Abstract
Ovarian cancer is the second most common gynaecological malignancy and was diagnosed in over 7,000 women in 2011 in the UK. There are currently no reliable biomarkers available for use in a regular screening assay for ovarian cancer and due to characteristic late presentation (78% in stages III and IV) ovarian cancer has a low survival rate (35% after 10 years). The mTOR pathway is a central regulator of growth, proliferation, apoptosis and angiogenesis; providing balance between available resources such as amino acids and growth factors, and stresses such as hypoxia, to control cellular behaviour accordingly. Emerging data links mTOR with the aetiopathogenesis of ovarian cancer. We hypothesised that mTOR inhibitors could play a therapeutic role in ovarian cancer treatment. In this study we began by validating the expression of four main mTOR pathway components, mTOR, DEPTOR, rictor and raptor, at gene and protein level in in vitro models of endometrioid (MDAH-2774) and clear cell (SKOV3) ovarian cancer using qPCR and ImageStream technology. Using a wound healing assay we show that inhibition of the mTOR pathway using rapamycin, rapalogues, resveratrol and NVP BEZ-235 induces a cytostatic and not cytotoxic response up to 18 h in these cell lines. We extended these findings up to 72 h with a proliferation assay and show that the effects of inhibition of the mTOR pathway are primarily mediated by the dephosphorylation of p70S6 kinase. We show that mTOR inhibition does not involve alteration of mTOR pathway components or induce caspase 9 cleavage. Preclinical studies including ovarian tissue of ovarian cancer patients, unaffected controls and patients with unrelated gynaecological conditions show that DEPTOR is reliably upregulated in ovarian cancer.
Keywords: ovarian cancer, rapalogues, resveratrol, S6K
Introduction
Ovarian carcinoma is the gynaecologic malignancy associated with the highest mortality in industrialised countries, with a reported 5-year survival rate of <30% (1). The prognosis for patients with ovarian cancer is determined by conventional factors such as surgical stage and histological grade and type. Nevertheless, no single molecular profile has helped identify the most aggressive tumours or aided in the guidance of suitable therapeutic strategies for specific patients. Due to the indefinite presentation of ovarian cancer, it is often not diagnosed until it has reached stage 3 or 4, at which point the 5-year survival rate drops significantly; the mortality rate has remained unchanged for >30 years. Despite this, due to the lack of a known, reliable biomarker for the disease, there is currently no ovarian cancer screening programme used by the National Health Service. Many carcinomas activate growth factor receptor signalling pathways that exhibit genetic alterations involving either the receptor, or other factors that drive the proliferation. Interestingly, several pathways converge on the highly conserved serine/threonine kinase mTOR (2).
The mTOR pathway is an integral hub of growth and proliferation; gauging external energy, growth factor and stress signals. It influences processes such as protein synthesis, lipid biogenesis and autophagy to find the balance between anabolic and catabolic mechanisms in relation to the whole-organism state. The PI3K/Akt/mTOR pathway is activated in advanced stage disease and inhibition of this pathway with inhibitors to Akt or its downstream effector, mTOR, increases chemo-sensitivity to paclitaxel in ovarian carcinoma cell lines (3). Cell signalling by mTOR plays a critical role in protein synthesis and proliferation of both normal and malignant cells. mTOR and one of its substrates, S6 kinase, is activated in ovarian cancer cells and inhibition of the mTOR pathways has anti-proliferative effects (4). Rapamycin inhibits the growth of a broad spectrum of malignancies including pancreatic cancer, leukaemia and B-cell lymphoma (5). Rapamycin (an mTOR inhibitor) may act as a substrate for the MDR transporter P-glycoprotein and consequently limit its utility in some tumours (6). In a transgenic mouse model of ovarian cancer, treatment with the rapamycin analogue everolimus (an inhibitor of mTOR) exhibited a delay in cancer progression (7). Deforolimus, another rapamycin analogue has been shown to inhibit sarcoma and endometrial cancer growth both in vitro and in vivo (8). In addition, the rapalogue temsirolimus has exhibited therapeutic benefit when administered to patients with clear cell carcinoma of the ovary (9).
Moreover, a limitation to successful cancer chemotherapy treatment is the acquisition of drug resistance. In advanced-stage ovarian cancer, mTOR pathway is upregulated, and inhibition of this pathway increases chemosensitivity in ovarian carcinoma cell lines. Previous data from our laboratory has revealed significant upregulation of DEPTOR in paclitaxel-resistant (TaxR) SKOV-3TaxR and PEO1TaxR cell lines. SKOV-3TaxR exhibited downregulation of RICTOR, RAPTOR and mTOR, whereas PEO1-TaxR showed down-regulation of RAPTOR and upregulation of RICTOR and mTOR (10).
In this study, we investigated the effects of rapalogues on ovarian cancer using two cell lines (SKOV3 and MDAH-2774) as in vitro experimental models. We expanded on these observations by mapping the expression of mTOR components (including DEPTOR, rictor, raptor and S6K) in tissue and peripheral blood of ovarian cancer patients.
Materials and methods
Ovarian cancer clinical samples
Gene expression of mTOR, Deptor, Rictor and Raptor were mapped in 12 clinical samples from ovarian cancer patients using qPCR. Clinical samples were of ovarian origin and obtained from patients at the 1st Department of Obstetrics and Gynecology, ‘Papageorgiou’ General Hospital, Medical School, Aristotle University, Thessaloniki, Greece. Ethical permission was obtained locally. The majority of ovarian cancers were deemed to be third grade (10 out of 12) and at stage 3 (11 out of 12).
RNA isolation, cDNA synthesis and quantitative RT-PCR
Ovarian tissue (40 mg) was lysed in a Qiagen Tissue Lyser II (Qiagen, Hilden, Germany) for 2 min with a 3-mm stainless steel ball bearing. RNA was extracted from tissue lysate using the GenElute™ mRNA MiniPrep kit (Sigma-Aldrich, MO, USA), a silica membrane/spin column method, and stored at −80°C until further use.
cDNA was synthesised from mRNA using Superscript II (Invitrogen, MA, USA). cDNA concentration was normalised using RNA concentrations determined by NanoDrop (Thermo Scientific, MA, USA) and was synthesised to a concentration of either 500 or 1,000 ng.
