Abstract
The Chemistry Development Kit (CDK) is a freely available open-source Java library for Structural Chemo-and Bioinformatics. Its architecture and capabilities as well as the development as an open-source project by a team of international collaborators from academic and industrial institutions is described. The CDK provides methods for many common tasks in molecular informatics, including 2D and 3D rendering of chemical structures, I/O routines, SMILES parsing and generation, ring searches, isomorphism checking, structure diagram generation, etc. Application scenarios as well as access information for interested users and potential contributors are given.
1. INTRODUCTION
Whoever pursues the endeavor of creating a larger software package in chemoinformatics or computational chemistry from scratch will soon be confronted with the Syssiphus task of implementing the standard repertoire of chemoinformatical algorithms and components invented during the last 20 or 30 years. The obvious workaround for this problem are commercially available chemoinformatics libraries that have been developed by companies such as MDL Information Systems, Inc., Daylight Chemical Information Systems, Inc., Advanced Chemistry Development, and certainly many others. A scientist in an academic environment, however, often feels obliged to openly share his results with the scientific community. Using proprietary components for software development makes it impossible to do so.
Generally, scientific software is too often closed source, leaving the user with a black box performing magical operations. Perceived as being counterproductive for the overall scientific progress, this trend fortunately seems to change. Sharing of ideas and results within communities is probably the most central paradigm in science. By publishing his results a scientist allows his colleagues to verify and build upon his results, thereby advancing the particular field as a whole [If I have seen further it is by standing on the shoulders of giants. - Isaac Newton]. One of the motivations for such contributions, besides the pure scientific curiosity, is, of course, the gain of social recognition and reputation among his peers.
In recent years the ideas sketched above have been part of the open-source revolution that took place in the world of software development, most widely recognized through the great success of the free Unix-like operating system GNU/Linux, a collaborative work of many individuals and organizations, including the Free Software Foundation lead by Richard Stallman and the Finish computer science student Linus Torvalds who started the project. According to several essays on this subject, open-source software, for which, by definition, the source code is always freely available to the public,1 has a number of intriguing benefits.
Most importantly, if the community of users is large enough and everyone can look at the sources and change them, it should not take too long until a particular software error is found and fixed. “Given enough eyeballs, all bugs are shallow”, as Eric Raymond put it in his widely recognized essay “The Cathedral and the Bazaar”,2 in which he analyses the mechanisms and principles of the open source movement.
Further, other scientists can easily build on existing results. Credit can still be given in the appropriate form, because open-source software is by no means freeware or in the public domain. Quite the contrary, the package as a whole as well as each piece of source code is labeled with a clear copyright notice, stating the name of the copyright holder and the nature of the license. This copyright notice must not be removed. Additional comments, however, regarding the changes and improvements made by others can, of course, be added. Substantial improvements to an existing piece of code by someone other than the copyright holder will usually lead to something like team formation, including appropriate copyright changes. This is especially important for academic scientists, who need to be able to point out their contributions to a particular field.
Considering the virtues of open-source software on one hand and the scientific tradition on the other hand, we started the CDK project under terms of a liberal open-source license.3 We use SourceForge,4 a Web based open-source development platform, for coordinating the contributions from about 10 developers from about five different countries. A greater number of people have subscribed to the developers mailing list and either listen silently or contribute by making feature requests or critical comments. SourceForge provides all the tools which are generally considered to be indispensable components for coordinating the contributions from developers and users in larger software projects, as there are Webspace, mailing lists, bug trackers, software versioning systems, release managers, etc.
This article is not only to describe the CDK project in scientific and software-technological terms but also to promote the underlying development model. The authors think that these principles form a paradigm for scientific software development where scientists can truly exploit the benefits of the Internet for a distributed collaboration that would not have been possible in pre-Internet times.
We are explicitly not claiming to give a general overview of chemical open source software. This will form an article of its own. However, we will give a synopsis on open source Java software in the following section instead.
The interested reader is cordially invited to visit the CDK project pages at http://cdk.sourceforge.net, get in touch with the developers, make use of the CDK package, and ultimately to extend its functionality.
2. OPEN SOURCE JAVA SOFTWARE IN CHEMISTRY
A number of libraries written in Java are freely available in binary form, but they do not include access to use and extend the source code.5–7 Libraries for other computer languages have been described in the literature but are, to our knowledge, not available to the public.8
To give an overview of the open source activitities in chemistry, we analyzed the open source projects registered at SourceForge.4 This Website has about 40 projects registered in the field of molecular chemistry, as found with a search on keywords such as molecule, molecular, chemistry, and chemical. Many projects are inactive: some are only registered but show no activity at all, and some showed activity in the past but never released software in binary form or source code. The number of active projects is about 25–30.
Of these projects 14 were found that use the Java programming language. Three of these are inactive for a long period and do not provide downloads. Two are succeeded by this project,9,10 and four are based on CDK.11–15 Four projects are interesting to note: MolMaster having a BSD license16 and including visualization of isosurfaces, jVisualizer having the GPL license17 for analyzing NMR couplings, CML having an Artistic License18 with tools around the Chemical Markup Language,19 and JOELib having the GPL license20 with an extensive file IO library based on OpenBabel21 and a library for molecular descriptors. Note that the first two are not really libraries but applications instead. CMLDOM and JOELib, however, are libraries with similar functionality for storing chemical content in memory.
