Abstract
Advanced prostate cancer (PCa) metastasizes to bone and lymph nodes, and currently available treatments cannot prevent the progression and metastasis of the disease. Therefore, an improved understanding of the molecular mechanisms of the progression and metastasis of advanced PCa using current genomic approaches is needed. Our miRNA expression signature in castration-resistant prostate cancer (CRPC) revealed that microRNA-320a (miR-320a) was significantly reduced in cancer tissues, suggesting that miR-320a may be a promising anticancer miRNA. The aim of this study was to investigate the functional roles of miR-320a in naïve PCa and CRPC cells and to identify miR-320a-regulated genes involved in PCa metastasis. The expression levels of miR-320a were significantly reduced in naïve PCa, CRPC specimens, and PCa cell lines. Restoration of mature miR-320a in PCa cell lines showed that miR-320a significantly inhibited cancer cell migration and invasion. Moreover, we found that lysosomal-associated membrane protein 1 (LAMP1) was a direct target of miR-320a in PCa cells. Silencing of LAMP1 using siRNA significantly inhibited cell proliferation, migration, and invasion in PCa cells. Overexpression of LAMP1 was observed in PCa and CRPC clinical specimens. Moreover, downstream pathways were identified using si-LAMP1-transfected cells. The discovery of tumor-suppressive miR-320a-mediated pathways may provide important insights into the potential mechanisms of PCa metastasis.
Keywords: microRNA, prostate cancer, castration-resistant prostate cancer, miR-320a, LAMP1, tumor suppressor, metastasis
Introduction
Prostate cancer (PCa) is the most frequently diagnosed cancer and the second leading cause of cancer-related death among men in developed countries (1). Most patients with PCa are initially responsive to androgen deprivation therapy; however, their cancers eventually become resistant to androgen-deprivation therapy and progress to castration-resistant prostate cancer (CRPC). Several novel treatments have recently been developed for patients with advanced PCa. These treatments, however, cannot completely control the progression and metastasis of PCa (2). Therefore, more effective treatment strategies, based on current genomic approaches, are required. Moreover, because it is difficult to obtain clinical specimens of CRPC, there are very few reports describing the genomic analysis of CRPC.
Small non-coding RNAs known as microRNAs (miRNAs) have been shown to regulate gene expression by repressing translation or cleaving RNA transcripts in a sequence-dependent manner (3). At present, a substantial amount of evidence has suggested that miRNAs are aberrantly expressed in many human cancers, including PCa, and play significant roles in human oncogenesis, metastasis, and drug resistance (4–7). Several reports have shown that miRNAs regulate ~30–60% (or more) of the protein-coding genes in the human genome by bioinformatics predictions (7,8). Thus, normal regulatory mechanisms of RNA networks can be disrupted by the aberrant expression of tumor-suppressive or oncogenic miRNAs in cancer cells. We propose that identification of aberrantly expressed miRNAs in clinical specimens is an important first step toward elucidating the details of RNA networks in cancer cells.
To identify novel RNA networks in PCa, we have previously constructed miRNA expression signatures using androgen-dependent PCa clinical specimens (9,10). Using this signature, we have identified tumor-suppressive miRNA-mediated PCa oncogenic pathways (11–16). More recently, we constructed miRNA expression signatures using CRPC clinical specimens and reported that the miRNA cluster miR-221/miR-222 functions as a tumor suppressor in PCa and CRPC cells (17). Moreover, miR-223 expression has been shown to be significantly reduced in PCa and CRPC specimens and to act as a tumor suppressor by targeting integrin-A3/B1 oncogenic signaling (18). The CRPC expression signature reported by our laboratory has provided important information needed to elucidate novel RNA networks in CRPC cells.
In this study, we focused on miR-320a because downregulation of miR-320a was observed in our two miRNA signatures for androgen-dependent PCa and CRPC clinical specimens (9,17). The aim of this study was to investigate the functional significance of miR-320a and to identify the molecular targets and pathways mediated by miR-320a in PCa and CRPC cells. Our data showed that restoration of mature miR-320a in PC3 and DU145 PCa cells significantly inhibited cancer cell migration and invasion. Direct regulation of lysosomal-associated membrane protein 1 (LAMP1) by miR-320a was observed in PCa cells. Silencing of LAMP1 using specific small interfering RNA (siRNA; si-LAMP1) significantly inhibited cell proliferation, migration, and invasion in PCa cells. The discovery of tumor-suppressive miR-320a-mediated molecular pathways provides new insights into the potential mechanisms of metastasis in PCa and suggests novel therapeutic strategies for the treatment of this disease.
Materials and methods
Clinical PCa specimens, cell lines, and RNA isolation
Clinical prostate specimens (PCa and normal prostate tissues) collected by needle biopsy or autopsy were obtained from patients admitted to Teikyo University Chiba Medical Center Hospital from 2008 to 2013. Twenty-nine prostate samples [PCa, n=15; non-cancerous prostate tissues (non-PCa), n=14] were obtained by transrectal prostate needle biopsy from patients with elevated serum prostate-specific antigen (PSA) levels, and eight metastatic PCa samples were obtained from patients who died of CRPC. The backgrounds of the patients are summarized in Table I. All patients provided written informed consent for tissue donation for research purposes. The protocol was approved by the Institutional Review Boards of Chiba University and Teikyo University.
Table I.
Patient characteristics.
| No. | Diagnosis | Age (years) | PSA (ng/ml) | Gleason score | cT | cN | cM |
|---|---|---|---|---|---|---|---|
| 1 | Non-PCa | 54 | 5.4 | - | - | - | - |
| 2 | Non-PCa | 60 | 5.6 | - | - | - | - |
| 3 | Non-PCa | 67 | 5.9 | - | - | - | - |
| 4 | Non-PCa | 67 | 8.1 | - | - | - | - |
| 5 | Non-PCa | 60 | 14 | - | - | - | - |
| 6 | Non-PCa | 69 | 6 | - | - | - | - |
| 7 | Non-PCa | 56 | 8.4 | - | - | - | - |
| 8 | Non-PCa | 61 | 8.6 | - | - | - | - |
| 9 | Non-PCa | 62 | 35.5 | - | - | - | - |
| 10 | Non-PCa | 57 | 5.2 | - | - | - | - |
| 11 | Non-PCa | 64 | 4.4 | - | - | - | - |
| 12 | Non-PCa | 60 | 5.7 | - | - | - | - |
| 13 | Non-PCa | 63 | 11.4 | - | - | - | - |
| 14 | Non-PCa | 65 | 13.2 | - | - | - | - |
| 15 | PCa | 65 | 277 | 4+5 | 4 | 1 | 1 |
| 16 | PCa | 73 | 478 | 4+3 | 3b | 0 | 1 |
| 17 | PCa | 75 | 1,000 | 4+5 | 4 | 1 | 1 |
| 18 | PCa | 79 | 63.2 | 4+5 | 3b | 1 | 1 |
| 19 | PCa | 69 | 95.6 | 4+4 | 4 | 1 | 1 |
| 20 | PCa | 70 | 248 | 3+4 | 3a | 1 | 0 |
| 21 | PCa | 66 | 36.1 | 4+5 | 3a | 1 | 0 |
| 22 | PCa | 81 | 1,338 | 4+5 | 4 | 0 | 1 |
| 23 | PCa | 72 | 102 | 4+4 | 3a | 0 | 0 |
| 24 | PCa | 65 | 212 | 4+4 | 4 | 1 | 1 |
| 25 | PCa | 81 | 11.4 | 4+4 | 2 | 0 | 0 |
| 26 | PCa | 75 | 22.7 | 4+4 | 3b | 0 | 0 |
| 27 | PCa | 73 | 467 | 4+4 | 3b | 1 | 0 |
| 28 | PCa | 58 | 482 | 4+4 | 3b | 1 | 0 |
| 29 | PCa | 73 | 76.4 | 4+4 | 3a | 0 | 0 |
| 30 | CRPC liver metastasis | 64 | 4,100 | 4+5 | 3b | 0 | 1b |
| 31 | CRPC lymph node metastasis | ||||||
| 32 | CRPC bone metastasis | ||||||
| 33 | CRPC bone metastasis | 75 | 4,690 | 4+5 | 3b | 0 | 0 |
| 34 | CRPC lung metastasis | ||||||
| 35 | CRPC liver metastasis | 78 | 3,850 | 3+5 | 4 | 0 | 1b |
| 36 | CRPC dural metastasis | ||||||
| 37 | CRPC bone metastasis |
PSA, prostate-specific antigen; PCa, prostate cancer; CRPC, castration-resistant prostate cancer.
