and summary graphs of the mEPSC frequency (left) and amplitude (right)).
Data are means ± SEM. Numbers of cells/cultures analyzed are shown in bars. Statistical significance was evaluated by either Student’s t-test (bar graphs) or Kolmogorov-Smirnov-test (cumulative probability plots), (**, p<0.01; ***, p<0.001).
Homozygous SHANK3 deletion aggravates SHANK3 haploinsufficiency phenotype
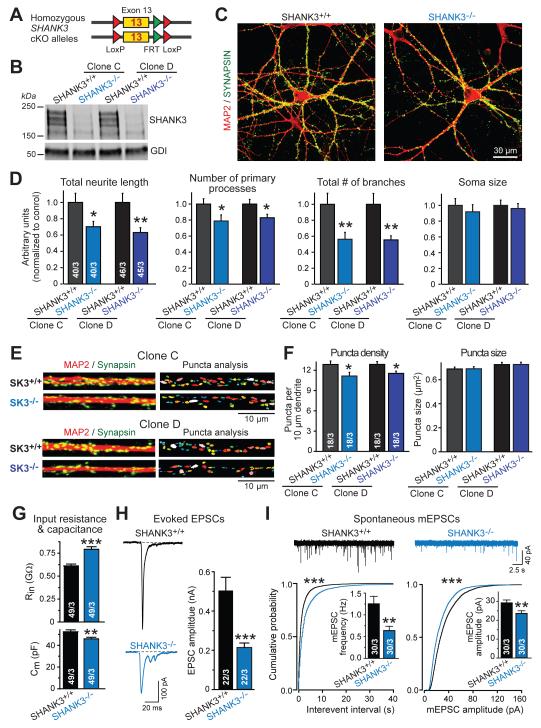
To inquire whether homozygous SHANK3 mutations produce phenotypes similar to those of heterozygous SHANK3 mutations, we generated mutant H1 ES cells with homozygous conditional SHANK3 mutations (22), and converted them into matching SHANK3+/+ or SHANK3−/− neurons by co-expression of Ngn2 and inactive or active Cre-recombinase, respectively (Fig. 3A, S4). Immunoblotting confirmed that the major SHANK3 protein isoforms were no longer expressed in SHANK3−/− neurons (Fig. 3B, S4E). Quantitative analyses uncovered a similar, but more severe phenotype in SHANK3−/− neurons than in SHANK3+/− neurons, with a significant decrease in dendritic arborization and in synapse density (Fig. 3C-3F, S5). Electrophysiologically, SHANK3−/− neurons also exhibited a large increase in input resistance, a massive decrease in evoked EPSC amplitude, and a significant decrease in mEPSC amplitude similar to SHANK3+/− neurons (Fig. 3G-3I). In addition, SHANK3−/− neurons displayed a decrease in capacitance and a notable decline in mEPSC frequency (Fig. 3G, 3I).
Figure 3. Homozygous SHANK3 deletion aggravates SHANK3 haploinsufficiency phenotype.
A) Diagram of homozygous SHANK3cKO/cKO alleles (see Supplemental Information for details).
B) SHANK3−/− neurons lack major SHANK3 proteins. Images depict immunoblots of matching SHANK3+/+ and SHANK3−/− neurons derived from two independent SHANK3cKO/cKO ES cell clones.
C) Representative images of matching SHANK3+/+ and SHANK3−/− neurons (day 21) stained by double immunofluorescence for MAP2 (red) and synapsin (green).
D) Homozygous SHANK3 deletion severely impairs dendritic arborization (summary graphs of indicated parameter [normalized to SHANK3+/+ controls] measured in matching SHANK3+/+ and SHANK3−/− neurons derived from two independent SHANK3cKO/cKO ES cell clones).
E) Representative images of dendrites stained for MAP2 (red) and synapsin (green) for analysis of synaptic puncta in SHANK3+/+ and SHANK3−/− neurons.
F) Homozygous SHANK3 deletion reduces synapse density (summary graphs of synaptic puncta density and size on proximal dendrites of isogenic SHANK3+/+ and SHANK3−/− neurons from two independent SHANK3cKO/cKO ES cell clones).
G) Homozygous SHANK3 deletion increases neuronal input resistance (Rin, top) and decreases capacitance (Cm, bottom).
H) Homozygous SHANK3 deletion reduces evoked EPSC amplitudes (left, representative traces; right, summary graphs of EPSC amplitudes).
I) Homozygous SHANK3 deletion decreases the frequency and amplitude of mEPSCs (top, representative traces; bottom, cumulative plots and summary graphs of mEPSC interevent intervals and frequency (bottom left) or mEPSC amplitudes (bottom right)).
Data in bar diagrams are means ± SEM. Numbers of cells/cultures analyzed are shown in bars. Statistical significance was evaluated by Student’s t-test (bar graphs) or Kolmogorov-Smirnov-test (cumulative probability plots), (*, p<0.05; **, p<0.01; ***, p<0.001). For additional data, see Fig. S3-S5.
Viewed together, our results suggest that conditional heterozygous and homozygous deletions of SHANK3 produce similar phenotypes in human neurons. Although the SHANK3-mutant neurons clearly exhibit synaptic impairments, their increased input resistance and decreased dendritic arborization cannot be readily explained in terms of a synaptic change, prompting us to search for other pathogenetic mechanisms.
