Abstract
The relationship between microRNA regulation and the specification of behaviour is only beginning to be explored. Here we find that mutation of a single microRNA locus (miR-iab4/8) in Drosophila larvae affects the animal’s capacity to correct its orientation if turned upside-down (self-righting). One of the microRNA targets involved in this behaviour is the Hox gene Ultrabithorax whose derepression in two metameric neurons leads to self-righting defects. In vivo neural activity analysis reveals that these neurons, the self-righting node (SRN), have different activity patterns in wild type and miRNA mutants whilst thermogenetic manipulation of SRN activity results in changes in self-righting behaviour. Our work thus reveals a microRNA-encoded behaviour and suggests that other microRNAs might also be involved in behavioural control in Drosophila and other species.
The regulation of RNA expression and function is emerging as a hub for gene expression control across a variety of cellular and physiological contexts including neural development and specification. Small RNAs such as microRNAs (miRNAs) have been shown to affect neural differentiation (1, 2) but their roles in the control of behaviour are only beginning to be explored.
Previous work in our laboratory focused on the mechanisms and impact of RNA regulation on the expression and neural function of the Drosophila Hox genes (3–6). These genes encode a family of evolutionarily conserved transcription factors that control specific programs of neural differentiation along the body axis (7–9) offering an opportunity to investigate how RNA regulation relates to the formation of complex tissues such as the nervous system.
Here we use the Hox gene system to investigate the roles played by a single miRNA locus (miR-iab4/8) (3, 10–14, 30) on the specification of the nervous system during early Drosophila development. This miRNA locus controls the embryonic expression of posterior Hox genes (3, 10–14). Given that we found no detectable differences in the morphological layout of the main components of the nervous system in late Drosophila embryos of wild type and miR-iab4/8 null mutants (herein ∆miR, (13)) (Fig S3B-F) we analysed early larval behaviour as a stratagem to probe the functional integrity of the late embryonic nervous system.
Most behaviours in early larva were unaffected by the miRNA mutation (Fig. S1, movie S1 and S2) except self-righting (SR) behaviour (Fig. 1A-C, movies S3–S4): miRNA mutant larvae were unable to return to their normal orientation at the same speed as their wild type counterparts.
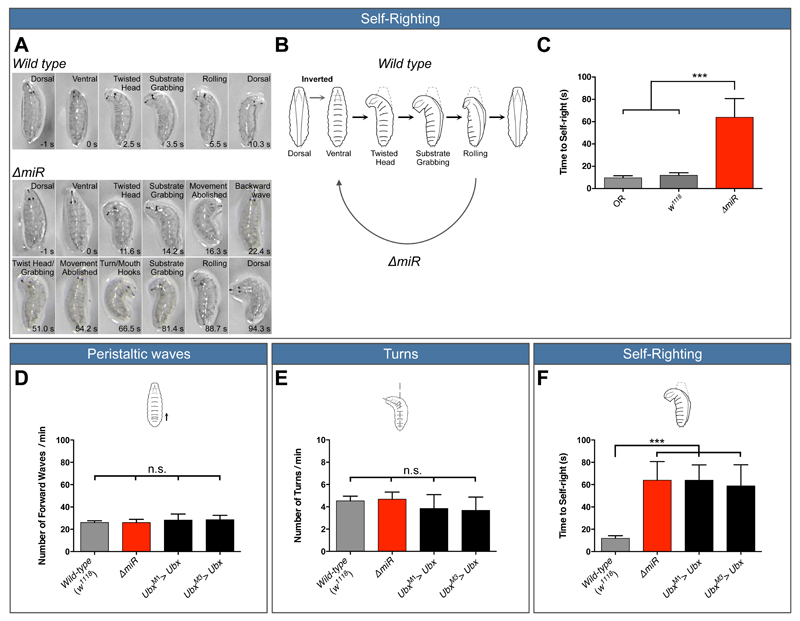
Figure 1. Both, removal of miR-iab4/iab8 and over-expression of Ubx disrupt a specific larval locomotor behaviour: self-righting.
