Abstract
Our knowledge of the processes involved in speciation of microalgae remains highly limited. In the present study, we investigated a potential role of ecological speciation processes in diversification of the filamentous green alga Klebsormidium. We examined 12 strains representing four different genotypes. The strains were collected from sandstone and limestone rocks and were cultivated at five different pH levels ranging from pH 4 to pH 8. We determined the responses of the 12 strains to the experimental pH conditions by (1) measuring the effective quantum yield of photosystem II, and (2) determining the growth rates after cultivation at different pH levels. Strong differences were found between the results obtained by these two methods. Direct counting of cells revealed a strong ecological differentiation of strains of Klebsormidium isolated from different substrate types. Strains isolated from limestone showed the highest growth rates at higher pH levels; whereas, the strains isolated from sandstone exhibited two distinct growth responses with optima at pH 5 and 6, respectively. In contrast, the effective quantum yield of photosystem II was always down-regulated at lower pH values, probably due to dissolved inorganic carbon limitation. In general, we determined distinct ecophysiological differentiation among distantly and closely related lineages, thereby corroborating our hypothesis that the sympatric speciation of terrestrial algae is driven by ecological divergence. We clearly showed that pH is a critical ecological factor that influences the diversity of autotrophic protists in terrestrial habitats.
Keywords: Diversity, Klebsormidium, pH, Protist, Speciation
Introduction
Protist species diversity has been the subject of considerable debate (Finlay 2002; Foissner 2006; Caron 2009). On one hand, some researchers think that diversity is very low because protists show a cosmopolitan distribution without endemic species. This ubiquity model predicts a very low probability of local extinction within protist populations (Fenchel & Finlay 2003). The consequence of this very low local extinction, coupled with extremely large population size, is high local protist diversity and low global diversity (Fenchel & Finlay 2003, 2004). Supporters of the ubiquity model estimate that approximately 20,000 protist species exist (Finlay & Fenchel 1999). Fenchel & Finlay (2003) argued that the distribution of the smallest organisms is dependent solely on habitat properties and not on contingencies of the evolutionary history, as is the case for multicellular organisms. On the other hand, other researchers think that diversity is very high because microorganisms also have a significant portion of endemic species. The existence of endemic species has been demonstrated through the example of flagship species, i.e. species with easily recognizable morphologies for which presence/absence can readily be demonstrated (Foissner 2006; Foissner et al. 2008). The considerable protist diversity is puzzling because the current paradigm holds that dispersal in microbes is ubiquitous, and therefore allopatric speciation must be strongly impeded by global gene flow. However, virtually nothing is known about alternative speciation mechanisms. Allopatric processes may play an important role in diatom speciation (Evans et al. 2009; Casteleyn et al. 2010). Moreover, some studies have demonstrated sympatric speciation (reviewed in Benton & Pearson 2001) based on fossil evidence. Nevertheless, it is clear that the distribution of some protists on the Earth’s surface is restricted by their habitat requirements. In addition, convergent morphological evolution has frequently led to the existence of simple morphotypes, which show extremely high phylogenetic diversity (Huss et al. 1999). Recently, many molecular studies have demonstrated the existence of very high cryptic diversity in different protistan taxa (de Vargas et al. 1999; von der Heyden et al. 2004; Škaloud & Peksa 2010; Škaloud et al. 2012).
Ecological adaptation is also known to influence diversity in algae (Huss et al. 2002; Logares et al. 2007; Rindi et al. 2008; Škaloud & Rindi 2013; Škaloud et al. 2014). For example, Rindi and Guiry (2002) reported that some species of Trentepohlia and Printzina formed perennial populations on different substrates in urban habitats in western Ireland. Similar ecological differentiation was reported for some species of Prasiola (Trebouxiophyceae) (Moniz et al. 2012). The influence of environmental factors was further demonstrated by Peksa & Škaloud (2011), who showed that specific lineages within the lichen photobiont genus Asterochloris exhibited clear environmental preferences, in relation to factors such as exposure to rain or sunlight, substrate type, and climate.
In comparison with the factors that determine the diversity of marine and freshwater algal communities (Machová-Černá & Neustupa 2009; Desrosiers et al. 2013; Svoboda et al. 2014), the critical factors that influence the distribution of aeroterrestrial algae remain unclear. The key factors that influence the community structure of terrestrial algae include light, humidity, temperature, nutrients, and pH (Hoffmann 1989). Pietrasiak et al. (2011) showed that in the absence of disturbance, several abiotic factors, particularly soil texture, pH, and electrical conductivity, seemed to be important for the development of biotic crusts. The pH seems to influence the dominance of the major groups of soil photoautotrophic organisms; cyanobacteria are known to prefer neutral and alkaline soils (Shields & Durell 1964; Brock 1973); whereas, green algae predominantly prefer acidic soils (Starks et al. 1981; Lukešová & Hoffmann 1995; Lukešová 2001). Bates et al. (2012) think that soil protistan richness and diversity were primarily influenced by climatic conditions that regulate annual moisture availability in soils (Bates et al. 2012). Similarly, climatic factors probably control the species composition of aerophytic algal communities growing on urban walls, e.g. Prasiolales-dominated assemblages in Atlantic parts of Europe vs Klebsormidium-dominated growths in continental and Mediterranean cities (Rindi & Guiry 2004; Rindi et al. 2007). Algal communities isolated from sandstone that was used as building blocks for a German castle differed markedly according to their exposure to direct sunlight (Hallmann et al. 2013).
In the present study, we investigated the genus Klebsormidium (Silva et al. 1972; Sluiman et al. 2008; Rindi et al. 2011) as a model to obtain a better insight into the ecologically driven speciation of microalgae. This genus comprises cosmopolitan filamentous green algae broadly distributed in various terrestrial and freshwater habitats (Rindi et al. 2008, 2011; Škaloud & Rindi 2013; Mikhailyuk et al. 2014, 2015; Ryšánek et al. 2015). The local distribution of Klebsormidium is generally influenced by different substrate preferences (Novis 2006; Rindi et al. 2008, 2011; Škaloud & Rindi 2013; Škaloud et al. 2014), which repeatedly originated during the evolution of the genus (Škaloud et al. 2014). Strains of Klebsormidium show a wide range of adaptation to unfavourable conditions, such as prolonged desiccation (Karsten & Holzinger 2012; Karsten et al. 2013; Holzinger et al. 2014; Herburger & Holzinger 2015; Herburger et al. 2016; Karsten et al. 2016), low temperature (Elster et al. 2008; Nagao et al. 2008), heavy metals (Gaysina et al. 2009), and ultraviolet (UV) radiation (Nagao et al. 2008; Kitzing et al. 2014).
The main objective of the present study was to understand whether ecological preferences may be drivers of sympatric speciation in Klebsormidium. We focused on pH as one of the major factors influencing the diversity of terrestrial photoautotrophs (see above) and investigated its influence on the effective quantum yield and growth rate of several strains of Klebsormidium isolated from two different substrates, sandstone and limestone. We posed the following questions: (1) Do strains isolated from different substrates show differences in their response to pH? (2) Do different genotypes that inhabit the same substrate show differences in their physiological and growth responses? (3) Do closely related strains isolated from different substrates show differences in their response to pH?
Material And Methods
Sampling sites and cultivation methods
We collected and isolated algal samples from rocks in the Czech Republic during the autumn seasons of 2012 and 2013. The strains isolated from sandstone (P05, P08, P09, AD31, AD32, and AD36) were collected in Labské pískovce (P05, P08, P09) (50°48′N, 14′14Έ) and Adršpach (AD31, AD32, AD36) (50°36′N, 16°7′E). The strains isolated from limestone (J06, J07, J11, MA12, MA16, and MA24) were collected near the village of Vápenná (J06, J07, J11) (50°16′N, 17°5′E) and in Moravský kras (MA12, MA16, MA24) (49°22′N, 16°43′E). The pH levels of the sandstone substrata from Labské pískovce and Adršpach were 5.05 and 5.17, respectively. The pH levels of the limestone substrata from Vápenná and Moravský kras were 6.97 and 7.06, respectively. The pH was measured by WTW pH-330 set with a flathead electrode (WTW SenTix Sur, Weilheim, Germany). The pH was measured on 10 different sides of rock, at each locality. All strains of Klebsormidium were isolated by cultivating samples from rock on 1.5% agar supplemented with Bold’s basal medium (BBM) (Starr & Zeikus 1993). The selected algal filaments were transferred repeatedly to fresh Petri dishes. After three changes of each isolate to fresh Petri dishes, the obtained cultures were observed to be unialgal by examination under an Olympus CX 31 light microscope (Olympus Corp., Tokyo, Japan). Unialgal stock cultures of Klebsormidium were maintained in BBM at 20°C under white fluorescent illumination of 30–50 μmol photons·m−2·s−1 provided by 18 W cool tubes (Philips TLD 18W/33, Amsterdam, the Netherlands), with a light:dark (L:D) cycle of 14:10 hours. We examined the morphology of 5-week-old cultures during the exponential growth phase.