Heterozygous and homozygous SHANK3 mutations impair Ih-currents
The input resistance of neurons depends at least in part on their ionic conductances and ion channels. Thus, we first asked whether changes in voltage-gated Na+-channels or K+-channels (delayed rectifier) which generate action potentials and determine neuronal excitability could be responsible for the increased input resistance of SHANK3-mutant neurons, prompted in part by binding of SHANK3 to K+-channels (26). However, we detected no changes in Na+- or K+-currents in SHANK3-mutant neurons (Fig. S6). We next tested whether decreased ionotropic glutamate receptor activation caused by impaired synaptic transmission in SHANK3-mutant neurons might be responsible. However, blocking ionotropic glutamate receptors had no effect on input resistance in SHANK3+/+ or SHANK3+/− neurons (Fig. 4A).
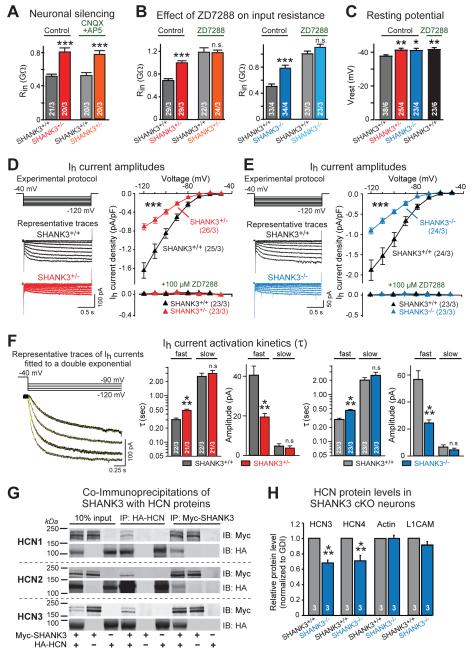
Figure 4. SHANK3 deletions impair Ih-currents in human neurons, and SHANK3 proteins interact with HCN channels.
A) Neuronal silencing by blocking glutamate receptors with CNQX (20 μM) and AP5 (50 μM) does not alter neuronal input resistance (Rin).
B) Blocking Ih-currents with ZD7288 (100 μM) increases the input resistance (Rin) of wild-type neurons, abolishing the difference between wild-type and mutant neurons.
C) SHANK3+/− and SHANK3−/− neurons exhibit a more negative resting potential (Vrest) than SHANK3+/+ neurons; inhibition of Ih-currents in SHANK3+/+ neurons with ZD7288 (5 μM) abolishes the difference.
D & E) SHANK3+/− (D) and SHANK3−/− neurons (E) exhibit decreased Ih-current amplitudes compared to matching SHANK3+/+ neurons (left, experimental protocol and sample traces; right, current/voltage relation of Ih-currents, which are validated by inhibition with ZD7288).
F) Kinetics of Ih-current activation is impaired in SHANK3+/− and SHANK3−/− neurons (left, representative capacity- and leak current-subtracted Ih-current recordings fitted with a double-exponential function [yellow line superimposed on traces]; center and right, summary graphs of time constants and amplitudes of fast and slow components of Ih-current activation at a test potential of −120 mV).
G) SHANK3 protein binds to HCN-channels mediating Ih-currents. Representative immunoblots document co-immunoprecipitation of Myc-tagged SHANK3 with HA-tagged HCN1, HCN2, and HCN3 proteins co-expressed in transfected HEK293T cells; cells expressing only one or the other protein serve as negative controls.
H) Levels of endogenous HCN3 and HCN4 proteins are decreased in SHANK3−/− neurons as determined by quantitative immunoblotting (for representative blots, see Fig. S10).
Data are means ± SEM. Numbers of cells/cultures analyzed are shown in bars or brackets. Statistical significance was evaluated by Student’s t-test (bar graphs in F, H), or two-way ANOVA (bar graphs in A-C) and two-way repeated measure ANOVA (I-V plots in D, E) followed by Bonferroni’s post hoc test, (*, p<0.05; **, p<0.01; ***, p<0.001).
A third candidate for an impaired membrane conductance as a cause of the increased input resistance are Ih-currents that are mediated by HCN-channels. In mammals, HCN-channels are encoded by four genes (HCN1-HCN4) and are expressed, like SHANKs, in neuronal and non-neuronal cells (27-30; Fig. S7). HCN-channels mediate hyperpolarization-activated Ih-currents that depolarize membranes towards the action-potential threshold and reduce membrane resistance. Ih-currents control neuronal excitability, membrane resting potentials, dendritic integration of synaptic potentials, and rhythmic oscillation of neurons (29,30). Giving their multifaceted functions, impairments of Ih-currents can have profound consequences for neuronal network activity (e.g., see 31-33).
Strikingly, we found that the Ih-current inhibitor ZD7288 (34,35) increased the input resistance of wild-type neurons dramatically but had only a small effect on SHANK3-mutant neurons, thereby abolishing the difference in input resistance between wild-type and SHANK3-mutant neurons (Fig. 4B). Moreover, SHANK3-deficient neurons displayed an increased resting membrane potential, and addition of ZD7288 to wild-type neurons increased their resting potential to that of SHANK3-deficient neurons (Fig. 4C).
