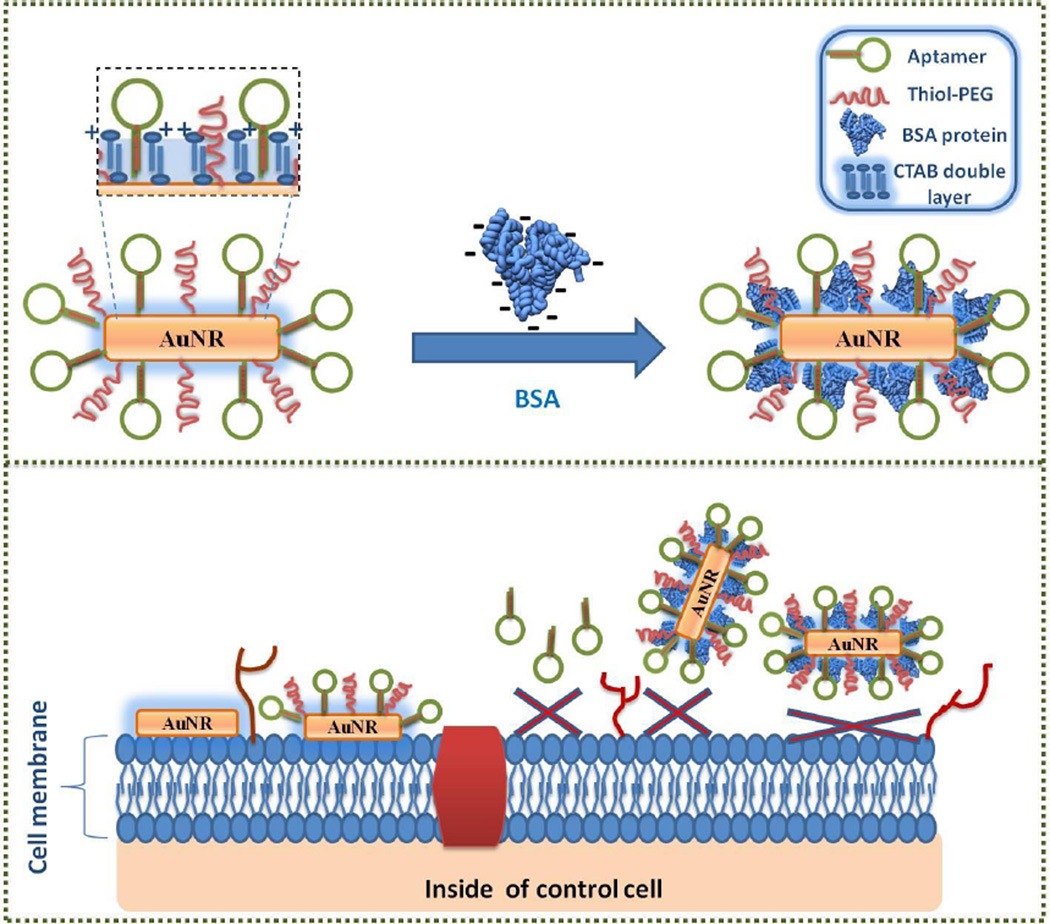
Abstract
Aptamer-conjugated gold nanorods (AuNRs) are excellent candidates for targeted hyperthermia therapy of cancer cells. However, in high concentrations of AuNRs, aptamer conjugation alone fails to result in highly cell-specific AuNRs due to the presence of positively charged cetyltrimethylammonium bromide (CTAB) as a templating surfactant. Besides causing nonspecific electrostatic interactions with the cell surfaces, CTAB can also be cytotoxic, leading to uncontrolled cell death. To avoid the nonspecific interactions and cytotoxicity triggered by CTAB, we report the further biologically inspired modification of aptamer-conjugated AuNRs with bovine serum albumin (BSA) protein. Following this modification, interaction between CTAB and the cell surface was efficiently blocked, thereby dramatically reducing the side effects of CTAB. This approach may provide a general and simple method to avoid one of the most serious issues in biomedical applications of nanomaterials: nonspecific binding of the nanomaterials with biological cells.
Graphical Abstract
Introduction
The emergence of gold nanorods (AuNRs) has attracted considerable scientific interest because of their high absorption cross sections and tunable absorption maxima in the NIR region.1 For instance, the extinction cross-section coefficients of AuNRs are more than double those of gold nanoshells (AuNSs)2, the first example of gold nanoparticles (AuNPs) able to absorb in the NIR region. High absorption cross sections facilitate efficient energy absorption in the NIR, thus allowing equally efficient conversion of this absorbed energy to thermal energy. The heating rate per gram of gold for AuNRs is at least six times faster than that for AuNSs.3 Therefore, AuNRs are excellent candidates as photothermal therapy (PTT) agents. Active targeting and destruction of cancer cells can be achieved by combining these intrinsic properties of AuNRs with the recognition capabilities of aptamers, single-stranded DNA or RNA oligonucleotides that can bind to their specific cell membrane proteins with affinities similar to those of antibodies.4, 5
Gold nanorods can be synthesized in high yields via a seed-mediated method, which employs cetyltrimethylammonium bromide (CTAB), a cationic surfactant known to form rod-like micelles in aqueous solutions.6 Thus, AuNRs synthesized with the seed-mediated method owe their distinct optoelectronic properties to this shape confining cationic surfactant (CTAB), present as a double layer on the gold surface.7–9 Besides confining the shape, cationic CTAB can also stabilize AuNRs in colloidal dispersions.10 However, despite these beneficial roles, CTAB can also cause side effects at elevated concentrations in cell experiments. Indeed, it has been demonstrated that CTAB can be toxic to many types of cells,11–15 and both the released monomers and the double-layer structure of CTAB can be responsible for this toxicity.11, 12, 14, 16 Moreover, since CTAB is positively charged, it can nonspecifically bind to negatively charged cell surfaces by electrostatic interactions.10 In fact, these interactions are the main cause of the toxicity. When CTAB interacts with the cell membrane, it forms blebs and defects (holes) on the membrane, eventually leading to cell death.10
A variety of strategies has been reported to reduce the toxic effects of CTAB. One of these involves coating the CTAB bilayer with polyelectrolytes. This coating can be achieved either with a single polyelectrolyte or layers of different polyelectrolytes [PSS (polystyrene sulfonate), PDDAC (poly(diallyldimethyl ammonium chloride), etc.] through electrostatic interactions.10, 11, 14 In this way, the bilayer structure of CTAB is encapsulated and cannot interact with the cell membrane. Another strategy avoids the release of CTAB monomers by fixing them via polymerization.17 As another surface coating strategy, silica-coating can also reduce the toxicity while using CTAB as a template to form a silica layer on the surface of AuNRs.18 Besides coating the bilayer structure of CTAB or making the monomers of CTAB more stable, thiol-terminated ligands can displace CTAB from the AuNR surface and thereby reduce cytotoxicity.13, 15 Even though there should be batch-to-batch variations for the CTAB quantity on the surface of AuNRs, surface coating strategies have comparable efficiencies in reducing the toxicity of CTAB, but not all of the CTAB can be replaced by the ligand-exchange strategies.15, 19 Thiol-terminated poly(ethylene glycol) (mPEG-SH) is the most commonly used ligand to remove the CTAB from the gold surface and maintain the stability of gold nanorods in aqueous solutions.20 However, when this additional surface modification step is carried out alone, it not only involves further longer incubation times (usually overnight incubations) and washing steps to remove as much CTAB as possible, but it can also reduce the potential surface density of the recognition elements to be immobilized on the gold nanorod surface. This is an important trade-off considering the presence of recognition elements with low binding affinities towards their target.