(A, B) Description of larval self-righting behaviour. (A) Time-lapses of larval self-righting behaviour. (A top) Wild type larvae were placed in an inverted position (ventral up), twisted their heads, grabbed the substrate with the mouth hooks and rolled their bodies onto their ventral surface (dorsal up). In contrast, (A bottom) ∆miR larvae displayed problems in self-righting their bodies. (B) Diagram of the self-righting behavioural response. (C) Quantification of the time required for the successful completion of the self-righting behaviour (mean ± SEM; N =27 to 29 larvae per genotype) in the two wild type controls (OR, w1118, light and dark grey respectively) and ∆miR larvae (red). (D-F) Quantification of larval behaviour in Ubx overexpression lines (UbxM1>Ubx and UbxM3>Ubx). Quantification of (D) number of forward peristaltic waves per minute, (E) larval turning per minute and (F) time to self-right in: wild-type (w1118, grey), ∆miR (red), UbxM1> Ubx and UbxM3> Ubx (black) (mean ± SEM; N =15 to 29 larvae per genotype). A non-parametric Mann-Whitney U test was performed to compare treatments; p > 0.05 (non-significant; n.s); p < 0.001 (***).
By means of selective target over-expression followed by SR phenotype analyses we identified the Drosophila Hox gene Ultrabithorax (Ubx) (17, 18) as a miRNA target implicated in the genetic control of SR behaviour (Fig. 1F). Notably, overexpression of Ubx did not affect any larval behavior tested except self-righting in remarkable agreement with the effects observed in miRNA mutants (Fig. 1D and E). Analysis of Ubx 3’UTR fluorescent reporter constructs expressed in the Drosophila CNS (Fig. S2) indicates that the interaction between miR-iab4/8 and Ubx is direct, in line with prior observations in other cellular contexts (10–12).
