Abstract
Here I take a somewhat personal perspective on signalling control, focusing on the rhomboid-like superfamily of proteins that my group has worked on for almost 20 years. As well as describing some of the key and recent advances, I attempt to draw out signalling themes that emerge. One important message is that the genetic and biochemical perspective on signalling has tended to underplay the importance of cell biology. There is clear evidence that signalling pathways exploit the control of intracellular trafficking, protein quality control and degradation, and other cell biological phenomena, as important regulatory opportunities.
Introduction
Signalling between cells ultimately controls most cellular decisions in a multicellular organism: differentiation, division, cell movement, physiological responses, cell death – all depend on the sending and receiving of signals. Clearly, these central controllers need themselves to be tightly regulated, and it is no surprise that the ultimate cause of many diseases is disruption of signalling pathways. The two most productive general approaches to understanding signalling in the last 50 years or more have been genetics and biochemistry: both have contributed hugely to the current high level of understanding of both the components and regulation of growth factor, cytokine, morphogen and other pathways, to the extent that there are now successful drugs derived from this knowledge [1, 2]. Nevertheless, the fundamentals that we don’t understand greatly outweigh what we do, and practical applications like novel drugs are still in their very early stages. Here I focus on the rhomboid-like superfamily of proteins but, as well as describing some of the key recent advances, I attempt to draw out themes that emerge about how they control signalling. One important message is that the genetic and biochemical perspective on signalling has tended to underplay the importance of cell biology. There is clear evidence that signalling pathways are regulated by cell biological phenomena such as intracellular trafficking, protein quality control and degradation.
Discovery of rhomboid proteases
The superfamily of rhomboid-like genes came to prominence when Drosophila Rhomboid-1 was discovered to be the first of a new class of enzyme: an intramembrane serine protease [3]. Rhomboid proteases were soon found to exist in almost all eukaryotes and prokaryotes [3, 4], and the family subsequently expanded to a superfamily when it became clear that there are many distantly related proteins that have lost protease activity during evolution [5–7] (Figure 1). The hallmark of this wider rhomboid-like superfamily is a conserved six-transmembrane domain (TMD) module which, in most cases, is extended by an extra N- or C-terminal TMD and/or extramembrane domains. The discovery of Drosophila Rhomboid-1 as a serine protease was reported in 2001 [3] and in the intervening 15 years much has been learned about some of the rhomboid-like proteins, although we still lack a clear view of the core function of the rhomboid-like domain. Nevertheless, both rhomboid proteases and their non-proteolytic cousins act in a very wide range of biologically significant cellular processes, including growth factor and cytokine signalling, mitochondrial function, intracellular protein quality control and trafficking, and bacterial protein export; in parallel, rhomboid-like proteins have also been implicated in a similarly broad span of human diseases including cancer, inflammation, and metabolic disorders [8].
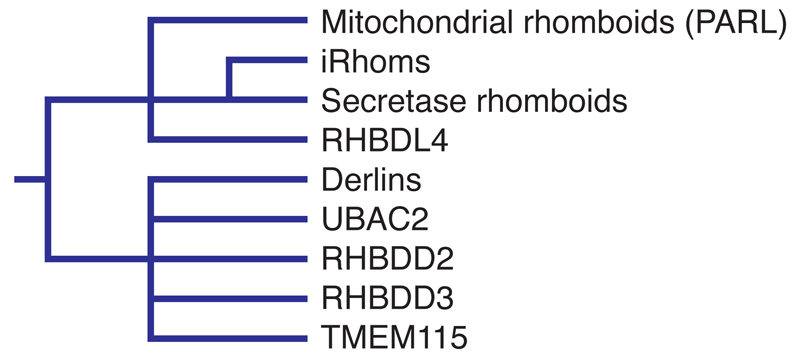
Figure 1.
Classification of members of the rhomboid-like superfamily in mammals. This shows the degree of similarity of rhomboid-like genes in mammals. It is not an evolutionary tree.