Molecular analyses
The DNA was isolated according to the protocol of Ryšánek et al. (2015) and stored at −20°C. We used 12 microcolonies of Klebsormidium for subsequent molecular analyses. For molecular screening of the isolated strains we used partial sequences of the plastid-encoded rbcL gene (the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase). The rbcL sequences were obtained by polymerase chain reaction (PCR) with a Touchgene Gradient cycler (Techne, Cambridge, United Kingdom) using the primers KF590 (5′-GAT GAA AAC GTA AAC TCT CAG C-3′) and rbcL-KR2 (5′-GGT TGC CTT CGC GAG CTA-3′; Škaloud & Rindi 2013). Both primers were designed specifically to amplify species of Klebsormidium. Each 20-μl reaction for PCR was composed as described by Ryšánek et al. (2015). The PCR protocol followed that of Škaloud & Rindi (2013). Sequencing reads were assembled and edited by using the SeqAssem software (Hepperle 2004).
For phylogenetic analyses, we used the newly obtained rbcL sequences of Klebsormidium and a selection of rbcL sequences of Klebsormidiales available in GenBank to produce an alignment. The final alignment of 632 base pairs (bp) was constructed by using ClustalW (Thompson et al. 1994) with MEGA 5.05 (Tamura et al. 2011). The aligned data set was analysed by using maximum parsimony with Phylogenetic Analysis Using Parsimony (PAUP 4.0b10; Swofford 2002), maximum likelihood with the Genetic Algorithm for Rapid Likelihood Inference (Zwickl 2006, unpublished Ph.D. dissertation), and Bayesian inference (BI) analysis with MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001). The evolutionary model was determined by using PAUP/MrModeltest 2.3 (Nylander 2004). The model selected under the Akaike information criterion was GTR + I + G. The BI analysis was performed by using the priors set as default in MrBayes. The robustness of the tree topologies was assessed by bootstrapping the data set as described by Škaloud & Rindi (2013).
Effective quantum yield in liquid medium
Six strains of Klebsormidium (limestone strains J06, J07, J11 and sandstone strains P05, P08, P09) were selected to evaluate their physiological performances in liquid medium. The exponentially growing strains were inoculated into 50ml Erlenmeyer flasks containing fresh BBM medium. The strains were grown at five different pH levels (pH 4, pH 5, pH 6, pH 7, and pH 8). Liquid BBM medium was buffered to pH 4, pH 5, or pH 6 with 1 mM 2-(N-morpholino) ethanesulphonic acid and to pH 7 or pH 8 with 1 mM N-(2-hydroxyethyl) piperazine-N′-2-ethanesulfonic acid hemisodium salt. The pH was checked and adjusted at 3-day intervals by using 0.1 M NaOH or 0.1 M HCl (InoLab pH/conductometer 720, WTW). The strains were cultivated at an optimum growth temperature of 20°C under continuous white fluorescent illumination of 20 μmol photons·m−2·second−1 for 8 days. Cultures of Klebsormidium (each with an approximate volume of 100 μl four times) were harvested daily and were concentrated on Whatman GF/F glass fibre filters (Whatman, Seattle, Washington USA). The filters were saturated with BBM at a similar pH as in harvested bottles and were maintained for 2 hours in Petri dishes at ambient room temperature (~22°C) under continuous white fluorescent illumination of 20–25 μmol photons·m−2·s−1. The effective quantum yield (ΔF/Fm′) of photochemistry was determined at regular intervals by using a pulse-amplitude modulated fluorimeter (PAM 2500; Heinz Walz GmbH, Pfullingen, Germany). The physiological state of the photosynthetic apparatus was determined by measuring the effective quantum yield of photosystem II photochemistry in a dark-acclimated state (ΔF/Fm′). The measurements were made after 2 and 3 hours, with four replicates per strain. In total, we performed eight measurements for each pH level.
Effective quantum yield on rock substrate
Eight strains of Klebsormidium (limestone strains MA12, MA24, J06, J07 and sandstone strains AD31, AD32, AD36, P05) were selected to investigate their performances when growing directly on rocky substrata. Small pieces of rock (c. 1 cm in diameter) were transferred to Petri dishes and were stabilized by using 1.5% solidified agar (in distilled water). Each strain was cultivated on sandstone (n = 4) and limestone (n = 4) rocks (Fig. S1) at an optimum growth temperature of 20°C under continuous white fluorescent illumination of ~20 μmol photons·m−2 second−1 for 10 days. The effective quantum yield (ΔF/Fm′) of photochemistry was determined daily by using a pulse-amplitude modulated fluorimeter (PAM 2500; Heinz Walz GmbH), with four replicates per strain.
Growth rate estimations
All 12 cultivated strains were used for growth rate measurements. After 2–3 weeks, approximately 1–1.5 ml of the experimental cultures growing in 50-ml Erlenmeyer flasks with fresh BBM medium were harvested into 2-ml tubes (Eppen-dorf, Hamburg, Germany) containing 2–3 glass balls, each with a diameter of 0.5 mm (Sigma-Aldrich, St Louis, Missouri USA). The tubes were inserted into a mill for grinding plant material (Retsch MM400, Haan, Germany) to fragment filaments into single cells; the mill was operated for 2–3 minutes at 18–24 frequencies/second. Next, approximately 80–120 μl of the solution (containing filament fragments of differing sizes) were pipetted onto Petri dishes (diameter 8 cm). To monitor the subsequent growth of single-cell fragments, we pipetted very low cell densities (c. 2600 cell fragments/ml). All strains were grown at different pH levels (pH 4, pH 5, pH 6, pH 7, and pH 8) in Petri dishes containing 1.5% agar supplemented with BBM. The solidified BBM medium was buffered in the same manner as described above for the experiment in liquid medium. The pH was measured at 2-day intervals by using a WTW 330/SET1 with SenTix Sur electrode. Strains were cultivated at an optimum growth temperature of 20°C under continuous white fluorescent illumination of 20 μmol photons·m−2·second−1 for 4 days. The culture time was relatively short because the pH increased slowly, and we were unable to adjust the pH of the agar plates. At the start of the experiment, we selected approximately 30–40 single cells for cultivation at each pH level, and we subsequently monitored the growth of each single cell over four consecutive days. Each day, we recorded the length of the filaments (number of cells) grown from these single cells; we used these measurements to determine the growth rates based on 30–40 replicates. We counted the cells by direct observation using an Olympus CX 31 light microscope.
Data analysis
The effect of different pH values and rock surfaces on effective quantum yield (ΔF/Fm′) was evaluated by multisample nonparametric Friedman two-way analysis of variance (ANOVA) tests by ranks, in connection with the Statistica software (StatSoft Inc, Tulsa, Oklahoma USA). Post hoc nonparametric multiple comparisons were computed by applying the ‘Post Hoc For Friedman.svb’ macro, comparing the absolute values of the differences for all analyzed pairs. Pair differences in mean ranks were displayed in R (R Core Team 2016), using the package corrplot. Growth response to different pH values was tested by one-way ANOVA tests with the Tukey’s pairwise comparisons in the program PAST 2.17c (Hammer et al. 2001). The graphs were created in SigmaPlot. The significance was tested to three levels: ***P < 0.001, **P < 0.01, *P < 0.05.