To identify the cellular basis for SR control we systematically over-expressed Ubx within sub-populations of neurons (Fig. S4). Increased levels of Ubx within the pattern of Cha(7.4kb)-Gal4, which largely targets cholinergic sensory and interneurons, phenocopied the miRNA SR anomalies (Fig. S4). Further analysis identified two metameric neurons as the minimal node required for the SR behaviour (Self-righting node, SRN) (Fig. 2A and B).
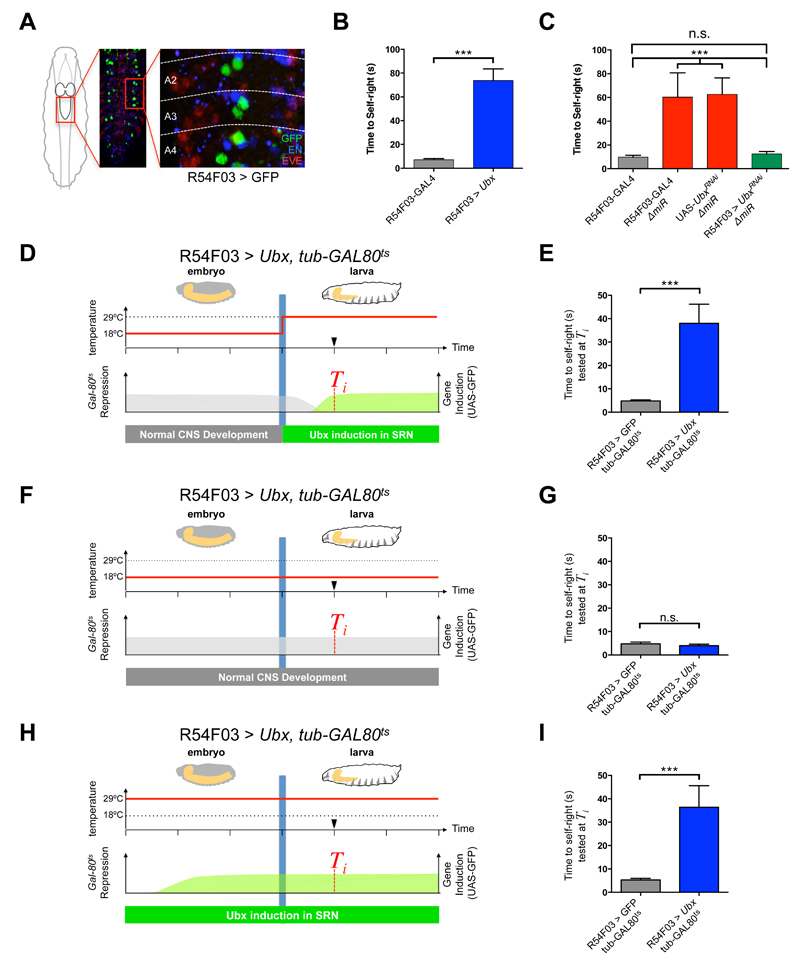
Figure 2. miRNA-dependent Ubx regulation in SRN cells underlies SR behaviour.
(A) R54F03-GAL4 expression (GFP, green) in the larval abdominal CNS (Even-skipped protein in red; Engrailed protein in blue; A2 refers to abdominal segment 2). (B) Artificial increase of Ubx expression in two metameric neurons driven by the R54F03-GAL4 promoter (mean ± SEM; N = 60 per genotype). (C) Artificial decrease of Ubx expression with UbxRNAi within SRN cells in ∆miR larvae (R54F03-GAL4, ∆miR/ UAS-UbxRNAi, ∆miR, green) (mean ± SEM; N = 20 to 23 per genotype). (D-I) Conditional increase of Ubx expression during embryonic and early larval development with tub-Gal80ts (Gal80ts represses GAL4 activity at 18°C) within SRN cells: R54F03 > Ubx, tub-GAL80ts (UAS-Ubx /+ ; R54F03-GAL4/ tub-Gal80ts). Controlled increase of Ubx expression in SRN cells in early larvae (D and E; mean ± SEM; N = 20 per genotype) and from mid-embryogenesis to early larvae (H and I; mean ± SEM; N = 15 per genotype). (F and G) Repressed increase of Ubx expression in SRN cells throughout embryogenesis and early larvae (mean ± SEM; N = 15 per genotype). A non-parametric Mann-Whitney U test was performed to compare treatments; p>0.05 (non-significant; n.s.); p<0.001 (***).
Several lines of evidence confirm the role of miRNA-dependent Ubx regulation within the SRN as a determinant of SR. First, both Ubx and miRNA transcripts (i.e. miR-iab4) derived from the miR-iab4/8 locus are detected within the SRN (Fig. 3A-C). Second, in the context of miRNA mutation, Ubx protein expression is increased within SRN (Fig. 3D-F). Third, reduction of Ubx (i.e. Ubx RNAi) specifically enforced within SRN cells is able to ameliorate or even rescue the SR phenotype observed in miRNA mutants (Fig. 2C).