3. THE ORIGIN OF THE CDK
The CDK originated as a support project for a couple of different chemoinformatics software packages, namely a structure editor,11 a Web database for organic compounds and their NMR chemical shifts,14 a program for computer assisted structure elucidation,22 and a 3D structure viewer and analyzer,13 which is still being ported to the CDK.
The authors of these programs generally agree on the benefits of the programming language Java, as there are as follows: clear object-oriented design, platform-independency, and the fact that it has become an important standard for client- and server-side applications on the Web. Since most of the scientifically interesting applications in chemistry have a computationally demanding kernel, they benefit from a client/server architecture because the server part can then be run on a powerful machine, while a user-friendly (Web-) interface can be used on whatever client machine the user chooses. These demands can be met much easier if one can still resort to a single programming language for the implementation and so we consider Java to be the programming language of choice not only for chemoinformatics and computational chemistry but also for scientific applications in general.
Concerns are frequently raised with respect to the performance of Java. However, the language structure itself, compared for example with C++, provides no good reason for Java having a generally lower performance than other languages more frequently used in high performance computing. Indeed, great efforts have been made to increase Java runtime performance and so, today, given a proper implementation and using the right runtime environment, server-side Java code does not need to be slower than C++ with the same scope. We would like to point the reader’s attention to a whole issue of the IBM systems journal dedicated to the subject of high performance computing in Java.23
4. DEVELOPMENT MODEL
To participate in CDK development, the interested individual needs to register with SourceForge (SF) to receive a free SF account and subscribe to the developers mailing list cdk-devel@lists.sourceforge.net. He or she then contacts one of the project administrators, who then adds the new member to the project’s developers list. Besides good Java programming skills, a working knowledge for the Concurrent Versions System (CVS) is needed. CVS is the most widely used system for version management in the Open Source community, which greatly facilitates the coordination of multiple developers working on the same source tree.
It is quite common in computer science to write a requirements specification before coding is started. Such a specification describes the intended behavior of the software (classes in this case) and can be used by developers to check the implementation and by users to see how those classes can be used. When the CDK was designed, such specification was only partly made using Unified Modeling Language (UML) diagrams.24 Currently we use Requests For Comment (RFC) documents for proposing a new specification to which the CDK library must conform. These RFC, which are a long time Internet standard for decision making, are discussed on the developers mailing list after which they are marked as final after majority voting.
5. PROJECT CONVENTIONS
In Java, source code is organized in so-called packages, which often (but not neccessarily) follow a naming scheme of something like an inverted Internet address. Putting a class such as Atom into a uniquely named package prevents class name collisions in cases where another library, used together with the CDK, also contains an Atom class with different function. Since the CDK is part of the OpenScience project,25 the CDK source tree is organized in packages under the org.openscience.cdk root package. Frequently, a new developer is interested in adding a particular functionality to the CDK, for example the capability for isomorphism and automorphism checking. He discusses the implications of his endeavor with the others CDK developers on the mailing list. Taking into account the suggestions, caveats, etc., of his codevelopers, he would then create a new subpackage org.openscience.cdk.isomorphism and add his contribution under this part of the source tree.
An important part of the CDK development effort is Unit Testing, which is based on the idea of writing easily repeatable tests for smallest units of the software package in question. Whenever a programmer adds a new module with new functionality to the CDK source tree, he is expected to add a test to the org.openscience.cdk.tests package, adhering to a particular naming convention. The unit testing itself is based on the JUnit package,26 which makes it easy to run a fully unattended test for the whole CDK package. This has proven to be of great value for such a distributed programming effort like the CDK. Especially if a developer changes something within the CDK core classes, a full JUnit test run of the CDK tests will show him within a few seconds whether his changes broke something or not. Further, each of these little test snippets is an instructive example on how to use a particular CDK module.
Indispensable for a library is documentation. The CDK is documented using the JavaDoc system—an integral part of the Java programming language. Using special tags, the code is documented directly in the source code, from which documentation can be produced automatically in various formats, most importantly as Web pages. We are using source code metrics to constantly measure the amount of documented source code statements, and we try to keep this percentage as high as possible. In addition to the JavaDoc API documentation, the user is guided by a few introductory manuals.
It should also be mentioned that the CDK’s software architecture has been independently chosen as subject of an M.Sc. thesis at the Technion (Israel Institute of Technology),27 focusing on automated methods for code inspection and review. This is a common industrial process by which source code is usually read manually to find errors, potential improvements, dependencies, etc. The thesis focuses on automizing the formal concept analysis using concept lattices28 for the review of individual java classes. Concept analysis is a mathematical classification technique, which is used for different problems in software research. This methodology is applied in three stages: (1) understanding the public interface of the class for use as a black box, (2) trying to reason about the design and possible errors in the class based on its lattice, and (3) inspecting actual source code. The first two stages are done without even having the source code: the methods and fields are determined by reverse engineering of the compiled class files. We have already received valuable input from this related project which will help us to resolve design flaws in our library.