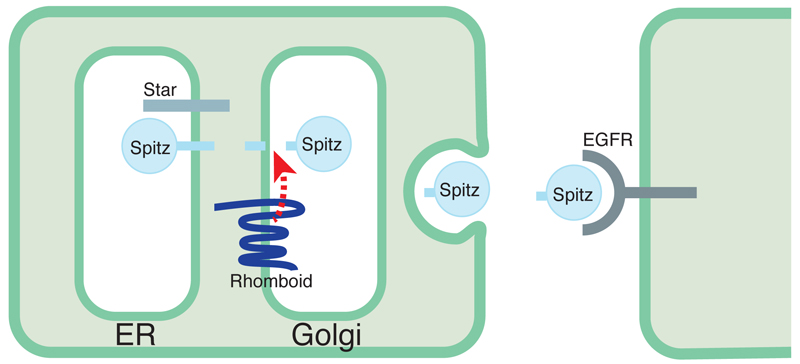
Rhomboids were discovered in Drosophila (and their name derives from an early mutation that gave rise to a pointy head skeleton in the fly embryo [9]). The path that led to understanding their molecular function started when rhomboid-1 mutations were discovered to affect EGF receptor signalling. Indeed, it became apparent that they were the primary regulators of this growth factor pathway in flies [10–12]. A convergence of genetic and functional approaches led to my group showing that Rhomboid-1 triggered EGFR activity by stimulating the release of active EGF ligand from its membrane tethered precursor [13]; we then showed that this was achieved by Rhomboid-1 being a protease that cuts the precursor in its TMD, thereby releasing the extracellular growth factor [3]. Our genetic screens also identified a protein, Star, that indicated a further complexity: the EGF precursor was confined to the endoplasmic reticulum (ER) until it encountered Star, which promoted its trafficking to the Golgi apparatus, the location of Rhomboid-1 [13]. This implied that the control of EGFR signalling in flies was largely effected at the level of membrane trafficking of the EGF ligand (Figure 2). Although not obvious at the time, it has subsequently become clear that this work illustrated two important themes: first, the production of signals (i.e. the pathway upstream of binding to a receptor) is an important but rather overlooked aspect of signal regulation; and second, since signalling occurs within and between cells, the cell biology of the signalling molecules is an important control point. I will return to these themes towards the end of this review.
Figure 2.
Rhomboid and Star regulate EGFR signalling in Drosophila. Star is needed for the EGF-like ligand, Spitz, to traffic from the ER to the Golgi apparatus. Once in the Golgi, the membrane tethered Spitz precursor is cleaved by Rhomboid, and is then free to leave the cell as a soluble and active growth factor
The wider rhomboid-like superfamily
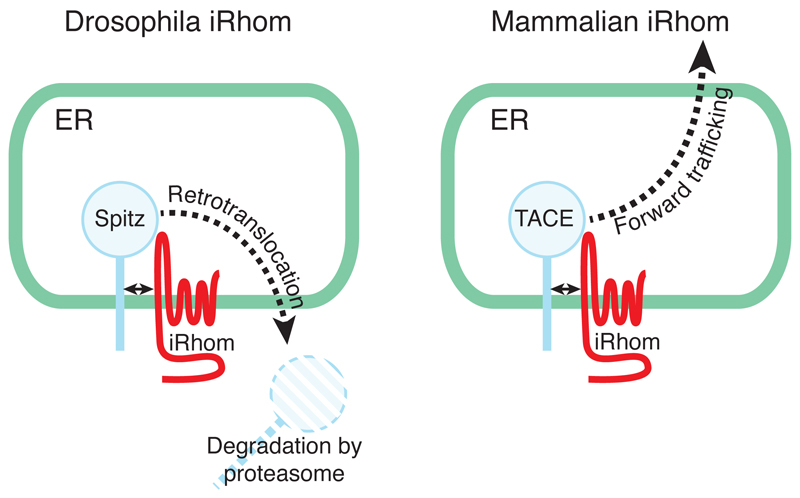
The search for rhomboid homologues led to early recognition that as well as many close relatives, there were also more distant family members that lacked key catalytic residues, implying that unlike Drosophila Rhomboid-1 they were not proteases [7, 14]. The potential importance of these proteins was implied by their evolutionary conservation but there was no indication of their function. Pursuing this puzzle, we returned to fly genetics to investigate the closest inactive homologue of the rhomboid proteases in Drosophila, which we named iRhom (reflecting its proteolytic inactivity, and no doubt also somewhat influenced by the prevailing fashion for ‘i’ as a prefix for cool new things). Again, a combined genetic and cellular approach was productive, showing that, like active Rhomboid-1, iRhom also interacts with EGF family ligands but instead of activating them, it degrades them before they can be released from the signal emitting cell, thereby inhibiting EGFR signalling [15]. At a functional level, iRhom acted in opposition to Rhomboid-1, inhibiting rather than activating EGFR signalling, but mechanistically there was little obvious relationship between the protease and the inactive homologue. iRhom, primarily localised to the ER, binds to and promotes ER associated degradation (ERAD) of EGF ligands (Figure 3).Although full mechanistic details remain unclear, the evolutionary relationships suggest that the binding between iRhom and its client EGF ligand may mimic the interaction between a rhomboid protease and its EGF ligand substrate, but in the case of the iRhom this promotes ERAD rather than proteolytic cleavage.