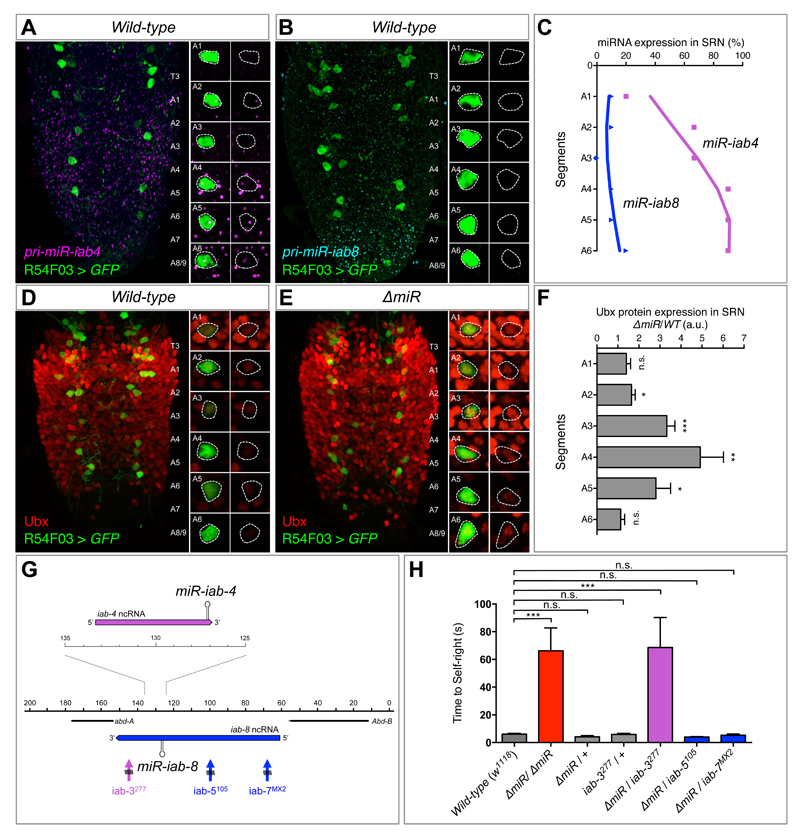
Figure 3. Regulation of Ubx protein expression in SRN cells by miR-iab4/iab8.
(A) Wild type expression of precursor miR-iab4 transcripts (RNA-FISH, magenta) in SRN cells (R54F03>GFP, green) of the ventral nerve cord (VNC) of first-instar Drosophila larvae. (B) Wild type expression of precursor miR-iab8 transcripts (RNA-FISH, cyan) in SRN cells (R54F03>GFP, green) of the VNC of first-instar Drosophila larvae. (C) Percentage of SRN cells expressing miR-iab4 (purple, square) and miR-iab8 (blue, triangle) precursors across A1 to A6 (N = 10). (D and E) Ubx protein expression (red) in SRN cells of wild type (D) and ∆miR (E) first-instar larvae VNCs. (F) Quantification of Ubx protein expression ratio of ∆miR over wild type within the SRN cells (red) by fluorescent intensity (N = 8 per genotype; arbitrary units, a.u.). (G) Diagram of a sub-region of the bithorax complex based on (13) showing iab-4 (purple) and iab-8 (blue) non-coding RNAs (ncRNA), and rearrangement breakpoints affecting miR-iab-4 (iab-3277, purple) and miR-iab-8 (iab-5105 and iab-7MX2, blue). (H) Genetic complementation tests to determine the involvement of miR-iab4 or miR-iab8 in SR behaviour using trans-heterozygote larvae for ∆miR and different chromosomal rearrangement breakpoints that disrupt the bithorax complex (mean ± SEM; N = 17 to 20 per genotype). A non-parametric Mann-Whitney U test was performed to compare treatments; p>0.05 (non-significant; n.s.); p < 0.05 (*); p < 0.01 (**); p<0.001 (***).
Two plausible scenarios arise to explain the effects of miR-iab4/8 in regards to SR behaviour. One is that miRNA input is required for the late embryonic development of the neural networks underlying SR arguing for a ‘developmental’ role of the miRNA; another, is that miRNA repression affects normal physiological/behavioural functions largely without disrupting neural development in line with a ‘behavioural’ role. Two independent experiments support that the primary roles of miR-iab4/8 are behavioural. First, anatomical analysis of SRN cells in wt, ∆miR and R54503>Ubx (i.e. SRN-driver line) show no significant differences in total numbers of SRN cells (Fig. S5B) nor in SRN cell body size (Fig. S5C); furthermore, analysis of wt, ∆miR and R54503>Ubx show indistinguishable SRN-projection patterns (Fig. S5D and E). Second, Gal-80ts mediated conditional expression experiments show that SRN-specific Ubx overexpression after embryogenesis is sufficient to trigger the SR behaviour (Fig. 2D-E).
