Abstract
Precision medicines exert selective pressure on tumor cells that leads to the preferential growth of resistant subpopulations, necessitating the development of next generation therapies to treat the evolving cancer. The PIK3CA/AKT/mTOR pathway is one of the most commonly activated pathways in human cancers1, which has led to the development of small molecule inhibitors that target various nodes in the pathway. Among these agents, first generation mTOR inhibitors (rapalogs) have caused responses in so-called “N-of-1” cases and second generation mTOR kinase inhibitors (TORKi) are currently in clinical trials2–4. We sought to delineate the likely resistance mechanisms to existing mTOR inhibitors as a guide for next generation therapies. The mechanism of resistance to the TORKi was unusual in that intrinsic kinase activity of mTOR was increased, rather than a direct active site mutation interfering with drug binding. Indeed, the identical drug resistant mutations have been also identified in drug-naïve patients4, suggesting that tumors with activating mTOR mutations will be intrinsically resistant to second generation mTOR inhibitors. Here, we report the development of a new class of mTOR inhibitors which overcomes resistance to existing first and second generation inhibitors. The third generation mTOR inhibitor exploits the unique juxtaposition of two drug binding pockets to create a bivalent interaction that allows inhibition of these resistant mutants.
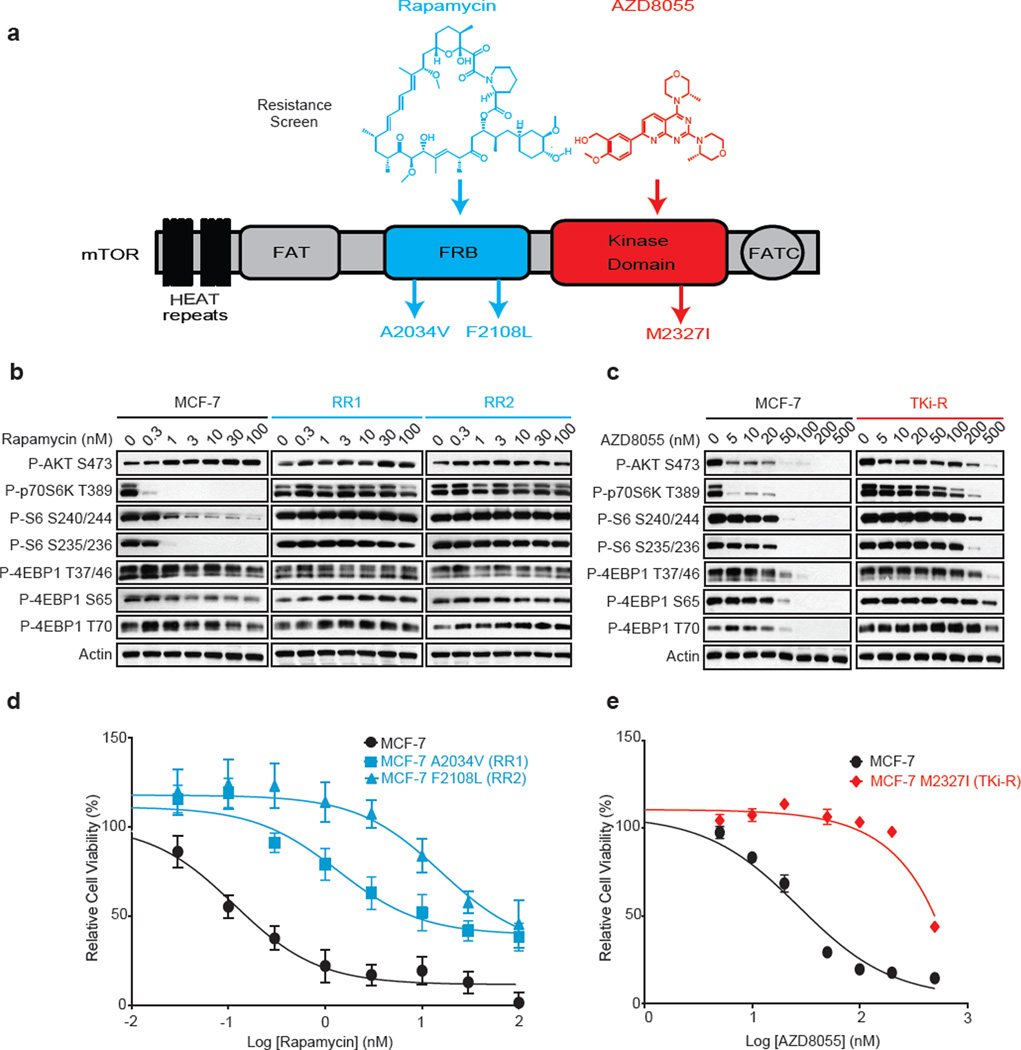
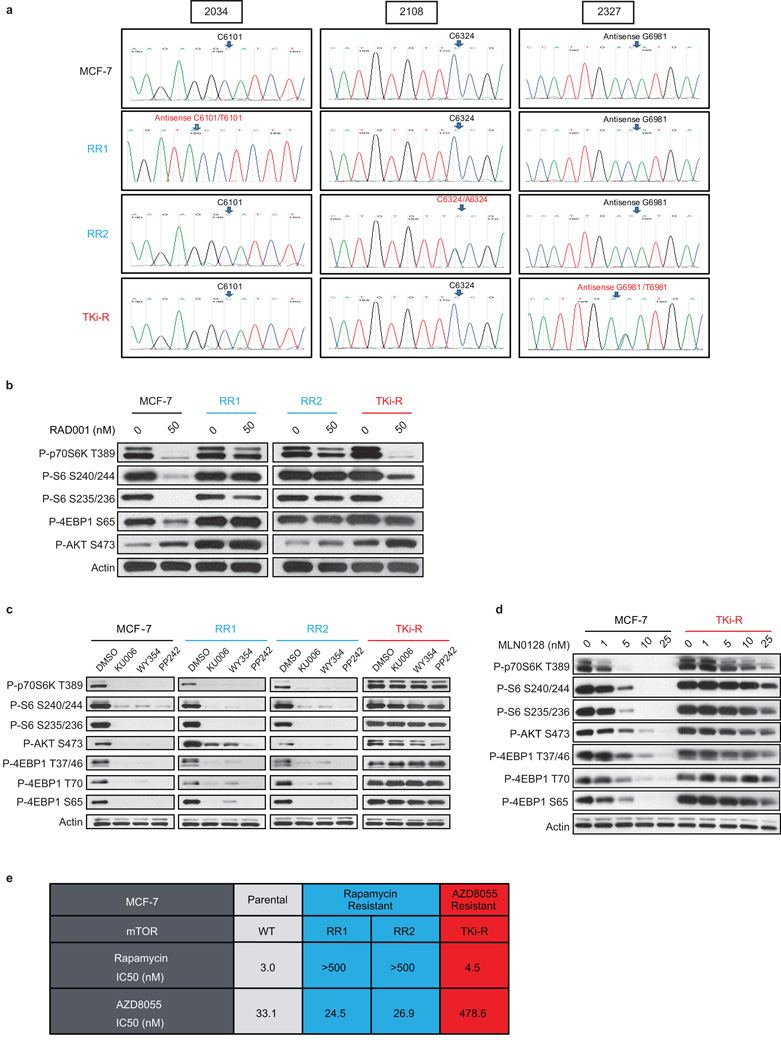
The MCF-7 breast cancer cell line was exposed to high concentrations of either a first generation mTORC1 inhibitor, rapamycin or a second generation mTOR ATP competitive inhibitor AZD8055 (a TORKi) for 3 months, until resistant colonies emerged. Deep sequencing revealed that the AZD8055-resistant (TKi-R) clones harbored an mTOR mutation located in the kinase domain at the M2327I position (Figure 1a, Extended Data Figure 1a) while two rapamycin-resistant (RR) clones contained mutations located in the FKBP12-rapamycin binding domain (FRB domain) at positions A2034V (RR1 cells) and F2108L (RR2 cells). The clinical relevance of these mutations is supported by a case report of a patient who acquired the identical F2108L mTOR mutation after relapse under everolimus treatment5 (Extended Data Table 1).