In real biological systems, when naked or, in other words unmodified, nanoparticles are introduced into biological fluids, they are usually covered with biomolecules such as proteins to form a corona around them that interacts with cells. This corona constitutes the identity of those nanoparticles in such a way that cells determine either to pursue or avoid the interactions with those nanoparticles.21 This assembly of biomolecules is still valid to some extent, even if the nanoparticles are modified with recognition elements such as antibodies, proteins and peptides or dispersion elements such as PEG.22–24 Among these biomolecules, mostly serum proteins are responsible for such constructs to avoid cellular interactions.21 The adsorbed serum proteins on the surface of nanoparticles limit the nonspecific interactions between the nanoparticle surface and cell membrane, and thus the cellular uptake of the nanoparticles is reduced.25–28 In order to mimic this biological machinery, in this work, Bovine Serum Albumin (BSA) is used to mimic the most abundant protein in human plasma, albumin. Therefore, CTAB on the surface of gold nanorods was passivated by BSA protein coating, effectively blocking interaction between the cell surface and CTAB. Also, instead of passive targeting as with the other strategies mentioned above, active targeting was achieved by aptamer conjugation. CTAB can be encapsulated or replaced on the surface of AuNRs with the strategies mentioned above, but these can reduce the possibility of cellular uptake by decreasing electrostatic interactions with the cell surface in the absence of recognition element modification.
It has been reported that serum proteins can interact with CTAB through electrostatic interactions.11 Therefore, positively charged CTAB can easily interact with the negatively charged BSA to form a layer on CTAB. BSA is one of the most common proteins used to reduce nonspecific binding in immunoassays,29 because it blocks the unoccupied sites of the solid capturing platform to prevent nonspecific protein binding and decrease the background signal. In general, blockers based on proteins are more efficient than synthetic blockers. Also it has been proven that BSA modified gold nanoparticles can be highly stable even under harsh environmental conditions where the salt (NaCl) concentration is well beyond the concentration in human blood.30 Thus, BSA modification holds great potential in biomedicine to mimic the natural biological machinery. Therefore, instead of polyelectrolytes, the natural stabilizing molecule BSA can be used to encapsulate CTAB to avoid nonspecific interactions and toxicity prior to photothermal therapy. This work demonstrates the optimization of the BSA modification of aptamer conjugated AuNRs to avoid undesired nonspecific interactions that cause cytotoxicity.
Materials and Methods
Cell Culturing
The cancer cell lines were obtained from ATCC (American Type Culture Collection). The selected cancer cell lines, CCRF-CEM (CCL-119 T-cell, human acute lymphoblastic leukemia), as target cells, and Ramos (CRL-1596, B-cell line, human Burkitt’s lymphoma), as control cells, were cultured at 37 °C under a 5% CO2 atmosphere in a solution consisting of RPMI 1640 medium (ATCC), 10% heat inactivated fetal bovine serum, FBS (Invitrogen, Carlsbad, CA, USA) and penicillin-streptomycin (100 IU/mL, GIBCO).