The results presented above suggest that miRNA-dependent Hox regulation within the SRN must somehow modify the normal physiology of SRN cells so that when the miRNA is mutated these neurons perform different functions than those in wild type animals. To test this hypothesis we used genetically-encoded calcium sensors [GCaMP6, (24)] specifically expressed in SRN cells and tracked down spontaneous profiles of neural activity. SRN cells in miRNA mutants produce activity traces that are significantly different from those observed in wild type SRN cells (Fig. 4B-C, Fig. S6A). Quantification of maximal amplitude and proportion of active cells in each genotype also reveal significant differences (Fig. 4D, Fig. S6B) in SRN function across the genotypes, but no change in cell viability is observed (Fig S6C). Neural activity differences across genotypes are significant within regions of expression of miR-iab4 (Fig. 4E) suggesting that this miRNA (and not miR-iab8) might be the main contributor to SR control. Analysis of mutations selectively affecting miR-iab4 or miR-iab8 (13–14, 25–26) strongly suggests that miR-iab4 is the key regulator of SR (Fig. 3G-H).
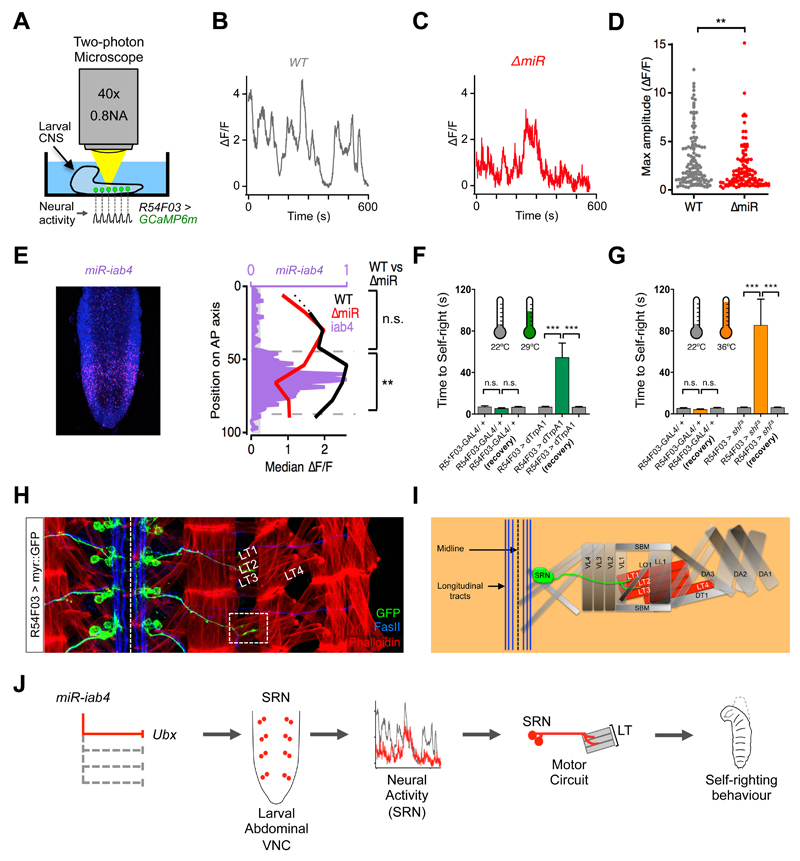
Figure 4. ∆miR mutants have abnormal patterns of neural activity in SRN cells.