6. DESCRIPTION OF THE LIBRARY’S FUNCTIONALITY
6.1. The Core Classes
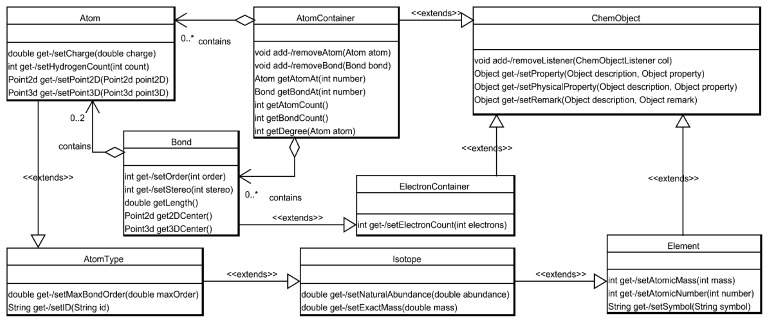
The classes contained in the root section of the CDK’s package hierarchy are all formalized representations of basic chemical concepts such as atoms, bonds, molecules, etc. Figure 1 shows an UML diagram explaining the inheritance hierarchy and the dependencies between the fundamental classes of the CDK. The UML diagrams shown in this article depict the relationship of only the core classes. They are thus edited and do only show a subset of their true interclass relationships. They show the central role of the ChemObject class, which is the superclass of all other classes and provides methods for storing even complex properties for any derived CDK object.
Figure 1.
UML diagram, showing the inheritance hierarchy and the dependencies of the fundamental classes within the CDK.
The first and probably most obvious inheritance chain to be mentioned in the core classes it that of Atom extending AtomType extending Isotope extending Element. This is not only logical from a chemical point of view but also provides the basis for a simple mechanism for the creation of Atoms, AtomTypes, Isotopes, and Elements based on subclasses of a single IsotopeFactory tool class, which will be discussed below. Placing the Atom in a long chain of inheritance provides central access points to the different levels of information. While the Element, for example, provides access to the symbol or the atomic number, some AtomType can further distinguish between the state of hybridization of an Atom or some other distinction a force field might need.
A further level of abstraction is incorporated by the AtomContainer and the ElectronContainer. The Electron-Container forms the base for constructs such as Bonds and Orbitals, whereas the AtomContainer is the envisioned storage for Atoms together with their Bonds and is the superclass for Rings, Molecules, and Substructures.
To support higher level concepts such as molecular ensembles or reactions, the CDK core is complemented by classes which group molecules into higher order constructs, like SetOfMolecules, ChemSequence, ChemModel, and ChemFile.
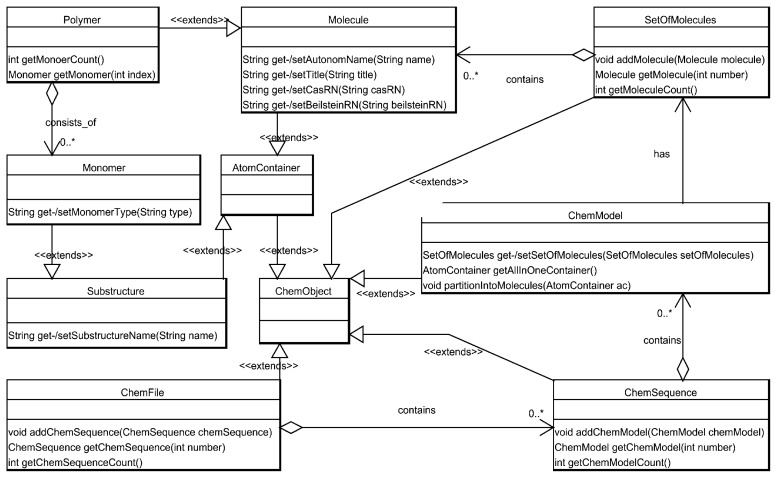
For clarity, the relationship of ChemObject and the AtomContainer has been moved to an additional UML diagram shown in Figure 2.
Figure 2.
UML diagram, showing the inheritance hierarchy and the dependencies of classes group based on the AtomContainer concept.
It shows how Molecules are contained in a SetOfMolecules, which is part of a ChemModel. ChemModels are meant to store the molecular information of the state of a chemical systems at a given point in time. To allow for the modeling of changes in time, we introduced the possibility of arranging various ChemModels into a ChemSequence. The ChemFile class is designed as the top level container, which can contain all the concepts stored in a chemical document among which one or more ChemSequences.
The Polymer class extends Molecule and provides convenient access to the Monomers it consists of. The Monomer itself is implemented as an AtomContainer. A subclass of Polymer is the BioPolymer used for representing protein and DNA molecules. The Polymer design allows BioPolymers to treat each amino acid as an AtomContainer.