Synthesis and Characterization of AuNRs
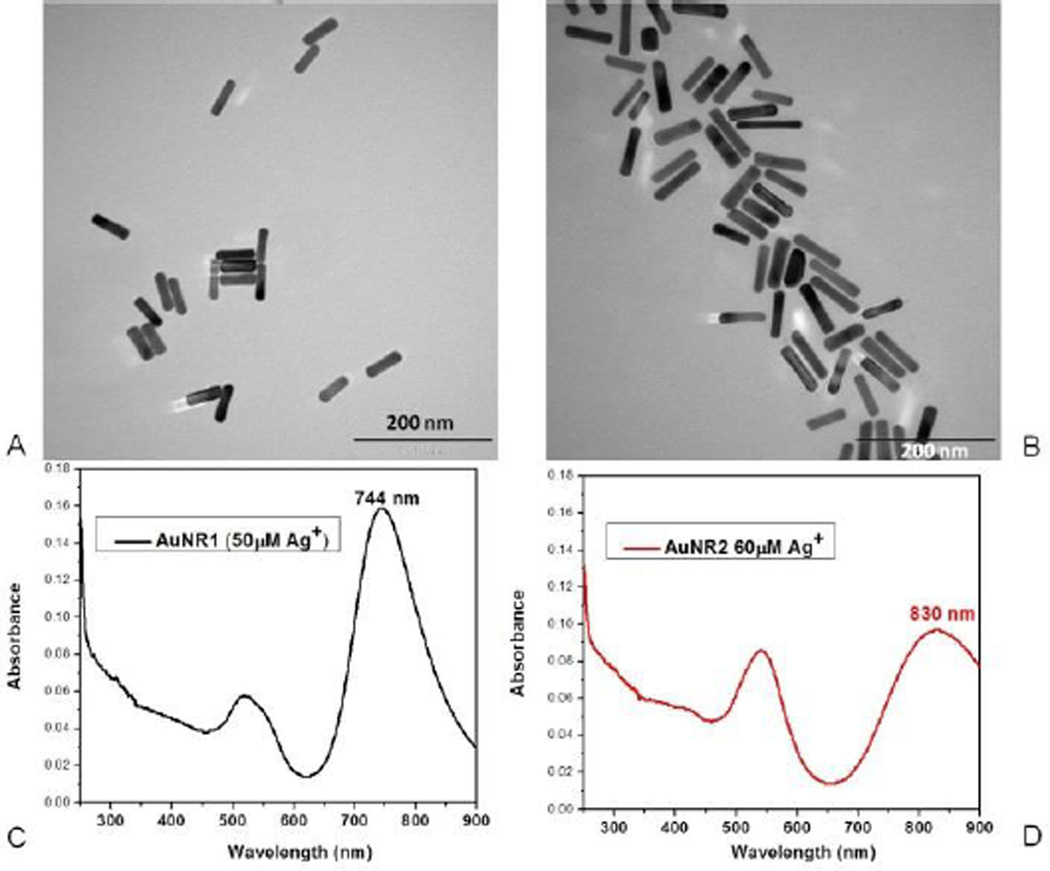
In this work, gold nanorods were synthesized and characterized by the same protocol that was adopted from the previous work.31 However, while preparing the growth solution, instead of 3.25 mL, 3.00 mL or 2.50 mL 0.004 M AgNO3 (60 or 50 µM Ag+ as the total concentration) was used to tune the size of AuNRs. The synthesized AuNRs were characterized by obtaining their UV/Vis spectra and TEM images using Cary Bio-300 (Varian, Walnut Creek, CA) and JEOL TEM 2010F, respectively. The sizes of gold nanorods prepared with 50 µM silver cation were determined to be 52 ± 4 nm and 14 ± 1 nm in length and width, respectively, by FemtoScan software. The sizes of those prepared with 60 µM silver cation were 70 ± 9 nm and 14 ± 1 nm in length and width, respectively.
Synthesis of Aptamer: sgc8c
The aptamer selected to target the CCRF-CEM cell line is sgc8c: 5`-ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GA-3`. The selected aptamer was modified with disulfide and fluorescein dye (FITC, FAM) at its 5`-terminus and 3`-terminus, respectively, using the same protocol that was previously reported.31
Conjugation of Aptamers on AuNR Surface
After reducing the disulfide groups to thiol groups at the 5`-terminus of the aptamer by using tris(2-carboxyethyl)phosphine (TCEP)31, 0.1 nM gold nanorod solution was incubated with 0.1 mM thiol-PEG (MW 5000) and 20 nM of the TCEPylated sgc8c in DNA grade water for 12 hours at room temperature. Next, the reaction solution was centrifuged at 14000 rpm at 25°C for 5 minutes to remove the excess aptamers and SH-PEG as the supernatant and to concentrate the gold nanorod solution. Finally, the precipitate was resuspended in DI or DNA grade water and briefly sonicated. The concentration of the aptamer-conjugated gold nanorods was evaluated by measuring the absorbance of the final solution via a Cary Bio-300 UV spectrometer (Varian, Walnut Creek, CA).
BSA Modification of Aptamer-conjugated AuNRs
Aptamer-conjugated AuNRs were incubated with 1, 3, 5 or 20 mg/mL of BSA in 100 µL binding buffer free of BSA for 2 hours at room temperature. After incubation, the suspensions were washed twice with washing buffer (Dulbecco’s PBS) by centrifuging at 14000 rpm and 25°C for 5 minutes to remove excess BSA. Finally, the precipitate was resuspended in binding buffer free of BSA for cell binding assays.
Cell Incubation and Flow Cytometry Analysis
The binding affinities of sgc8c-conjugated AuNRs (untreated and treated with BSA) were determined by incubating Ramos or CCRF-CEM (1×106 cells/mL) cells with AuNRs-sgc8c at 4°C for 30 min or at 37°C for 2h in 200 µL binding buffer free of BSA consisting of Dulbecco’s PBS with calcium chloride and magnesium chloride (Sigma), glucose (4.5 g/L), MgCl2 (5 mM) and yeast tRNA (Sigma, 0.1 mg/mL). Then cells were washed twice with 0.5 mL washing buffer consisting of Dulbecco’s PBS with calcium chloride and magnesium chloride (Sigma), glucose (4.5 g/L) and MgCl2 (5 mM). Finally, the cells were suspended in 200 µL binding buffer free of BSA and subjected to flow cytometry analysis by counting 10,000 events on a FACScan cytometer (Becton Dickinson Immunocytometry Systems, CA, USA), while using channel 1.
Cytotoxicity Assay
The cytotoxicity of AuNRs-sgc8c (untreated and treated with BSA) to CCRF-CEM and Ramos cells was evaluated by incubating 1×106 cells/mL of CCRF-CEM or Ramos cells with AuNRs-sgc8c in 200 µL binding buffer free of BSA at 37 °C for 2 h under 5% CO2 atmosphere. Following this step, cells were washed twice with washing buffer and then incubated at 37 °C for another 48 h in a 5% CO2 atmosphere. After washing twice, cells were suspended in BSA-free binding buffer and propidium iodide, PI (Invitrogen, Carlsbad, CA), at room temperature for 15 min to test cell viability. In this test, dead cells accumulated the dye and showed red fluorescence, which was analyzed by flow cytometry using channel 3.
Results and Discussion
Tuning the Size of AuNRs
Gold nanorods were prepared by the seed-mediated method.32–35 Their sizes were tuned by changing the concentration of the silver nitrate in the growth solution. Since low concentrations of silver cation allow less growth8, 32, the sizes of the gold nanorods prepared with the growth solution containing a lower concentration of silver cation were smaller in terms of length. Previous reports showed that the optimum size for efficient cellular uptake of gold nanoparticles is around 50 nm.36 Therefore, in this study, gold nanorods with the dimensions 52 ± 4 nm and 14 ± 1 nm in length and width, respectively, were used (Figure 1A).
Fig. 1.