Primers
Relative expression of mTOR, DEPTOR, rictor and raptor (Table I) were assessed by quantitative PCR (Q-PCR) on an xxpress® (BJS Biotechnologies, Middlesex, UK) thermal cycler using Kapa SYBR Fast Universal Mastermix (Kapa Biosystems, MA, USA). According to MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines (11), an assessment of the most stably expressed reference genes specific to the samples used must be carried out prior to any qPCR experiment. In light of this, a selection of 8 ovarian clinical samples were assessed using the geNorm human 12 gene kit (Primer Design, Southampton, UK) according to the manufacturer's instructions. Reference gene expression stability was analysed using qbaseplus software (Biogazelle, Zwijnaarde, Belgium). Primers for mTOR, Deptor, Rictor and Raptor were used as previously described (10). qPCR data were analysed using the ΔCq method whereby the Cq of the endogenous control was subtracted from the Cq of the gene of interest and an RQ (relative quantity) value was calculated by finding 2−ΔCq (11,12). Where more than one reference gene was used, the RQ values were averaged. A Student's t-test was used to calculate statistical significance.
Table I.
The primer sequences for the mTOR, Deptor, Rictor and Raptor genes used in qPCR experiments for the clinical samples and the in vitro experiments.
| Name | Product length (bases) | Strand | Size | Sequence (bases) |
|---|---|---|---|---|
| mTOR | 135 | Forward | 20 | tgccaactaccttcggaacc |
| Reverse | 20 | gctcgcttcacctcaaattc | ||
| Deptor | 202 | Forward | 20 | caccatgtgtgtgatgagca |
| Reverse | 20 | tgaaggtgcgctcatacttg | ||
| Rictor | 117 | Forward | 20 | ggaagcctgttgatggtgat |
| Reverse | 20 | ggcagcctgttttatggtgt | ||
| Raptor | 170 | Forward | 20 | actgatggagtccgaaatgc |
| Reverse | 20 | tcatccgatccttcatcctc |
Cell lines and treatments
SKOV3 (human ovarian clear cell adenocarcinoma) and MDAH-2774 (human ovarian endometrioid adenocarcinoma) cell lines were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA) and were used as models for epithelial ovarian cancer. Both cell lines were cultured at 37°C/5% CO2 in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin and 1% L-glutamine (all Gibco, MA, USA). SKOV3 and MDAH-2774 cells were treated with specific mTOR inhibitors (Table II) at two concentrations for three lengths of time, 24, 48 and 72 h.
Table II.
Details of the mTOR pathway inhibitory agents used in this study.
| Inhibitor | Mode of action | Concentrations | |
|---|---|---|---|
| Rap | Allosteric mTOR inhibitor | 20 nM | 100 nM |
| Eve | 20 nM | 100 nM | |
| Def | 100 nM | 1,000 nM | |
| Tem | 10 nM | 100 nM | |
| Res | Dual mTOR (via DEPTOR) and PI3 kinase (via ATP binding site) inhibitor | 25 μM | 50 μM |
| BEZ | Dual mTOR and PI3 kinase inhibitor via ATP binding sites of both | 10 nM | 100 nM |
Western blotting
Proteins extracted from cultured cells following treatments were assessed for cleaved and total caspase 9 and 3 levels, hallmarks of apoptosis, and phospho-p70S6K. Proteins were separated by mass by SDS-PAGE in a 12.5% resolving gel. The separated proteins were then electrophoretically transferred onto a nitrocellulose membrane (Thermo Scientific) in wet-transfer buffer. The membrane was probed using primary antibody for caspases 3 and 9 and phospho (Thr389) p70S6K (Cell Signaling Technology, MA, USA) and an anti-rabbit HRP-conjugated secondary antibody diluted in 5% bovine serum albumin/TBS Tween. The developed western blots were analysed densitometrically using ImageJ software (National Institutes of Health, MD, USA) and cleaved caspase 9 was normalised internally against uncleaved caspase 9. The data were analysed statistically using Student's t-test.
Ovarian tissue microarray
Unstained paraffin-embedded tissue micro-array slides containing 70 ovarian cancers were obtained from US Biomax (MD, USA). The paraffin-embedded slides were deparaffinised and rehydrated by a series of washes in reducing concentrations of ethanol (100, 95, 70 and 50%) followed by rinsing in tap water for 10 min. Antigen retrieval was accomplished by boiling slides in sodium citrate (pH 6.0) for 20 min in a microwave. Slides were washed in 0.4% of PBS Tween for 5 min and then incubated for 15 min in PBS containing 0.3% H2O2 to eliminate endogenous peroxidase activity. Blocking was carried out with 5% goat serum, followed by 48-h incubation with primary antibody (p70S6K). After several washes with PBS, slides were incubated with HRP conjugated secondary antibody for 60 min. Further washing in PBS Tween was carried out for 20 min before performing staining. Slides were then subjected to DAB staining, counterstained with haematoxylin and washed with 0.1% sodium bicarbonate. Slides were analysed for immunoreactivity of p70S6K by a light microscope and positive results were measured by the percentage of positive tumour cells.
Proliferation assay
Proliferation, death and viability of the cells were assessed after treatment using a Countess Automated Cell Counter (Invitrogen) and trypan blue stain. Media was aspirated from the cells which were then incubated with 200 μl of trypLE™ Express (Thermo Scientific) per well and manually agitated to detach the cells. The cells were resuspended in 800 μl of appropriate media to make 1 ml of cell suspension in total. An equal volume of cell suspension was mixed thoroughly with trypan blue stain (0.4%, trypan blue is selectively absorbed by dead cells) and applied to a Countess™ cell counting chamber slide (Invitrogen). Three readings were taken per sample and an average value calculated.
Wound healing assay
Cells were assessed for their ability to close an artificially created gap in cell growth area. One confluent T-75 cell culture flask was seeded per three 6-well plates with 2 ml appropriate media per well and grown until there was a confluent monolayer. A 20-μl pipette tip was used to create a ‘scratch’ in growth area perpendicular to a line drawn on the underside of the well in marker pen. The media was then aspirated and replaced with treated media. Cells were placed under a Zeiss Axiovert 200M microscope and images were recorded at 0, 6, 12 and 18 h. The perpendicular line of marker was used as a landmark to ensure that an image of the same area was taken at each time-point.
ImageStream
Protein expression and localisation of mTOR, DEPTOR, rictor and raptor was investigated using ImageStream high resolution flow cytometry (Merck Millipore, Darmstadt, Germany). Media was aspirated from the cells which were then incubated with TrypLE™ Express (Invitrogen). Washes in PBS were performed to remove debris. The cells were resuspended and incubated in 4% paraformaldehyde (PFA, Sigma, MO, USA) for 7 min and the PFA removed. Further washed in PBS were performed and stored ice-cold at −20°C until further use. The cells were incubated in blocking buffer (5% bovine serum in PBS) for 30 min followed by the appropriate primary antibody (diluted in blocking buffer) overnight at 4°C. Washes were performed with PBS and incubated in secondary antibody (diluted in blocking buffer) for 30 min. After secondary antibody incubation the cells were washed again in PBS to remove any remaining antibody. PBS was removed and the cells were resuspended in 100 μl Accumax (Innovative Cell Technologies, CA, USA) to dissociate any cellular aggregates. Draq5 nuclear stain (1 μl) was added before visualisation on ImageStream.