6.2. 2D Structure Graphical Handling
The ability to display and manipulate 2D drawings of chemical structures is one of the most important features of any chemoinformatics-related program. This includes the capability of generating coordinates for those chemical structures which have for example been generated by structure generator as coordinateless, chemical graphs. The details for this latter step are discussed in Section 6.4.
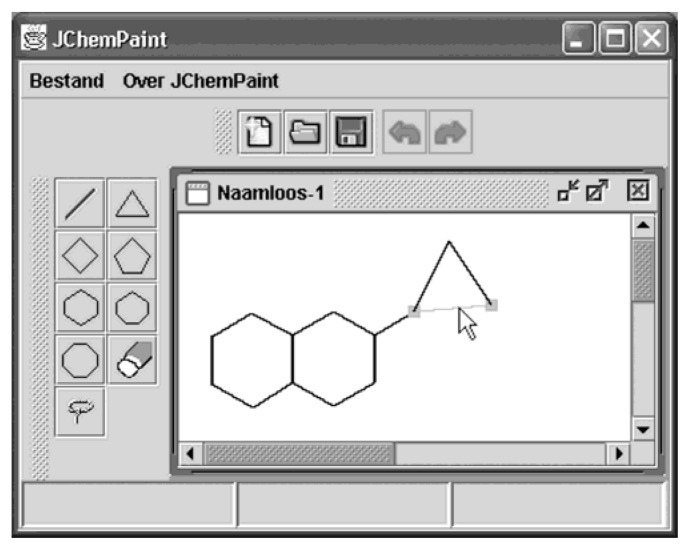
The Model-View-Controller paradigm (see for example ref 29) is used in the CDK library design wherever applicable. The classes for 2D structure graphical handling, for example, work on top of a ChemModel whose content they display and manipulate. A Renderer2D class produces a 2D drawing comparable to those produced by the major commercially available products. This view can be customized by altering the standard settings of a Renderer2DModel object. If the pure display is to be complemented by an option to manipulate the drawing, a Controler2D can be added to the setup. Its settings, again, are determined by a Controler2DModel and can be altered, for example, by using setDrawNumbers(true) in order to display atom numbers annotated to the structure. The Controler2D is an adapter to the available input devices, typically mouse and keyboard, and translates input into changes to the underlying models, which again are reflected by changes in the view produced by the Renderer2D. A simple resulting application is shown in Figure 3.
Figure 3.
Renderer2D and Controller2D cooperating in a simple, CDK-based version of JChemPaint. JChemPaint supports internationalization, with this example showing a dutch interface.
6.3. 3D Structure Handling
To provide high performance 3D graphics, the Java3D API is used within the CDK. This, however, makes CDK-based 3D applications no longer platform independent. This dependency originates from Java3D API relying on OpenGL or DirectX for the sake of higher performance. SUN microsystems does only provide the Java3D for Windows (both OpenGL and DirectX), Solaris and SGI IRIX, whereas a Linux version is developed by Blackdown30 and available for a variety of architectures.
In regard to loosing the platform independency the CDK does also contain classes for 3D rendering which are not based upon the Java3D API. Together with the separation of the rendering classes, due to the Model-View-Controller paradigm, this leads to the following four fundamental classes for 3D rendering: Renderer3D, Renderer3DModel, AcceleratedRenderer3D, and AcceleratedRenderer3DModel, the latter two based upon Java3D.
6.4. Structure Diagram Layout
Key fields of chemoinformatics, like virtual combinatorial chemistry, virtual screening, or computer-assisted structure elucidation, frequently handle chemical structures as one-dimensional graphs. These graphs are, for example, products of structure generators which use graph theoretical techniques to exhaustively and irredundantly generate all constistitutional isomers which are in agreement with a given molecular formula. In any of these programs, however, comes the point where, after a selection during a virtual screening, for example, the successful candidate structure(s) needs(s) to be presented to a chemist. At this point, a tool is needed that generates 2D or 3D coordinates to produce the kind of depiction a chemist is used to. This process has been termed Structure Diagram Generation.31 While 3D model builders such as CORINA32 are on our wishlist for the future and have not yet been implemented, the CDK features a 2D structure diagram generator, which has been written from scratch and which can easily be seen as one of the finest and most useful parts of the CDK, since most of its applications require structure diagram generation at several stages.
6.5. Graph Invariants
This package contains a few classes for the computation of graph invariants such as Wiener Indices,33 Morgan’s extended connectivity (EC) indices,34 and others.35 Morgan’s EC indices are, for example, used for canonical labeling of compounds. This package is likely to be one of the hot spot for future developments, since many chemoinformatics applications, like (quantitative) structure activity relationship ((Q)SAR) computations, do often rely on calculating various combinations of graph invariants of different types.
6.6. Structure Generators
This package holds some simple structure generators which are used by the SENECA system for computer-assisted structure elucidation.22 The class SingleRandomStructureGenerator can be used to generate a totally random structure from the constitutional space given by a certain molecular formula. Based on this randomly generated structure one can then use RandomGenerator to make small, random moves in constitution space, based on an algorithm suggested by Faulon.36 If such a generator is combined with a target function and simulated annealing protocol, one can effectively search constitution space for structures with certain desired properties, provided that these properties can be reliably backcalculated from a given constitutional formula.