Human prostate cancer cells (PC3 and DU145 cells) and normal prostate cells (RWPE-1) were obtained from the American Type Culture Collection (Manassas, VA, USA). PC3 and DU145 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 and 95% air at 37°C. RWPE-1 cells were cultured in keratinocyte serum-free medium containing 5 ng/ml epidermal growth factor and 50 μg/ml bovine pituitary extract.
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol as described previously (14,17,18).
Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR)
The expression levels of miR-320a were analyzed by TaqMan quantitative real-time PCR (assay ID: 0000542; Applied Biosystems, Foster City, CA, USA) and normalized to the expression of RNU48 (assay ID: 001006; Applied Biosystems). TaqMan probes and primers for LAMP1 (P/N: Hs00174766_m1; Applied Biosystems), GAPDH (the internal control; P/N: Hs02758991_m1; Applied Biosystems) and GUSB (the internal control; P/N: Hs00939627_m1; Applied Biosystems) were assay-on-demand gene expression products. The procedure for PCR quantification was described previously (14,17,18).
Transfection with mature miRNA and siRNA
The following mature miRNA species were used in this study: mature miRNA and Pre-miR miRNA Precursor (hsa-miR-320a; P/N: 4427975; Applied Biosystems). The following siRNAs were used: Stealth Select RNAi siRNA, si-LAMP1 (cat no. HSS180593, HSS180594; Invitrogen) and negative control miRNA/siRNA (P/N: AM17111; Applied Biosystems). Transfection procedures and transfection efficiencies of miRNA in PC3 and DU145 cells were described previously (14,17,18).
Cell proliferation, migration, and invasion assays
PC3 and DU145 cells were transfected with 10 nM miRNAs or siRNAs by reverse transfection. Cell proliferation was determined by XTT assay using a Cell Proliferation Kit II (Roche Applied Sciences, Tokyo, Japan). Cell migration activity was analyzed using uncoated Transwell polycarbonate membrane filters. Cell invasion was evaluated using modified Boyden chambers containing Transwell-precoated Matrigel membrane filter inserts. These assays were performed as described previously (14,17,18).
Selection of putative target genes regulated by miR-320a in PCa cells
To identify miR-320a target genes, we used in silico analysis and genome-wide gene expression analysis. First, we used TargetScan Release 7.0 (http://www.targetscan.org/) to search for genes containing the miR-320a seed sequence in the 3′-untranslated region (UTR). Next, to identify upregulated genes in clinical PCa specimens, we analyzed a publicly available gene expression dataset in the GEO database (GEO accession no. GSE29079). Finally, we attempted to identify miR-320a target genes using miR-320a-transfected PC3 cells. A SurePrint G3 Human GE 60K v3 microarray (Agilent Technologies, Santa Clara, CA, USA) was used for genome-wide expression analysis of miR-320a transfectants compared with mock-transfected PC3 cells (GEO accession no. GSE77790). As a result, genes fulfilling the three following conditions were listed: containing putative miR-320a target sites, upregulated in PCa clinical specimens, and downregulated by miR-320a restoration.
Immunohistochemical staining and scoring
A tissue microarray containing a total of 71 prostate samples, 51 PCa specimens, 10 prostatic intraepithelial neoplasia (PIN) samples, and 10 normal prostate samples was obtained from Provitro (Berlin, Germany; cat. no. 4012209, lot no. 146.1P.020212.27). Table III shows the characteristics of patients included in the tissue microarray. Four specimens were used as CRPC tissues (Table I, nos. 31 and 34–36). Tissue specimens were immunostained following the manufacturer's protocol with the Ultra-Vision Detection System (Thermo Scientific, Fremont, CA, USA). Primary rabbit polyclonal antibodies against androgen receptor (AR; 1:50, ab9474; Abcam, Cambridge, UK), antibodies against PSA (1:500, HPA000764; Sigma-Aldrich, St. Louis, MO, USA), and antibodies against LAMP1 (1:1,000; #9091; Cell Signaling Technology, Danvers, MA, USA) were used for immunochemistry. The slides were treated with biotinylated goat antibodies (Histofine SAB-PO kit; Nichirei, Tokyo, Japan). The IHC score was the sum of the score of the weighted intensity and extension of cancerous area. The intensity of staining was graded from 0 to 3 as follows: 0, negative staining; 1, mild staining; 2, moderate staining; and 3, intense staining. The area of staining in the cancerous area was graded from 0 to 3 as follows: 0, no staining of cells in any microscopic field; 1, <30% of malignant cells stained; 2, 30–60% of malignant cells stained; and 3, >60% of malignant cells stained. The procedure was carried out as described previously (14,17,18).
Table III.
Characteristics of patients included in the tissue microarray analysis.