Characterization of AuNRs. TEM images of gold nanorods prepared with (A) 50 µM Ag+, (B) 60 µM Ag+ with the dimensions 52 ± 4 nm, 14 ± 1 nm and 70 ± 9 nm, 14 ± 1 nm in length and width, respectively. Absorption spectra of gold nanorods prepared with (C) 50 µM Ag+, (D) 60 µM Ag+ with longitudinal bands at 744 nm and 830 nm, respectively.
Quantification of Aptamers Immobilized on AuNR Surface
The average number of aptamers immobilized on the surface per AuNR was determined by dividing the concentration of aptamers immobilized on the AuNR surface by the concentration of the AuNRs used in conjugation.31, 37, 38 In order to find the concentration of the surface-bound aptamers, the concentration of the unbound aptamers was subtracted from the overall aptamer concentration used in the modification. Aptamer concentrations were evaluated using a fluorescence standard calibration curve for standard solutions of the sgc8c aptamer modified with FITC dye (Figure S-1A). According to these calculations, approximately 160 sgc8c aptamers were immobilized on each gold nanorod surface. Also, the aptamer immobilization could be monitored by the zeta potential change of AuNRs before and after aptamer immobilization. Since CTAB is a highly cationic surfactant, the surface of the AuNRs was highly positively charged before aptamer immobilization (Figure S-1B). After aptamer conjugation, the surface of the AuNRs became negative due to the negative charges on the aptamers (Figure S-1B).
Specificity Test of sgc8c and sgc8c-conjugated AuNRs
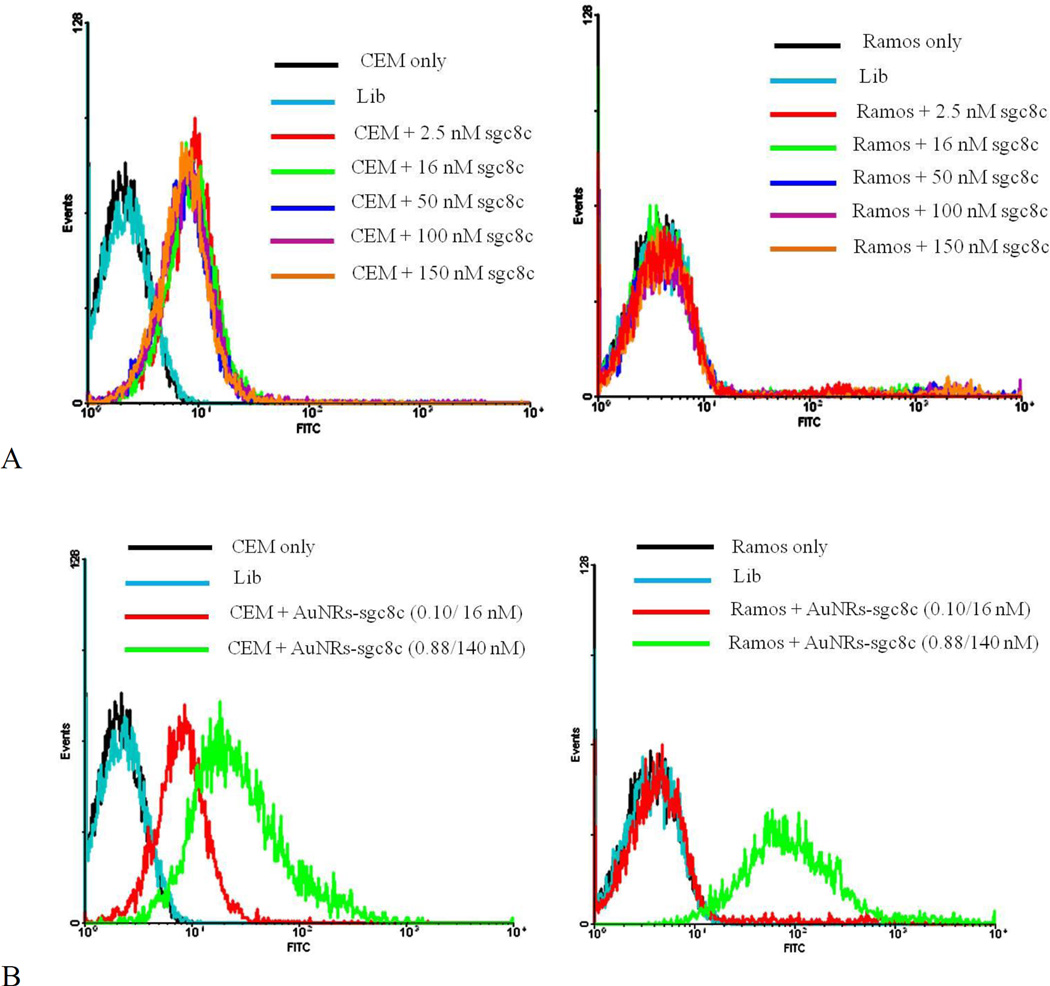
Sgc8c aptamer can specifically bind to the CEM cancer cell line4, but since its biomarker protein is absent from the Ramos cancer cell line, it cannot bind to Ramos cells. Different concentrations of FITC-modified sgc8c aptamer were incubated with both cancer cell lines at 4°C, and no nonspecific binding to Ramos cells was observed (Figure 2A). However, when the concentration of AuNRs conjugated with sgc8c aptamer was increased, an obvious nonspecific binding to Ramos cells was observed (Figure 2B). This nonspecific binding did not occur with similar concentrations of free sgc8c aptamers incubated with Ramos cells; therefore, it was caused by the increased concentrations of AuNRs. The increased concentration of AuNRs results in a corresponding increase in positively charged CTAB content, which, in turn, increases the possibility of electrostatic interactions between the positively charged CTAB and negatively charged cell membrane, even though the overall surface charge of the AuNRs is negative after aptamer immobilization.
Fig. 2.
Specificity test of (A) sgc8c aptamer only and (B) sgc8c-conjugated AuNRs under different concentrations to CEM and RAMOS cell lines. In B, the first concentration is for AuNRs, and the second concentration is for aptamers. Lib stands for “library” and it is the random DNA sequence with the same length as sgc8c aptamer to show the specificity of sgc8c.