To be able to build a structure generator for chemical graphs based on evolutionary algorithms (like the well-known genetic algorithm), we also included a CrossOverMachine, which accepts two chemical graphs in the form of Atom-Containers and produces two offsprings. Genetic Algorithms are population based methods which produce new offsprings for the next generation by a carefully chosen combination of mutation and crossover procedures, applied to the current population. The CrossOverMachine does thus complement the mutation operation used in the RandomGenerator class.
6.7. Ring Searches
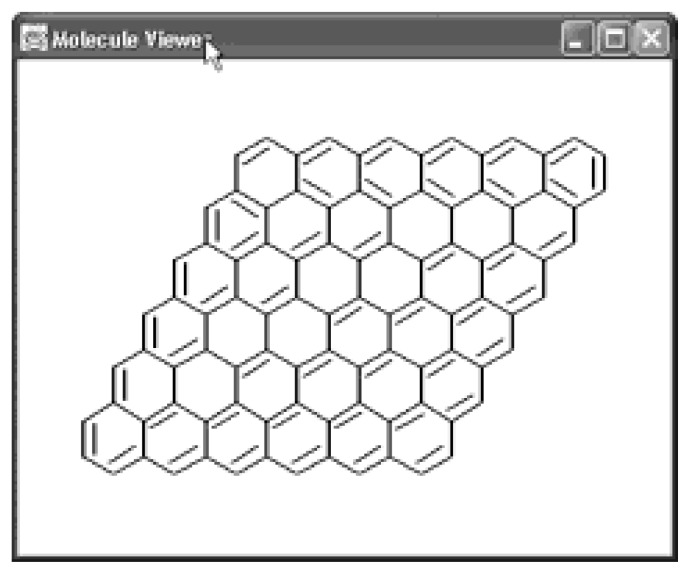
John Figueras’ fast algorithm for finding the Smallest Set of Smallest Rings (SSSR) has been implemented and is used for example by the structure diagram generation package.37 Especially large condensed ring systems, for which the process of coordinate generation could take up to a minute due to a slow depth first ring perception algorithm in older systems,38 can now be layed out within fractions of a second as shown in Figure 4. Further this package contains a class for partioning a given ring systems into AtomContainers, one for each ring.
Figure 4.
A rings system parsed from a SMILES, analyzed by Figueras’ SSSR algorithm and displayed by the MoleculeViewer class. The process takes 300 ms on a 600 MHz Pentium with Windows XP and JDK 1.3.1.
In other applications, like aromaticity detection, for example, it is essential to compute the Set of All Rings (SAR). While procedures have been published to produce the SAR from a SSSR, it is computationally more efficient to use specialized algorithms for this purpose. The CDK contains an implementation of a fast and efficient algorithm given by Hanser et al.39
6.8. Aromaticity Detection
There are various definitions of aromaticity and at least as many ways of detecting aromaticity according to these definitions. This package is the intended container for all of them and does currently hold an implementation of a HueckelAromaticityDetector class. Based on the SAR detection algorithm by Hanser et al. (see section 6.7) this class starts with the largest detected ring, counts the number of alternating double or triple bond electrons, and does also take into account free electron pairs of heteroatoms. It then checks whether the ring contains 4n + 2 π-electrons, according to the well-known Hückel rule. The ring, all its atoms, and bonds are marked as aromatic, and the search continues with the remaining rings of equal or smaller size, leaving out those rings that are completely part of an already detected larger aromatic system.
6.9. Isomorphism
Being able to determine if two chemical structures are identical or whether one structure is a subgraph of another structure is one of the most important capabilities of a chemoinformatics library. The Isomorphism subpackage contains a versatile module for Maximum Common Substructure (MCSS) Searches. Since MCSS determination is the most general case of graph matching, it can be used to determine structure identity and to do subgraph matching and maximum common substructure searches.
6.10. File Input/Output
File input and output is generalized in CDK. All file i/o classes implement either ChemObjectReader or ChemObjectWriter. Each file format is represented by two separate classes implementing one of these interfaces.
CDK currently supports IO classes for XYZ, MDL molfile,40 PDB,41 and CML.42 The latter format was developed by Murray-Rust and Rzepa as the first XML based file format for chemical content. The CDK contains both an input and output class for this format. The CML input reader uses an alternative to Murray-Rust’s DOM approach and is based on SAX.43
6.11. Interaction with other Java Libraries
Besides file i/o, CDK supports a second method to exchange data with other programs and libraries. The interface to other libraries makes it possible to combine methods from both libraries giving access to a larger set of functionality. CDK provides direct conversion of CDK classes to JOELib20 classes. Support for CMLDOM19 is planned.
6.12. SMILES
Simplified Molecular Line Entry Specification (SMILES) provides string representations of molecular constitutions.44 Due to their compactness and relative simplicity they are now widely used as an interchange format for coordinateless molecular structures. Based on a specification for unique (canonical) SMILES,45 it is also possible to perform graph isomorphism checks. The CDK features a generator for canonical SMILES, written to comply with the rules published by the Daylight Inc. founders. While the SMILES generator implements all of the published SMILES standard including chirality, the SMILES parser in the CDK package only complies to the (slightly extended) Super Simplified SMILES specification46 which is sufficient to code most organic structures.