| No. | Diagnosis | Age (years) | Gleason score | Stage pT | Stage pN |
|---|---|---|---|---|---|
| 1 | PCa | 64 | 4+3 | 3b | 0 |
| 2 | PCa | 67 | 3+4 | 2b | 0 |
| 3 | PCa | 58 | 3+4 | 2b | 0 |
| 4 | PCa | 63 | 7 | 3b | 0 |
| 5 | PCa | 65 | 3+3 | 2b | 0 |
| 6 | PCa | 61 | 4+4 | 3b | x |
| 7 | PCa | 62 | 3+4 | 2b | x |
| 8 | PCa | 66 | 4+4 | 2b | x |
| 9 | PCa | 61 | 3+4 | 3a | x |
| 10 | PCa | 74 | 4+3 | 2b | x |
| 11 | PCa | 54 | 3+4 | 2c | x |
| 12 | PCa | 68 | 3+4 | 3a | 0 |
| 13 | PCa | 58 | 3+4 | 3a | 0 |
| 14 | PCa | 65 | 3+3 | 2a | 0 |
| 15 | PCa | 77 | 3+4 | 4 | 0 |
| 16 | PCa | 58 | 3+4 | 3a | 0 |
| 17 | PCa | 50 | 4+3 | 2b | 0 |
| 18 | PCa | 53 | 3+3 | 2b | 0 |
| 19 | PCa | 59 | 4+5 | 3a | 0 |
| 20 | PCa | 70 | 2+3 | 2b | 0 |
| 21 | PCa | 65 | 5+4 | 3a | 0 |
| 22 | PCa | 57 | 3+5 | 2b | 0 |
| 23 | PCa | 68 | 4+4 | 2b | 0 |
| 24 | PCa | 58 | 3+3 | 2b | 0 |
| 25 | PCa | 63 | 3+4 | 2b | 0 |
| 26 | PCa | 56 | 3+4 | 2b | 0 |
| 27 | PCa | 63 | 5+3 | 3a | 0 |
| 28 | PCa | 64 | 3+5 | 3a | 0 |
| 29 | PCa | 60 | 3+4 | 2b | 0 |
| 30 | PCa | 60 | 3+3 | 3a | 0 |
| 31 | PCa | 57 | 3+2 | 2b | 0 |
| 32 | PCa | 50 | 3+3 | 2a | 0 |
| 33 | PCa | 68 | 3+3 | 3a | 0 |
| 34 | PCa | 65 | 3+4 | 3b | 1 |
| 35 | PCa | 69 | 5+5 | 3a | 1 |
| 36 | PCa | 51 | 2+3 | 2b | 0 |
| 37 | PCa | 62 | 3+3 | 3a | 0 |
| 38 | PCa | 61 | 3+4 | 3a | 0 |
| 39 | PCa | 53 | 4+4 | 3b | 1 |
| 40 | PCa | 56 | 4+3 | 2b | 0 |
| 41 | PCa | 59 | 2+3 | 2b | 0 |
| 42 | PCa | 61 | 3+4 | 2b | 0 |
| 43 | PCa | 62 | 3+4 | 3b | 1 |
| 44 | PCa | 66 | 3+3 | 3a | 0 |
| 45 | PCa | 62 | 3+3 | 2b | 0 |
| 46 | PCa | 56 | 3+3 | 2b | 0 |
| 47 | PCa | 58 | 3+3 | 3a | 0 |
| 48 | PCa | 66 | 5+4 | 3a | 0 |
| 49 | PCa | 55 | 3+4 | 3a | 0 |
| 50 | PCa | 67 | 2+3 | 2b | 0 |
| 51 | PCa | 61 | 3+5 | 2b | 0 |
| 52 | PIN | 59 | - | - | - |
| 53 | PIN | 58 | - | - | - |
| 54 | PIN | 62 | - | - | - |
| 55 | PIN | 51 | - | - | - |
| 56 | PIN | 58 | - | - | - |
| 57 | PIN | 68 | - | - | - |
| 58 | PIN | 64 | - | - | - |
| 59 | PIN | 56 | - | - | - |
| 60 | PIN | 61 | - | - | - |
| 61 | PIN | 51 | - | - | - |
| 62 | Normal | 70 | - | - | - |
| 63 | Normal | 63 | - | - | - |
| 64 | Normal | 62 | - | - | - |
| 65 | Normal | 81 | - | - | - |
| 66 | Normal | 67 | - | - | - |
| 67 | Normal | 76 | - | - | - |
| 68 | Normal | 66 | - | - | - |
| 69 | Normal | 69 | - | - | - |
| 70 | Normal | 63 | - | - | - |
| 71 | Normal | 71 | - | - | - |
PCa, prostate cancer; PIN, prostatic intra-epithelial neoplasia.
Western blotting
Cells were harvested 72 or 96 h after transfection, and lysates were prepared. Cell lysates (20 μg protein) were separated on Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA, USA) and transferred to PVDF membranes. Immunoblotting was carried out with rabbit anti-LAMP1 antibodies (1:2,000; #9091; Cell Signaling Technology); anti-GAPDH antibodies (1:4,000; ab8245; Abcam) were used as an internal loading control. Membranes were washed and incubated with anti-rabbit IgG horseradish peroxidase (HRP)-linked antibodies (7074; Cell Signaling Technology). Complexes were visualized with Clarity Western ECL Substrate (Bio-Rad). The procedure was performed as described previously (14,17,18).
Plasmid construction and dual-luciferase reporter assays
The partial wild-type sequence of the LAMP1 3′-UTR or that with deletion of the miR-320a target site was inserted between the XhoI-PmeI restriction sites in the 3′-UTR of the hRluc gene in the psiDHECK-2 vector (C8021; Promega, Madison, WI, USA). The procedure for the dual-luciferase reporter assay was described previously (14,17,18).
Statistical analysis
The relationships between 2 groups and the numerical values obtained by RT-PCR were analyzed using Mann-Whitney U tests. The relationships among more than three variables and numerical values were analyzed using Bonferroni-adjusted Mann-Whitney U tests. All analyses were performed using Expert StatView (version 5; SAS Institute Inc., Cary, NC, USA).
Results
Expression levels of miR-320a in PCa specimens and cell lines
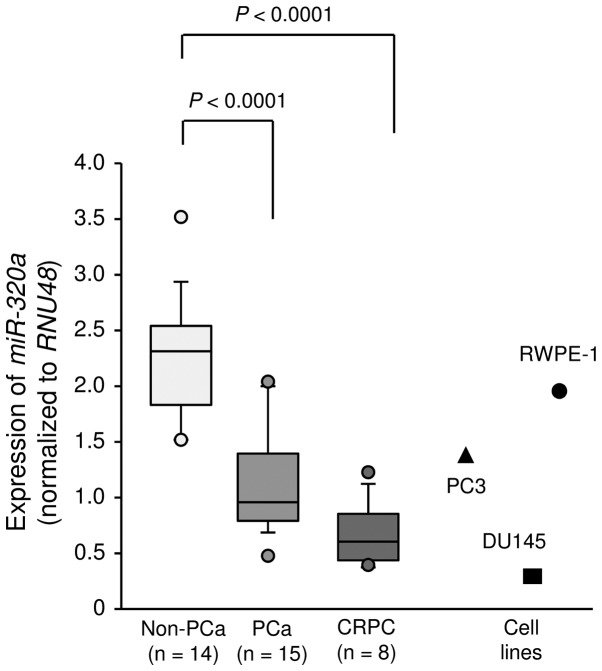
First, we evaluated the expression of miR-320a in clinical prostate specimens and cell lines. The median PSA level of patients with normal prostate tissues was 7.05 ng/ml (range, 4.4–35.5 ng/ml), and that of patients with PCa was 212 ng/ml (range, 11.4–1338 ng/ml). Of the patients with PCa, only 4 patients had organ-confined disease (Table I). The expression levels of miR-320a were significantly lower in cancer tissues than in non-cancerous tissues (P<0.0001; Fig. 1). In PC3 and DU145 cells, expression levels of miR-320a were relatively low compared with those in clinical specimen and were lower than those in normal prostate cells (RWPE-1 cells; Fig. 1).
Figure 1.
Expression levels of miR-320a in clinical prostate cancer specimens and cell lines. Expression levels of miR-320a were determined by RT-PCR. RNU48 was used for normalization.