Because these results suggest that the changed electrical properties of SHANK3-mutant neurons may be due to an impairment of Ih-currents, we next directly measured Ih-currents in precisely matching SHANK3+/+ and SHANK3+/− or SHANK3−/− neurons using whole-cell recordings (Fig. 4D, 4E, S8). Ih-currents were readily activated by brief (2 sec) hyperpolarizing voltage steps, exhibited a reversal potential of −32 mV, and were inhibited by extracellular Cs+ and ZD7288 (Fig. S8). Compared to SHANK3+/+ neurons, both SHANK3+/− and SHANK3−/− neurons exhibited a severe decrease in Ih-current density (Fig. 4D, 4E) and a deceleration of Ih-current activation (Fig. 4F). However, other basic properties of Ih-currents, such as their half-maximal activation potential (V50), were not significantly altered (Fig. S8). Furthermore, depolarizing voltage sag responses that are evoked by brief injections of hyperpolarizing currents in current-clamp mode (36, 37), were also significantly impaired in SHANK3+/− and SHANK3−/− neurons; these impairments again were occluded by ZD7288, suggesting that these phenotypes are also due to a specific alteration in Ih-channel function (Fig. S8).
Viewed together, these data suggest that SHANK3 mutations cause Ih-channel dysfunction in human neurons. To test the possibility that SHANK3 interacts with HCN-channels, we co-expressed SHANK3 with HCN1, HCN2, or HCN3 channels in HEK293T cells. Co-immunoprecipitations revealed that SHANK3 bound to all three isoforms of HCN-channels in this assay (Fig. 4G). Mapping of the interaction domains using glutathione S-transferase (GST)-pulldowns suggested that the SHANK3 ankyrin repeats directly bind to HCN-channels (Fig. S9). Finally, measurements of the levels of two human HCN isoforms to which antibodies were available, HCN3 and HCN4, demonstrated that the SHANK3 deletion significantly decreased the levels of endogenous HCN proteins in human neurons, consistent with a direct interaction (Fig. 4H, S10).
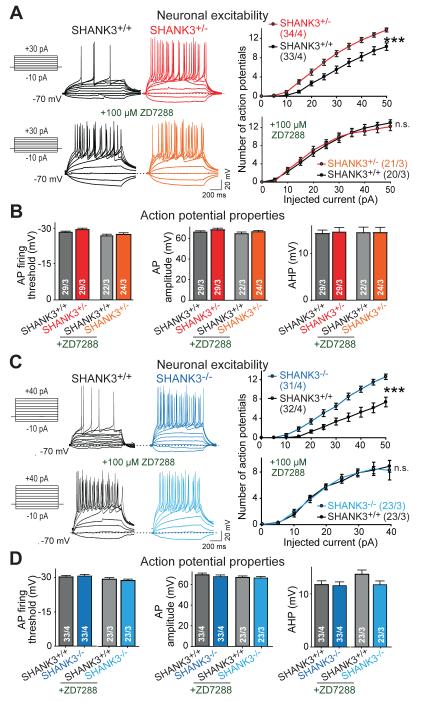
Because Ih-currents are major determinants of neuronal excitability (29, 30), we examined the effects of SHANK3 mutations on action potential generation (Fig. 5). Consistent with the increased input resistance, SHANK3+/− and SHANK3−/− neurons fired significantly more action potentials than SHANK3+/+ neurons in response to depolarizing current injections; this phenotype was abolished by addition of ZD7288 and thus was Ih-current dependent (Fig. 5). Moreover, because sustained rhythmic oscillations are a hallmark of neuronal circuits in various brain regions that overlap with highly enriched regions of SHANK3 expression (14,29,38), we monitored the spontaneous spiking activity of SHANK3-mutant neurons. When neurons were held at the resting membrane potential, a large proportion of wild-type cells (~60%) fired action potentials spontaneously and regularly (Fig. S11). In contrast, SHANK3-mutant neurons fired fewer action potentials; again, this phenotype was reversed by addition of the Ih-current inhibitor ZD7288 (Fig. S11).
Figure 5. SHANK3 mutations render neurons hyperexcitable by impairing Ih-currents.
A) SHANK3+/− mutant neurons reach action potential (AP) firing threshold earlier than matching SHANK3+/+ human neurons and exhibit a steeper input-output relationship as assessed by the number of APs elicited by increasing current injections (from −10 pA to +50 pA, 1 s, 5 pA increments) during current-clamp recordings. Ih-channel channel inhibition with ZD7288 abolishes the difference (left, experimental protocol and representative traces; right, plots of the mean action potential number vs. injected current).
B) Summary graphs of active electrical properties of matching SHANK3+/+ and SHANK3+/− neurons (measured without or with ZD7288 application). From left to right: AP firing threshold, AP amplitude, and AP after-hyperpolarization amplitude (AHP).
C) Same as A, but for matching SHANK3−/− and SHANK3+/+ neurons.
D) Same as B, but for matching SHANK3−/− and SHANK3+/+ neurons.
Data are means ± SEM. Numbers of cells/cultures analyzed are shown in bars or brackets. Statistical significance was evaluated by two-way ANOVA (bar graphs), or two-way repeated measure ANOVA followed by Bonferroni’s post hoc test (input-output plots), (n.s., not significant; *, p<0.05; **, p<0.01; ***, p<0.001).