Statistical analysis
Changes observed in experiments were assessed for statistical significance using the Student's t-test. An assessment for homoscedasticity (variance) of data from each category was made using the F-test. If homoscedasticity was proven, an unpaired, two-tailed Student's t-test was performed to assess significance in all cases as no matched pairs of samples were used. If data were not homoscedastic, an unpaired, two-tailed Student's t-test with Welch's correction was performed to account for variance. All statistical tests were performed using GraphPad Prism® software (GraphPad Software Inc., CA, USA). P-values were denoted on graphs if significant (p<0.05).
Results
Expression of mTOR components in ovarian cancer cell lines
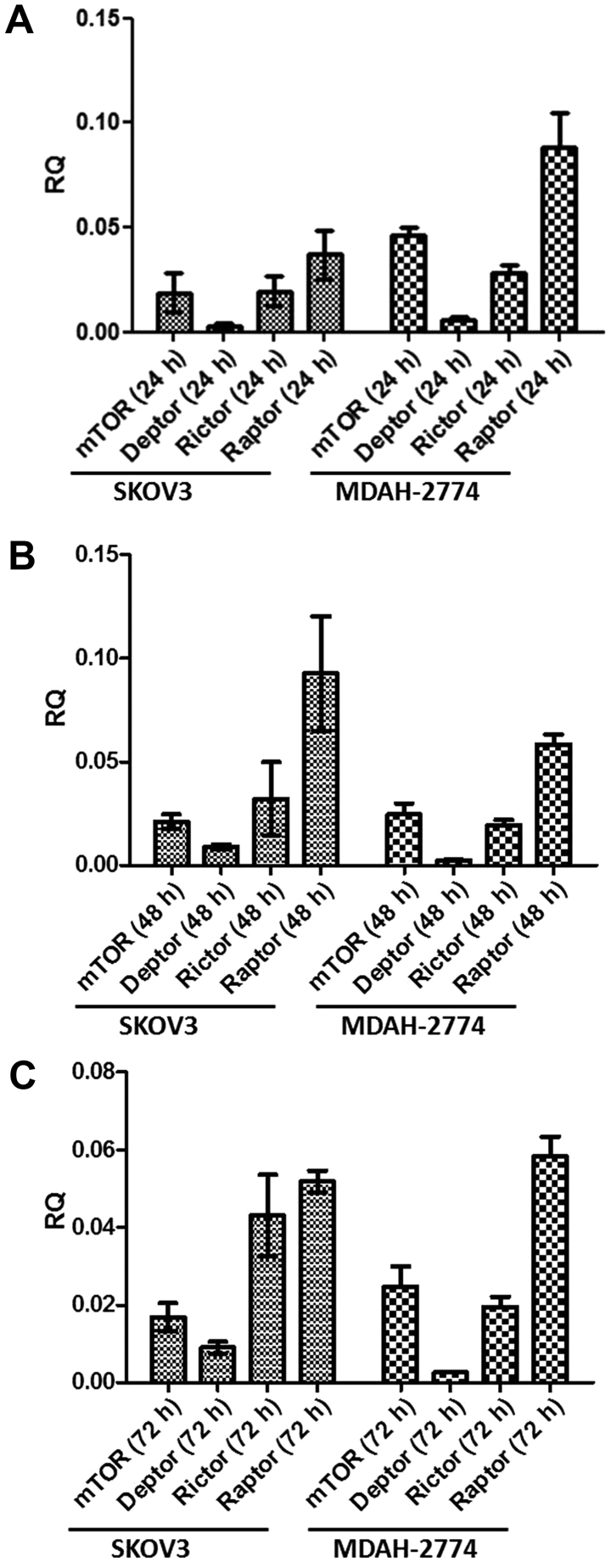
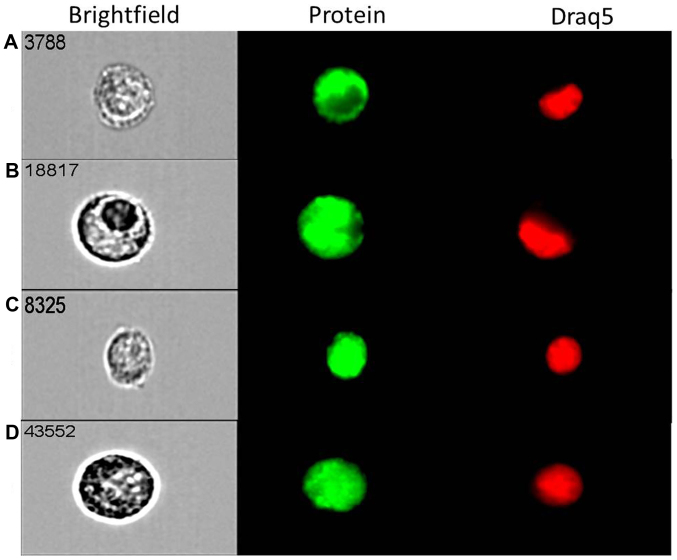
Prior to studying the effects of mTOR inhibitors, we assessed the basal expression of mTORC1 and mTORC2 components in both cell lines over 72 h. All key components, i.e., mTOR, DEPTOR, rictor and raptor are expressed in both cell lines in a similar fashion over the course of 72 h (Fig. 1). The predominant component was raptor, with DEPTOR demonstrating minimal expression in both cell lines under basal conditions. Following qPCR analysis, we used the state-of-the-art ImageStream technology that combines flow cytometry with cell imaging for parallel quantification and visualisation of single cells. Strong cytoplasmic and nuclear expression for both mTOR (Fig. 2A) and DEPTOR (Fig. 2B) was evident in most of ovarian cancer (MDAH-2774) single cells studied. The expression of rictor (Fig. 2C) and raptor (Fig. 2D) was primarily nuclear/cytoplasmic. Similar cellular distribution of these key components of both mTORC1 and mTORC2 complexes was evident in SKOV3 cells (data not shown).
Figure 1.
Gene expression analysis of mTOR pathway components, mTOR, DEPTOR, rictor and raptor, in SKOV3 and MDAH-2774 cells. SKOV3 and MDAH-2774 cells were seeded in 6-well plates at 100,000 cells per well and cultured under basal conditions over a 72-h period. qPCR was performed in cDNA synthesised from extracted mRNA after 24 (A), 48 (B) and 72 h (C) using primers for mTOR, DEPTOR, rictor and raptor corrected with YWHAZ (reference gene). Error bars depict standard deviation.
Figure 2.