Effects of miR-320a restoration on the proliferation, migration, and invasion of PCa cells
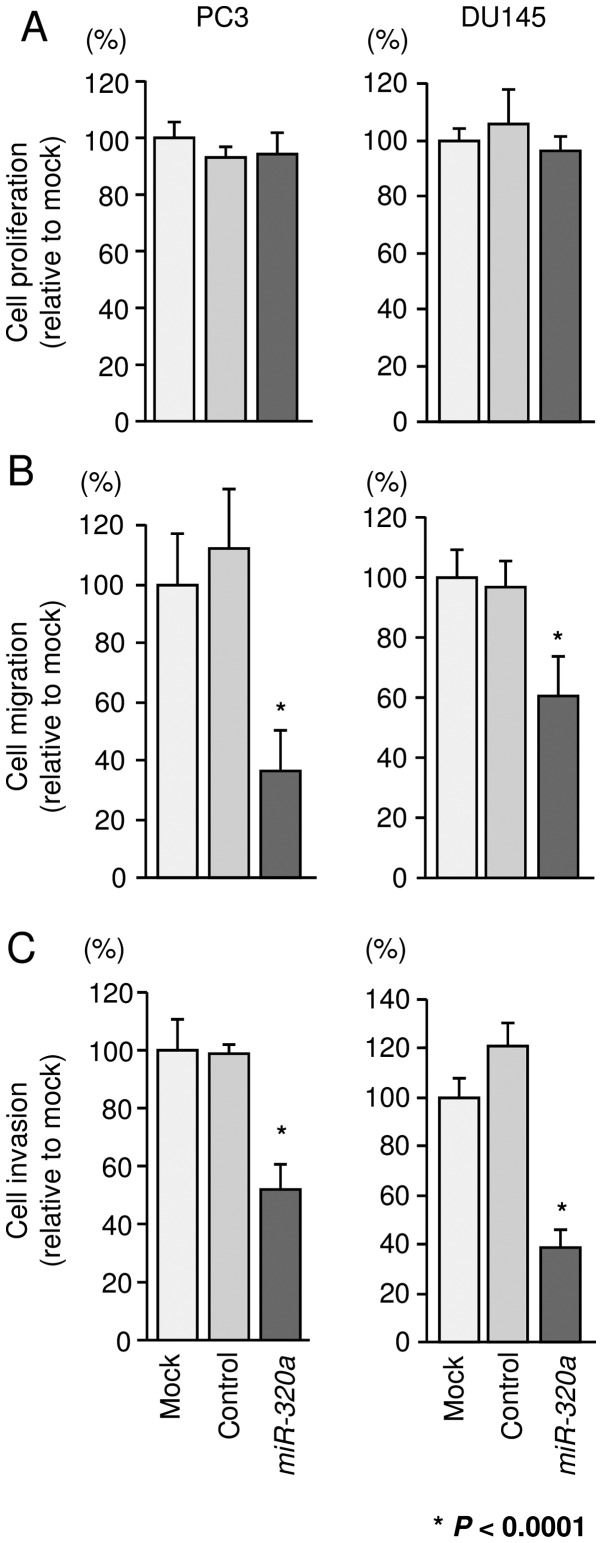
To investigate the functional effects of miR-320a, we performed gain-of-function studies using miRNA transfection into PC3 and DU145 cells. XTT assays revealed that cell proliferation was not inhibited in miR-320a transfectants in comparison with that in mock- or miR-control-transfected PC3 and DU145 cells (Fig. 2A). However, cell migration and invasion activities were significantly inhibited in miR-320a transfectants in comparison with those in mock- or miR-control-transfectants (P<0.0001; Fig. 2B and C).
Figure 2.
Effects of miR-320a restoration on the proliferation, migration, and invasion of PCa cells. (A) Cell proliferation was determined 72 h after transfection using XTT assays. (B) Cell migration activity was determined 48 h after transfection using Boyden chamber migration assays. (C) Cell invasion activity was determined 48 h after transfection using Matrigel invasion assays. *P<0.0001. Experiments were performed triplicate. Bars indicate SDs.
Identification of candidate target genes of miR-320a in PCa cells
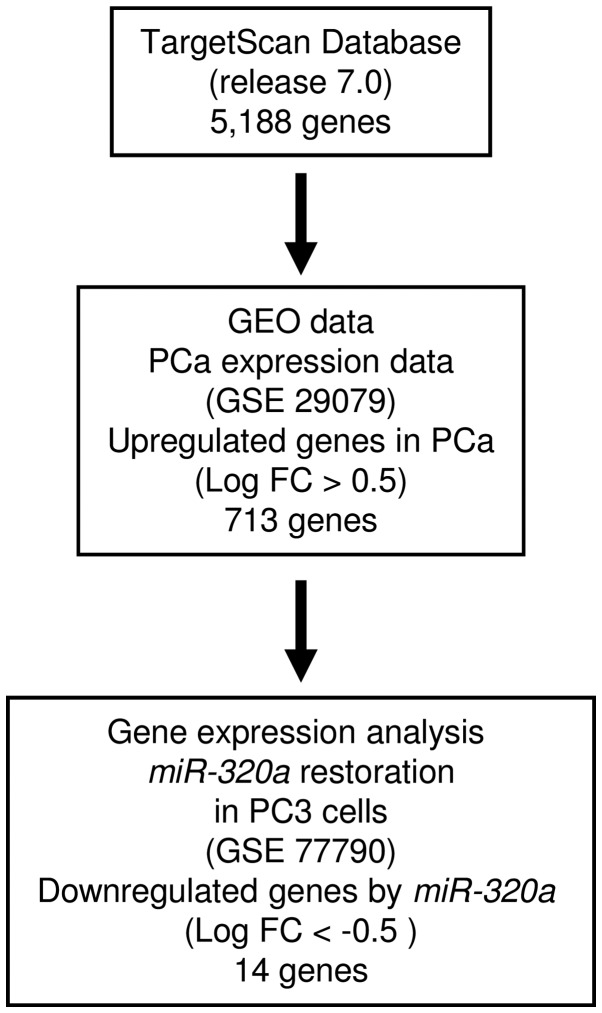
To identify target genes regulated by miR-320a, we performed a combination of in silico analysis and genome-wide gene expression analysis. First, we screened genes having putative 3′-UTR sites matching the seed sequence of miR-320a. Using the TargetScan online program, 5,188 genes with putative target sites for miR-320a were selected. Next, to choose upregulated genes in PCa clinical specimens compared with normal prostatic tissue, the genes were analyzed with available gene expression data from GEO (accession no. GSE29079), which compared 48 normal and 47 PCa tissue samples. As a result, 713 genes upregulated (log2 ratio >0.5) in PCa tissue were selected. To gain further insight into which genes were affected by tumor-suppressive miR-320a in PCa, we carried out genome-wide gene expression analysis using a microarray comparing miR-320a-transfected and mock-transfected PC3 cells (deposited in the GEO database, accession no. GSE77790). Consequently, a total of 14 candidate genes were identified as target genes of miR-320a (Table II). Of these, we focused on the LAMP1 gene for further analyses because the functional significance of LAMP1 in PCa had not been reported. A flow chart outlining our strategy for identification of candidate target genes of miR-320a is shown in Fig. 3.
Table II.
Candidate target genes regulated by miR-320a in PCa.
| Gene symbol | Gene name | PC3 miR-320a transfectant | No. of conserved sites | No. of poorly conserved sites | GEO fold change |
|---|---|---|---|---|---|
| CNOT6 | CCR4-NOT transcription complex, subunit 6 | −1.4261212 | 1 | 4 | 0.539992 |
| VDAC1 | Voltage-dependent anion channel 1 | −1.2843819 | 1 | 0 | 0.609059 |
| LAMP1 | Lysosomal-associated membrane protein 1 | −0.8793476 | 1 | 0 | 0.623045 |
| TPD52 | Tumor protein D52 | −0.8678817 | 1 | 0 | 0.606609 |
| SERBP1 | SERPINE1 mRNA binding protein 1 | −0.6758766 | 1 | 0 | 0.709015 |
| HELZ | Helicase with zinc finger | −0.65864474 | 2 | 1 | 0.675884 |
| MYEF2 | Myelin expression factor 2 | −0.6576734 | 1 | 1 | 0.597199 |
| FUS | Fused in sarcoma | −0.6464454 | 1 | 1 | 0.568564 |
| NEDD4L | Neural precursor cell expressed, developmentally downregulated 4-like | −0.6353359 | 1 | 0 | 1.241135 |
| FAM117B | Family with sequence similarity 117, member B | −0.62850374 | 1 | 4 | 0.729023 |
| NUFIP2 | Nuclear fragile X mental retardation protein interacting protein 2 | −0.60588837 | 3 | 1 | 0.775047 |
| ZC3H7B | Zinc finger CCCH-type containing 7B | −0.5742502 | 2 | 1 | 0.571856 |
| RANBP2 | RAN binding protein 2 | −0.55872875 | 1 | 1 | 0.562369 |
| FBXO28 | F-box protein 28 | −0.52747154 | 1 | 0 | 0.591344 |
Figure 3.
Strategy for identification of candidate target genes of miR-320a. TargetScan Release 7.0 was used to identify genes containing the miR-320a seed sequence in the 3′-UTR. Next, we analyzed the gene expression dataset using the GEO database (GEO accession no. GSE29079). Finally, we attempted to identify miR-320a target genes using miR-320a-transfected PC3 cells (GEO accession no. GSE77790).