Figure 3.
iRhom triggers ERAD in flies but forward trafficking in mammals. In Drosophila the primary known function of iRhom is to trigger degradation of the EGF-like ligand Spitz by ERAD; it is not know if this function also occurs in mammalian cells. The known function in mammals is to promote the forward trafficking from the ER to the Golgi apparatus of TACE, the enzyme that activates TNFα and several EGFR ligands. In both flies and mammals, iRhom binds directly to its client protein, which has a single TMD.
A similar genetic and cellular approach in mammals demonstrated that mammalian iRhoms have a different primary function [16, 17], although there are similarities: mammalian iRhoms (of which there are two) appear primarily located in the ER, and they too bind to a single pass TMD protein, thereby affecting its subsequent fate. But in this case, instead of directing their client to degradation, they license its onward trafficking from the ER to the Golgi apparatus. In the absence of iRhom, the client cannot leave the ER (Figure 3). So far, the only confirmed client of mammalian iRhoms is TACE (aka ADAM17), a membrane tethered metalloprotease of the ADAM family [18]. TACE is the TNFα converting enzyme, named because one of its main functions is to release active TNFα from its membrane tethered precursor, at the surface of inflammatory cells. TNFα is sometime called the primary inflammatory cytokine; accordingly, iRhom2 mutant mice are defective in their inflammatory response [16, 17, 19]. Indeed, since TNFα is the target of very successful anti-inflammatory therapy, used extensively for rheumatoid arthritis (RA) but also in other diseases, the discovery that iRhom2 is a physiologically important new regulator of TNFα activity, and its consequent involvement in RA models, has caused significant pharmaceutical interest.
Much of the focus of mammalian iRhom function has been associated with its TNFα and inflammatory function, but the iRhom client TACE is also responsible for shedding multiple other cell surface proteins. Although not all of these are known to be physiologically important, TACE is definitely the primary activator of several EGF receptor ligands in mammals (with a similar role as Rhomboid-1 in flies: releasing the active growth factor from a membrane tethered precursor) [20]. Consistent with this, iRhoms also affect aspects of EGFR signalling in mammals, although the mechanistic details are less well understood than in the case of TNFα. What is clear is that iRhoms can indeed modulate signalling by the TACE-dependent EGFR ligands – TGFα, HB-EGF, amphiregulin, and epiregulin [21]. Intriguingly, iRhom is reported to control the selectivity of TACE for different EGFR ligands [22], as well as its ER exit, but the relationship between these functions is not known. Consistent with a role in EGFR signalling, mutations in mouse iRhom2 leads to wound healing and skin/hair follicle defects, although the full chain of cause and effect is not yet clear [23, 24]. Importantly, iRhom2 is also implicated in human disease. The rare autosomal dominant disease Tylosis with oesophageal cancer (TOC) is caused by mutations in the cytoplasmic domain of iRhom2 [25], leading to a variety of symptoms including palmoplantar hyperkeratosis and oesophageal cancer. Whether this is caused by defects in inflammatory signalling via TNFα, or EGFR signalling, or by other mechanisms, is not yet resolved, although a recent report shows that mutations in the cytoplasmic domain enhances cancer susceptibility in a mouse model [23], and another suggests that cell death may be aberrant due to elevated TNFR shedding from TOC cells [26]. Although TOC is a very rare disease, cells from these patients provide a valuable genetic resource for learning more about iRhom function in humans.