The same trend of nonspecific binding also occurred when the AuNR-sgc8c conjugate was incubated with Ramos cells (control cells) at 37°C for 2 h. The nonspecific binding was evident when the Ramos cells were incubated with 0.3 nM AuNR-sgc8c, and it increased along with the increasing concentration of AuNRs (Figure 3A).
Fig. 3.
Cytotoxicity and nonspecificity test of AuNR-sgc8c to Ramos cells. (A) Flow cytometric assay to monitor the binding of different concentrations of AuNR-sgc8c with Ramos cells (control cells). The first concentration is for AuNRs, and the second concentration is for aptamers. (B) Cell viability of Ramos cells after incubation with AuNR-sgc8c at 37°C for 2 h.
Cytotoxicity of AuNRs
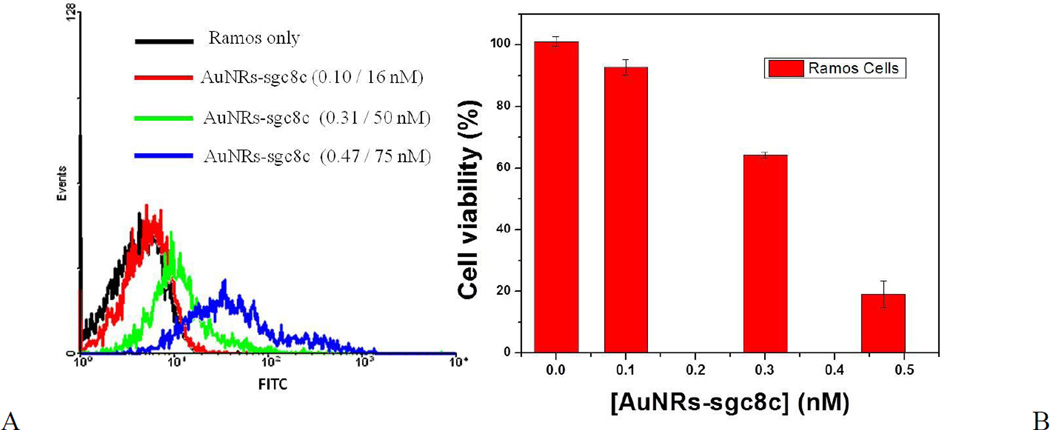
When the concentration of AuNRs increases, the CTAB content also increases, thus exposing both target and control cells to further cytotoxic effects. As shown in Figure S-2, when the incubation concentration of AuNR-sgc8c was increased beyond 0.3 nM, the cytotoxic effects were evident by the low cell count, as determined by flow cytometry. Similar cytotoxic effects were observed when AuNR-sgc8c was incubated with Ramos cells (control cells) at 37°C for 2 h (Figure 3), as a result of the nonspecific binding of AuNRs. In this case, when the concentration of AuNRs was 0.3 nM and higher, the percentage of Ramos cell viability decreased dramatically and went down to 19 % for 0.47 nM AuNRs, as shown by PI staining (Figure 3B).
BSA Modification of AuNRs to Avoid Cytotoxicity and Nonspecific Interactions
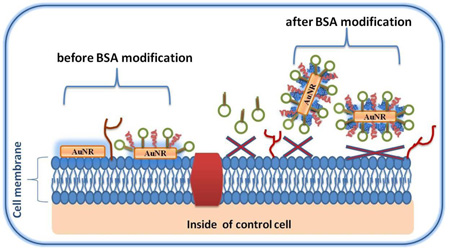
Nonspecific binding of CTAB results primarily from interactions between the positively charged CTAB and negatively charged cell surface caused by the presence of proteins and phospholipids.10 Thus, cytotoxic effects of CTAB can be minimized if the interaction between the cell surface and CTAB can be blocked. In this study, BSA (bovine serum albumin) protein was used to coat the CTAB bilayer on the surface of the AuNRs through electrostatic interactions (Figure 4), because BSA (pI = 4.7) is negatively charged, and CTAB is positively charged at pH 7.4 (binding buffer).11, 39
Fig. 4.
Scheme illustrating avoidance of nonspecific binding to control cell lines using BSA modification of AuNR-sgc8c (Note that the schematic is not drawn to scale).
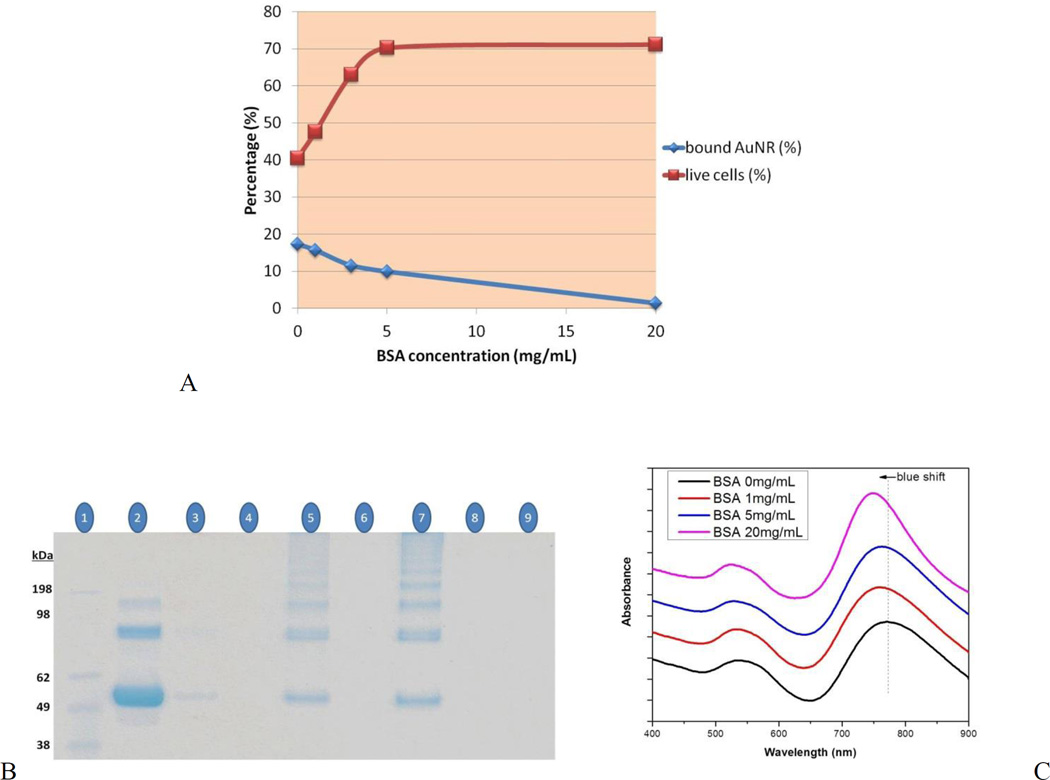
In order to optimize the BSA concentration required for coating the AuNR surface to minimize the interactions between the CTAB and the cell surface, CEM cells were incubated with AuNRs (0.47 nM) treated with different concentrations of BSA at 37°C for 2 h. After incubation, according to the results of PI staining, AuNRs treated with 5 and 20 mg/mL BSA showed much lower toxicity compared to the untreated AuNRs and AuNRs treated with 1 and 3 mg/mL BSA (Figure 5A). However, the cell viabilities were similar for cells incubated with AuNRs treated with 5 or 20 mg/mL BSA. Thus, 5 mg/mL was chosen as the optimal concentration of BSA for coating the AuNR surface. As shown in Figure 5A, toxicity decreased when the percentage of the surface-bound AuNRs decreased, indicating that the interaction between CTAB and the cell surface is the main cause of toxicity.