6.13. Fingerprints
Fingerprinting is nowadays an indispensable tool for judging molecular similarity, as a prefilter for isomorphism checking and thus for structure searching in databases. Here as well as in the case of SMILES an own subpackage for this class of algorithms is justified because there are various ways of computing fingerprints. By allowing the addition of different fingerprinters instead of just having one monolithic org.openscience.cdk.tools.Fingerprinter we give the user the freedom of choosing whatever methods yields the best performance for his case. The Fingerprinter class in the CDK produces Daylight-type fingerprints.47 It works by running a breadth-first search, starting at each atom in the molecule, thereby producing string representations of paths up to the length of six atoms. For each of theses SMILES-like strings, hash codes are computed, using the standard string hashing algorithm provided by the Java language. With these hash codes, a pseudorandom number generator with a default working range of [0–1023] is seeded and the first random number is retrieved. This number indicates a position in a fingerprint bitstring of length 1024, which is then set to “1”. Based on the entirety of all computed paths from the molecule, a molecular fingerprint is obtained in the form of this bitstring.
6.14. Tools
The tools package contains utility classes for all those cases that did not justify the creation of a dedicated package. The IsotopeFactory, for example, can return pre-configured instances of Elements and Isotopes for a given element symbol or a given atomic mass.
The ConnectivityChecker class tests whether a given chemical graph is connected, i.e., whether there is a bond path between every possible pair of atoms in the graph and, in the case of a nonconnected graph, it can return a Vector with the disjunct pieces of the graph, stored in AtomContainer objects. Related to ConnectivityChecker is the Path-Tools class which, for example, provides methods for finding the shortest path between to given atoms in a molecule.
The MFAnalyser class has methods of returning the molecular formula of a given Molecule object and for creating an unbonded AtomContainer object from a given molecular formula string. The HOSECodeGenerator produces HOSE codes48 for each atom in a given AtomContainer. By feeding these HOSE codes into the BremserOneSphere-HOSECodePredictor class, one can predict expectation ranges for carbon-13 NMR chemical shifts.49
7. RESULTS
The CDK is now the basis for a number of software projects. The chemical editor JChemPaint11 which takes advantage of the CDK and for which the CDK’s Model-View-Controller mechanisms have been implemented is again just a support tool for higher level applications such as the Web database NMRShiftDB for organic compounds and their NMR chemical shifts, or SENECA, a program for computer assisted structure elucidation.22
While allowing the fast assembly of such large monolithic applications such as SENECA or NMRShiftDB, the true strength of the CDK lies in its ability to serve as a chemoinformatician’s workbench. By just writing a few lines of code, one can quickly test new ideas or modify existing CDK based applications to make them suit other needs.
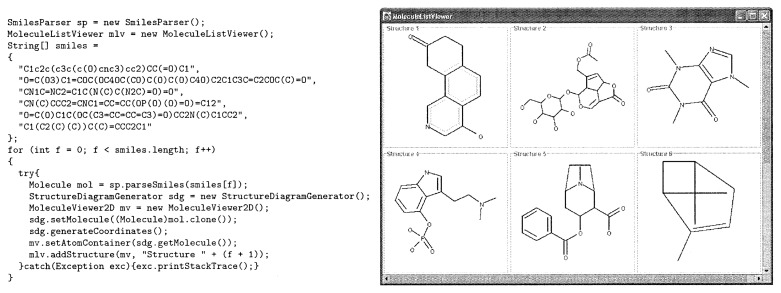
The following code snippet illustrates how one can quickly parse a list of SMILES strings into AtomContainers, produce 2D coordinates, and display the results in a MoleculeList-Viewer.
8. CONCLUSION
We have presented details of a new open-source Java library facilitating the implementation of software packages in chemoinformatics. The CDK is freely available50 under the terms of the GNU Lesser General Public License (LGPL)3. The source code may thus be downloaded and improved or adapted for specific needs. In contrast to the famous GNU General Public License (GPL)51 the LGPL allows for the use of the CDK in proprietary software packages. While any use of the CDK for proprietary and closed-source project is thus welcome, we also highly appreciate feedback and any potential backflow. Companies are using the CDK for commercial projects, such as SafeBase, a theragenomics knowledge management system on adverse drug reactions.52 At the IBM Germany Development Lab in Böblingen an Extreme Blue internship project group has been started to write a CDK-based open source 2D/3D editor for chemical structures. The company IXELIS, situated in Strasbourg, France, is working on a global semantic information system applied to scientific knowledge and has contributed the MCSS code, which came into existence during their work with the CDK.
Further, our chemoinformatics software kit is the basis for other open-source projects, like the SENECA system for computer-assisted structure elucidation22 and NMRShiftDB,14 a free database of organic chemicals and their NMR data.