ImageStream high resolution flow cytometry analysis of mTOR (A), DEPTOR (B), rictor (C) and raptor (D) protein expression in MDAH-2774 cells grown under basal conditions. MDAH-2774 cells were fixed in paraformaldehyde and stained with antibodies for mTOR, DEPTOR, rictor and raptor and an Alexa Fluor® conjugated secondary antibody before visualisation. mTOR, DEPTOR, rictor and raptor were expressed at protein level in this cell line.
mTOR inhibitors compromise migratory capacity and cell proliferation of ovarian cancer cells in vitro
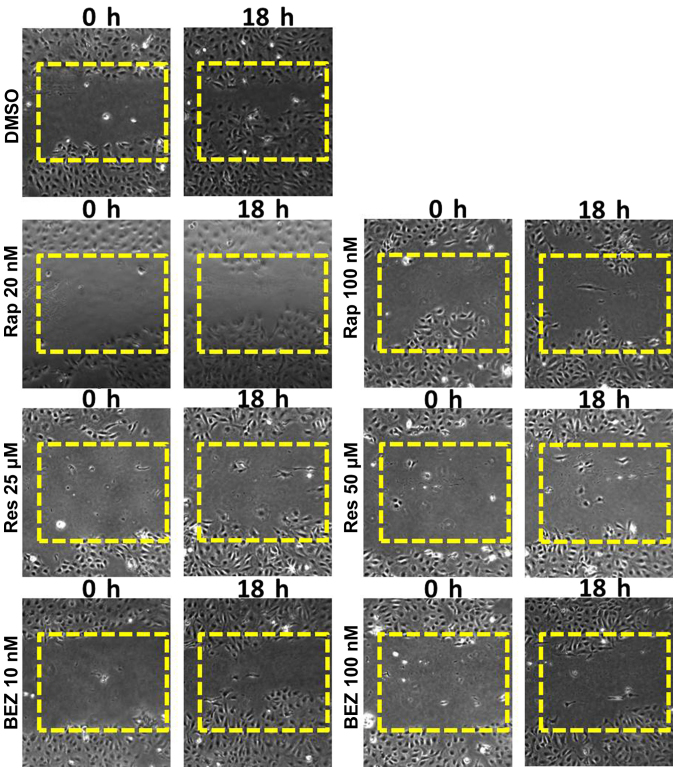
Wound healing assays can give a visual indication of cell proliferative and migratory capacity spatially in vitro. Rapamycin (20 and 100 nM) was selected from the rapalogues (since they have a similar chemical structure) and resveratrol (25 and 50 μM) and BEZ (10 and 100 nM) were used as they are mechanistically different to the rapalogues and one another. SKOV3 cells showed a marked growth into the wound which was not shared by any of the inhibitor-treated cells (Fig. 3). In MDAH-2774 cells, a more modest response was recorded using the same inhibitors (data not shown).
Figure 3.
Wound healing analysis of SKOV3 cells treated with rapamycin (Rap), resveratrol (Res) and NVP BEZ-235 (BEZ). SKOV3 cells were seeded in 6-well plates and grown to a confluent monolayer. A scratch was created in the growth area with a p200 pipette tip and the cells were cultured in media containing either Rap (20 or 100 nM), Res (25 or 50 μM) or BEZ (10 or 100 nM). The ‘wound’ was monitored over an 18-h period and images recorded at 0, 6, 12 and 18 h. mTOR pathway inhibition induced retardation of growth into the created gap.
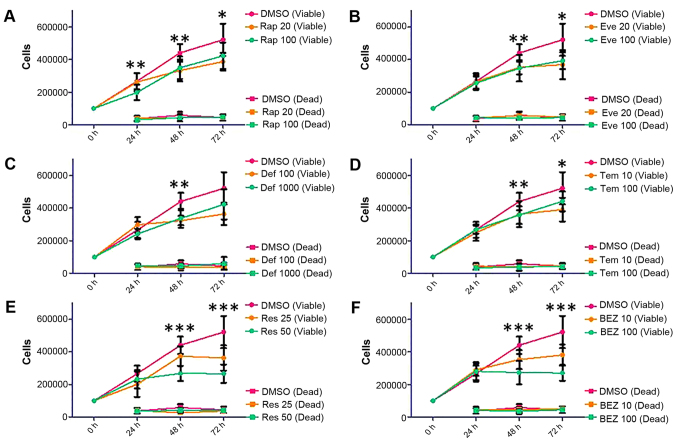
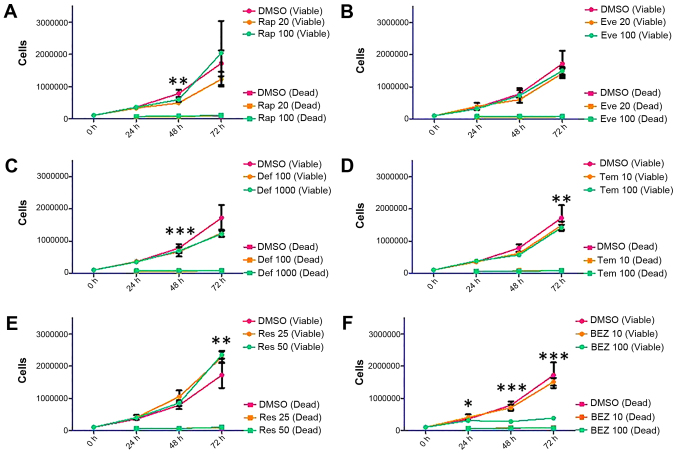
Based on the wound healing data, we have decided to expand on the repertoire of rapalogues; therefore, trypan blue staining was used to make an assessment of the proliferative capacity of SKOV3 and MDAH-2774 cells up to 72 h following treatment with the following inhibitors: rapamycin (Rap, 20 and 100 nM), everolimus (Eve, 20 and 100 nM), deforolimus (Def, 100 and 1,000 nM), temsirolimus (Tem, 10 and 100 nM), resveratrol (Res, 25 and 50 μM), and NVP BEZ-235 (BEZ, 10 and 100 nM). Treatment of the SKOV3 cell line with Rap and the rapalogues showed a non-dose-dependent decrease in viable cells (Fig. 4, Rap 100 nM: p=0.0355 at 72 h, Eve 100 nM: p=0.0047 at 72 h, Tem 100 nM: p=0.0446 at 72 h, Def 1,000 nM did not reach statistical significance: p=0.0597). The decrease in viable cells shown by Res and BEZ was dose-dependent (Fig. 4, Res 50 μM: p<0.0001 at 72 h, BEZ 100 nM: p<0.0001 at 72 h).
Figure 4.