Fig. 5.
(A) Cell viability of CEM cells incubated (37°C for 2 h) with AuNRs only (0.47 nM) or AuNRs treated with different concentrations of BSA (1, 3, 5 and 20 mg/mL). Cell viability was analyzed by PI staining. Surface-bound AuNRs were analyzed by absorption measurements. (B and C) Monitoring BSA modification on AuNRs: (B) Gel electrophoresis of captured BSA: lane 1, protein marker; lane 2, BSA protein standard (10 µg); lanes 3, 5, 7, BSA captured, respectively via 0.1, 0.3 and 0.5 nM AuNRs incubated with 100 µg of BSA; lane 9, 0.5 nM AuNRs only. Nothing was loaded in lanes 4, 6 and 8. (C) UV-vis spectra of AuNRs treated with different concentrations of BSA.
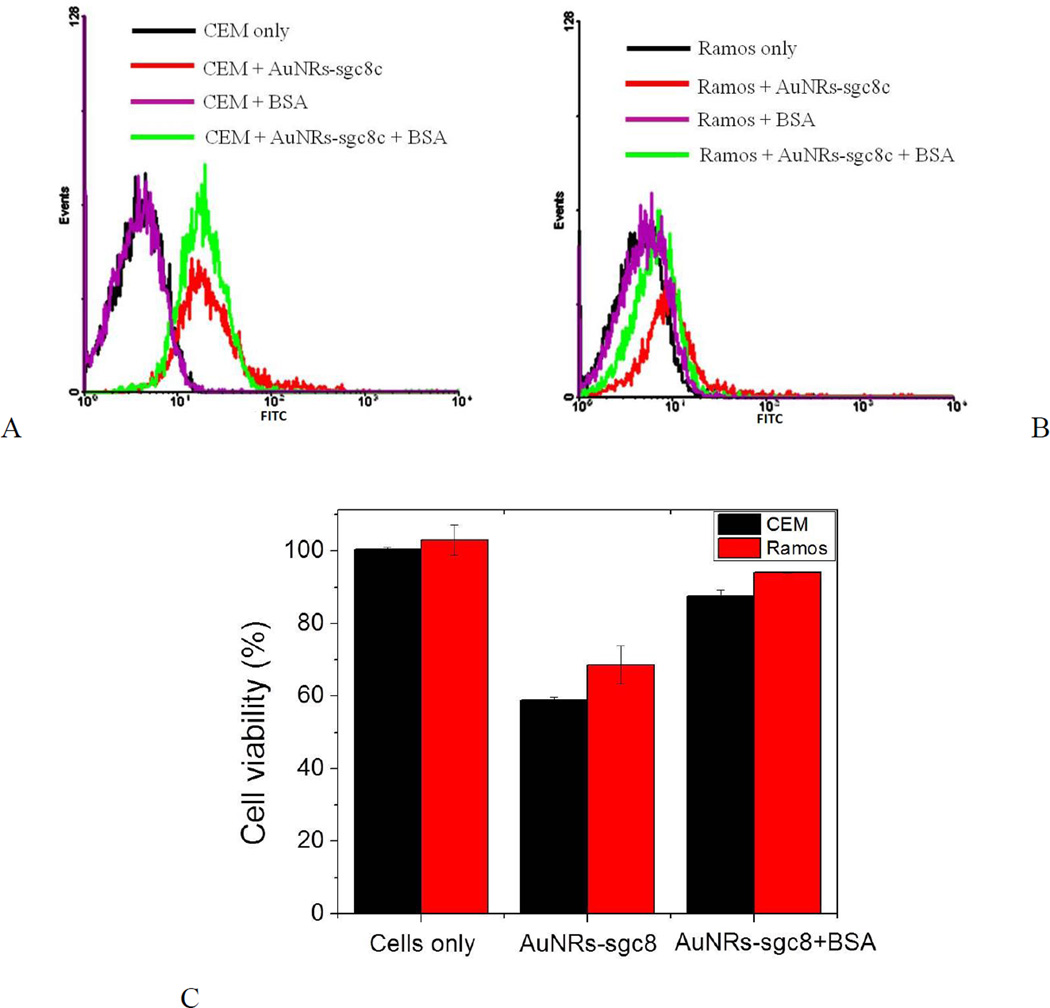
Since 0.3 nM and higher concentrations of AuNRs showed an evident cytotoxicity against bound cells after incubation at 37°C for 2 h (Figures S–2, 3 and 5), concentrations of 0.3 and 0.47 nM AuNRs were chosen for BSA treatment. When CEM cells (target cells) were incubated with AuNR-sgc8c (0.47 nM) treated with 1, 5, 20 mg/mL BSA, cell viability increased dramatically and similarly for the nanorods treated with 5 and 20 mg/mL BSA (Figure S-3A, C). These results correlate with those obtained for the AuNRs without aptamer modification (Figure 5A). The cell viabilities of both CEM and Ramos cells decreased dramatically after incubation with untreated AuNR-sgc8c at 0.47 nM (Figure S-3). However, the cell viabilities of both the CEM and Ramos cells were recovered almost completely if they were incubated with 5 mg/mL BSA-treated AuNR-sgc8c at 0.47 nM (Figure S-3C). The same trend occurred when CEM and Ramos cells were incubated with 5 mg/mL BSA-treated AuNR-sgc8c at 0.3 nM and incubated at 37°C for 2 h (Figure 6). Since the concentration of the AuNRs was lower (0.3 nM), toxicity was not as severe as it was with 0.47 nM AuNRs (Figure 6C). Toxicity decreased as the concentration of AuNRs decreased, because the CTAB concentration also decreased.