Results
Strain morphology and molecular analyses
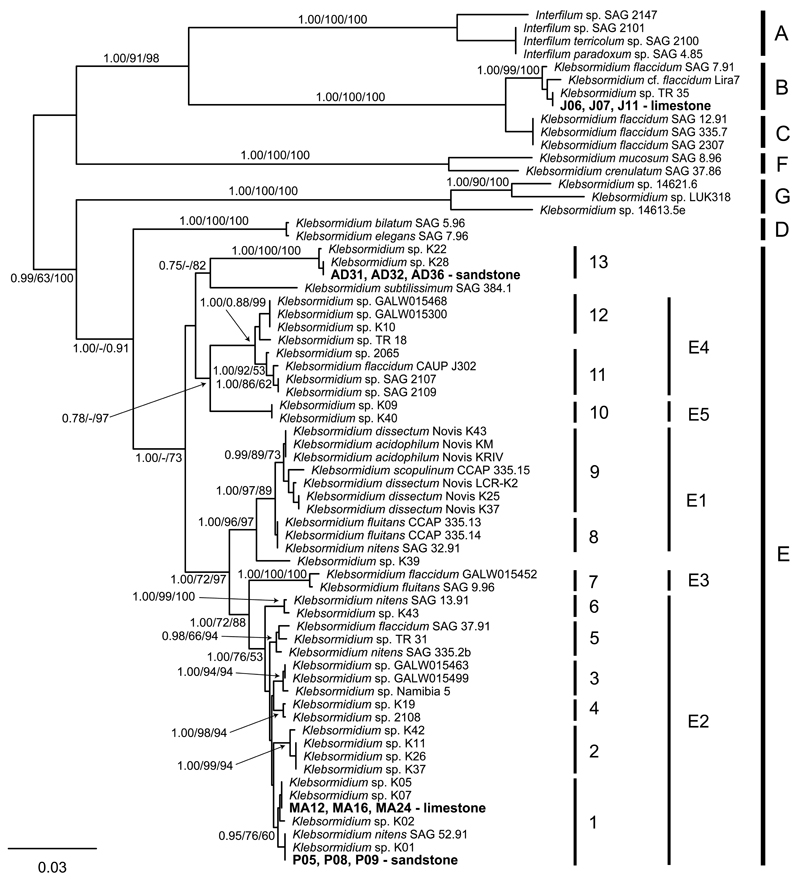
The results of our molecular analyses revealed that the isolated strains were representative of four different genotypes, which were specific to the sampled localities (Fig. 1). These four genotypes were inferred within three distinct clades. Two genotypes (sandstone strains P05, P08, and P09 and limestone strains MA12, MA16, and MA24) were inferred within clade E1 sensu Škaloud & Rindi (2013); sandstone strains AD31, AD32, and AD36 were inferred within clade E13 sensu Škaloud & Rindi (2013); and limestone strains J06, J07, and J11 were shown to belong to superclade B sensu Rindi et al. (2011). The rbcL sequences of the 12 investigated strains of Klebsormidium were deposited in GenBank under accession numbers KU528666–KU528677. With the exception of superclade B, in which small differences between strains were observed, the strain morphology was relatively uniform within and among clades. In most of the strains, the filaments were short, 4–6 μm wide, and tended to segregate into shorter filaments or individual cells. Cells were 1.4–3.2 times longer than wide; the chloroplast covered 50% of the cell wall and usually possessed a small pyrenoid. With the exception of strain J11, H-shaped pieces were not observed.
Fig 1.
Phylogenetic tree obtained from Bayesian analysis based on rbcL dataset, showing the position of investigated strains of Klebsormidium and their relatives. Values at the nodes indicate statistical support estimated by MrBayes posterior probabilities (left), maximum likelihood bootstrap (middle), and maximum parsimony bootstrap (right). The clade numbering (A–G, E1–E6) follows Rindi et al. (2011) and clades (1–13) are according to Škaloud and Rindi (2013).
Response to pH
Effective Quantum Yield In Liquid Medium
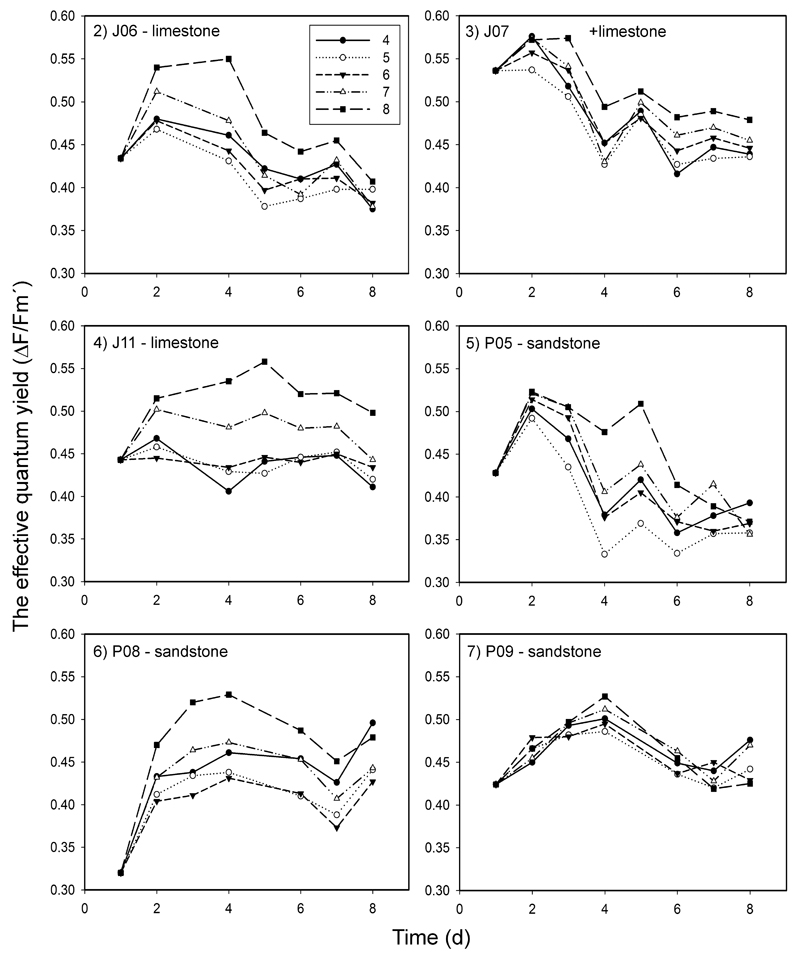
In our first approach to evaluate the organisms’ response to different pH values, we determined the physiological performance of three limestone strains (J06, J07, and J11) and three sandstone strains (P05, P08, and P09) in liquid media buffered to distinct pH values. We used a pulse-amplitude modulated fluorimeter to measure the effective quantum yield of photosystem II (Figs 2–7; Tables S1–S2). In all investigated strains, the effective quantum yield (ΔF/Fm′) showed different responses to pH: ΔF/Fm′ was significantly higher at pH 8 than at lower pH values (Figs 8–13), as shown by the two-way Friedman ANOVA tests, with the single exception of strain P09 (Fig. 13). In the strain P08 (Fig. 12), the effective quantum yield was significantly higher at pH 4 than at pH 6, as well. Throughout the experiments, the lowest ΔF/Fm′ values were measured at pH 5 (Fig. 8).
Fig 2–7.
The effective quantum yield (ΔF/Fm′) of PS II measured for three limestone (J06, J07, J11) and three sandstone (P05, P08, P09) strains of Klebsormidium at five different pH levels (4–8). The values plotted represent means of four replicated measurements. Differences were evaluated by post hoc comparisons of nonparametric Friedman two-way ANOVA tests (see Figs 8–13). Standard deviations of those measurements are given in Tables S1–S2.
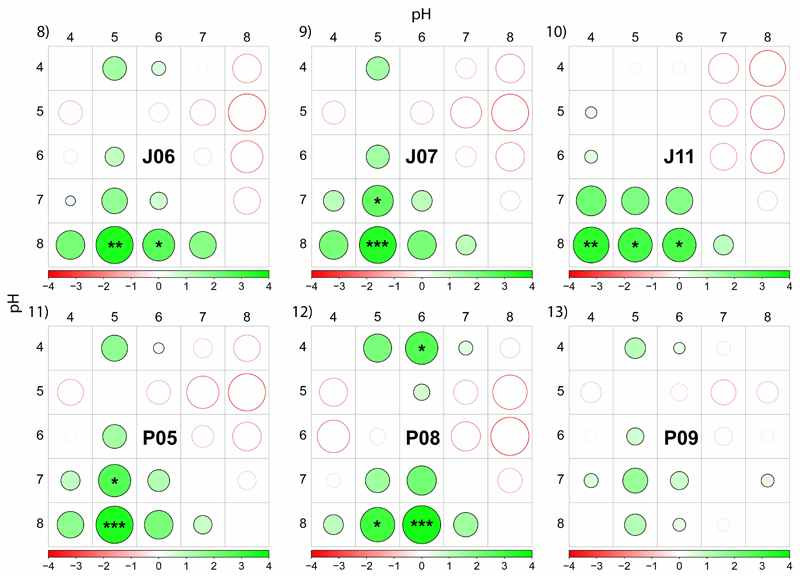
Fig 8–13.