Most vertebrates have two iRhoms, and the work described above has focused on iRhom2, which is primarily expressed in macrophages. Because of this tissue specificity, iRhom2 null mutant mice appear healthy apart from macrophage driven inflammatory defects. The phenotype of iRhom1 mutant mice is less certain, in that that there are two different papers describing different defects. Our group has published that loss of iRhom1 leads to multiple organ defects and death within 2-4 weeks of birth [27]. We found that iRhom1/2 double mutants die in utero, around embryonic day 10. In contrast, the group of Carl Blobel subsequently reported that another iRhom1 mutation led to no obvious phenotype, and that the iRhom1/2 double mutant were born at normal Mendelian ratio but with eyes open, and die perinatally [28]. The eyes open phenotype is significant because it is associated with loss of TACE, the iRhom client [29]. The reasons for this discrepancy are not yet clear: the two mutations are caused by different lesions so although both are expected to be null, it is possible they have different functional consequences. The one we produced has a larger genomic deletion, making it possible that it disrupts a control element of a neighbouring gene in addition to iRhom1. Finally, the genetic backgrounds of the two mutations are not identical, which could affect the phenotype. Further work is ongoing to resolve this issue, which is important not just for understanding the physiopathology of the mutants but also mechanistically. We know that iRhom1 and iRhom2 are largely interchangeable in their ability to regulate TACE trafficking and selectivity: they appear to be mechanistically similar [27, 28]. If the double knockout closely resembles a TACE knockout, that would suggest that TACE is the primary or only client of the iRhoms. If, however, the double knockout is significantly worse, there is an implication of other, as yet unknown clients or functions.
To date more has been discovered about the physiology and genetics of iRhom mutations than about their mechanistic cell biology. There are major gaps and puzzles to solve. How does iRhom in Drosophila promote ERAD? And in mammals, how does it promote ER to Golgi trafficking and TACE substrate selectivity? Is TACE really the only client in mammals? – it seems a priori unlikely but, on the other hand, TACE is an unusually powerful and multifunctional signalling activator, so it is possible that the iRhoms have evolved as specialised regulators. Another important issue is how the same protein in flies and mammals can do apparently different things, a question made more intriguing by the result that, like Drosophila iRhom, mammalian iRhom can also promote ERAD [15], although it is not yet clear whether this is physiologically meaningful. An unexpected recent piece of molecular insight, albeit not one that immediately resolves any of the open questions, is a report that iRhom1 can regulate the activity of proteasomes, probably by interaction with the proteasome chaperones PAC1 and 2 [30]. Overall, there are more questions than answers about the molecular mechanisms by which iRhoms act to control inflammatory and growth factor signalling, but all current evidence points to them being key regulators of major signalling pathways, so there are strong incentives to learn more.
Emerging themes
Our knowledge of the rhomboid-like superfamily is still patchy, making it hard to develop clear mechanistic or functional themes that connect all its members. Most work has focused on the rhomboid proteases, particularly on their enzymology and structure [31], and on the iRhoms, the class of proteolytically inactive rhomboid-like proteins that are closest to the active rhomboids. More distant members of the family include the derlins, which are important components of the ERAD pathway but whose molecular mechanism remains controversial [5]; UbaC2, an ER protein reported to be involved in the regulation of lipid droplet biosynthesis [32, 33]; RHBDD2, genetically associated with cancer but mechanistically unexplored [34–36]; RHBDD3 an inflammatory regulator of natural killer and dendritic cells, functioning at least in part by interaction with ubiquitin [37, 38]; and TMEM115, a Golgi protein reported to bind to the conserved oligomeric Golgi (COG) complex and to regulate retrograde transport [39]. The phylogenetic tree of rhomboid-like proteins suggests that they may have all evolved from an ancient rhomboid that was a protease, with only one branch having retained that protease function [4, 6].