LAMP1 was strongly expressed in clinical PCa/CRPC specimens
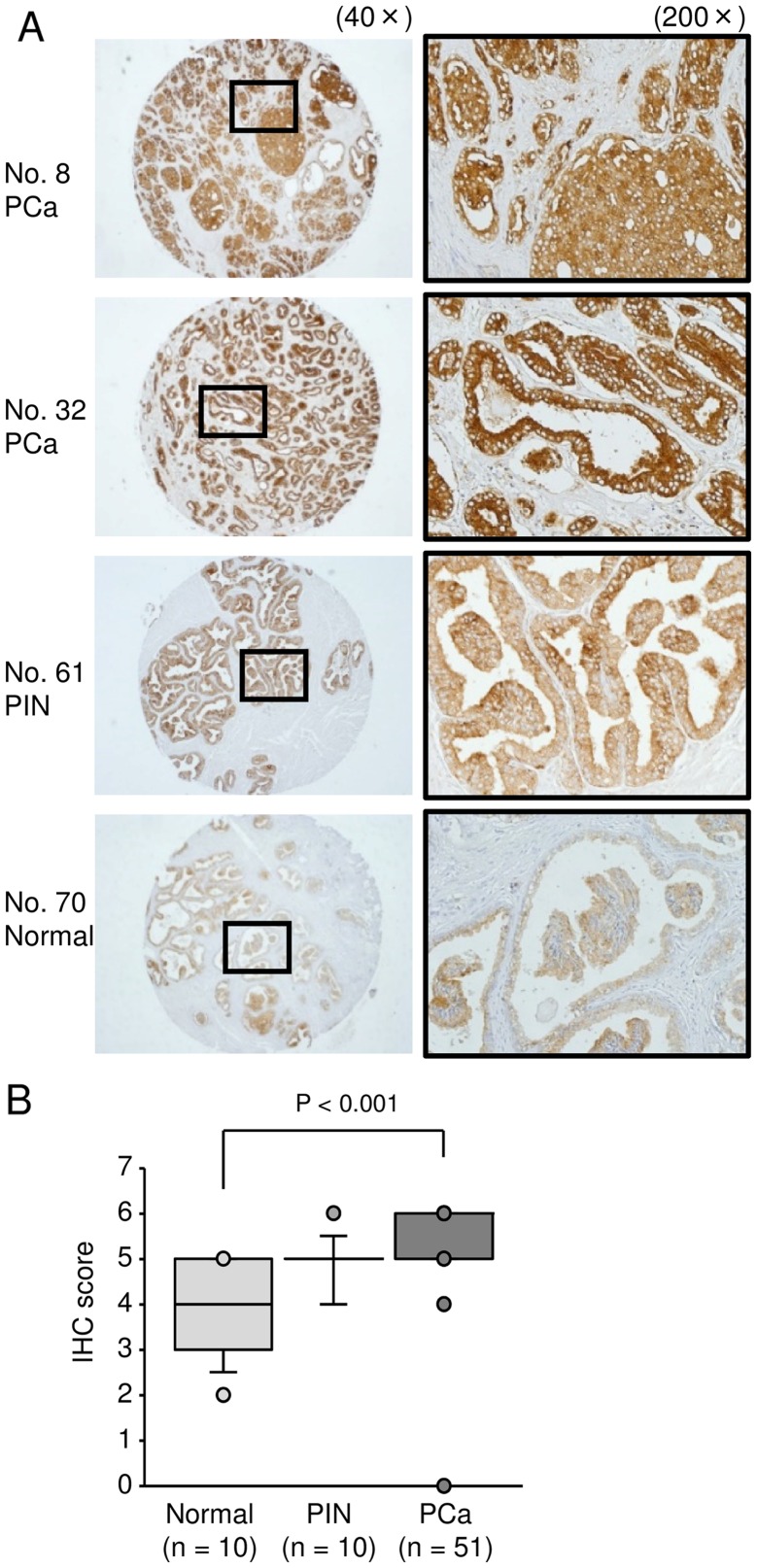
To analyze whether LAMP1 was upregulated in PCa clinical specimens, we carried out immunohistochemical staining of PCa, PIN, prostatic hyperplasia (non-PCa), and CRPC tissues. Using a tissue microarray, we confirmed that LAMP1 expression was significantly increased in PCa specimens compared with that in non-PCa specimens (Fig. 4B; P<0.0001). Representative images of IHC staining for LAMP1 in the tissue microarray are shown in Fig. 4A. In CRPC specimens, we also found strong expression of LAMP1 (Fig. 5).
Figure 4.
Expression of LAMP1 in clinical PCa specimens using a tissue microarray. (A) Representative immunohistochemical staining for LAMP1 in a prostate cancer tissue microarray (Table III; nos. 8, 32, 61 and 70). LAMP1 was strongly expressed in PCa. (B) Comparison of immunohisto-chemical staining of LAMP1 in prostate cancer tissues using IHC scores. LAMP1 expression was significantly higher in PCa tissues than in normal prostate tissues.
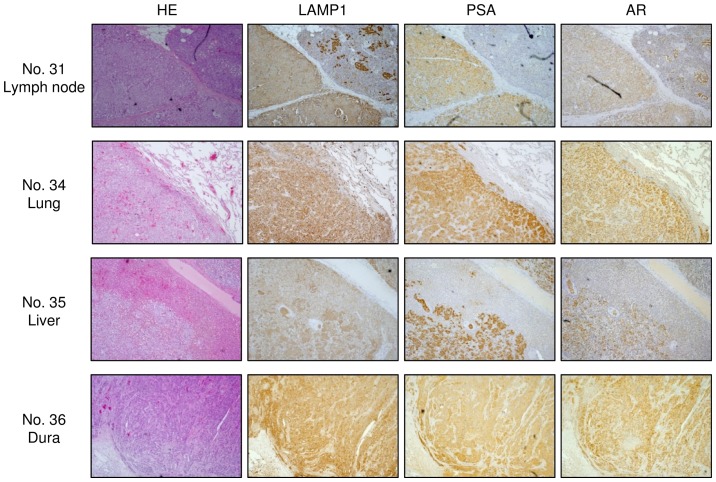
Figure 5.
Expression of LAMP1 in clinical CRPC specimens. Representative immunostaining for LAMP1 in CRPC clinical specimens. PSA and AR were used to confirm prostate cancer metastasis.
miR-320a directly regulated LAMP1 in PCa cells
Next, we performed quantitative RT-PCR and western blotting to analyze whether restoration of miR-320a in PC3 and DU145 cells reduced the expression of LAMP1. As shown in Fig. 6A and B, the expression levels of LAMP1 were significantly repressed by miR-320a transfection in comparison with those in mock- or miR-control-transfected cells.
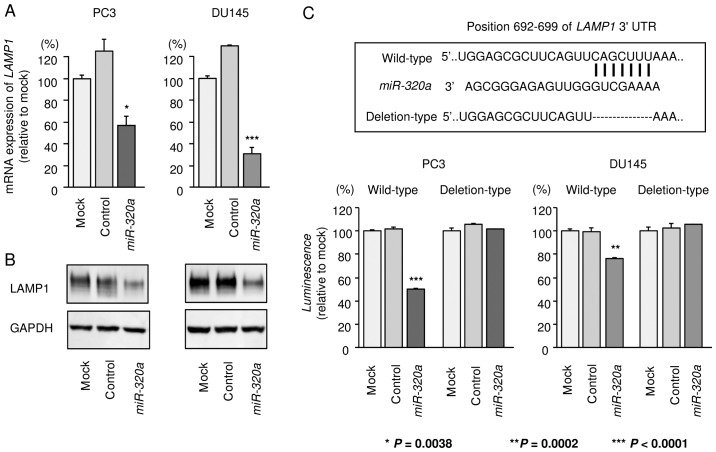
Figure 6.