Differences in the effective quantum yield (ΔF/Fm′) of PS II measured at five different pH levels (4–8) on six selected strains (see Figs 2–7 for measured data). Differences were evaluated by post hoc comparisons of nonparametric Friedman two-way ANOVA tests. First, repeated ΔF/Fm′ measurements at different pH values were ordered from highest to lowest. Then, mean ranks were calculated for every pH level. Within each row (a particular pH level), positive and negative differences in ΔF/Fm′ mean ranks are displayed by a symbol size and shading. Filled and empty circles display positive and negative differences in mean ranks, respectively, varying from −4 to 4 (see the colour shade legend). For example, in the strain J06 mean rank in ΔF/Fm′ at pH 4 was higher relative to pH 5 and 6 but lower relative to pH 8. Significant differences as determined by Friedman two-way ANOVA tests are indicated by asterisks (***P < 0.001, **P < 0.01, *P < 0.05).
Effective Quantum Yield On Rock Substrate
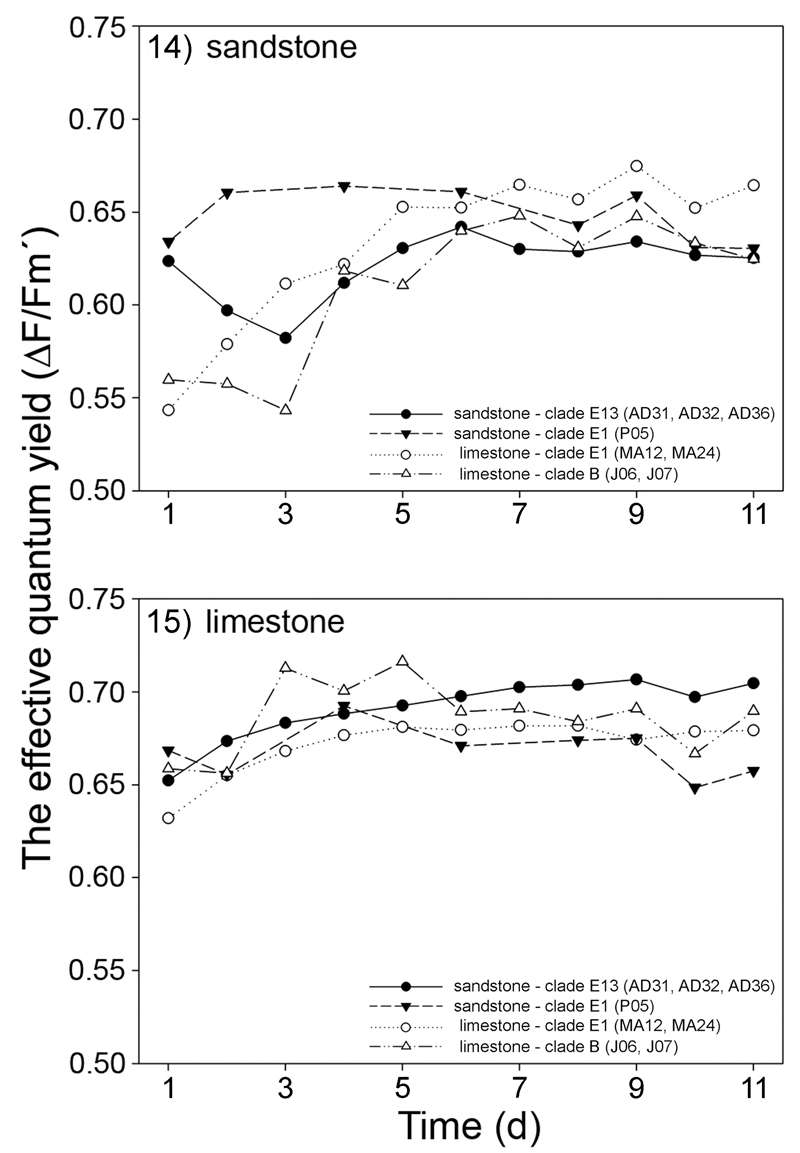
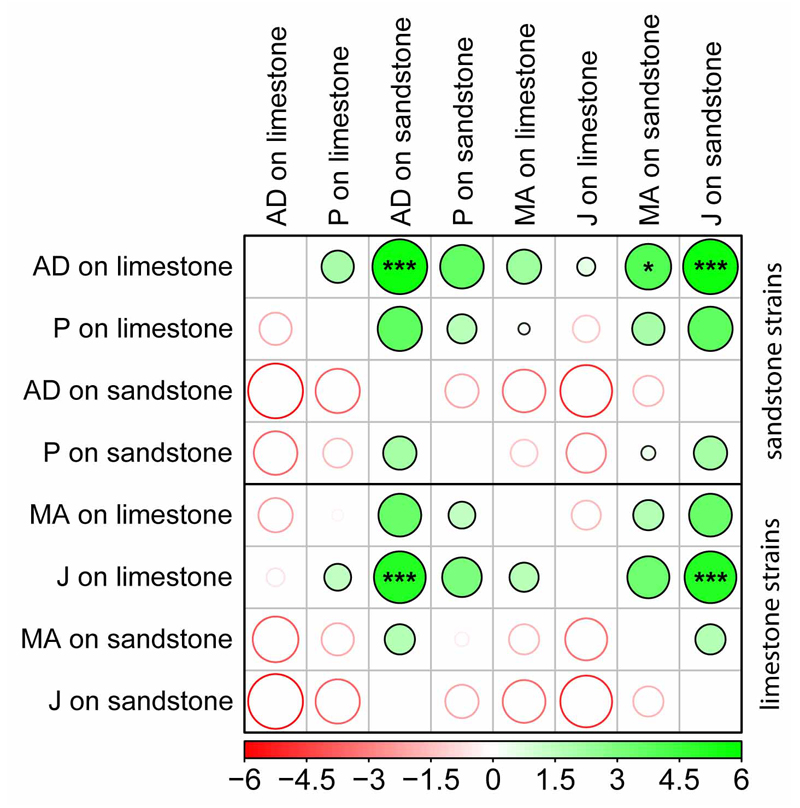
In our second approach, we cultivated four sandstone (AD31, AD32, AD36, and P05) and four limestone (MA12, MA24, J06, and J07) strains, respectively, directly on pieces of sandstone and limestone. As with the previous experiments, the physiological performance of investigated strains was evaluated by measuring the effective quantum yield of photosystem II (ΔF/Fm′). We found no significant differences in physiological performances of limestone and sandstone strains when they were cultivated on the same type of rock (Figs 14–15; Tables S3–S4). However, Friedman two-way ANOVA tests revealed significant differences in the pairwise comparisons of the physiological performance of some strains (Fig. 16). ΔF/Fm′ was significantly higher on the strains cultivated on limestone, as determined by two-way Friedman ANOVA tests. Interestingly, the ΔF/Fm′ of the limestone strains were inhibited during the first 4 days of cultivation on sandstone (Fig. 14). After this acclimation period, their physiological performance was, however, comparable to the sandstone strains.
Fig 14–15.
The effective quantum yield (ΔF/Fm′) of PS II measured for four limestone (MA12, MA24, J06, J07) and four sandstone (AD31, AD32, AD36, P05) strains when growing directly on sandstone (Fig. 14) and limestone (Fig. 15) rock substrate. The values plotted represent means of four replicated measurements. Genetically identical strains were displayed by a single value for better clarity. Differences were evaluated by post hoc comparisons of nonparametric Friedman two-way ANOVA tests (see Fig. 16). Standard deviations of the measurements are given in Tables S3–S4.
Fig 16.
Differences in the effective quantum yield (ΔF/Fm′) of PS II measured at two different natural substrate (sandstone and limestone) on eight selected strains (see Figs 14–15 for measured data). Differences were evaluated by post hoc comparisons of nonparametric Friedman two-way ANOVA tests (see Figs 8–13 legend for further explanation). Filled and empty circles display positive and negative differences in mean ranks, respectively, varying from −6 to 6 (see the colour shade legend). Significant differences as determined by Friedman two-way ANOVA tests are indicated by asterisks (***P < 0.001, **P < 0.01, *P < 0.05).