(A) Schematic of the lateral larval CNS expressing GCaMP6m in SRN cells (R54F03> GCaMP6m, green) imaged in a two-photon microscope. (B) Examples of spontaneous activity recorded over 10 mins from wild type (WT: UAS-GCaMP6m/+; R54F03-GAL4/+) and (C) ∆miR mutants (UAS-GCaMP6m/+; R54F03-GAL4, ∆miR/∆miR) in SRN cells. (D) Maximum amplitude of spontaneous activity in SRN cells: WT (median ∆F/F=1.91; N = 120) and in ∆miR mutants (median ∆F/F=1.27; N = 115) (** p<0.01, Mann-Whitney U test). (E) Expression pattern of miR-iab4 (purple) and DAPI (blue) in the VNC of a freshly hatched larva (left panel). Median ∆F/F in SRN cells of WT (black line) and ∆miR (red line) larval VNCs, and relative expression of miR-iab4 (purple) along the anterior-posterior (A-P) axis. Median ∆F/F of WT (median of 2.132, n=73) and ∆miR (median of 1.122, n=68) in regions of high miR-iab4 expression (** p<0.01, Mann-Whitney U test). Regions of low miR-iab4 expression have a median ∆F/F of 1.763 in WT (N = 47) and 1.749 (N = 47) in ∆miR specimens (n.s., p>0.05; Mann-Whitney U test). (F and G) Thermogenetic manipulation of neural activity in SRN cells. Activation (F, R54F03>dTrpA1) and inhibition (G, R54F03>shits) of SRN neural activity (*** p<0.0001) [29°C (green) for activation (H) and 36°C (orange) for inhibition (I)]. (H) Wild-type motor axonal projections of SRN cells (UAS-myr::GFP/ UAS-myr::GFP; R54F03-GAL4/ R54F03-GAL4, green) into muscles (phalloidin, red) lateral transverse 1 and 2 (LT1 and LT2) in late embryos (stage 17)(Fasciclin II, FASII, blue).). Scale bars (white bars) represent 10μm (I) Diagram of SRN cells projecting to the LT1 and LT2 muscles. (J) A model that summarises the data reported in this study. Mutation of miR-iab4 (left) leads to Ubx de-repression in the SRN node affecting SRN neural activity patterns and triggering an anomalous self-righting behaviour (right).
To demonstrate that the changes in SRN neural activity were causal to SR behaviour we artificially activated (Fig. 4F) or inhibited (Fig. 4G) SRN cells (27, 28) and showed that this triggered the aberrant SR phenotype. This suggested that activation of SRN cells in larvae placed ‘right-side up’ might be sufficient to ‘evoke’ actions reminiscent of a self-righting response. We developed an optogenetic system where we activated SRN cells by means of R54F03-driven channelrhodopsin 2 (ChR2) in trans-retinal fed larvae. Under blue light stimulation larvae performed an atypical bending movement, frequently adopting a “lunette” position (Fig. S7 and movie S5). Neither parental line R54F03-Gal4 or UAS-Ch2R showed similar reactions to stimulation confirming the specificity of this effect (Fig. S7, movies S6 and S7).
To study the links between SRN neurons and the SR movement we labelled SRN projections with myr-GFP and discovered that SRN cells innervate two of the lateral transverse (LT) muscles and that they can be co-labelled with anti-Fasciclin 2 (Fas2) (Fig. 4H) demonstrating these to be motorneurons. LT muscles are innervated by Bar-H1+ motorneurons (Fig. S8A) so we used Bar-H1-Gal4 as a second driver to demonstrate that appropriate Ubx levels in these cells are required for normal self righting behavior (Fig. S8B) establishing the SRN cells as the LT-MNs.
We have therefore shown that miRNA-dependent Hox gene repression within a distinct group of motorneurons (SRN/LT-MNs) is required for the control of a specific locomotor behaviour in the early Drosophila larva. Our finding that Hox gene post-transcriptional regulation is involved in SR control suggests that other RNA-based regulatory processes affecting Hox gene expression might also impinge on specific neural outputs; we are currently investigating this possibility with especial regard to the roles of the Hox genes in the specification of neural lineages with axial-specific architectures and systematically testing the roles of other miRNAs on behaviour.