Shank3 deletions impair Ih-currents in developing mouse neurons
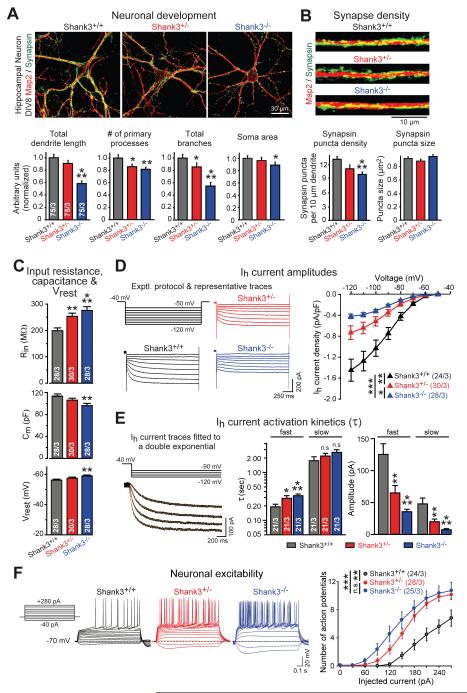
To test whether Shank3-mutant mice also exhibit an impairment in Ih-currents, we cultured hippocampal neurons from newborn littermate Shank3+/+, Shank3+/−, and Shank3−/− mice. In the Shank3-mutant mice, exons encoding the PDZ-domain of SHANK3 are deleted similar to our conditionally SHANK3-mutant human neurons (14). Strikingly, we observed an overall very similar phenotype in developing mouse neurons as in human neurons (Fig. 6, S12, S13). Specifically, quantitative imaging revealed that at 8 days in culture (DIV8), homo- but not heterozygous Shank3 mutations caused a significant impairment in dendritic arborization and synapse density similar to human SHANK3 deletions (Fig. 6A, 6B, S12). Electrophysiological recordings showed that although the total input resistance of mouse neurons was lower than that of human neurons, both hetero- and homozygous Shank3 mutations produced a dramatic increase in input resistance (Fig. 6C). Homozygous Shank3 deletions also significantly decreased cell capacitance and increased the resting membrane potential similar to human mutations. We then directly measured Ih-currents, and detected a massive impairment in both hetero- and homozygous Shank3-deficient mouse neurons (Fig. 6D, 6E, S12). The Ih-current amplitude was decreased more than two-fold by Shank3 mutations, the Ih-current voltage sag was reduced, and the Ih-current activation kinetics was significantly decelerated. In all of these phenotypes, the homozygous mutation was more deleterious than the heterozygous mutation. These changes closely resemble those observed in human SHANK3-deficient neurons, and also led to a markedly increased excitability in mouse Shank3-deficient neurons (Fig. 6F).
Figure 6. Developing hippocampal neurons from Shank3 knockout mice reproduce phenotype of human SHANK3-mutant neurons.
A & B) Dendritic arborization (A) and synapse formation (B) are impaired in developing Shank3-deficient mouse neurons (top, representative images of hippocampal neurons (A) and dendritic segments (B); bottom, summary graphs of the indicated dendritic, cellular and synaptic parameters). Neurons cultured from littermate Shank3+/+, Shank3+/− or Shank3−/− mice were stained at 8 days in vitro (DIV8) for MAP2 (red) and synapsin (green).
C) Hetero- and homozygous Shank3 deletions increase neuronal input resistance (top), decrease cell capacitance (center), and enhance the resting membrane potential (bottom) in hippocampal neurons cultured from littermate Shank3+/+, Shank3+/−, and Shank3−/− mice and analyzed at DIV8-9.
D) Hetero- and homozygous Shank3 deletions impair neuronal Ih-currents in hippocampal neurons at DIV8-9 (left, experimental protocol and sample traces; right, summary graph of the current/voltage relation).
E) Hetero- and homozygous Shank3 deletions decelerate Ih-current activation (left top, experimental protocol; left bottom, capacitance- and leak current-subtracted representative traces fitted with the sum of two exponential functions shown superimposed on the traces in yellow; right, summary graphs of activation time constants (τ) and component amplitudes obtained at a test potential of −120 mV).
F) Hetero- and homozygous Shank3 deletions render hippocampal neurons hyperexcitable (left, experimental protocol and representative traces of stepwise depolarizing current injections; right, summary plots of the action potential number vs. injected current during current-clamp recordings of neurons analyzed at DIV8-9).
Data are means ± SEM. Numbers of cells/cultures analyzed are shown in bars or brackets. Statistical significance was evaluated by one-way ANOVA followed by Tukey’s post hoc test (bar graphs), or two-way repeated measure ANOVA followed by Bonferroni’s post hoc test (I-V plot), (*, p<0.05; **, p<0.01; ***, p<0.001). For additional data, see Fig. S12-S13.
Chronic Ih-current inhibition impairs neuronal development similar to SHANK3 mutations
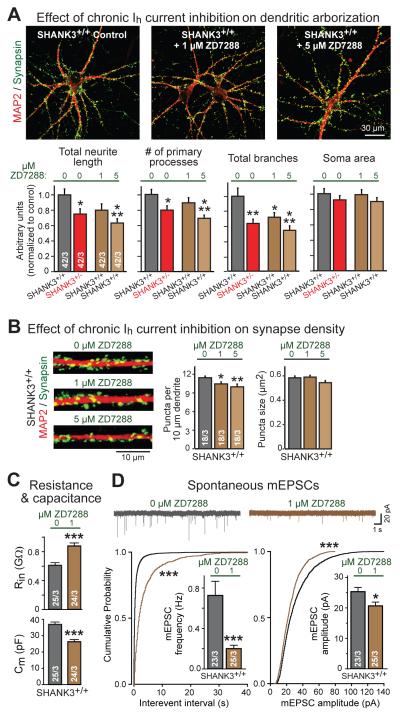
A decrease in Ih-currents by SHANK3 mutations likely accounts for the electrophysiological changes of SHANK3-mutant neurons, but can it also account, at least in part, for their morphological and synaptic changes? To investigate this question, we chronically inhibited Ih-channels in wild-type neurons by continuous application of low concentrations of ZD7288 (1 and 5 μM) during neuronal differentiation.