Fig. 6.
Specificity and cytotoxicity test of AuNR-sgc8c (0.3 nM) to CEM and Ramos cells. Flow cytometric assay to monitor the binding of AuNR-sgc8c with (A) CEM and (B) Ramos after 5 mg/mL BSA treatment. Incubation with cells was performed at 37°C for 2 h. (C) Cell viability of CEM and Ramos cells analyzed by PI staining.
As shown in Figure 5A, AuNRs that were not modified with aptamers could bind to CEM cells as a result of the electrostatic interactions between the CTAB and cell surface. This nonspecific binding was also observed in the confocal images of control Ramos cells incubated with AuNR-sgc8c (0.5 nM) at 37°C for 2 h (Figure S-4A). The green signal results from the emission of the FITC dye on the sgc8c aptamers. (The signal is weak because confocal microscopy only collects the emission from a certain area, while flow cytometry integrates the emission signal generated by each cell until the pre-set cell number is reached.) On the other hand, when Ramos cells were incubated with BSA-treated AuNR-sgc8c (0.5 nM) at 37°C for 2 h, no signal was observed, indicating the absence of nonspecific binding (Figure S-4B).
BSA modification of the AuNRs was also monitored by SDS-PAGE (Figure 5B). After incubating the AuNRs (0.1, 0.3 and 0.5 nM) with 100 µg of BSA for 2 h, the excess BSA was removed, and the captured BSA (including a standard BSA (10 µg) sample) were loaded in a 12% Bis-Tris SDS-polyacrylamide gel for electrophoresis. Since AuNRs cannot move in the gel (Figure 5B, lane 9), the captured BSA was eluted by heating the AuNR-BSA at 95°C for 5 minutes before loading into the gel. As expected, the band intensities of the captured BSA increased as the concentration of the incubated AuNRs increased (Figure 5B lanes 3, 5 and 7). If the second BSA band from the bottom was taken as a reference (excluding the polymerized BSA bands and the contaminant band, first from the bottom, that were mainly the artifacts of the heat-shock preparation process of albumins), the percentage of the BSA capture (compared to standard BSA band) is 1, 5 and 11% for 0.1, 0.3 and 0.5 nM AuNRs, respectively (Figure S-5). Furthermore, according to the zeta potential measurements (Figure S-1B), BSA modified AuNRs-sgc8c (−18 mV) were more negatively charged compared to the AuNRs-sgc8c (−11 mV), which is another proof of BSA modification. On the other hand, BSA modification did not affect the morphology of the AuNRs adversely, because the transverse and more importantly longitudinal absorption bands could still be resolved well upon BSA treatment in different concentrations (Fig. 5C). However, a blue SPR shift was observed in the longitudinal absorption band of AuNRs when the incubation concentration of BSA increased, which can be considered as another proof of the BSA modification of the AuNR surface.
In fact, there are serum protein receptors on the cell membrane that are responsible for serum protein recognition. For instance, there are albumin receptors on the cell membrane to recognize albumin protein as nutrition for the cells. However, it has been reported that BSA that is adsorbed on the cationic nanoparticles via electrostatic or hydrophobic interactions undergoes a structural change.40, 41 Thus, this denatured form of BSA cannot be recognized by the albumin receptors present on the cell membrane.
In view of these results, it can be concluded that BSA modification of AuNRs minimized CTAB/cell surface interaction. This step circumvents the toxic effects of CTAB, even if high concentrations of AuNRs are employed. Consequently, this procedure can facilitate the progress of biological applications, especially in vivo applications, where high concentrations of AuNRs are needed. Lower concentrations of AuNRs can generate sufficient heat to kill cells (Figure S-6). However, even if all these AuNRs bind to the target tumor, there is no guarantee that the amount will be sufficient to destroy the entire tumor tissue by elevating its temperature. Therefore, higher concentrations of AuNRs are preferred.
Conclusions
This study emphasizes the vital role of surface modification of gold nanorods for biomedical applications. Despite the high specificity of aptamers and high NIR absorption of gold nanorods, their conjugation is not sufficient to achieve efficient targeted photothermal therapy of cancer cells as a result of CTAB nonspecificity and cytotoxicity. These toxic effects occur with high concentrations of gold nanorods and should be controlled before laser treatment is undertaken. In this study, with the inspiration of the biological machinery, BSA encapsulation of CTAB on the surface of AuNRs minimized CTAB/cell surface interaction such that nonspecific binding to cells was dramatically decreased, thus limiting the toxic effects of CTAB on both control and target cells before laser treatment. The efficiency of BSA encapsulation in reducing the toxicity is also comparable to that reported for polyelectrolyte coatings. Thus, aptamer-conjugated BSA-modified AuNRs are efficient and highly selective PTT agents with negligible cytotoxicity.
Supplementary Material
Acknowledgments
This work is supported by grants awarded by the National Institutes of Health (GM079359 and CA133086). We would like to thank Dr. Kathryn Williams from UF for her help in revising this paper, and Dr. Martin Kurnik from UCSB for the BSA protein scheme.