Combined with what we know about the function of the proteases and the iRhoms it is attractive to speculate that a common function of the rhomboid-like domain may be to recognise and bind TMDs – either to cleave them, in the case of the active proteases, or to handle them in other ways. This is consistent with the core 6 TMD domain being almost entirely membrane embedded, but it is too early to be confident that a TMD binding function extends to the more distant members of the clan. Rhomboid-like proteins exist in many different membranes – for examples organelles of the secretory pathway, plasma membrane, mitochondria – but they all have significant cytoplasmic domains. Interestingly, these are not well conserved, suggesting that they may determine some of the functions distinct to different rhomboids. Many rhomboid-like proteins also have significant luminal/periplasmic/extracellular domains, again not well conserved, but there is less evidence so far about their functional importance.
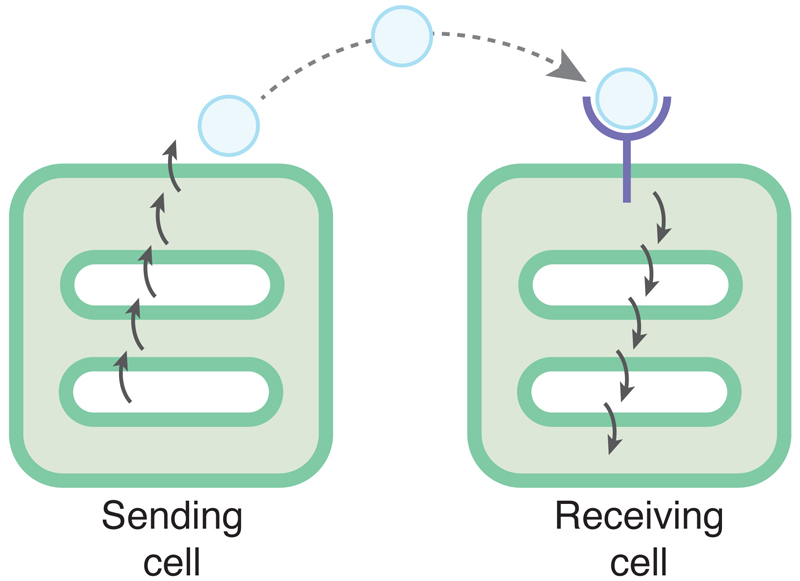
A broader signalling theme is illustrated by rhomboid research: there is much control of signalling between cells at the level of signal production. This is important because historically a greater focus has been placed on understanding signal transduction pathways, that is, the pathways from the receptor to the nucleus in the receiving cell (Figure 4). This imbalance of effort probably derives from the early biochemical approaches that founded much of the field: it is much easier to study what happens autonomously within the receiving cell. Genetic approaches are not restricted in this way, and it is notable that genetic screens have identified signalling regulators that act to control signal emission. An even more general message emerges from studying the genetics of signalling, as alluded to at the beginning of this review: although both classical genetics and biochemistry tend to produce maps of signalling pathways that show proteins linked to each other by arrows, often depicted non-specifically floating inside (or outside) cells, signalling pathways are actually hugely influenced by cell biology. As exemplified by the signalling associated with the rhomboid-like clan, many key signalling components are membrane proteins and therefore subject to regulated membrane trafficking as well as quality and protein stability control. Moreover, the cytoskeleton influences the intracellular movement of signalling machinery, and membrane lipid composition also can affect signal production and reception. These are just examples of the myriad cell biological factors that can act as important regulators of signalling pathways. This relatively poorly studied interface between signalling and cell biology will be productive and illuminating at both a fundamental and translational level: if rational drug design is to live up to its huge promise, we need to understand the detailed mechanisms of the pathways in which we aim to intervene.
Figure 4.
Signal sending and receiving. To date, most signalling research has focused on signal pathways in the signal receiving cell. It is becoming apparent from genetic and cell biology appraoches that there is at least as much important regulation in the signal sending cell.
Finally, one even broader issue arises from the study of iRhoms: the potential functional significance of inactive enzyme homologues or pseudoenzymes [40–42]. Browsing the EC enzyme database it is clear that most enzymes, across all classes, have inactive cognates. Where these are conserved, it must be assumed that there is selective pressure for their retention and there that they are functional, despite not having enzyme activity. The pseudokinases have been most studied. They comprise about 10% of the human ‘kinome’. No other class has been studied as systematically, but what does emerge from many of the cases that have been investigated, including the iRhoms, is that, having lost enzyme activity, they have evolved regulatory relationships with the pathways in which their cognate enzymes act. This is perhaps not surprising: despite loss of a catalytic residue or an active site, the overall architecture of the protein is likely to remain similar when mutated so that it may still bind to cofactors or substrates, and it may retain expression in the same place and time as the enzyme from which it evolved. This provides a solid platform from which to evolve regulatory factors. It should be stressed that this will not be a universal phenomenon, and the nature and function of the evolved regulator can be very diverse, but it seems like a safe prediction that many pseudoenzymes will be found to regulate the process in which their cognate enzyme acts, and that a chain of evolutionary logic will exist that links the two functions.