Chronic inhibition of Ih-channels dramatically impaired neuronal morphology in a manner indistinguishable from that of the SHANK3 haploinsufficiency (Fig. 7A, S14). Specifically, chronic application of ZD7288 caused a dose-dependent decrease in neurite outgrowth, number of primary processes, and dendritic branching. Moreover, ZD7288 decreased the synapse density, again suggesting that major phenotypes of SHANK3 mutations may represent indirect effects of an Ih-current impairment (Fig. 7B). In addition, chronic inhibition of Ih-channels in human neurons increased the neuronal input resistance and impaired spontaneous synaptic transmission, measured after washout of ZD7288 used to inhibit Ih-currents (Fig. 7C, 7D). These results suggest that long-term inhibition of Ih-currents has dramatic effects on multiple facets of neuronal development.
Figure 7. Chronic inhibition of Ih-currents in human neurons mimics SHANK3 haploinsufficiency phenotype.
A) Chronic partial inhibition of Ih-currents in wild-type SHANK3+/+ neurons causes dendritic arborization defects similar to SHANK3+/− haploinsufficiency. Matching SHANK3+/+ and SHANK3+/− neurons were differentiated from SHANK3+/cKO ES cells; SHANK3+/+ neurons were treated with low-dose ZD7288 (1 μM or 5 μM) from day 3-21 of neural induction (top, representative images of neurons double-labeled for MAP2 and synapsin; bottom, summary graphs of indicated parameters normalized to the untreated SHANK3+/+ control).
B) Chronic inhibition of Ih-currents in SHANK3+/+ neurons decreases synapse numbers. Experiments were performed as described for panel A (left, representative images of dendritic segments double labeled for MAP2 and synapsin; right, summary graphs of density and size of synaptic puncta).
C) Chronic inhibition of Ih-currents in SHANK3+/+ neurons significantly increases input resistance (Rin, top) and decreases cell capacitance (Cm, bottom). Experiments were performed as described for panel A; recordings were performed after washout of ZD7288.
D) Chronic inhibition of Ih-currents in SHANK3+/+ neurons with ZD7288 (1 μM) reduces the frequency and amplitude of spontaneous mEPSCs (top, representative traces; bottom left, summary plots of the interevent interval and summary graph of the mEPSC frequency; bottom right, same for the mEPSC amplitude). Experiments were performed as described for panel A; recordings were performed after washout of ZD7288.
Data are means ± SEM. Numbers of cells/cultures analyzed are shown in the bars. Statistical significance was evaluated by Student’s t-test (bar graphs in C and D), one-way ANOVA followed by Tukey’s post hoc test (bar graphs in B), or two-way ANOVA followed by Bonferroni’s post hoc test (bar graphs in A) or Kolmogorov-Smirnov-test (cumulative probability plots in D), (*, p<0.05; **, p<0.01; ***, p<0.001).
Summary
Many mutations are thought to predispose to idiopathic ASDs by causing primary impairments in synaptic transmission (1-5). Our data show that SHANK3 haploinsufficiency impairs synaptic function, but also demonstrate that SHANK3 haploinsufficiency decreases Ih-channel function as a primary impairment, which in turn produces major changes in intrinsic neuronal properties and secondarily affects synaptic function. Indeed, the fact that several salient phenotypes of hetero- and homozygous SHANK3-mutant human neurons were reproduced by pharmacologic inhibition of Ih-channels suggests that Ih-channel dysfunction is a major effect of SHANK3 haploinsufficiency. Moreover, the very similar phenotypes produced by Shank3 mutations in developing mouse neurons, which also severely impaired Ih-currents, indicates a general role for SHANK3 in scaffolding HCN-channels during neuronal development at a period coinciding with the manifestation of ASDs. A role for SHANK3 as a scaffolding protein for HCN-channels is plausible given the broad expression of SHANK3 in nonneuronal cells that also express HCN-channels, and SHANK3 may function to enrich HCN-channels at postsynaptic sites together with other proteins (31,32). Thus, we would like to suggest that an Ih-current impairment is a major pathogenetic force of SHANK3 mutations in predisposing to ASDs and in Phelan-McDermid syndrome. Changes in Ih-channel function can conceivably be influenced pharmacologically, suggesting that pharmacological manipulation of Ih-channels may be therapeutically beneficial.
HCN-channel mutations have been linked clinically with human neurological disorders including epilepsy, sleep disorder and impaired learning, which are commonly associated with enhanced neuron firing and/or aberrant neuronal firing patterns and are also observed in mice with HCN-channel mutations (29,30,38-41). The symptoms resulting from impaired HCN-channels agree well with the hypothesized involvement of SHANK3 deletions in ASDs and Phelan-McDermid syndrome that are also commonly associated with intellectual disability, impaired learning and memory, and epilepsy (1-5). Therefore, our data collectively suggest that impairment of HCN-channel function may contribute to the manifestations of ASDs in patients with SHANK3 mutations and Phelan-McDermid syndrome.
Methods Summary
See the supplementary materials for full details of the materials and methods (22).