Besides its proven usability in research and production quality scientific software, the CDK has also become a valuable tool for teaching chemoinformatics. At least one of our authors (C.S.) is using the software package in lectures to demonstrate many standard chemoinformatics algorithms on the functionality level as well as on the source code level. Due to the inherent modularization of the object oriented language Java, most of the classes and methods are concise and easy to understand.
It should be mentioned that we have experienced, albeit on a smaller scale than the large open-source projects, the benefits and the fascination of the principles mentioned in the Introduction. Based on this experience, this article is also supposed to promote these ideas and to attract further contributors for our project. The inspiring experience is that as soon as a certain amount of material has accumulated and a certain amount of publicity has been gained, an open-source project becomes something like a self-runner, contributors start adding their own subprojects, and new ideas are integrated which would probably never have been borne in mind if the CDK were created by a single organization and even individual. Of course, such a development model also has disadvantages. It is probably much more difficult to adhere to certain quality standards, to respond to deadlines (but on the other hand, there rarely are any in such small projects), and to do strategic planning. It has been shown, however, that these problems can be overcome.
Figure 5.
A CDK code snippet illustrating the use of SmilesParser and StructureDiagramGenerator is shown followed by its output.
ACKNOWLEDGMENT
The authors would like to thank all members of the CDK project for their contributions, corrections, and helpful comments.
REFERENCES AND NOTES
- 1.The Open Source Initiative (OSI) [accessed on Aug 2002]. 2002. http://www.opensource.org.
- 2.Raymond ES. The Cathedral and the Bazaar: Musings on Linux and Open Source by an Accidental Revolutionary. O’Reilly and Associates; Sebastopol, CA: 1999. [Google Scholar]
- 3.GNU Lesser General Public License – GNU Project – Free Software Foundation (FSF) [accessed on Aug 2002]. 2002. http://www.gnu.org/licenses/lgpl.html.
- 4.SourceForge.net. [accessed on Aug 2002]. 2002. http://www.sf.net/
- 5.Rzepa H, Tonge A. VChemLab: A Virtual Chemistry Laboratory. The Storage, Retrieval, and Display of Chemical Information Using Standard Internet Tools. J Chem Inf Comput Sci. 1998;38:1048–1053. [Google Scholar]
- 6.Csizmadia F. JChem: Java Applets and Modules Supporting Chemical Database Handling from Web Browsers. J Chem Inf Comput Sci. 2000;40:323–324. doi: 10.1021/ci9902696. [DOI] [PubMed] [Google Scholar]
- 7.Blauch D. Java Classes for Managing Chemical Information and Solving Generalized Equilibrium Problems. J Chem Inf Comput Sci. 2002;42:143–146. doi: 10.1021/ci010074+. [DOI] [PubMed] [Google Scholar]
- 8.Bauerschmidt S, Gasteiger J. Overcoming the limitations of a connection table description: A universal representation of chemical species. J Chem Inf Comput Sci. 1997;37:705–714. [Google Scholar]
- 9.The CompChem libraries. [accessed on Aug 2002]. 2002. http://compchem.sourceforge.net/
- 10.The JMDraw Structure Diagram Generation Engine. [accessed on Aug 2002]. 2002. http://jmdraw.sourceforge.net/
- 11.Steinbeck C, Krause S, Willighagen E. JChemPaint – Using the Collaborative Forces of the Internet to Develop a Free Editor for 2D Chemical Structures. Molecules. 2000;5:93–98. [Google Scholar]
- 12.The JChemPaint Structure Editor. [accessed on Aug 2002]. 2002. http://jmdraw.sourceforge.net/
- 13.The Jmol 3D Molecular Visualization Software. [accessed on Aug 2002]. 2002. http://jmol.sourceforge.net/
- 14.Kuhn S, Krause S, Steinbeck C. NMRShiftDB – An Open-Access, Open-Submission, Open-Source Database for Organic Structures and their NMR data. 2002 Manuscript in preparation. [Google Scholar]
- 15.The NMRShiftDB NMR Database. [accessed on Aug 2002]. 2002. http://www.nmrshiftdb.org/
- 16.The MolMaster Molecular Visualization Package. [accessed on Aug 2002]. 2002. http://molmaster.sourceforge.net/
- 17.The JVisualizer NMR Analysis Package. [accessed on Aug 2002]. 2002. http://jvisualizer.sourceforge.net/
- 18.The Chemical Markup Language Supporting Software Pages. [accessed on Aug 2002]. 2002. http://cml.sourceforge.net/
- 19.Murray-Rust P, Rzepa H. Chemical Markup XML, and the Worldwide Web. 2. Information Objects and the CMLDOM. J Chem Inf Comput Sci. 2001;41:1113–1123. doi: 10.1021/ci000404a. [DOI] [PubMed] [Google Scholar]