Growth Rate Estimations
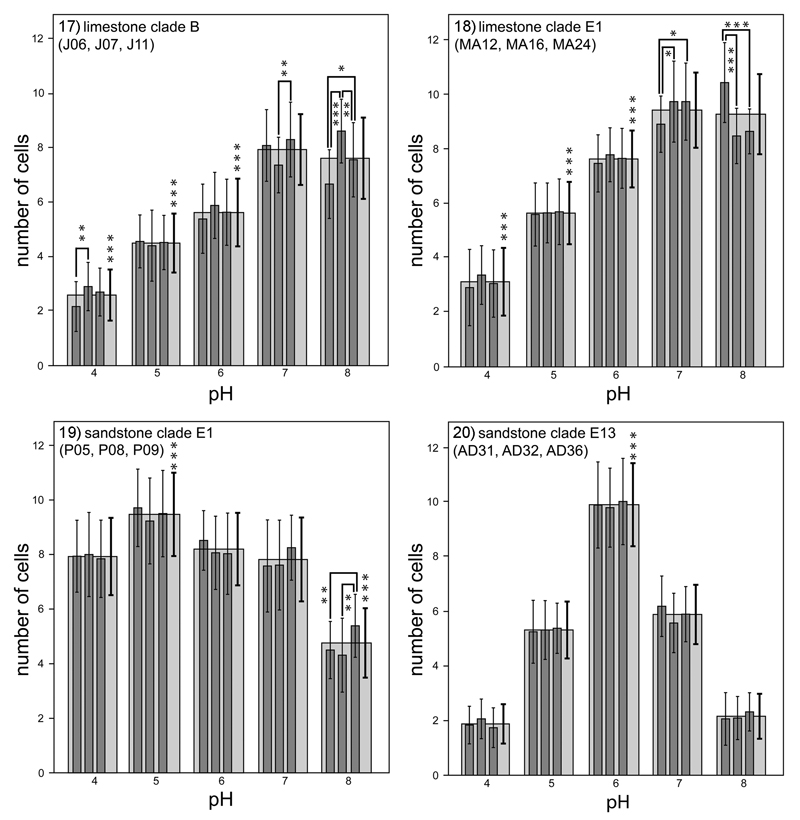
Finally, we evaluated the influence of pH on the specific growth rates of all 12 cultivated strains of Klebsormidium. We found that all strains isolated from limestone (MA12, MA16, MA24, J06, J07, and J11) exhibited similar responses to pH; these strains showed increases in growth rate as the pH of the medium increased (Figs 17–18). After 4 days of cultivation, we identically counted only three cells at pH 4 but >10 cells at pH 7 and pH 8. However, strains J06, J11, MA16, and MA24 showed the highest growth rates at pH 7; whereas, strains J07 and MA12 showed the highest growth rates at pH 8. Strains MA12, MA16, and MA24 grew slightly faster than did strains J06, J07, and J11. The statistical tests revealed no significant differences in growth rates between pH 7 and 8 but the growth rates at other pH values were significantly different (P < 0.001), as determined by one-way ANOVA tests.
Fig 17–20.
Growth response of four lineages of Klebsormidium (limestone clades B and E1; sandstone clades E1 and E13) to different pH levels (4–8). The graphs display the total number of cells in young filaments grown from single cells after 4 days of cultivation on agar plates. Each lineage is represented by three investigated strains. Dark grey bars display mean number of cells determined for each studied strain, light grey bars display the overall means for the lineage. The mean values were calculated from 30–40 replicates, and the standard deviations are displayed for each measurement. Significant differences as determined by one-way ANOVAs Tukey’s pairwise comparisons are indicated by asterisks (***P < 0.001, **P < 0.01, *P < 0.05).
We observed different responses to pH between the two sandstone genotypes (Figs 19–20). Strains belonging to clade E13 (AD31, AD32, and AD36; Fig. 19) showed significantly different growth patterns (P < 0.001) at the investigated pH levels. These strains exhibited the highest growth rates at pH 6; at this pH, they produced an average of 10 cells after 4 days of cultivation. In contrast, at pH 4 and pH 8, these strains grew more slowly and produced only approximately two cells after 4 days. Strains belonging to clade E1 (Fig. 20) grew well at all investigated pH levels; however, they showed higher growth rates at pH 5 than at pH 8 (c. 9 cells vs c. 4–6 cells). For E1 sandstone strains, the statistical tests supported the significant differentiation of growth rates at pH 5 and 8, respectively (P < 0.001). The highest growth rate of E13 sandstone strains at pH 6 was also significantly supported (P < 0.001).
Discussion
In this study, we applied two different methods to monitor the response of strains of Klebsormidium to different pH levels. First, we used a pulse-amplitude modulated fluorimeter to measure the effective quantum yield (ΔF/Fm′) of photosystem II. The main advantage of this method is that it enables the maintenance of cultures in liquid medium, thereby allowing pH adjustment during cultivation. This in turn enables experiments to be continued for prolonged periods (up to 8 days in the present study). Second, we used direct counting of cells on agar plates. This method is straightforward and provides an accurate estimate of cell growth rates at different pH levels. However, it is almost impossible to adjust the pH during the experiment. After 4 days in the present study, the pH had increased by c. 0.5 pH units. This increase in pH during cultivation experiments is caused by the photosynthetic activity of phototrophic organisms (Shiraiwa et al. 1993). Therefore, this technique should be used only for algal strains with relatively high growth rates.
Interestingly, we found clear differences between the results obtained by these two methods. Direct counting of cells revealed a strong ecological differentiation of strains of Klebsormidium isolated from different substrate types. Strains isolated from limestone showed the highest growth rates at pH 7 and pH 8 and had significantly lower growth rates at pH 4 and pH 5. The strains isolated from sandstone exhibited distinct growth responses. While strains belonging to clade E1 showed generally similar growth rates at all investigated pH levels, the strains inferred within clade E13 showed the highest growth rate at pH 6.
In contrast, the effective quantum yield (ΔF/Fm′) of photosystem II did not show any differentiation between the strains isolated from different rock types. Instead, all investigated strains showed the highest ΔF/Fm′ at pH 8, and its down-regulation at lower pH values. Such coincident responses indicate the presence of a common mechanism inducing an increase of photosynthetic efficiency at higher pH values.
Since the intracellular pH of photoautotrophs is usually maintained fairly constant over a wide range of external pH values (Lane & Burris 1981), we hypothesize that a strong positive effect of pH to ΔF/Fm′ was probably caused by dissolved inorganic carbon (DIC) limitation at lower pH values. In aqueous solutions, the dissolved CO2 dissociates into bicarbonate (HCO3−), and carbonate (CO32−), maintaining a certain ratio depending on pH, ion concentrations, and salinity (Falkowski & Raven 2007). At high pH values, HCO3− is the dominant carbon species. However, Rubisco reacts only with dCO2, not bicarbonate or carbonate ions (Baba & Shiraiwa 2012). The majority of microbial phototrophs use dCO2 when it is freely available (Reynolds 1984). However, some Cyanobacteria and green algae use both dCO2 and HCO3− in photosynthesis due to the extra- and intracellular activity of carbonic anhydrase (Allen & Spence 1981). Although the use of different DIC components has not been studied in Klebsormidium, we consider the stimulation of photosynthesis by HCO3− as the most likely explanation of the positive correlation of ΔF/Fm′ and pH in investigated strains (Shelp & Canvin 1980). In general, our results clearly show that, at least in our experimental system, ΔF/Fm′ does not reflect the overall fitness of studied organisms but rather indicates the efficiency of PSII under different DIC conditions. We therefore consider growth rate responses to different pH as the only measured data useful for assessing the ecological adaptation of the studied strains.
The tolerance of the investigated strains of Klebsormidium to a wide range of pH levels is in accordance with the results of previous investigations of the ecophysiology of this genus (Karsten et al. 2014). In general, strains of Klebsormidium are relatively tolerant to various physiological stresses such as UV radiation (Nagao et al. 2008; Kitzing et al. 2014; Kitzing & Karsten 2015), desiccation (Karsten et al. 2010, 2016; Karsten & Holzinger 2014), and osmotic stress (Kaplan et al. 2012). The genomic machinery required for adaptation to terrestrial environments was detected in the genome of Klebsormidium flaccidum (Kützing) P.C.Silva, K.R.Mattox & W.H.Blackwell (Hori et al. 2014), which is a member of clade E5 sensu Škaloud & Rindi (2013). This machinery includes genes involved in the signalling pathways for the phytohormones cytokinin, ABA, and ethylene, which was recently confirmed in the desiccation transcriptome of Klebsormidium crenulatum (Holzinger & Becker 2015). This is interesting, as the receptors of, e.g. cytokinin signalling show a strictly pH-dependent ligand binding in vascular plants (Lomin et al. 2015).