Direct regulation of LAMP1 by miR-320a. (A) LAMP1 mRNA expression was determined at 96 h after transfection of miR-320a. GAPDH was used as an internal control. (B) LAMP1 protein expression 72 h after transfection with miR-320a. GAPDH was used as a loading control. (C) Luciferase reporter assays using a vector encoding the putative miR-320a target site in the LAMP1 3′-UTR (wild-type and deletion constructs). *P=0.0038, **P=0.0002, and ***P<0.0001. Experiments were performed in triplicate. Bars indicate SDs.
Next, to determine whether LAMP1 mRNA was directly regulated by miR-320a, we performed luciferase reporter assays. We used vectors encoding either the partial wild-type sequence of the 3′-UTR of LAMP1, including the predicted miR-320a target site (position 692–699 of the LAMP1 3′-UTR), or deletion vectors lacking this position. We found that the luminescence intensities were significantly reduced by transfection with miR-320a and the vector carrying the wild-type 3′-UTR of LAMP1, whereas transfection with the deletion vector blocked the decrease of luminescence in PC3 and DU145 cells. These data suggested that miR-320a bound directly to this site in the 3′-UTR of LAMP1 (Fig. 6C).
Effects of silencing LAMP1 on cell proliferation, migration, and invasion in PCa cell lines
To investigate the functional role of LAMP1, we performed loss-of-function studies using si-LAMP1 transfectants. First, we evaluated the knockdown efficiency of si-LAMP1 transfection in PC3 and DU145 cells. Real-time PCR and western blotting indicated that two siRNAs (si-LAMP1-1 and si-LAMP1-2) could effectively reduce the expression of LAMP1 in PC3 and DU145 cells (Fig. 7A and B).
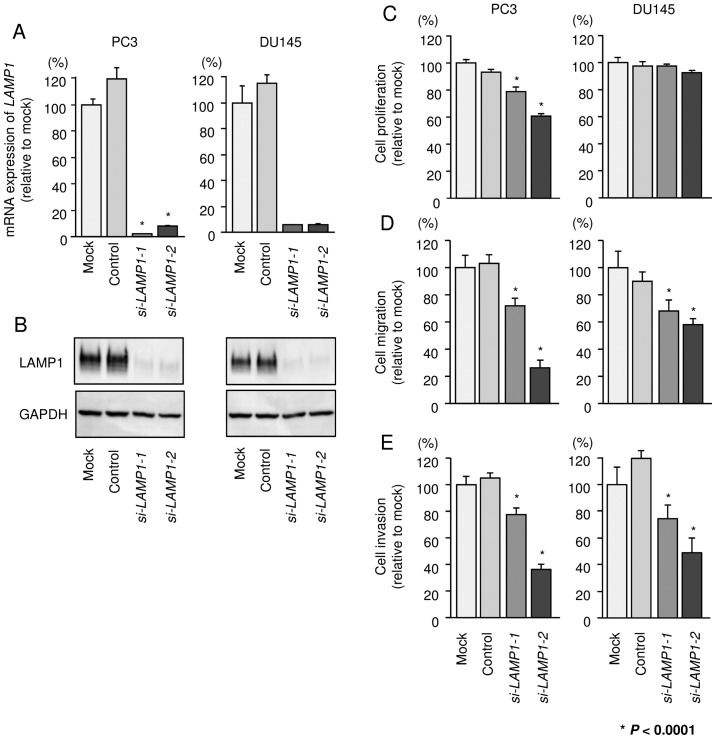
Figure 7.
Effects of silencing LAMP1 on cell proliferation, migration, and invasion in PCa cell lines. (A) LAMP1 mRNA expression was determined at 72 h after transfection with si-LAMP1. GAPDH was used as an internal control. (B) LAMP1 protein expression was evaluated by western blotting at 72 h after transfection with si-LAMP1. GAPDH was used as a loading control. (C) Cell proliferation was determined by XTT assays. (D) Cell migration activity was determined by Boyden chamber migration assays. (E) Cell invasion activity was determined by Matrigel invasion assays. *P<0.0001. Experiments were performed in triplicate. Bars indicate SDs.
Using these two siRNAs, we carried out functional analyses. XTT assays demonstrated that cell proliferation was inhibited only in PC3 cells by si-LAMP1 transfection (Fig. 7C). Cell migration and invasion activities were significantly inhibited in si-LAMP1 transfectants in both PC3 and DU145 in comparison with mock or negative control transfectants (Fig. 7D and E).
Downstream genes affected by silencing of LAMP1 in PCa cells
To further investigate which genes are modulated by LAMP1 signaling, we performed genome-wide gene expression analysis using si-LAMP1 in PC3 cells. A SurePrint G3 Human GE 60K v3 microarray was used for genome-wide expression analysis. The raw data were submitted to the GEO database (accession no. GSE77790). In this study, we selected significantly downregulated genes by si-LAMP1-1 or si-LAMP1-2 transfection [Log2(si-LAMP1/mock) <-2.0]. LAMP1 was the most significantly downregulated gene, indicating the reliability of this array analysis. Genes significantly downregulated by silencing of LAMP1 are shown in Table IV.
Table IV.
Genes downregulated by silencing of LAMP1 in PC3 cells.
| Gene symbol | Gene name | Log2(si-LAMP1/mock) |
|---|---|---|
| LAMP1 | Lysosomal-associated membrane protein 1 | −5.447 |
| ANLN | Anillin, actin binding protein | −2.699 |
| KIF2C | Kinesin family member 2C | −2.610 |
| KIF14 | Kinesin family member 14 | −2.595 |
| AXL | AXL receptor tyrosine kinase | −2.594 |
| CCNA2 | Cyclin A2 | −2.581 |
| SPC25 | SPC25, NDC80 kinetochore complex component | −2.543 |
| NEK2 | NIMA-related kinase 2 | −2.507 |
| ESCO2 | Establishment of sister chromatid cohesion N-acetyltransferase 2 | −2.478 |
| MYL10 | Myosin, light chain 10, regulatory | −2.464 |
| HMGB2 | High mobility group box 2 | −2.440 |
| HIST2H3A | Histone cluster 2, H3a | −2.432 |
| MZT1 | Mitotic spindle organizing protein 1 | −2.416 |
| CCNE2 | Cyclin E2 | −2.404 |
| NUF2 | NUF2, NDC80 kinetochore complex component | −2.403 |
| CENPF | Centromere protein F, 350/400 kDa | −2.378 |
| RRM2 | Ribonucleotide reductase M2 | −2.377 |
| CDK1 | Cyclin-dependent kinase 1 | −2.319 |
| SKA1 | Spindle and kinetochore associated complex subunit 1 | −2.301 |
| PBK | PDZ binding kinase | −2.293 |
| TOP2A | Topoisomerase (DNA) IIα 170 kDa | −2.274 |
| CASC5 | Cancer susceptibility candidate 5 | −2.233 |
| lnc-AKR1C2-3 | lnc-AKR1C2-3:2 | −2.224 |
| NUSAP1 | Nucleolar and spindle associated protein 1 | −2.212 |
| CEP55 | Centrosomal protein 55 kDa | −2.201 |
| UTS2 | Urotensin 2 | −2.188 |
| SKA3 | Spindle and kinetochore associated complex subunit 3 | −2.185 |
| HIST1H3B | Histone cluster 1, H3b | −2.184 |
| G3BP2 | GTPase activating protein (SH3 domain) binding protein 2 | −2.174 |
| SPC248 | SPC24, NDC80 kinetochore complex component | −2.170 |
| CENPA | Centromere protein A | −2.135 |
| GNB4 | Guanine nucleotide binding protein (G protein), β polypeptide 4 | −2.131 |
| MKI67 | Marker of proliferation Ki-67 | −2.119 |
| TK1 | Thymidine kinase 1, soluble | −2.118 |
| MKI67 | Marker of proliferation Ki-67 | −2.115 |
| GLUD2 | Glutamate dehydrogenase 2 | −2.102 |
| TYMS | Thymidylate synthetase | −2.099 |
| E2F8 | E2F transcription factor 8 | −2.087 |
| LOC255187 | Uncharacterized LOC255187 | −2.076 |
| KIF4A | Kinesin family member 4A | −2.073 |
| CREG2 | Cellular repressor of E1A-stimulated genes 2 | −2.073 |
| FAM83D | Family with sequence similarity 83, member D | −2.068 |
| HIST1H1D | Histone cluster 1, H1d | −2.063 |
| DTL | Denticleless E3 ubiquitin protein ligase homolog (Drosophila) | −2.062 |
| FAM72D | Family with sequence similarity 72, member D | −2.054 |
| KIF15 | Kinesin family member 15 | −2.042 |
| ANXA1 | Annexin A1 | −2.022 |
| FAM72A | Family with sequence similarity 72, member A | −2.020 |
| NMU | Neuromedin U | −2.019 |
| BIRC5 | Baculoviral IAP repeat containing 5 | −2.019 |
| BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | −2.016 |
| E2F2 | E2F transcription factor 2 | −2.015 |
| RAB31 | RAB31, member RAS oncogene family | −2.010 |
Discussion
Metastatic PCa initially responds to androgen-deprivation therapy; however, the cancer cells gradually acquire resistance to first-line androgen-deprivation therapy and consequently progress to CRPC. There are no effective treatments for preventing progression of metastasis in patients with CRPC, and this condition significantly affects the survival rates of men with advanced PCa (2,19). Therefore, developing a deeper understanding of the molecular mechanisms of metastatic pathways in advanced PCa will facilitate the development of novel treatment options for the disease. Recently, many studies have shown that aberrantly expressed miRNAs disrupt normally regulated RNA networks in cancer cells, and these events trigger cancer development, progression, and metastasis (4,10). Based on this background, we have identified tumor-suppressive miRNAs that specifically block cancer cell migration and invasion in PCa cells (9,11–18,20).
