Abstract
Centrosomes are components of the mitotic spindle responsible for organizing microtubules and establishing a bipolar spindle for accurate chromosome segregation. In budding yeast, Saccharomyces cerevisiae, the centrosome is called the spindle pole body, a highly organized tri-laminar structure embedded in the nuclear envelope. Here we describe a detailed protocol for the purification of fluorescently labeled spindle pole bodes from S. cerevisiae. Spindle pole bodies are purified from yeast using a TAP-tag purification followed by velocity sedimentation.
This highly reproducible TAP-tag purification method improves upon previous techniques and expands the scope of in vitro characterization of yeast spindle pole bodies. The genetic flexibility of this technique allows for the study of spindle pole body mutants as well as the study of spindle pole bodies during different stages of the cell cycle. The ease and reproducibility of the technique makes it possible to study spindle pole bodies using a variety of biochemical, biophysical, and microscopic techniques.
Keywords: Spindle pole body (SPB), purification, mitosis, centrosome, TAP-tag
1. INTRODUCTION
The mitotic spindle ensures accurate segregation of genetic material in a dividing cell. The centrosome is a crucial component of this macromolecular machine, nucleating and organizing microtubules. Because components of the mitotic spindle are highly conserved throughout eukaryotes, the yeast spindle serves as an excellent model.
In yeast, the centrosome equivalent is the spindle pole body. The spindle pole body is a highly organized laminar structure, consisting of three plaques. The core spindle pole body components form the central plaque, which is embedded in the nuclear envelope throughout the cell cycle (1, 2). The inner plaque sits on the nuclear side of the central plaque and is the site of nuclear microtubule nucleation. The inner plaque contains the γ-tubulin small complex, which is essential for microtubule nucleation, and Spc110, a linker protein that binds the γ-tubulin small complex to the core of the spindle pole body (3, 4, 5). The outer plaque is on the cytoplasmic side of the central plaque. This structure also contains the γ-tubulin small complex, which is bound to the spindle pole body via interactions with Spc72 (6).
Yeast spindle pole bodies (SPBs) have historically been studied by thin section electron microscopy of yeast cells (1, 2, 7) or purified to identify components, determine structure, and examine function of the SPB in vitro (3, 8, 9, 10, 11). A method to purify SPBs out of the lesser characterized yeast S. uvarum generated high yields of purified SPBs, but was limited in the ability to perform genetic analyses and the SPBs could only be visualized by electron microscopy (3). More recently, SPBs were co-purified from S. cerevisiae with a TAP-tagged nuclear pore component, Mlp2 (12). Expanding on this method, we discovered that TAP-tagging Spc97, a component of the spindle pole body, increases yield. Adding fluorescent tags to spindle pole body components helps to both visualize the SPBs and quantify yields. Here we offer a detailed purification protocol for growing, harvesting, and lysing cells to purify spindle pole bodies by TAP-tag purification followed by velocity sedimentation.
2. MATERIALS
2.1 Growing, Harvesting, and Lysing Cells
S. cerevisiae strain expressing SPC97-TAP::kanMX with SPC42-mCherry::hphMX or S. cerevisiae strain expressing SPC97-TAP::kanMX with SPC42-mCherry::hphMX and TUB4-GFP::kanMX
YPD media: 1% yeast extract, 2% peptone, 2% glucose
Resuspension buffer: 20 mM sodium-HEPES buffer (pH 7.4), 1.2% polyvinylpyrrolidone (average MW 40,000), 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride, 4 μg/ml aprotinin, 4 μg/ml chymostatin, 4 μg/ml leupeptin, 4 μg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate
PM 100 ball mill grinder (Retsch)
Stainless steel 125 ml planetary ball mill grinding jar with 20 mm diameter mill grinding balls (Retsch)
Liquid nitrogen
18G needle
2.2 Spindle Pole Body Purification
Extraction Buffer with 300 mM NaCl (EB1 w/ 300 mM NaCl): 20 mM sodium-HEPES buffer (pH 7.4), 300 mM NaCl, 0.5% Triton X-100, 2 mM MgCl2, 100 μM GTP, 1 mM ATP, 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride, 4 μg/ml aprotinin, 4 μg/ml chymostatin, 4 μg/ml leupeptin, 4 μg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 5% glycerol
Extraction Buffer with 200 mM NaCl (EB1 w/ 200 mM NaCl): 20 mM sodium-HEPES buffer (pH 7.4), 200 mM NaCl 0.5% Triton X-100, 2 mM MgCl2, 100 μM GTP, 1 mM ATP, 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride, 4 μg/ml aprotinin, 4 μg/ml chymostatin, 4 μg/ml leupeptin, 4 μg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 5% glycerol
TEV cleavage buffer: 40 mM sodium-HEPES buffer (pH 7.4), 200 mM NaCl, 2 mM MgCl2, 1 mM GTP, 1 mM ATP, 1 mM EDTA (pH 8), 1 mM dithiothreitol, 5% glycerol
PCU-2-110 Homogenizer (Kinematica)
M-270 Epoxy Dynabeads (Invitrogen) conjugated to rabbit IgG (MP Biomedicals) according to manufacturer’s protocol
Magnetic stands for 50 ml Falcon tube and 1.7 ml microcentrifuge tubes
TEV protease (stored in 50 mM Tris buffer (pH 7.5), 1 mM EDTA, 5 mM DTT, 50% glycerol, 0.1% Triton X-100)
2.3 Velocity Sedimentation
Sucrose solutions: 10%, 20%, 30%, 40%, and 2.5 M sucrose in 10 mM Bis-Tris buffer (adjust to pH 6.5 with HCl), 0.1 mM MgCl2
Thick wall polycarbonate tubes: 11 × 34 mm (Beckman Coulter)
Ultracentrifuge Rotor TLS-55 (Beckman Coulter)
3. METHODS
3.1 Growing, Harvesting, and Lysing Cells
This protocol has been adapted from Michael Rout’s lab (http://lab.rockefeller.edu/rout/protocols). The lysis technique with the Retsch PM 100 ball mill grinder increases the percent of cells lysed and decreases protein degradation, resulting in higher yields.