Generation and analysis of human ES cells with heterozygous and homozygous SHANK3 conditional KO (cKO) alleles
SHANK3–mutant ES cells were generated from H1 ES cells (passage 40; WiCell, http://www.wicell.org) using recombinant adeno-associated virus- (AAV−) mediated homologous recombination (Fig. 1A; S1A)(24,25). AAVs used for gene targeting contained a puromycin-resistance cassette surrounded by FRT sites (for Flp-recombinase mediated deletion). The cassette was inserted into the 3′ intron of exon 13 (76 bp) of the human SHANK3 gene, exon 13 was flanked by LoxP sites (for Cre-recombinase mediated deletion), and 5′ and 3′ DNA homology arms were added. The same gene targeting template was also used in the generation of homozygous SHANK3 cKO ES cells, except that a CRISPR/Cas9 targeting method was used to specifically target the second wild-type allele of the SHANK3 gene and not the already targeted first SHANK3 cKO allele. Heterozygous and homozygous SHANK3 cKO ES cell clone mutations were confirmed by PCR, and the puromycin-resistance cassette was removed by Flp-recombinase. To generate isogenic control and SHANK3-mutant human neurons, hetero- or homozygous SHANK3 cKO ES cells were transdifferentiated to neurons using forced expression of Ngn2 as described (23). Lentiviruses encoding active (Cre) or inactive Cre-recombinase (ΔCre) were applied at one day before induction of neuron differentiation. Neurons were analyzed at day 21-23 in most experiments. For the experiments of chronic Ih-current inhibition with low-dose ZD7288 (Fig. 7), wild-type human neurons were treated with 1 μM or 5 μM ZD7288 from day 3 to day 21.
Generation and analysis of Shank3-mutant hippocampal mouse neurons
Breedings of heterozygous Shank3-mutant mice with deletion of the PDZ domain (exon 13-16; ref. 14; B6.129-Shank3tm2Gfng/J; Jackson Labs stock No. 017688) were used to produce littermate wild-type, heterozygous and homozygous Shank3-mutant mice. Hippocampal neurons were cultured from newborn mice (42,43), and analyzed as immature developing neurons at 8-9 days in vitro (DIV8-9). Mouse genotyping was performed by PCR using the Jackson Labs protocol.
Morphological analyses
Dendritic arborizations were analyzed in neurons that were sparsely transfected with an EGFP to obtain fluorescent images of individual neurons. Confocal images were analyzed unbiasedly using the “neurite outgrowth” application on MetaMorph software. For synapse morphology analyses, fixed neurons were stained by double immunofluorescence with antibodies to MAP2 (to stain for dendrites) and synapsin (to label pre-synaptic terminals) or HOMER1 (to label post-synaptic specializations), and images were again quantified using MetaMorph software (Molecular Devices) (44).
Electrophysiological recordings
Whole-cell patch-clamp recordings were performed essentially as described (23,42). Excitatory postsynaptic currents (EPSCs) were pharmacologically isolated with picrotoxin (PTX, 50 μM) and recorded at a −70 mV holding potential in voltage-clamp mode in response to extracellular stimulation with a concentric bipolar electrode (43). Spontaneous miniature EPSCs (mEPSCs) were monitored in the presence of tetrodotoxin (1 μM). Recordings of the intrinsic and active membrane properties were generally recorded in human neurons in the presence of 50 μM PTX (unless otherwise stated), and in hippocampal mouse neurons in the presence of CNQX (6-cyano-7-nitroquinoxaline-2,3-dione; 20 μM), AP5 (2-Amino-5-phosphonopentanoic acid; 50 μM) and PTX (50 μM).
Input resistance (Rin) was calculated as the slope of linear fits of current-voltage plots generated from a series of increasing current injection steps in current-clamp. Ih-channel activity was measured in voltage-clamp mode as the amplitude of the slowly activating inward current component elicited by 2 s voltage steps from −50 mV to −120 mV in 10 mV increments from a holding potential of −40 mV with 2 mM 4-Aminopyridine (4-AP) and 0.5 mM BaCl2 in the bath solution. Depolarizing voltage-sag responses were evoked by brief injections of hyperpolarizing currents in current-clamp mode. Voltage-dependent Na+ (INa) and K+ (IKD) currents were recorded in voltage-clamp mode at a holding potential of −70 mV in the presence of 2 mM 4-AP; voltage steps ranging from −90 mV to +40 mV were delivered at 10 mV increments. Intrinsic action potential (AP) firing properties of neurons were recorded in current-clamp mode. To assess neuronal excitability, first minimal currents were introduced to hold membrane potential around −65 to −70 mV, then increasing amounts of depolarizing currents were injected for 1s in stepwise manner. For spontaneous AP firing, cells were held at their resting membrane potential (Vrest), no current was injected. The Vrest obtained during spontaneous firing was determined as the mean steady-state voltage recorded during interspike intervals. All experiments were performed at room temperature.
Protein-protein interaction analyses were performed by co-immunoprecipitation of Myc-tagged full length Shank3 protein with HA-tagged full length HCN proteins expressed in HEK293T cells. Co-immunoprecipitations of truncated Shank3 proteins with full-length HCN proteins were also performed to map the responsible interaction domain on Shank3. In order to further demonstrate the interaction between Shank3 and HCN proteins, a GST pull-down assay was performed using purified Shank3 ankyrin repeats domain and full length HCN1 protein.