This ability to adapt to a wide range of conditions typical of terrestrial environments may explain the cosmopolitan distribution of the genus Klebsormidium (Ryšánek et al. 2015). Nevertheless, the relatively high abundance of this genus in different types of localities might simply be a sampling artefact. In other words, strains that are common and geographically widespread have been discovered and studied; whereas, rarer strains (e.g. such as those belonging to clade E13) have either not yet been studied or have been studied only sporadically. In the present study, strains belonging to clade E13 grew optimally in a very narrow range of pH levels; they showed the highest growth rates at pH 6 but grew very slowly at pH 4 and pH 8. This finding might imply that clade E13 is a specialist rather than a generalist, and this in turn may reflect its ecologically restricted occurrence. Generalists have a wider range of suitable habitats and are therefore discovered more frequently; in addition, they are geographically more widespread than are specialists (Finlay et al. 2002).
In the present study, we showed clear differences in growth responses between strains isolated from sandstone (lower pH preference) and limestone (higher pH preference) substrates. Our results are similar to those of Lowe et al. (2007), who reported a relatively important influence of pH on the structure of an algal community growing on a wet wall. Indeed, acidic conditions strongly increase the chemical solubility, and thus mobility of metals, resulting in high concentrations of heavy metals such as Fe, Cu, Pb, Al, and Zn in soils and waters with low pH levels (Gross 2000; Aguilera et al. 2007; Novis & Harding 2007). High concentrations of heavy metals can therefore substantially influence the diversity of terrestrial microalgae growing on acidic substrates.
Our results suggest that different lineages of Klebsormidium are adapted to the substrate on which they originally occur, independently of their evolutionary distance. We found that closely related lineages differed ecophysiologically to the same extent as unrelated clades. Our findings may indicate the widespread existence of sympatric speciation in Klebsormidium through ecological divergence, and we hypothesize that this situation is probably common among other taxa of terrestrial algae. The mechanisms of genetic differentiation are not yet fully understood. Specializations to habitats (Gächter & Weisse 2006; Logares et al. 2007), selection pressures (Vanormelingen et al. 2009), and/or persistent founder effects (De Meester et al. 2002) have been hypothesized as important factors contributing to the structure of protist populations. Ecological differentiation facilitates allopatric (our data; de Vargas et al. 1999) and sympatric (e.g. Amato et al. 2007; Weisse 2008; Vanelslander et al. 2009) speciation of protistan cryptic species. Congruent with the recent studies of Fontaneto et al. (2007) and Birky et al. (2010), our data suggest the existence of distinct species units and sympatric speciation in asexual protists.
In addition, we found that adaption to specific substrates has originated many times during the evolutionary history of Klebsormidium. Strains belonging to clade E13 grew optimally in a very narrow range of pH levels. Moreover, two closely related genotypes inferred within clade E1 showed clear ecophysiological adaptation to the substrate from which they were originally sampled. These genotypes differed in their ecology and ecophysiology but were genetically similar. An analogous situation was reported by Logares et al. (2007), who investigated two dinophytes that had identical ribosomal DNAs but which differed from each other ecologically, physiologically, and even phenetically. Such clear differentiation has been attributed to rapid adaptive evolution, which has not yet been reflected in the ribosomal divergence.
In summary, in the present study, we showed that all strains isolated from sandstone and limestone were able to grow over the range of investigated pH levels but to differing extents. Strains isolated from limestone showed the highest growth rates at pH 7 and pH 8; these strains grew very slowly at pH 4 and pH 5. Strains isolated from sandstone exhibited two different growth responses. Strains from one of the investigated genotypes showed the highest growth rate at pH 6; whereas, strains of the other genotype had almost identical growth rates at all of the investigated pH levels. We conclude that pH is a critical ecological factor that influences the diversity of Klebsormidium in terrestrial habitats. Moreover, our data highlighted distinct ecophysiological differentiation among distantly and closely related lineages, thereby corroborating our hypothesis that the common sympatric speciation of terrestrial algae is driven by ecological divergence. However, further research will be necessary to provide support to this general conclusion.
Supplementary Data
Supplementary data associated with this article can be found online at 10.2216/15-110.1.s1
Acknowledgements
We would like to thank Prof. Ulf Karsten, University of Rostock, Germany, for several helpful suggestions and discussion on the experimental design of the study. This study was supported by AKTION ‘Austria-Czech Republic’ (http://www.oead.at), Project 65 p5 to Martina Pichrtová and A.H., Charles University Prague, and by a grant from the Charles University Science Foundation (GAUK n. 1544214). Moreover the study was supported by the Austrian Science Fund (FWF) Project P 24242-B16 and FWF project I 1951-B16 to AH and by The Czech Science Foundation grant 15-34645 L to Martina Pichrtová, Charles University, Prague.
References
- Aguilera A, Zettler E, Gomez F, Amaral-Zettler L, Rodriguez N, Amils R. Distribution and seasonal variability in the benthic eukaryotic community of RioTinto (SW, Spain), an acidic, high metal extreme environment. Systematic and Applied Microbiology. 2007;30:531–546. doi: 10.1016/j.syapm.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Allen ED, Spence DHN. The differential ability of aquatic plants to utilize the inorganic carbon supply in fresh waters. New Phytologist. 1981;87:269–283. [Google Scholar]
- Amato A, Kooistra WHCF, Ghiron JHL, Mann DG, Pröschold T, Montresor M. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Baba M, Shiraiwa Y. High-CO2 response mechanisms in microalgae. In: Najafpour Mohammad., editor. Advances in photosynthesis-fundamental aspects. InTech; Rijeka, Croatia: 2012. pp. 299–320. [Google Scholar]
- Bates ST, Clemente JC, Flores GE, Walters WA, Parfrey LW, Knight R, Fierer N. Global biogeography of highly diverse protistan communities in soil. The ISME Journal. 2012;7:1–8. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Pearson PN. Speciation in the fossil record. Trends in Ecology & Evolution. 2001;16:405–411. doi: 10.1016/s0169-5347(01)02149-8. [DOI] [PubMed] [Google Scholar]
- Birky CW, Jr, Adams J, Gemmel M, Perry J. Using population genetic theory and DNA sequences for species detection and identification in asexual organisms. PLoS One. 2010;5(5):e10609. doi: 10.1371/journal.pone.0010609. 101371/journal.pone.0010609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TD. Lower pH limit for existence of blue-green algae: evolutionary and ecological implications. Science. 1973;179:480–483. doi: 10.1126/science.179.4072.480. [DOI] [PubMed] [Google Scholar]
- Caron DA. Past president’s address: protistan biogeography: why all the fuss? Journal of Eukaryotic Microbiology. 2009;56:105–112. doi: 10.1111/j.1550-7408.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- Casteleyn G, Leliaert F, Backeljau T, Debeer AE, Kotaki Y, Rhodes L, Lundholm N, Sabbe K, Vyverman W. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12952–12957. doi: 10.1073/pnas.1001380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester L, Gomez A, Okamura B, Schwenk K. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecologica. 2002;23:121–135. [Google Scholar]
- De Vargas C, Norris R, Zaninetti L, Gibb SW, Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers C, Leflaive J, Eulin A, Ten-Hage L. Bioindicators in marine waters: benthic diatoms as a tool to assess water quality from eutrophic to oligotrophic coastal ecosystems. Ecological Indicators. 2013;32:25–34. [Google Scholar]
- Elster J, Degma P, Kováčík L, Valentová L, Šramková K, Pereira AB. Freezing and desiccation injury resistance in the filamentous green alga Klebsormidium from the Antarctic, Arctic and Slovakia. Biologia. 2008;63:843–851. [Google Scholar]
- Evans KM, Chepurnov VA, Sluiman HJ, Thomas SJ, Spears BM, Mann DG. Highly differentiated populations of the fresh water diatom Sellaphora capitata suggest limited dispersal and opportunities for allopatric speciation. Protist. 2009;160:386–396. doi: 10.1016/j.protis.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Raven JA. Aquatic photosynthesis. Princeton University Press; Princeton, New Jersey: 2007. p. 488. [Google Scholar]
- Fenchel T, Finlay BJ. Is microbial diversity fundamentally different from biodiversity of larger animals and plants? European Journal of Protistology. 2003;39:486–490. [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Fenchel T. Divergent perspective on protist species richness. Protist. 1999;150:229–233. doi: 10.1016/S1434-4610(99)70025-8. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Monaghan EB, Maberly SC. Hypothesis: the rate and scale of dispersal of fresh water diatom species is a fiction of their global abundance. Protist. 2002;153:261–273. doi: 10.1078/1434-4610-00103. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozoologica. 2006;45:111–136. [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodiversity and Conservation. 2008;17:235–242. [Google Scholar]
- Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. Independently evolving species in asexual bdelloid rotifers. PLoS Biology. 2007;5:e87. doi: 10.1371/journal.pbio.0050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gächter E, Weisse T. Local adaptation among geographically distant clones of the cosmopolitan freshwater ciliate Meseres corlissi. I. Temperature response. Aquatic Microbial Ecology. 2006;45:291–300. [Google Scholar]
- Gaysina LA, Purina ES, Safiullina LM, Bakieva GR. Resistance of Klebsormidium flaccidum (Streptophyta) to heavy metals. Plant Sciences. 2009;3:39–41. [Google Scholar]
- Gross W. Ecophysiology of algae living in highly acidic environments. Hydrobiologia. 2000;433:31–37. [Google Scholar]
- Hallmann C, Stannek L, Fritzlar D, Hause-Reitner D, Friedl T, Hoppert M. Molecular diversity of phototrophic biofilms on building stone. FEMS Microbiology Ecology. 2013;84:355–372. doi: 10.1111/1574-6941.12065. [DOI] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD. PAST: Paleontological Statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4(1):9. [Google Scholar]
- Hepperle D. SeqAssem©. A sequence analysis tool, contig assembler and trace data visualisation tool for molecular sequences, version 09/2004. 2004 http://www.sequentix.de.