Grow 10 ml YPD culture of cells overnight and maintain in log phase growth to inoculate 2 L culture.
Grow 2 L YPD culture of cells overnight to 4.5 × 107 cells/ml (still in log phase)
Pellet cells at 4000xg for 10 minutes at 4°C in 1 L bottles.
Resuspend cell pellet in 25 ml cold dH2O on ice. Transfer cells to 50 ml Falcon tubes and pellet cells at 2600xg for 5 minutes at 4°C.
Combine 2 cell pellets and resuspend in 15 ml cold dH2O. Spin down at 2600xg for 5 minutes at 4°C.
Resuspend pellet in 15 ml cold resuspension buffer. Pellet at 2600xg for 15 minutes at 4°C. Decant the supernatant.
Pellet again at 2600xg for 15 minutes at 4°C to remove excess buffer. At this point, the pellet should be a relatively dry, thick paste.
Cool a 50 ml Falcon tube and fill with liquid nitrogen. Using an 18G needle, poke holes in the tube lid to allow liquid nitrogen vapor to escape.
With a spatula, scoop out the cell paste and transfer to a 20 ml syringe. Press the cell paste into the liquid nitrogen in the Falcon tube, creating noodles.
When all cells have been frozen, loosely cap tube to allow liquid nitrogen to escape. Store at −80°C.
To lyse cells, cool a 125 ml stainless steel grinding jar and stainless steel grinding balls in liquid nitrogen.
Empty the contents of one 50 ml Falcon tube of frozen cell noodles into the grinding jar and fill the jar with grinding balls.
Ensure the counterweight is properly set.
Start grinding program: 3 × 1 min grinding cycles at 400 rpm, reversing directions with each cycle.
At the end of the grinding program, cool the grinding jar in liquid nitrogen.
Repeat the grinding program and cooling 7 more times, or until you get satisfactory lysis.
When lysis is complete, scoop out the lysed cell dust and transfer to the cooled 50 ml Falcon tube (see Note 1). Store lysed cells at −80°C.
3.2 Spindle Pole Body Purification
Previous purification protocols were modified to increase the yield and stability of purified spindle pole bodies. Prior techniques were laborious and time-intensive, whereas this protocol has reduced the purification to a few hours.
Resuspend 4 g of lysed cells in 20 ml of cold Extraction Buffer with 300 mM NaCl.
Homogenize for 30 seconds at speed 5 using PCU-2-110 Homogenizer (Kinematica).
Clear lysate at 2000xg for 10 minutes at 4°C.
While clearing the lysate, prepare IgG magnetic beads. In a 1.7 ml microcentrifuge tube, take 250 μl of Dynabeads conjugated to rabbit IgG, and place in a magnetic stand to magnetize and collect the beads. Remove the supernatant and resuspend in 250 μl EB1 w/ 300 mM NaCl. Magnetize and repeat wash twice. (We do our own conjugations by following the manufacturer’s instructions.)
Transfer cleared cell lysate to a clean 50 ml Falcon tube. Add 250 μl of IgG conjugated Dynabeads.
Incubate on a Nutator (Clay Adams) to prevent beads from settling for 30 minutes at 4°C.
Magnetize beads and resuspend in 100 μl EB1 w/ 200 mM NaCl and transfer to 1.7 ml low retention microcentrifuge tube.
Incubate 2 minutes on a Nutator. Magnetize beads and repeat wash with EB1 w/ 200 mM NaCl twice.