Immunoblotting
All immunoblots were visualized by fluorescently labeled secondary antibodies, and quantified on Odyssey CLx Infrared Imager and Odyssey software (LICOR Biosciences). Signals were normalized to human GDI as the neuronal loading control.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Y. Zhang, S. Maxeiner, and S.J. Lee for advice, Dr. G. Feng (MIT) for reagents, and Dr. V. Sebastiano (Stanford) for sharing instruments. This work was supported by grants from the NIH (MH092931 to M.W.; NS077906 to T.C.S.) and a postdoctoral fellowship from Vetenskapscrdet, Sweden (to S.C.B.).
Footnotes
AUTHOR CONTRIBUTIONS
F.Y. performed the ES cell, molecular biology, expression, and cell-biology experiments, T.D. the electrophysiological experiments, S.C.B. the protein chemistry experiments, C.P. the immunoblotting experiments, and C.H.P. the gene-expression experiments. All authors planned the experiments, analyzed data, and edited the paper written by T.C.S.
REFERENCES and NOTES
- 1.Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genetics. 2014;10:e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betancur C, Buxbaum JD. SHANK3 haploinsufficiency: a "common" but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Molecular Autism. 2013;4:17. doi: 10.1186/2040-2392-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosti RO, Sadek AA, Vaux KK, Gleeson JG. The genetic landscape of autism spectrum disorders. Dev. Med. Child Neurology. 2014;56:12–18. doi: 10.1111/dmcn.12278. [DOI] [PubMed] [Google Scholar]
- 4.Grabrucker AM, Schmeisser MJ, Schoen M, Boeckers TM. Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 2011;21:594–603. doi: 10.1016/j.tcb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Carbonetto S. A blueprint for research on Shankopathies: a view from research on autism spectrum disorder. Dev. Neurobiol. 2014;74:85–112. doi: 10.1002/dneu.22150. [DOI] [PubMed] [Google Scholar]
- 6.Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilmatre A, Huguet G, Delorme R, Bourgeron T. The emerging role of SHANK genes in neuropsychiatric disorders. Dev. Neurobiol. 2014;74:113–122. doi: 10.1002/dneu.22128. [DOI] [PubMed] [Google Scholar]
- 8.Han K, Holder JL, Jr., Schaaf CP, Lu H, Chen H, et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–77. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 11.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 12.Sheng M, Kim E. The Shank family of scaffold proteins. J. Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 13.Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Molecular Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peca J, Feliciano C, Ting JT, Wang W, Wells MF, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speed HE, Kouser M, Xuan Z, Reimers JM, Ochoa CF, et al. Autism-Associated Insertion Mutation (InsG) of Shank3 Exon 21 Causes Impaired Synaptic Transmission and Behavioral Deficits. J. Neurosci. 2015;35:9648–9665. doi: 10.1523/JNEUROSCI.3125-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, et al. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J. Neurosci. 2013;33:18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang M, Bozdagi O, Scattoni ML, Wohr M, Roullet FI, et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J. Neurosci. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drapeau E, Dorr NP, Elder GA, Buxbaum JD. Absence of strong strain effects in behavioral analyses of Shank3-deficient mice. Disease Models & Mechanisms. 2014;7:667–681. doi: 10.1242/dmm.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Human Mol. Genetics. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Chung C, Ha S, Lee D, Kim DY, et al. Shank3-mutant mice lacking exon 9 show altered excitation/inhibition balance, enhanced rearing, and spatial memory deficit. Front. Cell. Neurosci. 2015;9:94. doi: 10.3389/fncel.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uppal N, Puri R, Yuk F, Janssen WG, Bozdagi-Gunal O, et al. Ultrastructural analyses in the hippocampus CA1 field in Shank3-deficient mice. Molecular Autism. 2015;6:41. doi: 10.1186/s13229-015-0036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.For experimental procedures, see Supplementary Materials and Methods.
- 23.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pak C, Danko T, Zhang Y, Aoto J, Anderson G, et al. Human neuropsychiatric disease modeling using condition deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1. Cell Stem Cell. 2015;17:316–328. doi: 10.1016/j.stem.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patzke C, Han Y, Covy J, Yi F, Maxeiner S, Wernig M, Sudhof TC. Analysis of conditional heterozygous STXBP1 mutations in human neurons. The Journal of Clinical Investigation. 2015;125:3560–3571. doi: 10.1172/JCI78612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proepper C, Putz S, Russell R, Boeckers TM, Liebau S. The Kvbeta2 subunit of voltage-gated potassium channels is interacting with ProSAP2/Shank3 in the PSD. Neuroscience. 2014;261:133–143. doi: 10.1016/j.neuroscience.2013.10.045. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393:587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Wang J, Siegelbaum SA. Properties of hyperpolarization-activated pacemaker current defined by coassembly of HCN1 and HCN2 subunits and basal modulation by cyclic nucleotide. J. Gen. Phys. 2001;117:491–504. doi: 10.1085/jgp.117.5.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benarroch EE. HCN channels: function and clinical implications. Neurology. 2013;80:304–310. doi: 10.1212/WNL.0b013e31827dec42. [DOI] [PubMed] [Google Scholar]
- 30.Postea O, Biel M. Exploring HCN channels as novel drug targets. Nature Reviews Drug Discovery. 2011;10:903–914. doi: 10.1038/nrd3576. [DOI] [PubMed] [Google Scholar]
- 31.Nolan MF, Malleret G, Lee KH, Gibbs E, Dudman JT, et al. The hyperpolarization-activated HCN1 channel is important for motor learning and neuronal integration by cerebellar Purkinje cells. Cell. 2003;115:551–564. doi: 10.1016/s0092-8674(03)00884-5. [DOI] [PubMed] [Google Scholar]