Proliferation and survival analysis of SKOV3 cells following treatment with Rap [(A) 20 and 100 nM], everolimus [(B) Eve, 20 and 100 nM], deforolimus [(C) Def, 100 and 1,000 nM], temsirolimus [(D) Tem, 10 and 100 nM], Res [(E) 25 and 50 μM] and BEZ [(F) 10 and 100 nM]. SKOV3 cells were seeded in 6-well plates at a density of 100,000 cells per well and allowed to proliferate for 24 h before treatment. Cells were treated with mTOR pathway inhibitors for 24, 48 and 72 h. At each time-point, cells were detached and mixed with an equal volume of trypan blue. A count of viable and dead cells was made using a Countess automated cell counter (Invitrogen). Error bars depict standard deviation, *p=0.01–0.05, **p=0.001–0.009, ***p<0.009.
Viable cell count in MDAH-2774 cells was differentially affected by Rap (Rap 100 nM: p=0.0013 at 48 h), rapalogues (Def 1,000 nM: p=0.0004 at 48 h, Tem 100 nM: p=0.0065 at 72 h) or Res (Res 50 μM: p=0.0014 at 72 h) treatment (Fig. 5). BEZ treatment at 10 nM concentration did not have an effect on viable cell count, but at 100 nM, proliferation was almost entirely eliminated (Fig. 5, p<0.0001 at 48 and 72 h).
Figure 5.
Proliferation and survival analysis of MDAH-2774 cells following treatment with Rap [(A) 20 and 100 nM], everolimus [(B) Eve, 20 and 100 nM], deforolimus [(C) Def, 100 and 1,000 nM], temsirolimus [(D Tem, 10 and 100 nM), Res [(E) 25 and 50 μM] and BEZ [(F) 10 and 100 nM)]. MDAH-2774 cells were seeded in 6-well plates at a density of 100,000 cells per well and allowed to proliferate for 24 h before treatment. Cells were treated with mTOR pathway inhibitors for 24, 48 and 72 h. At each time-point, cells were detached and mixed with an equal volume of trypan blue. A count of viable and dead cells was made using a Countess automated cell counter (Invitrogen). Error bars depict standard deviation, *p=0.01–0.05, **p=0.001–0.009, ***p<0.009.
Effects of mTOR inhibitors on caspase 9 and phospho-p70S6K
Cleavage of caspase 9 could be seen after all treatments and time-points in both cell lines. When analysed densitometrically, SKOV3 cells showed a clear trend of highest caspase 9 cleavage at 24 h and lessening over time in each case (Fig. 6A). These changes did not reach statistical significance in comparison to controls; however, significant changes were found between time-points in Rap 20 nM (p=0.0069, 24–72 h), Rap 100 nM (p=0.00159, 24–72 h), Def 1,000 nM (p=0.0496, 24–48 h), Res 25 μM (p=0.0367, 24–48 h and p=0.0353, 24–72 h) and Res 50 μM (p=0.0483, 24–72 h) treatments, indicating that cleavage of caspase 9 does decrease significantly over time in these cases. However, MDAH-2774 cells showed no increase in caspase 9 cleavage upon treatment with mTOR inhibitors (Fig. 6B and C).
Figure 6.
Analysis of caspase 9 cleavage following mTOR pathway inhibition in SKOV3 (A) and MDAH-2774 (B) cells following treatment with Rap (20 and 100 nM), Eve (20 and 100 nM), Def (100 and 1,000 nM), Tem (10 and 100 nM), Res (25 and 50 μM) and BEZ (10 and 100 nM). SKOV3 and MDAH-2774 cells were treated with mTOR pathway inhibitors for 24, 48 and 72 h. At each time-point, cells were lysed and western blots were performed using an antibody recognising cleaved large fragment and total caspase 9. Developed western blots were analysed densitometrically using ImageJ software (National Institutes of Health). Caspase 9 cleavage was increased after 24 h in SKOV3 cells but was not maintained at 48 or 72 h. No increase in caspase 9 cleavage was seen in MDAH-2774 cells. Error bars depict standard deviation. Representative image shows extractions after 72 h (C).
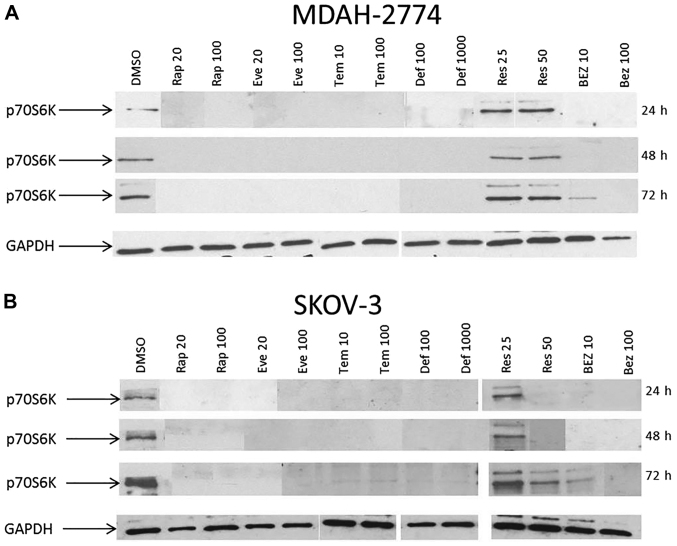
Phosphorylation status of p70S6K was examined to better understand the activity of the mTOR pathway after inhibition. At 24, 48 and 72 h, the basal phosphorylation of p70S6K was completely abolished in both MDAH-2774 and SKOV3 (Fig. 7) cells treated with rapamycin, all of the other rapalogues and at 100 nM of BEZ treatment. In SKOV3 cells treated with Res, dephosphorylation of p70S6K only occurred at 50 μM of Res at 24 and 48 h. Res treatment did not dephosphorylate p70S6K at 25 or 50 μM in MDAH-2774 cells.
Figure 7.
Analysis of phosphorylated p70S6K protein expression following mTOR pathway inhibition in MDAH-2774 (A) and SKOV3 (B) cells following treatment with Rap (20 and 100 nM), Eve (20 and 100 nM), Def (100 and 1,000 nM), Tem (10 and 100 nM), Res (25 and 50 μM) and BEZ (10 and 100 nM). SKOV3 and MDAH-2774 cells were seeded in 6-well plates at a density of 100,000 cells per well and allowed to proliferate for 24 h before treatment. Cells were treated with mTOR pathway inhibitors for 24, 48 and 72 h. At each time-point, cells were lysed and protein extracted in Laemmli buffer. Western blots were performed using an antibody recognising phosphorylated p70S6K (Thr389). GAPDH was used as a reference.