The fact that we could not detect any obvious neuro-anatomical changes in miRNA mutant embryos suggests these to be either very subtle or that the role of miRNA regulation may be primarily ‘behavioral’ in the sense of affecting the performance of a correctly wired neural system, rather than ‘developmental’ i.e. contributing to the development of the network (29). Given that miR-iab4/iab8 is involved in adult ovary innervation (30) it seems that miRNAs – much like ordinary protein-coding genes– can be involved in several distinct roles within the organism.
The results of this study contribute to the understanding of how complex innate behaviours are represented in the genetic program. Our data lead us to propose that other miRNAs might also be involved in the control of behaviour in Drosophila and other species.
Supplementary Material
Acknowledgements
We would like to thank Leon Lagnado for his support to this project and Sofia Pinho and Peter Reed for technical assistance. We also thank Rob White for antibodies, Welcome Bender, Ernesto Sánchez-Herrero and the Bloomington Stock Centre for Drosophila stocks, and Pedro Patraquim for bioinformatic support. This work was funded by a Wellcome Trust Investigator Award to C.R.A. [WT grant 098410/Z/12/Z] and a PhD studentship to J.P.O. by Fundação para a Ciência e a Tecnologia (Portugal) [FCT grant SFRH/BD/63312/2009]. J.B. is funded by a Sir Henry Dale Fellowship (Wellcome Trust and the Royal Society) Grant 105568/Z/14/Z and M.L. was supported by grants from the Biotechnology and Biological Sciences Research Council (UK) (BB/I022414/1) and the Wellcome Trust (092986/Z).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Li X, Jin P. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- 2.Sun AX, Crabtree GR, Yoo AS. Curr Opin Cell Biol. 2013;25:215–221. doi: 10.1016/j.ceb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Development. 2010;137:2951–2960. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- 4.Reed HC, et al. Genetics, genetics. 2010;109:112086. [Google Scholar]
- 5.de Navas LF, et al. Development. 2011;138:107–116. doi: 10.1242/dev.051409. [DOI] [PubMed] [Google Scholar]
- 6.Rogulja-Ortmann A, et al. Development. 2014;141:2046–2056. doi: 10.1242/dev.101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 8.Maeda RK, Karch F. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 9.Mallo M, Alonso CR. Development. 2013;140:3951–3963. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- 10.Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyler DM, et al. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark A, et al. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender W. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gummalla M, et al. PLoS genetics. 2012;8:e1002720. doi: 10.1371/journal.pgen.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso CR. Trends Genet. 2012;28:78–88. doi: 10.1016/j.tig.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Bridges CB, Morgan TH. The Third-Chromosome Group of Mutant Characters of Drosophila melanogaster. Carnegie Institution of Washington; 1923. [Google Scholar]
- 18.Sanchez-Herrero E, Vernós I, Marco R, Morata G. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- 19.Akam ME, Martinez-Arias A. EMBO J. 1985;4:1689–1700. doi: 10.1002/j.1460-2075.1985.tb03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White RA, Wilcox M. EMBO J. 1985;4:2035–2043. doi: 10.1002/j.1460-2075.1985.tb03889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall JC, Kankel DR. Genetics. 1976 [Google Scholar]
- 22.Pfeiffer BD, et al. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning L, et al. CellReports. 2012;2:1002–1013. [Google Scholar]
- 24.Chen T-W, et al. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gummalla M, Galetti S, Maeda RK, Karch F. Front Cell Neurosci. 2014;8:96. doi: 10.3389/fncel.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karch F, et al. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- 27.Hamada FN, et al. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitamoto T. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 29.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garaulet DL, et al. Developmental Cell. 2014;29:635–648. doi: 10.1016/j.devcel.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.