- 32.Nolan MF, Malleret G, Dudman JT, Buhl DL, Santoro B, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119:719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 33.DiFrancesco JC, DiFrancesco D. Dysfunctional HCN ion channels in neurological diseases. Front. Cell. Neurosci. 2015;6:174. doi: 10.3389/fncel.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.BoSmith RE, Briggs I, Sturgess NC. Inhibitory actions of ZENECA ZD7288 on whole-cell hyperpolarization activated inward current (If) in guinea-pig dissociated sinoatrial node cells. British J. of Pharmacology. 1993;110:343–349. doi: 10.1111/j.1476-5381.1993.tb13815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin KS, Rothberg BS, Yellen G. Blocker state dependence and trapping in hyperpolarization-activated cation channels: evidence for an intracellular activation gate. J. Gen. Phys. 2001;117:91–101. doi: 10.1085/jgp.117.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCormick DA, Pape HC. Properties of a hyperpolarization-activated cation current and its role in rhythmic oscillation in thalamic relay neurones. J. Physiology. 1990;431:291–318. doi: 10.1113/jphysiol.1990.sp018331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon JS, Nerbonne JM. Hyperpolarization-activated currents in isolated superior colliculus-projecting neurons from rat visual cortex. J. Physiology. 1993;462:393–420. doi: 10.1113/jphysiol.1993.sp019561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis AS, Chetkovich DM. HCN channels in behavior and neurological disease: too hyper or not active enough? Mol. Cell. Neurosci. 2011;46:357–367. doi: 10.1016/j.mcn.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips AM, Kim T, Vargas E, Petrou S, Reid CA. Spike-and-wave discharge mediated reduction in hippocampal HCN1 channel function associates with learning deficits in a genetic mouse model of epilepsy. Neurobiology of Disease. 2014;64:30–35. doi: 10.1016/j.nbd.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Vaidya SP, Johnston D. Temporal synchrony and gamma-to-theta power conversion in the dendrites of CA1 pyramidal neurons. Nature Neuroscience. 2013;16:1812–1820. doi: 10.1038/nn.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, Gamo NJ, Yang Y, Jin LE, Wang XJ, et al. Neuronal basis of age-related working memory decline. Nature. 2011;476:210–213. doi: 10.1038/nature10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maximov A, Sudhof TC. Autonomous function of synaptotagmin 1 in triggering synchronous release independent of asynchronous release. Neuron. 2005;48:547–554. doi: 10.1016/j.neuron.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Maximov A, Pang ZP, Tervo DG, Sudhof TC. Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J Neurosci Methods. 2007;161:75–87. doi: 10.1016/j.jneumeth.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Anderson GR, Aoto J, Tabuchi K, Foldy C, Covy J, et al. β-Neurexins control neural circuits by regulating synaptic endocannabinoid signaling. Cell. 2015;162:593–606. doi: 10.1016/j.cell.2015.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W, Morishita W, Buckmaster PS, Pang ZP, Malenka RC, Sudhof TC. Distinct neuronal coding schemes in memory revealed by selective erasure of fast synchronous synaptic transmission. Neuron. 2012;73:990–1001. doi: 10.1016/j.neuron.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franke B, Figiel M, Engele J. CNS glia are targets for GDNF and neurturin. Histochemistry and cell biology. 1998;110:595–601. doi: 10.1007/s004180050322. [DOI] [PubMed] [Google Scholar]
- 47.Pang ZP, Cao P, Xu W, Sudhof TC. Calmodulin controls synaptic strength via presynaptic activation of calmodulin kinase II. The Journal of neuroscience. 2010;30:4132–4142. doi: 10.1523/JNEUROSCI.3129-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic acids research. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Sudhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18120–18125. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chevaleyre V, Castillo PE. Assessing the role of Ih channels in synaptic transmission and mossy fiber LTP. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9538–9543. doi: 10.1073/pnas.142213199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Liao L, Liu X, Luo F, Yang T, Li C. Is ZD7288 a selective blocker of hyperpolarization-activated cyclic nucleotide-gated channel currents? Channels (Austin) 2012;6:438–442. doi: 10.4161/chan.22209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maccaferri G, McBain CJ. The hyperpolarization-activated current (Ih) and its contribution to pacemaker activity in rat CA1 hippocampal stratum oriens-alveus interneurones. The Journal of physiology. 1996;497(Pt 1):119–130. doi: 10.1113/jphysiol.1996.sp021754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson RB, Siegelbaum SA. Hyperpolarization-activated cation currents: from molecules to physiological function. Annual review of physiology. 2003;65:453–480. doi: 10.1146/annurev.physiol.65.092101.142734. [DOI] [PubMed] [Google Scholar]
- 56.Wahl-Schott C, Biel M. HCN channels: structure, cellular regulation and physiological function. Cellular and molecular life sciences: CMLS. 2009;66:470–494. doi: 10.1007/s00018-008-8525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neuhoff H, Neu A, Liss B, Roeper J. I(h) channels contribute to the different functional properties of identified dopaminergic subpopulations in the midbrain. The Journal of neuroscience. 2002;22:1290–1302. doi: 10.1523/JNEUROSCI.22-04-01290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Xu Q, Bey AL, Lee Y, Jiang YH. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Molecular Autism. 2014;5:30. doi: 10.1186/2040-2392-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biel M, Wahl-Schott C, Michalakis S, Zong X. Hyperpolarization-activated cation channels: from genes to function. Physiological reviews. 2009;89:847–885. doi: 10.1152/physrev.00029.2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.