Wash once with 100 μl TEV cleavage buffer. Magnetize beads and resuspend in 50 μl TEV cleavage buffer. Add 1 μg TEV.
Incubate on a rotator for 2 hours at 4°C (see Note 2).
Magnetize beads and take supernatant. Store supernatant on ice until velocity sedimentation (see Note 3).
3.3 Velocity Sedimentation
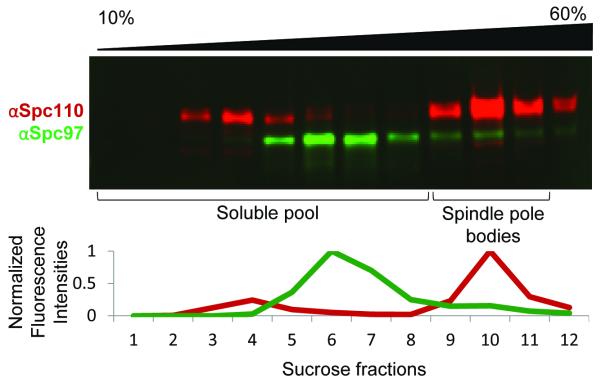
The soluble pool of γ-tubulin small complex and other spindle pole body components exist in the higher fractions of the sucrose gradient (fractions 1-7). Spindle pole bodies, as determined by western blot analysis and verified by mass spectrometry, migrate near the 2.0 M/2.5 M sucrose interface. Intact spindle pole bodies reproducibly fall in fractions 9-11 (out of 12). The velocity sedimentation effectively separates the soluble pool of spindle pole body components from the intact spindle pole bodies, resulting in a more homogeneous sample.
Using a wide-bore tip, load 200 μl of 2.5 M sucrose into a 1 ml thick wall polycarbonate tube for the TLS-55 rotor.
Carefully, to avoid mixing, use wide-bore tips to layer 40%, 30%, 20%, and 10% sucrose solutions in tube.
Let sit at 4°C for 2 hours to allow the gradient to equilibrate.
Load purified sample onto the sucrose gradient with a wide-bore tip and centrifuge at 50,000 rpm for 5 hours at 4°C in a TLS-55 swinging bucket rotor.
Stepwise remove 90 μl fractions from the top of the gradient using a wide-bore tip.
To identify which fractions of the sucrose gradient contain spindle pole bodies, spindle pole body components Spc110 and Spc97 can be probed via western blot analysis (see Figure 1).
Flash freeze 5 μl aliquots of the spindle pole body-containing fractions in liquid nitrogen and store at −80°C.
Figure 1.
Western blot analysis of sucrose gradient fractions. Twelve fractions (90 μl each) were taken from the top of the sucrose gradient, with sucrose concentration increasing from left (10%) to right (60%). Sucrose fractions were probed for Spc110 (in red) and Spc97 (in green) by western blot analysis. Integrated intensities were normalized to the peak intensity. The soluble pool of spindle pole body components migrate in fractions 1-7 while fully formed spindle pole bodies migrate in fractions 9-11. The presence of all spindle pole body components in fractions 9-11 was further confirmed by mass spectrometry. Note that the antibody against Spc110 is much more sensitive than the antibody against Spc97.
4. NOTES
The ease by which the lysed cells are removed from the grinding jar varies greatly. It helps to fill the jar entirely, with a full 50 ml Falcon tube of cell noodles, as well as thoroughly cool the jar between grinding programs. This will result in a loose powder that can easily be scooped out and transferred to a 50 ml Falcon tube for storage. Insufficient cooling between grinding programs can cause the cell dust to cake on the grinding jar walls, requiring substantial effort to scrape off.
At this step, incubation on a rotator is preferred over a Nutator. With the smaller volume during TEV cleavage, the Nutator is insufficient for preventing the Dynabeads from settling to the bottom of the tube.
At this step in the purification, you have spindle pole bodies at a high concentration, but contaminated with nucleoli, fragments of the nucleus and soluble γ-tubulin small complex. If spindle pole body concentration is of more importance than purity, the TEV eluate from this step can be used in downstream experiments. If purity is of high importance, the TEV eluate can be further purified by velocity sedimentation, however, the concentration of spindle pole bodies will decrease.
5. REFERENCES
- 1.Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae. J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16(7):1550–1564. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen T, Vinh DB, Crawford DK, Davis TN. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol Biol Cell. 1998;9(8):2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17(14):3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr., McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129(6):1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- 9.Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- 10.Hyams JS, Borisy GG. Nucleation of microtubules in vitro by isolated spindle pole bodies of the yeast Saccharomyces cerevisiae. J Cell Biol. 1978;78(2):401–414. doi: 10.1083/jcb.78.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89(7):1077–1986. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- 12.Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mpl2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005;170(2):225–235. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]