- Herburger K, Holzinger A. Localization and quantification of callose in the streptophyte green algae Zygnema and Klebsormidium: Correlation with desiccation tolerance. Plant and Cell Physiology. 2015;56:2259–2270. doi: 10.1093/pcp/pcv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herburger K, Karsten U, Holzinger A. Entransia and Hormidiella, sister lineages of Klebsormidium (Streptophyta), respond differently to light, temperature and desiccation stress. Protoplasma. 2016 doi: 10.1007/s00709-015-0889-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L. Algae of terrestrial habitats. The Botanical Review. 1989;55:77–105. [Google Scholar]
- Holzinger A, Becker B. Desiccation tolerance in the streptophyte green alga Klebsormidium: the role of phytohormones. Communicative and Integrative Biology. 2015;8(4):e1059978. doi: 10.1080/19420889.2015.1059978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A, Kaplan F, Blaas K, Zechmann B, Komsic-Buchmann K, Becker B. Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense. PLoS One. 2014;9(10):e110630. doi: 10.1371/journal.pone.0110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, Moriyama T, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huss VAR, Frank C, Hartmann EC, Hirmer M, Kloboucek A, Seidel BM, Wenzeler P, Kessler E. Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta) Journal of Phycology. 1999;35:587–598. [Google Scholar]
- Huss VAR, Ciniglia C, Cennamo P, Cozzolino S, Pinto G, Pollio A. Phylogenetic relationships and taxonomic position of Chlorella-like isolates from low pH environments (pH < 3.0) BMC Evolutionary Biology. 2002;26:2–13. doi: 10.1186/1471-2148-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Lewis LA, Wastian J, Holzinger A. Plasmolysis effects and osmotic potential of two phylogenetically distinct alpine strains of Klebsormidium (Streptophyta) Protoplasma. 2012;249:789–804. doi: 10.1007/s00709-011-0324-z. [DOI] [PubMed] [Google Scholar]
- Karsten U, Holzinger A. Light, temperature, and desiccation effects on photosynthetic activity, and drought-induced ultrastructural changes in the green alga Klebsormidium dissectum (Streptophyta) from a high alpine soil crust. Microbial Ecology. 2012;63:51–63. doi: 10.1007/s00248-011-9924-6. [DOI] [PubMed] [Google Scholar]
- Karsten U, Holzinger A. Green algae in alpine biological soil crust communities: acclimation strategies against ultraviolet radiation and dehydration. Biodiversity and Conservation. 2014;23:1845–1858. doi: 10.1007/s10531-014-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten U, Lütz C, Holzinger A. Ecophysiological performance of the aeroterrestrial green alga Klebsormidium crenulatum (Charophyceae, Streptophyta) isolated from an alpine soil crust with an emphasis on desiccation stress. Journal of Phycology. 2010;46:1187–1197. doi: 10.1111/j.1529-8817.2011.00980.x. [DOI] [PubMed] [Google Scholar]
- Karsten U, Pröschold T, Mikhailyuk T, Holzinger A. Photosynthetic performance of different genotypes of the green alga Klebsormidium sp. (Streptophyta) isolated from biological soil crusts of the Alps. Algological Studies. 2013;142:45–62. [Google Scholar]
- Karsten U, Herburger K, Holzinger A. Dehydration, temperature and light tolerance in members of the aeroterrestrial green algal genus Interfilum (Streptophyta) from biogeographically different temperate soils. Journal of Phycology. 2014;50:804–816. doi: 10.1111/jpy.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten U, Herburger K, Holzinger A. Living in biological soil crust communities of African deserts – physiological traits of green algal Klebsormidium species (Streptophyta) to cope with desiccation, light and temperature gradients. Journal of Plant Physiology. 2016 doi: 10.1016/j.jplph.2015.09.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing C, Karsten U. UV-induced effects on growth, photosynthetic performance and sunscreen contents in members of the genera Interfilum, Klebsormidium, Hormidiella and Entransia (Klebsormidiophyceae, Streptophyta) European Journal of Phycology. 2015;50:279–287. [Google Scholar]
- Kitzing C, Pröschold T, Karsten U. UV-induced effects on growth, photosynthetic performance and sunscreen contents in different populations of the green alga Klebsormidium fluitans (Streptophyta) from alpine soil crusts. Microbial Ecology. 2014;67:327–340. doi: 10.1007/s00248-013-0317-x. [DOI] [PubMed] [Google Scholar]
- Lane AE, Burris JE. Effects of environmental pH on the internal pH of Chlorella pyrenoidosa, Scenedesmus quadricauda and Euglena mutabilis. Plant Physiology. 1981;68:439–442. doi: 10.1104/pp.68.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Rengefors K, Kremp A, Shalchian-Tabrizi K, Boltovskoy A, Tengs T, Shurtleff A, Klaveness D. Phenotypically different microalgal morphospecies with identical ribosomal DNA: a case of rapid adaptive evolution? Microbial Ecology. 2007;53:549–561. doi: 10.1007/s00248-006-9088-y. [DOI] [PubMed] [Google Scholar]
- Lomin SN, Krivosheev DM, Steklov MY, Arkhipov DV, Osolodkin DI, Schmülling T, Romanov GA. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. Journal of Experimental Botany. 2015;66:1851–1863. doi: 10.1093/jxb/eru522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe RL, Furey PC, Ress JA, Johansen JR. Diatom biodiversity and distribution on wet walls in Great Smoky Mountains National Park. Southeastern Naturalist. 2007;6:135–152. [Google Scholar]
- Lukešová A. Soil algae in brown coal and lignite post-mining areas in central Europe (Czech Republic and Germany) Restoration Ecology. 2001;9:341–350. [Google Scholar]
- Lukešová A, Hoffmann L. Soil algae from acid rain impacted forest areas of Krušné hory mountains (Czech Republic). 2. Effects of pH on growth. Algological Studies. 1995;78:39–51. [Google Scholar]
- Machová-Černá K, Neustupa J. Spatial distribution of algal assemblages in a temperate lowland peat bog. International Review of Hydrobiology. 2009;94:40–56. [Google Scholar]
- Mikhailyuk T, Holzinger A, Mlassalski A, Karsten U. Morphology and ultrastructure of Interfilum and Klebsormidium (Klebsormidiales, Streptophyta) with special reference to cell division and thallus formation. European Journal of Phycology. 2014;49:395–412. doi: 10.1080/09670262.2014.949308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailyuk T, Glaser K, Holzinger A, Karsten U. Biodiversity of Klebsormidium (Streptophyta) from alpine biological soil crusts (Alps, Tyrol, Austria and Italy) Journal of Phycology. 2015;51:750–767. doi: 10.1111/jpy.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz MB, Rindi F, Novis PM, Broady PA, Guiry MD. Molecular phylogeny of antarctic Prasiola (Prasiolales, Trebouxiophyceae) reveals extensive cryptic diversity. Journal of Phycology. 2012;48:940–955. doi: 10.1111/j.1529-8817.2012.01172.x. [DOI] [PubMed] [Google Scholar]
- Nagao M, Matsui K, Uemura M. Klebsormidium flaccidum, a charophycean green alga, exhibits cold acclimation that is closely associated with compatible solute accumulation and ultrastructural changes. Plant, Cell & Environment. 2008;31:872–885. doi: 10.1111/j.1365-3040.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- Novis PM. Taxonomy of Klebsormidium (Klebsormidiales, Charophyceae) in New Zealand streams and the significance of low-pH habitats. Phycologia. 2006;45:293–301. [Google Scholar]
- Novis PM, Harding JS. Extreme acidophiles: freshwater algae associated with acid mine drainage. In: Seckbach J, editor. Extremophilic algae, cyanobacteria and non-photosynthetic protists: from prokaryotes to astrobiology. Springer Verlag; Dordrecht, the Netherlands: 2007. pp. 445–463. [Google Scholar]
- Nylander JAA. MrModeltest, version 2. 2004 http://www.abc.se/~nylander.