Expression of S6K in ovarian tissue array
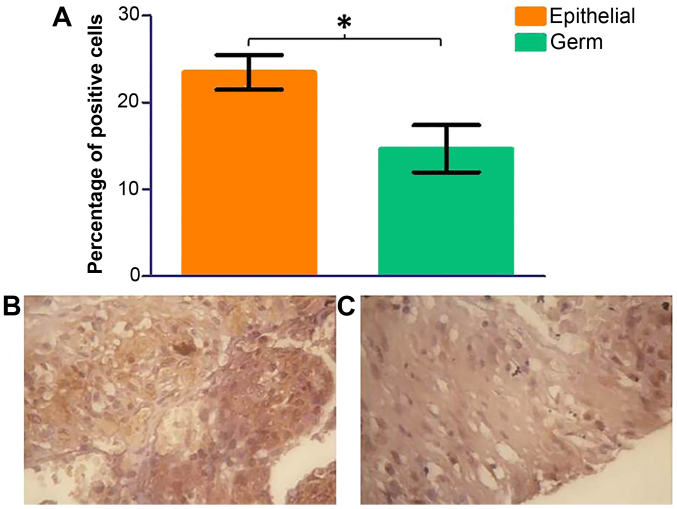
Based on the previous data on p70S6K, it is evident that the mTORC1 activity is compromised upon treatment with mTOR inhibitors. Given the significance that p70S6K has in moderating the activity of mTORC1, we studied the cellular distribution of this kinase on an ovarian tissue array. Ovarian cancer (n=70) tissue samples were probed with a p70S6K antibody by immunohistochemistry using DAB staining. Three areas of each tissue sample were selected at random and total cells and total positive cells were counted in each area. An average percentage of positive cells was calculated. There was a significant decrease in phospho-p70S6K protein expression in germ cell tumours in comparison to epithelial cell tumours (Fig. 8, p=0.0398). No significant change was seen in the grade of ovarian cancer (data not shown); however, phospho-p70S6 kinase shows a significant increase in later stages (Fig. 9).
Figure 8.
Immunohistochemistry for phosphorylated p70S6 kinase was performed on paraffin-embedded ovarian tissue clinical samples from a range of ovarian tumours. Three areas of tissue were analysed for percentage of positively stained cells. A significant decrease in phosphorylated p70S6K was detected in germ cell tumours in comparison to epithelial tumours (A). (B and C) Representative images of serous papillary carcinoma (epithelial) and a germ cell tumour, respectively. Error bars depict standard error.
Figure 9.
Immunohistochemistry for phosphorylated p70S6 kinase was performed on paraffin-embedded ovarian tissue clinical samples from a range of ovarian tumours. Three areas of tissue were analysed for percentage of positively stained cells. No significant change was seen between individual stage of disease; however a significant difference was detected between early and late stage disease [(p=0.0461 (A)]. (B and C) Representative images of stage I and stage IV serous papillary carcinoma, respectively. Error bars depict standard error.
Expression of mTOR components in ovarian cancer patients
It is becoming more widely accepted that an explorative test should be carried out for each set of samples to determine a suitable reference gene in each case (11,13). For this reason, we performed an analysis of reference gene stability in ovarian clinical tissue samples using the geNorm™ 12 gene kit. Eight clinical samples were selected to represent the whole cohort. The two most stable genes were RPL13A (ribosomal protein L13A; a gene that encodes a component of the 60S ribosomal subunit) and YWHAZ (a gene that encodes a 14-3-3 signal transduction protein). These reference genes were used in qPCR experiments involving clinical samples as they have been proven stable enough to provide robust normalisation.
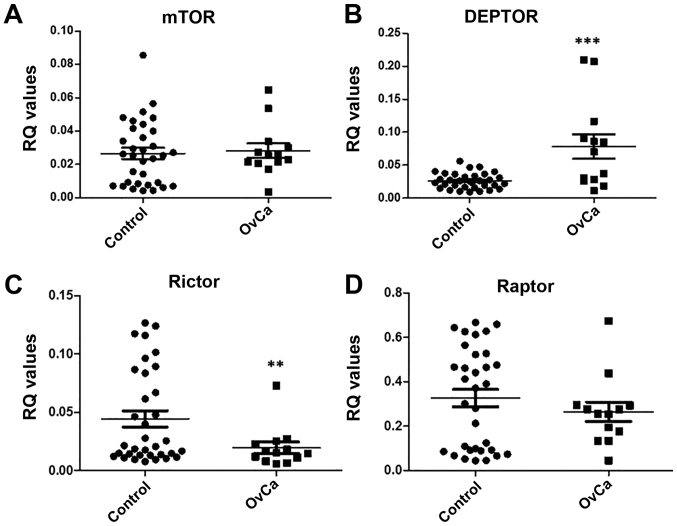
Therefore, qPCR for mTOR, DEPTOR, rictor and raptor was carried out using the reference genes RPL13A and YWHAZ on cDNA synthesised from the extracted RNA from ovarian tissue of ovarian cancer patients (n=13) and non-affected controls (n=34). Data were analysed using the ΔCq method and an RQ value calculated by 2−ΔCq. Results show a significant increase in DEPTOR expression (p=0.0007) and a significant decrease in rictor expression (p=0.0388) in comparison to control (Fig. 10).
Figure 10.
Gene expression analysis of mTOR pathway components, mTOR, DEPTOR, rictor and raptor in ovarian clinical samples. cDNA was synthesised from mRNA extracted from ovarian tissue obtained from ovarian cancer patients and unaffected controls. qPCR was performed using primers for mTOR, DEPTOR, rictor and raptor corrected with YWHAZ and RPL13A (reference genes). Error bars depict standard error.
Discussion
In this study, using two different ovarian cancer cell lines and clinical samples, we investigated the effect of rapalogues as well as dual kinase inhibitors in ovarian cancer. We have used SKOV3 and MDAH-2774 cell lines as models for epithelial ovarian cancer. The cell lines differed in their cells of origin, SKOV3 cells are clear cell derived, and MDAH-2774 are endometrioid. Using these cell lines we were able to study the two forms of malignancy most associated with endometriosis (14–16). In future, a useful addition to this study should be the inclusion of primary malignant and non-malignant ovarian surface epithelial cells. All key components of mTORC1 and mTORC2 complexes, i.e., mTOR, DEPTOR, rictor and raptor are expressed in both cell lines in a similar manner. The predominant component was raptor, with DEPTOR demonstrating minimal expression under basal (no treatment) conditions. Using high-resolution flow cytometry we demonstrate for the first time a cytoplasmic and nuclear expression of mTOR and DEPTOR in MDAH-2774. Similar localisation was evident in SKOV3 corroborating previous data from our laboratory (10). Rictor and raptor appeared to be expressed primarily in the cytoplasm (data not shown). It is evident therefore, that under basal conditions both mTOR complexes are expressed at mRNA and protein level.