Our recent studies indicated that the miR-221/miR-222 cluster is significantly downregulated in PCa and CRPC specimens and suppresses cancer cell aggressiveness by targeting Ecm29, a scaffold protein that links the 26S proteasome to motor proteins (17). Moreover, we showed that tumor-suppressive miR-223 inhibits cancer cell migration and invasion by targeting ITGA3 and ITGB1 (18). Interestingly, knockdown of ITGB1 significantly inhibits downstream oncogenic signaling pathways, including the focal adhesion kinase (FAK), AKT, and extracellular signal-regulated kinase (ERK) pathways (18). Thus, suppression of ITGA3/ITGB1 signaling in PCa cells may have applications in the development of novel therapies for PCa.
In this study, we focused on miR-320a because our miRNA signatures of hormone-naïve PCa and CRPC showed that miR-320a was significantly downregulated in cancer tissues (9,17). We validated the low expression of miR-320a in hormone-naïve PCa and CRPC clinical specimens. Restoration of miR-320a significantly inhibited cancer cell migration and invasion in both PC3 and DU145 cell lines, suggesting that miR-320a acted as a tumor-suppressive miRNA in PCa cells.
The anticancer effects of miR-320a have been reported in PCa and various other types of cancers (21–23). For example, overexpression of miR-320 inhibits β-catenin expression and the expression of cancer stem cell markers, such as CD44, Oct-4, and CD133, in PCa cells (21). β-catenin is a multi-functional protein that regulates cell adhesion and functions as a transcriptional coactivator by interacting with several transcriptional factors (21,22,24). Accumulating evidence has shown that β-catenin colocalizes with AR, and these events are more frequently observed in CRPC than in hormone-naïve PCa (25). Therefore, β-catenin/AR signaling may be potential therapeutic target for the management of CRPC. Another study demonstrated that miR-320a expression is reduced in primary salivary adenoid cystic carcinoma (SACC) with metastasis compared with that in SACC without metastasis (26). Overexpression of miR-320a inhibits the adhesion, invasion, and migration of SACC cells through regulation of ITGB3 (26). Chronic myeloid leukemia (CML) is a myeloproliferative disease involving the BCR/ABL fusion protein. Interestingly, a recent study showed that miR-320a expression is reduced in K562 cells and CML cancer stem cells. Additionally, restoration of miR-320a inhibits K562 cell migration, invasion, and proliferation and promotes apoptosis by targeting the BCR/ABL oncogene (27). These studies indicated that continuous analysis of tumor-suppressive miR-320a-regulated oncogenic pathways may provide insights into novel RNA networks in cancer cells.
To better understand PCa progression and metastasis, we identified miR-320a target genes using a combination of in silico and genome-wide gene expression analyses. Recent miRNA studies in our laboratory have utilized this strategy to identify novel molecular targets and pathways regulated by tumor-suppressive miRNAs in several cancers, including PCa (17,18). A total of 14 putative target genes of miR-320a were identified in this study. Among these genes, we focused on the LAMP1 gene because few reports have described the role of this target in PCa. Here, we found that downregulation of LAMP1 inhibited the migration and invasion of cancer cells. Additionally, overexpression of LAMP1 was observed in hormone-naïve PCa and CRPC clinical specimens.
LAMP1 is a heavily glycosylated lysosomal membrane protein that is expressed mainly in the endosome-lysosomal membrane of cells (28,29). High expression of LAMP1 on the plasma membrane has been observed in several metastatic cell lines, including human melanoma, colon carcinoma, fibrosarcoma, and myelomonocytic leukemia cell lines (30–33). Previous studies have indicated that cell surface glycosylation in cancer cells differs from that in normal cells (30). Additionally, the expression of β1,6 branched N-oligosaccharides on the cell surface in several human cancers has been shown to be correlated with the malignant potential of the cells (34). LAMP1 acts as a major carrier of β1,6 branched N-oligosaccharides in B16 melanoma cells (31). A recent study also showed that LAMP1 expression on the cell surface enhances lung metastasis by providing ligands for galectin-3 (Gal-3), a member of the multifunctional β-galactoside binding lectin family (31). Gal-3 has high affinity for β-1,6-N-acetylglucosamine branched glycans. This interaction mediates binding of the lectin to many glycoproteins in the cell membrane, such as cadherin, integrins, and growth factor receptors, and these events may enhance and promote cancer cell growth and metastasis (35). Several studies have demonstrated that Gal-3 plays pivotal roles in cancer progression and aggressiveness by regulating cell proliferation, apoptosis, invasion, and metastasis (35,36). Moreover, overexpression of Gal-3 promotes PCa cell progression, and its heterogeneous expression may be associated with different PCa subtypes (37). These studies indicate that cell surface expression of LAMP1 may affect Gal-3 activation and mediate multiple stages of cancer progression and metastasis.
In this study, we identified LAMP1-mediated cancer pathways by using genome-wide gene expression analysis of si-LAMP1-transfected cells. Our data showed that several genes known to contribute to cancer cell aggressiveness, including ANLN, AXL, CCNA2 and CENPF (16,38–40), were involved in LAMP1 downstream pathways. The identification of these novel molecular targets mediated by the miR-320a/LAMP1 axis may lead to a better understanding of PCa progression and metastasis.
In conclusion, downregulation of miR-320a was observed in PCa and CRPC clinical specimens, and this miRNA was shown to function as a tumor-suppressive miRNA in PCa cells. LAMP1 was directly regulated by tumor-suppressive miR-320a and contributed to cancer cell aggressiveness. The identification of novel molecular pathways regulated by the miR-320a/LAMP1 axis may lead to a better understanding of PCa progression and metastasis.