- Peksa O, Škaloud P. Do photobionts influence the ecology of lichens? A case study of environmental preferences in symbiotic green alga Asterochloris (Trebouxiophyceae) Molecular Ecology. 2011;20:3936–3948. doi: 10.1111/j.1365-294X.2011.05168.x. [DOI] [PubMed] [Google Scholar]
- Pietrasiak N, Johansen JR, Drenovsky RE. Geologic composition influences distribution of microbiotic crusts in the Mojave and Colorado Deserts at the regional scale. Soil Biology and Biochemistry. 2011;43:967–974. [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2016. version R 2.15.1 http://www.R-project.org/ [Google Scholar]
- Reynolds CS. The ecology of freshwater phytoplankton. Cambridge University Press; Cambridge, UK: 1984. p. 384. [Google Scholar]
- Rindi F, Guiry MD. Diversity, life history and ecology of Trentepohlia and Printzina (Trentepohliales, Chlorophyta) in urban habitats in western Ireland. Journal of Phycology. 2002;38:39–54. [Google Scholar]
- Rindi F, Guiry MD. Composition and spatial variability of terrestrial algal assemblages occurring at the bases of urban walls in Europe. Phycologia. 2004;43:225–235. [Google Scholar]
- Rindi F, Mcivor L, Sherwood AR, Friedl T, Guiry MD, Sheath RG. Molecular phylogeny of the green algal order Prasiolales (Trebouxiophyceae, Chlorophyta) Journal of Phycology. 2007;43:811–822. [Google Scholar]
- Rindi F, Guiry MD, López-Bautista JM. Distribution, morphology, and phylogeny of Klebsormidium (Klebsormidiales, Charophyceae) in urban environment in Europe. Journal of Phycology. 2008;44:1529–1540. doi: 10.1111/j.1529-8817.2008.00593.x. [DOI] [PubMed] [Google Scholar]
- Rindi F, Mikhailyuk TI, Sluiman HJ, Friedl T, LÓpez-Bautista JM. Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta) Molecular Phylogenetics and Evolution. 2011;58:218–231. doi: 10.1016/j.ympev.2010.11.030. [DOI] [PubMed] [Google Scholar]
- Ryšánek D, Hrčková K, Škaloud P. Global ubiquity and local endemism of free-living terrestrial protists: phylogeographic assessment of the streptophyte alga Klebsormidium. Environmental Microbiology. 2015;17:689–698. doi: 10.1111/1462-2920.12501. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Canvin DT. Utilization of exogenous inorganic carbon species in photosynthesis by Chlorella pyrenoidosa. Plant Physiology. 1980;65:774–779. doi: 10.1104/pp.65.5.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields LM, Durell LW. Algae in relation to soil fertility. The Botanical Review. 1964;30:92–128. [Google Scholar]
- Shiraiwa Y, Goyal A, Tolbert NE. Alkalization of the medium by unicellular green algae during uptake dissolved inorganic carbon. Plant and Cell Physiology. 1993;34:649–657. [Google Scholar]
- Silva PC, Mattox KR, Blackwell WH. The generic name Hormidium as applied to green algae. Taxon. 1972;21:639–645. [Google Scholar]
- Škaloud P, Peksa O. Evolutionary inferences based on ITS rDNA and actin sequences reveal extensive diversity of the common lichen alga Asterochloris (Trebouxiophyceae, Chlorophyta) Molecular Phylogenetics and Evolution. 2010;54:36–46. doi: 10.1016/j.ympev.2009.09.035. [DOI] [PubMed] [Google Scholar]
- Škaloud P, Rindi F. Ecological differentiation of cryptic species within an asexual protest morphospecies: a case study of filamentous green alga Klebsormidium (Streptophyta) Journal of Eukaryotic Microbiology. 2013;60:350–362. doi: 10.1111/jeu.12040. [DOI] [PubMed] [Google Scholar]
- Škaloud P, Kynčlová A, Benada O, Kofroňová O, Škaloudová M. Toward a revision of the genus Synura, section Petersenianae (Synurophyceae, Heterokontophyta): morphological characterization of six pseudo-cryptic species. Phycologia. 2012;51:303–329. [Google Scholar]
- Škaloud P, Lukešová A, Malavasi V, Ryšánek D, HrčKová K, Rindi F. Molecular evidence for the polyphyletic origin of low pH adaptation in the genus Klebsormidium (Klebsormidiophyceae, Streptophyta) Plant Ecology and Evolution. 2014;147:333–345. [Google Scholar]
- Sluiman HJ, Guihal C, Mudimu O. Assessing phylogenetic affinities and species deliminations in Klebsormidiales (Streptophyta): nuclear-encoded rDNA phylogenies and ITS secondary structure models in Klebsormidium, Hormidiella, and Entransia. Journal of Phycology. 2008;44:183–195. doi: 10.1111/j.1529-8817.2007.00442.x. [DOI] [PubMed] [Google Scholar]
- Starks TL, Shubert LE, Trainor FR. Ecology of soil algae: a review. Phycologia. 1981;20:65–80. [Google Scholar]
- Starr RC, Zeikus JA. UTEX – the Culture Collection of Algae at the University of Texas at Austin 1993 List of Cultures. Journal of Phycology. 1993;29:1–106. [Google Scholar]
- Svoboda P, Kulichová J, Šťastiný J. Spatial and temporal community structure of desmids on a small spatial scale. Hydrobiologia. 2014;722:291–303. [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and other methods) Sinauer Associates; Sunderland, Massachusetts: 2002. Version 4. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving sensitivity of progressive multiple sequence alignment through sequence weighting, position 86 specific gap penalties, and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanelslander B, Creach V, Vanormelingen P, Ernst A, Chepurnov VA, Sahan E, Sabbe K. Ecological differentiation between sympatric pseudocryptic species in the estuarine benthic diatom Navicula phyllepta (Bacillariophyceae) Journal of Phycology. 2009;45:1278–1289. doi: 10.1111/j.1529-8817.2009.00762.x. [DOI] [PubMed] [Google Scholar]
- Vanormelingen P, Vyverman W, De Bock D, Van der Gucht K, De Meester L. Local genetic adaptation to grazing pressure of the green alga Desmodesmus armatus in a strongly connected pond system. Limnology and Oceanography. 2009;54:503–511. [Google Scholar]
- Von der Heyden S, Chao E, Cavalier-Smith T. Genetic diversity of goniomonads: an ancient divergence between freshwater and marine species. European Journal of Phycology. 2004;39:343–350. [Google Scholar]
- Weisse T. Distribution and diversity of aquatic protists: an evolutionary and ecological perspective. Biodiversity and Conservation. 2008;17:243–259. [Google Scholar]
- Zwickl DJ. Dissertation. The University of Texas at Austin; Texas: 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion; p. 115. Available from Digital Repository, University of Texas Libraries. http://hdl.Handle.net/2152/2666. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.