Furthermore, we assessed the expression of these genes in ovarian cancer patients (n=13) and controls (n=34). We demonstrate a significant increase in DEPTOR expression and a significant decrease in rictor expression in comparison to controls. In order to assess mTOR pathway component gene expression in a large cohort of patients (>50), we utilised the in silico analysis method Oncomine™. mTOR, DEPTOR and raptor expression was analysed, but due to the small sample size, rictor data was not available. mTOR gene expression was significantly higher (1.166-fold) in data from the Bonome dataset in ovarian carcinoma (n=185) patients compared to controls (n=10). DEPTOR gene expression was significantly higher (1.683-fold) in patients with ovarian serous adenocarcinoma (n=43) compared to controls (n=10; Yoshihara dataset). There were no significant changes in the expression of raptor between ovarian serous adenocarcinoma patients (n=43) and controls (n=10; Yoshihara dataset).
As mentioned, this study investigated the effects of mTOR pathway inhibition in vitro. Western blot experiments indicated cleavage of caspase 9 after 24-h treatment which was then decreased by 48 h in SKOV3 cells. This indicates that initial response to mTOR inhibitor treatment is to induce apoptosis but this effect is rescued by 48 h of treatment. Caspase 9 is an initiator caspase which cleaves the pro-form of caspase 3 to induce apoptosis. However, there was no cleavage of caspase 3 in either of the cell lines following treatment with mTOR inhibitors (data not shown), suggesting that there is limited impact on apoptotic events. This data was further supported by proliferation assays showing no change in dead cell number over 72 h. Results showing no significant change in dead cell number over the course of the experiment in any case indicate that the effects of mTOR pathway inhibition are cytostatic as opposed to cytotoxic. This data is supported by previous research which shows that rapamycin and temsirolimus cause cell cycle arrest at G0/G1 phase (17), whereas Eve treatment reduced viable cell number, but was not able to induce apoptosis in SKOV3 cells (18).
The inhibitory effects of Res and BEZ on SKOV3 cells were significantly greater for these dual mTOR/PI3 kinase inhibitors than for the rapalogues alone. This suggests that additional targeting of PI3 kinase increases inhibitory efficacy in these ovarian cancer cells. In the more proliferative MDAH-2774 cell line, 100 nM BEZ treatment profoundly decreased the number of viable cells to almost basal level; however dual mTOR and PI3 kinase inhibitors were not more effective than inhibition by rapamycin or rapalogue in these cells. The lower efficacy of dual inhibitors observed in MDAH-2774 cells may be due to the negative feedback loop directed to PI3 kinase that is relieved on mTOR inhibition and can induce PI3 kinase signalling via Akt to cause upregulation in mTORC2 signalling (19). Dual mTOR and PI3 kinase, or mTORC1 and mTORC2 inhibitors are able to target both mTORC1 signalling and the after-effect of mTORC2 signalling and therefore provide better repression of the mTOR pathway (20,21). There is also evidence that a negative feedback loop exists from mTOR to the MAP kinase pathway (22) and inhibition of both the MAP kinase and PI3 kinase pathways has proven to be more effective than targeting solely PI3 kinase (20). Collectively, these data demonstrate that these inhibitors act in a cell-specific manner within the microenvironment of ovarian cancer.
Phosphorylation status of p70S6K was also assessed to obtain a better understanding of the effects of these inhibitors on mTORC1 activity. Rapamycin, rapalogues and BEZ (at high concentration in MDAH-2774) completely dephosphorylated p70S6K in both cell lines. In SKOV3 cells, treatment with 50 μM of Res also dephosphorylated p70S6K. However, Res did not dephosphorylate p70S6K in MDAH-2774 cells. These data corroborate previous findings on p70S6K using BEZ in neuroblastoma cells (23), temsirolimus and everolimus in EGFR mutant lung cancer cells, PC-9, and HCC827 (24). In one of our in vitro models, Res had little effect on the phosphorylation status of p70S6K. This again could be a cell-specific effect since Res inhibits p70S6K in other cell lines such as the osteoblast-like MC3T3-E1 cells (24), or the human nasopharyngeal carcinoma (NPC) cells (26) to name a few.
Given the importance of p70S6K in ovarian cancer, we mapped the protein expression of this phospho-p70S6K in 70 ovarian cancer patients and controls. We demonstrate for the first time a significant increase in staining for phospho-p70S6K with worsening stage. Since p70S6K can be a prerequisite of tumour metastasis (27), targeting this kinase particularly in patients with high stage tumours could be of therapeutic value. Recently it has been shown that phosphorylated p70S6K predicted tamoxifen resistance in postmenopausal breast cancer patients (28). Future studies should concentrate on immunostaining for phosphorylated p70S6K in a larger cohort of patients in order to define whether this kinase is of a diagnostic or prognostic value in ovarian cancer.
This study highlights the need for ‘tailor-made’ therapies against ovarian cancer depending on the gene expression profile of the patient. The non-apoptotic effects of the mTOR pathway inhibitors used in this study may exclude them from being effective single agent therapies in the treatment of malignancy. However, it has been shown that rapalogue treatment is able to potentiate the apoptotic effects of tamoxifen, doxorubicin, UCN-01 and cisplatin treatment in vitro, indicating that mTOR pathway inhibitors may be useful in combinatorial therapeutic approaches to cancer treatment (18,29–31). Using mTOR pathway inhibitors may therefore allow for lower doses of cytotoxic chemotherapeutic agents in cancer treatment and therefore a reduction in the harmful side-effects brought about by these drugs (31).