In conclusion, I have attempted two goals in this short review. First, I have outlined the history and current state of play of research into the rhomboid-like superfamily. Once it became clear that rhomboid-like proteins were much more widespread than just the rhomboid proteases, the field diversified and grew substantially. Then, as medical significance has begun to emerge for several members of the clan, interest has grown further and there are prospects of rapid progress on many fronts in the near future. My second goal has been to use the rhomboid-like field to draw out some important broader principles, mostly about how signalling pathways – the all-important regulators of most biology in multicellular organisms – are regulated by cell biological processes. I have also speculated that pseudoenzymes illustrate a profound evolutionary phenomenon of potential regualtory significance.
Acknowledgments
Work in my group has been primarily supported by the Wellcome Trust [grant number 101035/Z/13/Z], the Medical Research Council [programme number U105178780], and fellowships from EMBO, HFSP, and the Marie Curie programme of the European Union.
Footnotes
Novartis Medal Award Lecture
SA174: Organelle Crosstalk in Membrane Dynamics and Cell Signalling
References
- 1.Druker BJ, Lydon NB. Lessons learned from the development of an abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 3.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinch LN, Grishin NV. Bioinformatics perspective on rhomboid intramembrane protease evolution and function. Biochim Biophys Acta. 2013;1828:2937–2943. doi: 10.1016/j.bbamem.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman M. The Rhomboid-Like Superfamily: Molecular Mechanisms and Biological Roles. Annu Rev Cell Dev Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 9.Mayer U, Nüsslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 1988;2:1496–1511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- 10.Wasserman JD, Urban S, Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signalling. Genes Dev. 2000;14:1651–1663. [PMC free article] [PubMed] [Google Scholar]
- 11.Guichard A, Biehs B, Sturtevant MA, Wickline L, Chacko J, Howard K, Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development. 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- 12.Bang AG, Kintner C. Rhomboid and Star facilitate presentation and processing of the Drosophila TGF-alpha homolog Spitz. Genes Dev. 2000;14:177–186. [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 14.Lemberg MK, Freeman M. Cutting Proteins within Lipid Bilayers: Rhomboid Structure and Mechanism. Mol Cell. 2007;28:930–940. doi: 10.1016/j.molcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor- alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 19.Issuree PD, Maretzky T, McIlwain DR, Monette S, Qing X, Lang PA, Swendeman SL, Park-Min KH, Binder N, Kalliolias GD, et al. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J Clin Invest. 2013;123:928–932. doi: 10.1172/JCI66168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, et al. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem. 2002;277:12838–12845. doi: 10.1074/jbc.M112050200. [DOI] [PubMed] [Google Scholar]
- 21.Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP. Substrate Selectivity of Epidermal Growth Factor-Receptor Ligand Sheddases and their Regulation by Phorbol Esters and Calcium Influx. Mol Biol Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maretzky T, McIlwain DR, Issuree PD, Li X, Malapeira J, Amin S, Lang PA, Mak TW, Blobel CP. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1302553110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosur V, Johnson KR, Burzenski LM, Stearns TM, Maser RS, Shultz LD. Rhbdf2 mutations increase its protein stability and drive EGFR hyperactivation through enhanced secretion of amphiregulin. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1323908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leilei Y, Bing L, Yang L, Shaoxia W, Yuan X, Dongping W, Huahu Y, Shichen S, Guangzhou Z, Ruiyun P, et al. iRhom2 Mutation Leads to Aberrant Hair Follicle Differentiation in Mice. PLoS One. 2014;9:e115114. doi: 10.1371/journal.pone.0115114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaydon DC, Etheridge SL, Risk JM, Hennies HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr NK, et al. RHBDF2 mutations are associated with Tylosis, a familial esophageal cancer syndrome. Am J Hum Genet. 