Acknowledgements
This study was supported by the KAKENHI, grant numbers (C) 15K10801, (C) 15K20070, (C) 15K20071, and (B) 25293333.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Sridhar SS, Freedland SJ, Gleave ME, Higano C, Mulders P, Parker C, Sartor O, Saad F. Castration-resistant prostate cancer: From new pathophysiology to new treatment. Eur Urol. 2014;65:289–299. doi: 10.1016/j.eururo.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 10.Goto Y, Kurozumi A, Enokida H, Ichikawa T, Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol. 2015;22:242–252. doi: 10.1111/iju.12700. [DOI] [PubMed] [Google Scholar]
- 11.Goto Y, Kojima S, Nishikawa R, Enokida H, Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T, Seki N. The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget. 2014;5:7748–7759. doi: 10.18632/oncotarget.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishikawa R, Goto Y, Kojima S, Enokida H, Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y, et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol. 2014;45:401–410. doi: 10.3892/ijo.2014.2437. [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa R, Goto Y, Sakamoto S, Chiyomaru T, Enokida H, Kojima S, Kinoshita T, Yamamoto N, Nakagawa M, Naya Y, et al. Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci. 2014;105:802–811. doi: 10.1111/cas.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M, Goto Y, Matsushita R, Kurozumi A, Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, et al. MicroRNA-26a/b directly regulate La-related protein 1 and inhibit cancer cell invasion in prostate cancer. Int J Oncol. 2015;47:710–718. doi: 10.3892/ijo.2015.3043. [DOI] [PubMed] [Google Scholar]
- 15.Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, et al. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa R, Goto Y, Kurozumi A, Matsushita R, Enokida H, Kojima S, Naya Y, Nakagawa M, Ichikawa T, Seki N. MicroRNA-205 inhibits cancer cell migration and invasion via modulation of centromere protein F regulating pathways in prostate cancer. Int J Urol. 2015;22:867–877. doi: 10.1111/iju.12829. [DOI] [PubMed] [Google Scholar]
- 17.Goto Y, Kojima S, Nishikawa R, Kurozumi A, Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M, et al. MicroRNA expression signature of castration-resistant prostate cancer: The microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br J Cancer. 2015;113:1055–1065. doi: 10.1038/bjc.2015.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, et al. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2016;107:84–94. doi: 10.1111/cas.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: Emerging therapeutic strategies. Nat Rev Clin Oncol. 2011;8:357–368. doi: 10.1038/nrclinonc.2011.127. [DOI] [PubMed] [Google Scholar]
- 20.Goto Y, Nishikawa R, Kojima S, Chiyomaru T, Enokida H, Inoguchi S, Kinoshita T, Fuse M, Sakamoto S, Nakagawa M, et al. Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett. 2014;588:1973–1982. doi: 10.1016/j.febslet.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, Yeh SD, Hong TM, Chen YL. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 22.Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun. 2012;420:787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 23.Lü M, Ding K, Zhang G, Yin M, Yao G, Tian H, Lian J, Liu L, Liang M, Zhu T, et al. MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting ARPP-19 and ERRγ. Sci Rep. 2015;5:8735. doi: 10.1038/srep08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmans R, Städeli R, Basler K. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol. 2005;15:1207–1211. doi: 10.1016/j.cub.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama NN, Shao S, Hoang BH, Mercola D, Zi X. Wnt signaling in castration-resistant prostate cancer: Implications for therapy. Am J Clin Exp Urol. 2014;2:27–44. [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Liu B, Lin Z, Yao Y, Chen Y, Li Y, Chen J, Yu D, Tang Z, Wang B, et al. MiR-320a acts as a prognostic factor and Inhibits metastasis of salivary adenoid cystic carcinoma by targeting ITGB3. Mol Cancer. 2015;14:96. doi: 10.1186/s12943-015-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xishan Z, Ziying L, Jing D, Gang L. MicroRNA-320a acts as a tumor suppressor by targeting BCR/ABL oncogene in chronic myeloid leukemia. Sci Rep. 2015;5:12460. doi: 10.1038/srep12460. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Jensen SS, Aaberg-Jessen C, Christensen KG, Kristensen B. Expression of the lysosomal-associated membrane protein-1 (LAMP-1) in astrocytomas. Int J Clin Exp Pathol. 2013;6:1294–1305. [PMC free article] [PubMed] [Google Scholar]
- 29.Parkinson-Lawrence EJ, Dean CJ, Chang M, Hopwood JJ, Meikle PJ, Brooks DA. Immunochemical analysis of CD107a (LAMP-1) Cell Immunol. 2005;236:161–166. doi: 10.1016/j.cellimm.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Furuta K, Ikeda M, Nakayama Y, Nakamura K, Tanaka M, Hamasaki N, Himeno M, Hamilton SR, August JT. Expression of lysosome-associated membrane proteins in human colorectal neoplasms and inflammatory diseases. Am J Pathol. 2001;159:449–455. doi: 10.1016/S0002-9440(10)61716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal AK, Gude RP, Kalraiya RD. Regulation of melanoma metastasis to lungs by cell surface lysosome associated membrane protein-1 (LAMP1) via galectin-3. Biochem Biophys Res Commun. 2014;449:332–337. doi: 10.1016/j.bbrc.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 32.Sarafian V, Jadot M, Foidart JM, Letesson JJ, Vanden Brûle F, Castronovo V, Wattiaux R, Coninck SW. Expression of Lamp-1 and Lamp-2 and their interactions with galectin-3 in human tumor cells. Int J Cancer. 1998;75:105–111. doi: 10.1002/(SICI)1097-0215(19980105)75:1<105::AID-IJC16>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 33.Mane SM, Marzella L, Bainton DF, Holt VK, Cha Y, Hildreth JE, August JT. Purification and characterization of human lysosomal membrane glycoproteins. Arch Biochem Biophys. 1989;268:360–378. doi: 10.1016/0003-9861(89)90597-3. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan V, Bane SM, Kawle PD, Naresh KN, Kalraiya RD. Altered melanoma cell surface glycosylation mediates organ specific adhesion and metastasis via lectin receptors on the lung vascular endothelium. Clin Exp Metastasis. 2005;22:11–24. doi: 10.1007/s10585-005-2036-2. [DOI] [PubMed] [Google Scholar]
- 35.Fortuna-Costa A, Gomes AM, Kozlowski EO, Stelling MP, Pavão MS. Extracellular galectin-3 in tumor progression and metastasis. Front Oncol. 2014;4:138. doi: 10.3389/fonc.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed H, AlSadek DM. Galectin-3 as a potential target to prevent cancer metastasis. Clin Med Insights Oncol. 2015;9:113–121. doi: 10.4137/CMO.S29462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Nangia-Makker P, Tait L, Balan V, Hogan V, Pienta KJ, Raz A. Regulation of prostate cancer progression by galectin-3. Am J Pathol. 2009;174:1515–1523. doi: 10.2353/ajpath.2009.080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dachineni R, Ai G, Kumar DR, Sadhu SS, Tummala H, Bhat GJ. Cyclin A2 and CDK2 as novel targets of aspirin and salicylic acid: A potential role in cancer prevention. Mol Cancer Res. 2016;14:241–252. doi: 10.1158/1541-7786.MCR-15-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaltriti M, Elkabets M, Baselga J. Molecular pathways: AXL, a membrane receptor mediator of resistance to therapy. Clin Cancer Res. 2016;22:1313–1317. doi: 10.1158/1078-0432.CCR-15-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W, Wang Z, Shen N, Pi W, Jiang W, Huang J, Hu Y, Li X, Sun L. Knockdown of ANLN by lentivirus inhibits cell growth and migration in human breast cancer. Mol Cell Biochem. 2015;398:11–19. doi: 10.1007/s11010-014-2200-6. [DOI] [PubMed] [Google Scholar]