Footnotes
Electronic Supplementary Information (ESI) available: [Figures S-1 to S-5 are included]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Yasun E, Kang H, Erdal H, Cansiz S, Ocsoy I, Huang YF, Tan W. Interface Focus. 2013;3:20130006. doi: 10.1098/rsfs.2013.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu M, Chen JY, Li ZY, Au L, Hartland GV, Li XD, Marquez M, Xia YN. Chem. Soc. Rev. 2006;35:1084–1094. doi: 10.1039/b517615h. [DOI] [PubMed] [Google Scholar]
- 3.von Maltzahn G, Park JH, Agrawal A, Bandaru NK, Das SK, Sailor MJ, Bhatia SN. Cancer Res. 2009;69:3892–3900. doi: 10.1158/0008-5472.CAN-08-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J Proteome. Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Mol. Cell. Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Jana NR, Gearheart L, Murphy CJ. J Phys. Chem. B. 2001;105:4065–4067. [Google Scholar]
- 7.Murphy CJ, Sau TK, Gole AM, Orendorff CJ, Gao J, Gou L, Hunyadi SE, Li T. J Phys. Chem. B. 2005;109:13857–13870. doi: 10.1021/jp0516846. [DOI] [PubMed] [Google Scholar]
- 8.Perezjuste J, Pastorizasantos I, Lizmarzan L, Mulvaney P. Coord. Chem. Rev. 2005;249:1870–1901. [Google Scholar]
- 9.Nikoobakht B, El-Sayed MA. Langmuir. 2001;17:6368–6374. [Google Scholar]
- 10.Wang L, Jiang X, Ji Y, Bai R, Zhao Y, Wu X, Chen C. Nanoscale. 2013;5:8384–8391. doi: 10.1039/c3nr01626a. [DOI] [PubMed] [Google Scholar]
- 11.Hauck TS, Ghazani AA, Chan WC. Small. 2008;4:153–159. doi: 10.1002/smll.200700217. [DOI] [PubMed] [Google Scholar]
- 12.Alkilany AM, Nagaria PK, Hexel CR, Shaw TJ, Murphy CJ, Wyatt MD. Small. 2009;5:701–708. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- 13.Niidome T, Yamagata M, Okamoto Y, Akiyama Y, Takahashi H, Kawano T, Katayama Y, Niidome Y. Journal of controlled release : official journal of the Controlled Release Society. 2006;114:343–347. doi: 10.1016/j.jconrel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 14.Qiu Y, Liu Y, Wang L, Xu L, Bai R, Ji Y, Wu X, Zhao Y, Li Y, Chen C. Biomaterials. 2010;31:7606–7619. doi: 10.1016/j.biomaterials.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 15.Vigderman L, Manna P, Zubarev ER. Angew Chem Int Edit. 2012;51:636–641. doi: 10.1002/anie.201107304. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Liu Y, Li W, Jiang X, Ji Y, Wu X, Xu L, Qiu Y, Zhao K, Wei T, Li Y, Zhao Y, Chen C. Nano Lett. 2011;11:772–780. doi: 10.1021/nl103992v. [DOI] [PubMed] [Google Scholar]
- 17.Alkilany AM, Nagaria PK, Wyatt MD, Murphy CJ. Langmuir. 2010;26:9328–9333. doi: 10.1021/la100253k. [DOI] [PubMed] [Google Scholar]
- 18.Hu X, Gao X. Phys. Chem. Chem. Phys. 2011;13:10028–10035. doi: 10.1039/c0cp02434a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oyelere AK, Chen PC, Huang X, El-Sayed IH, El-Sayed MA. Bioconjugate chemistry. 2007;18:1490–1497. doi: 10.1021/bc070132i. [DOI] [PubMed] [Google Scholar]
- 20.Liao H, Hafner JH. Chem. Mater. 2005;17:4636–4641. [Google Scholar]
- 21.Monopoli MP, Aberg C, Salvati A, Dawson KA. Nature Nanotech. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari M. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 23.Stephan MT, Moon JJ, Um SH, Bershteyn A, Irvine DJ. Nature medicine. 2010;16:1035–1041. doi: 10.1038/nm.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HR, Andrieux K, Delomenie C, Chacun H, Appel M, Desmaele D, Taran F, Georgin D, Couvreur P, Taverna M. Electrophoresis. 2007;28:2252–2261. doi: 10.1002/elps.200600694. [DOI] [PubMed] [Google Scholar]
- 25.Lesniak A, Fenaroli F, Monopoli MP, Aberg C, Dawson KA, Salvati A. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Li WX, Li QN, Li YG, Li YF, Zhang XY, Huang Q. Carbon. 2009;47:1351–1358. [Google Scholar]
- 27.Bajaj A, Samanta B, Yan H, Jerry DJ, Rotello VM. J Mater. Chem. 2009;19:6328. doi: 10.1039/b718745a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunov O, Syrovets T, Loos C, Beil J, Delacher M, Tron K, Nienhaus GU, Musyanovych A, Mailander V, Landfester K, Simmet T. ACS Nano. 2011;5:1657–1669. doi: 10.1021/nn2000756. [DOI] [PubMed] [Google Scholar]
- 29.Buchwalow I, Samoilova V, Boecker W, Tiemann M. Scientific reports. 2011;1:28. doi: 10.1038/srep00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez-Medina S, Blankenburg J, Olson J, Landes CF, Link S. ACS Sustain Chem Eng. 2013;1:833–842. doi: 10.1021/sc400042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasun E, Gulbakan B, Ocsoy I, Yuan Q, Shukoor MI, Li C, Tan W. Anal. Chem. 2012;84:6008–6015. doi: 10.1021/ac300806s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikoobakht B, El-Sayed MA. Chem. Mater. 2003;15:1957–1962. [Google Scholar]
- 33.Johnson CJ, Dujardin E, Davis SA, Murphy CJ, Mann S. J Mater. Chem. 2002;12:1765–1770. [Google Scholar]
- 34.Gole A, Murphy CJ. Chem. Mater. 2004;16:3633–3640. [Google Scholar]
- 35.Gao J, Bender CM, Murphy CJ. Langmuir. 2003;19:9065–9070. [Google Scholar]
- 36.Chithrani BD, Ghazani AA, Chan WC. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 37.Huang YF, Sefah K, Bamrungsap S, Chang HT, Tan W. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 38.Huang YF, Chang HT, Tan W. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 39.Peng ZG, Hidajat K, Uddin MS. J Colloid Interface Sci. 2005;281:11–17. doi: 10.1016/j.jcis.2004.08.086. [DOI] [PubMed] [Google Scholar]
- 40.Fleischer CC, Payne CK. Acc Chem Res. 2014;47:2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam S, Mukhopadhyay A. J Phys Chem C. 2014;118:27459–27464. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.