References
- 1.De Cecco L, Marchionni L, Gariboldi M, Reid JF, Lagonigro MS, Caramuta S, Ferrario C, Bussani E, Mezzanzanica D, Turatti F, et al. Gene expression profiling of advanced ovarian cancer: Characterization of a molecular signature involving fibroblast growth factor 2. Oncogene. 2004;23:8171–8183. doi: 10.1038/sj.onc.1207979. [DOI] [PubMed] [Google Scholar]
- 2.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 3.Kim SH, Juhnn YS, Song YS. Akt involvement in paclitaxel chemoresistance of human ovarian cancer cells. Ann NY Acad Sci. 2007;1095:82–89. doi: 10.1196/annals.1397.012. [DOI] [PubMed] [Google Scholar]
- 4.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci USA. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang S, Houghton PJ. Inhibitors of mammalian target of rapamycin as novel antitumor agents: From bench to clinic. Curr Opin Investig Drugs. 2002;3:295–304. [PubMed] [Google Scholar]
- 6.Kurmasheva RT, Huang S, Houghton PJ. Predicted mechanisms of resistance to mTOR inhibitors. Br J Cancer. 2006;95:955–960. doi: 10.1038/sj.bjc.6603353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mabuchi S, Altomare DA, Connolly DC, Klein-Szanto A, Litwin S, Hoelzle MK, Hensley HH, Hamilton TC, Testa JR. RAD001 (Everolimus) delays tumor onset and progression in a transgenic mouse model of ovarian cancer. Cancer Res. 2007;67:2408–2413. doi: 10.1158/0008-5472.CAN-06-4490. [DOI] [PubMed] [Google Scholar]
- 8.Squillace RM, Miller D, Cookson M, Wardwell SD, Moran L, Clapham D, Wang F, Clackson T, Rivera VM. Antitumor activity of ridaforolimus and potential cell-cycle determinants of sensitivity in sarcoma and endometrial cancer models. Mol Cancer Ther. 2011;10:1959–1968. doi: 10.1158/1535-7163.MCT-11-0273. [DOI] [PubMed] [Google Scholar]
- 9.Takano M, Kikuchi Y, Kudoh K, Goto T, Furuya K, Kikuchi R, Kita T, Fujiwara K, Shiozawa T, Aoki D. Weekly administration of temsirolimus for heavily pretreated patients with clear cell carcinoma of the ovary: A report of six cases. Int J Clin Oncol. 2011;16:605–609. doi: 10.1007/s10147-010-0177-z. [DOI] [PubMed] [Google Scholar]
- 10.Foster H, Coley HM, Goumenou A, Pados G, Harvey A, Karteris E. Differential expression of mTOR signalling components in drug resistance in ovarian cancer. Anticancer Res. 2010;30:3529–3534. [PubMed] [Google Scholar]
- 11.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Jacob F, Guertler R, Naim S, Nixdorf S, Fedier A, Hacker NF, Heinzelmann-Schwarz V. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS One. 2013;8:e59180. doi: 10.1371/journal.pone.0059180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, Nagle CM, Doherty JA, Cushing-Haugen KL, Wicklund KG, et al. Ovarian Cancer Association Consortium. Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case-control studies. Lancet Oncol. 2012;13:385–394. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinton LA, Sakoda LC, Sherman ME, Frederiksen K, Kjaer SK, Graubard BI, Olsen JH, Mellemkjaer L. Relationship of benign gynecologic diseases to subsequent risk of ovarian and uterine tumors. Cancer Epidemiol Biomarkers Prev. 2005;14:2929–2935. doi: 10.1158/1055-9965.EPI-05-0394. [DOI] [PubMed] [Google Scholar]
- 16.Merritt MA, Green AC, Nagle CM, Webb PM Australian Cancer Study (Ovarian Cancer); Australian Ovarian Cancer Study Group. Talcum powder, chronic pelvic inflammation and NSAIDs in relation to risk of epithelial ovarian cancer. Int J Cancer. 2008;122:170–176. doi: 10.1002/ijc.23017. [DOI] [PubMed] [Google Scholar]
- 17.Fagone P, Donia M, Mangano K, Quattrocchi C, Mammana S, Coco M, Libra M, McCubrey JA, Nicoletti F. Comparative study of rapamycin and temsirolimus demonstrates superimposable anti-tumour potency on prostate cancer cells. Basic Clin Pharmacol Toxicol. 2013;112:63–69. doi: 10.1111/j.1742-7843.2012.00923.x. [DOI] [PubMed] [Google Scholar]
- 18.Treeck O, Wackwitz B, Haus U, Ortmann O. Effects of a combined treatment with mTOR inhibitor RAD001 and tamoxifen in vitro on growth and apoptosis of human cancer cells. Gynecol Oncol. 2006;102:292–299. doi: 10.1016/j.ygyno.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zitzmann K, Rüden J, Brand S, Göke B, Lichtl J, Spöttl G, Auernhammer CJ. Compensatory activation of Akt in response to mTOR and Raf inhibitors - a rationale for dual-targeted therapy approaches in neuroendocrine tumor disease. Cancer Lett. 2010;295:100–109. doi: 10.1016/j.canlet.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Hisamatsu T, Mabuchi S, Matsumoto Y, Kawano M, Sasano T, Takahashi R, Sawada K, Ito K, Kurachi H, Schilder RJ, et al. Potential role of mTORC2 as a therapeutic target in clear cell carcinoma of the ovary. Mol Cancer Ther. 2013;12:1367–1377. doi: 10.1158/1535-7163.MCT-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durbas M, Horwacik I, Boratyn E, Kamycka E, Rokita H. GD2 ganglioside specific antibody treatment downregulates PI3K/Akt/mTOR signaling network in human neuroblastoma cell lines. Int J Oncol. 2015;47:1143–1159. doi: 10.3892/ijo.2015.3070. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa D, Takeuchi S, Nakagawa T, Sano T, Nakade J, Nanjo S, Yamada T, Ebi H, Zhao L, Yasumoto K, et al. mTOR inhibitors control the growth of EGFR mutant lung cancer even after acquiring resistance by HGF. PLoS One. 2013;8:e62104. doi: 10.1371/journal.pone.0062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo A, Otsuka T, Kuroyanagi G, Yamamoto N, Matsushima-Nishiwaki R, Mizutani J, Kozawa O, Tokuda H. Resveratrol inhibits BMP-4-stimulated VEGF synthesis in osteoblasts: Suppression of S6 kinase. Int J Mol Med. 2014;33:1013–1018. doi: 10.3892/ijmm.2014.1626. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Zhou X, Zhou K. Resveratrol inhibits human nasopharyngeal carcinoma cell growth via blocking pAkt/p70S6K signaling pathways. Int J Mol Med. 2013;31:621–627. doi: 10.3892/ijmm.2013.1237. [DOI] [PubMed] [Google Scholar]
- 27.Ip CK, Yung S, Chan TM, Tsao SW, Wong AS. p70 S6 kinase drives ovarian cancer metastasis through multicellular spheroid-peritoneum interaction and P-cadherin/b1 integrin signaling activation. Oncotarget. 2014;5:9133–9149. doi: 10.18632/oncotarget.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beelen K, Opdam M, Severson TM, Koornstra RH, Vincent AD, Wesseling J, Muris JJ, Berns EM, Vermorken JB, van Diest PJ, et al. Phosphorylated p-70S6K predicts tamoxifen resistance in postmenopausal breast cancer patients randomized between adjuvant tamoxifen versus no systemic treatment. Breast Cancer Res. 2014;16:R6. doi: 10.1186/bcr3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avellino R, Romano S, Parasole R, Bisogni R, Lamberti A, Poggi V, Venuta S, Romano MF. Rapamycin stimulates apoptosis of childhood acute lymphoblastic leukemia cells. Blood. 2005;106:1400–1406. doi: 10.1182/blood-2005-03-0929. [DOI] [PubMed] [Google Scholar]
- 30.Hahn M, Li W, Yu C, Rahmani M, Dent P, Grant S. Rapamycin and UCN-01 synergistically induce apoptosis in human leukemia cells through a process that is regulated by the Raf-1/MEK/ERK, Akt, and JNK signal transduction pathways. Mol Cancer Ther. 2005;4:457–470. doi: 10.1158/1535-7163.MCT-04-0137. [DOI] [PubMed] [Google Scholar]
- 31.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]