2012;90:340–346. doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maney SK, McIlwain DR, Polz R, Pandyra AA, Sundaram B, Wolff D, Ohishi K, Maretzky T, Brooke MA, Evers A, et al. Deletions in the cytoplasmic domain of iRhom1 and iRhom2 promote shedding of the TNF receptor by the protease ADAM17. Sci Signal. 2015;8:ra109. doi: 10.1126/scisignal.aac5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christova Y, Adrain C, Bambrough P, Ibrahim A, Freeman M. Mammalian iRhoms have distinct physiological functions including an essential role in TACE regulation. EMBO Rep. 2013;14:884–890. doi: 10.1038/embor.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Maretzky T, Weskamp G, Monette S, Qing X, Issuree PD, Crawford HC, McIlwain DR, Mak TW, Salmon JE, et al. iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1505649112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 30.Lee W, Kim Y, Park J, Shim S, Lee J, Hong SH, Ahn HH, Lee H, Jung YK. iRhom1 regulates proteasome activity via PAC1/2 under ER stress. Sci Rep. 2015;5:11559. doi: 10.1038/srep11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks CL, Lemieux MJ. Untangling structure-function relationships in the rhomboid family of intramembrane proteases. Biochim Biophys Acta. 2013;1828:2862–2872. doi: 10.1016/j.bbamem.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd SJ, Raychaudhuri S, Espenshade PJ. Subunit architecture of the Golgi Dsc E3 ligase required for sterol regulatory element-binding protein (SREBP) cleavage in fission yeast. J Biol Chem. 2013;288:21043–21054. doi: 10.1074/jbc.M113.468215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci U S A. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacunza E, Rabassa ME, Canzoneri R, Pellon-Maison M, Croce MV, Aldaz CM, Abba MC. Identification of signaling pathways modulated by RHBDD2 in breast cancer cells: a link to the unfolded protein response. Cell Stress Chaperones. 2013 doi: 10.1007/s12192-013-0466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmedli NB, Gribanova Y, Njoku CC, Naidu A, Young A, Mendoza E, Yamashita CK, Ozgul RK, Johnson JE, Fox DA, et al. Dynamics of the Rhomboid-Like Rhbdd2 Expression in Mouse Retina and Involvement of its Human Ortholog in Retinitis Pigmentosa. J Biol Chem. 2013 doi: 10.1074/jbc.M112.419960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacunza E, Canzoneri R, Rabassa ME, Zwenger A, Segal-Eiras A, Croce MV, Abba MC. RHBDD2: a 5-fluorouracil responsive gene overexpressed in the advanced stages of colorectal cancer. Tumour Biol. 2012 doi: 10.1007/s13277-012-0503-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Han C, Xie B, Wu Y, Liu S, Chen K, Xia M, Zhang Y, Song L, Li Z, et al. Rhbdd3 controls autoimmunity by suppressing the production of IL-6 by dendritic cells via K27-linked ubiquitination of the regulator NEMO. Nat Immunol. 2014 doi: 10.1038/ni.2898. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Liu S, Xia M, Xu S, Wang C, Bao Y, Jiang M, Wu Y, Xu T, Cao X. Rhomboid domain-containing protein 3 is a negative regulator of TLR3-triggered natural killer cell activation. Proc Natl Acad Sci U S A. 2013;110:7814–7819. doi: 10.1073/pnas.1220466110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ong YS, Tran TH, Gounko NV, Hong W. TMEM115 is an integral membrane protein of the Golgi complex involved in retrograde transport. J Cell Sci. 2014;127:2825–2839. doi: 10.1242/jcs.136754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pils B, Schultz J. Inactive enzyme-homologues find new function in regulatory processes. J Mol Biol. 2004;340:399–404. doi: 10.1016/j.jmb.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 41.Todd AE, Orengo CA, Thornton JM. Sequence and structural differences between enzyme and nonenzyme homologs. Structure. 2002;10:1435–1451. doi: 10.1016/s0969-2126(02)00861-4. [DOI] [PubMed] [Google Scholar]
- 42.Adrain C, Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13:489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]