- 20.JOELib – a java based computational chemistry package. [accessed on Aug 2002]. 2002. http://joelib.sourceforge.net/
- 21.The OpenBabel Chemical File Format Conversion Package. [accessed on Aug 2002]. 2002. http://openbabel.sourceforge.net/
- 22.Steinbeck C. SENECA: A Platform-Independent, Distributed and Parallel System for Computer-Assisted Structure Elucidation in Organic Chemistry. J Chem Inf Comput Sci. 2001;41:1500–1507. doi: 10.1021/ci000407n. [DOI] [PubMed] [Google Scholar]
- 23.IBM Systems Journal – Java Performance. 2000. [Google Scholar]
- 24.Stevens P, Pooley R. Using UML: software engineering with objects and components. (Object Technology Series Addison-Wesley). 1999 Updated edition for UML1.3: first published 1998 (as Pooley and Stevens) [Google Scholar]
- 25.The OpenScience Project. [accessed on Aug 2002]. 2002. http://www.openscience.org/
- 26.JUnit, Testing Resources for Extreme Programming. [accessed on Aug 2002]. 2002. http://www.junit.org/
- 27.Dekel U. Personal Communication; 2002. [Google Scholar]
- 28.Ganter B, Wille R. Concept Analysis: Mathematical Foundations. Springer-Verlag: Berlin-Heidelberg; 1999. [Google Scholar]
- 29.Krasner G, Pope S. A Cookbook for using the Model-View-Controller User Interface Paradigm in Smalltalk-80. JOOP. 1988:29–49. [Google Scholar]
- 30.Java-Linux. [accessed on Aug 2002]. 2002. http://www.blackdown.org/
- 31.Helson H. Structure Diagram Generation. Rev Comput Chem. 1999;13:313–398. [Google Scholar]
- 32.Gasteiger J, Rudolph C, Sadowski J. Automatic Generation of 3D-Atomic Coordinates for Organic Molecules. Tetrahedron Comput Method. 1990;4:537–547. [Google Scholar]
- 33.Wiener H. Correlation of Heat of Isomerization and Difference in Heat of Vaporization of Isomers Among Paraffin Hydrocarbons. J Am Chem Soc. 1947;69:17–20. [Google Scholar]
- 34.Morgan HL. The Generation of a Unique Machine Description for Chemical Structures – A Technique Developed at Chemical Abstracts Service. J Chem Doc. 1965;5:107–113. [Google Scholar]
- 35.Hu CY, Lu L. On highly discriminating molecular topological index. J Chem Inf Comput Sci. 1996;36:82–90. [Google Scholar]
- 36.Faulon J-L. Stochastic Generator of Chemical Structure. 2. Using Simulated Annealing To Search the Space of Constitutional Isomers. J Chem Inf Comput Sci. 1996;36:731–740. [Google Scholar]
- 37.Figueras J. Ring Perception Using Breadth-First Search. J Chem Inf Comput Sci. 1996;36:986–991. [Google Scholar]
- 38.Bley K, Brandt J, Dengler A, Frank R, Ugi I. Constitutional Formulae generated from Connectivity Information: the Program MDRAW. J Chem Res (M) 1991:2601–2689. [Google Scholar]
- 39.Hanser T, Jauffret P, Kaufmann G. A new algorithm for exhaustive ring perception in a molecular graph. J Chem Inf Comput Sci. 1996;36:1146–1152. [Google Scholar]
- 40.Description of Several Chemical Structure File Formats Used by Computer Programs Developed at Molecular Design Limited, 1992 An updated online version of this document can be found on http://www.mdli.com/downloads/literature/ctfile.pdf.
- 41.Protein Data Bank Atomic Coordinate and Bibliographic Entry Format Description. 1985. [Google Scholar]
- 42.Murray-Rust P, Rzepa H. Chemical Markup XML, and the Worldwide Web. 1. Basic Principles. J Chem Inf Comput Sci. 1999;39:928–942. [Google Scholar]
- 43.Willighagen E. Processing CML conventions in Java. Internet J Chem. 2001;4:4. [Google Scholar]
- 44.Weininger D. SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J Chem Inf Comput Sci. 1988;28:31–36. [Google Scholar]
- 45.Weininger D, Weininger A, Weininger J. SMILES. 2. Algorithm for Generation of Unique SMILES Notation. J Chem Inf Comput Sci. 1989;29:97–101. [Google Scholar]
- 46.SMILES Home Page. [accessed on Aug 2002]. 2002. http://www.daylight.com/dayhtml/smiles/
- 47.James CA, Weininger D, Delany J. Daylight Theory Manual. [accessed on Aug 2002]. 2000. http://www.daylight.com/dayhtml/doc/theory/theory.toc.html.
- 48.Bremser W. HOSE – A Novel Substructure Code. Anal Chim Act. 1978;103:355–365. [Google Scholar]
- 49.Bremser W. Expectation Ranges of 13-C NMR Chemical Shifts. Magn Reson Chem. 1985;23:271–275. [Google Scholar]
- 50.The Chemical Development Kit. [accessed on Aug 2002]. 2002. http://cdk.sf.net/
- 51.GNU General Public License – GNU Project – Free Software Foundation (FSF) [accessed on Aug 2002]. 2002. http://www.gnu.org/licenses/gpl.html.
- 52.TheraSTrat – Taking drug safety a step further. [accessed on Aug 2002]. 2002. http://www.therastrat.com/