Figure 1. Single amino acid mutation accounts for acquired resistance to mTOR inhibitors.
a, Graphic representation of mTOR domains and site mutagenesis isolated in rapamycin- and AZD8055-resistant cells. b, The effects of rapamycin or AZD8055 (c) on mTOR signaling was assessed in MCF-7, RR1 and RR2 cells (or in TKi-R cells (c)) by immunoblotting 4 hours after treatment. For gel source data, see Supplemental Figure 1. d, Dose-dependent cell growth inhibition curves of MCF-7 and rapamycin-resistant MCF-7 A2034V (RR1) and MCF-7 F2108L (RR2) cells treated with rapamycin at day 3 or e, MCF-7 and AZD8055-resistant MCF-7 M2327I (TKi-R) cells treated with AZD8055. Each dot and error bar on the curves represents mean ± SD (n=8). All experiments were repeated at least three times.
To verify that the mutations altered the efficacy of their respective drugs and were not simply passenger mutations, we analyzed the phosphorylation of effectors downstream of mTOR in several cellular systems. In the RR cells, phosphorylation of the normally rapamycin sensitive sites on S6K (T389) and S6 (S240/244 and S235/236) were unaffected even at high rapalog concentrations (100 nM) (Figure 1b, Extended Data Figure 1b). Phosphorylation of the key mTOR effector 4EBP-1 is normally unaffected by rapamycin but strongly reduced by TORKi6–8. In the TKi-R cells, however, 4EBP-1 phosphorylation was significantly less sensitive to a variety of TORKi (Figure 1c, Extended Data Figures 1c, d). Consistent with this weakened signaling inhibition, the RR and TKi-R clones were significantly less sensitive to their respective drugs in a 72h proliferation assay when compared to the parental line (Figures 1d, e, Table in SI). To determine if the RR and TKi-R mTOR mutations were directly responsible for the drug-resistance phenotype, each mutant was expressed in another model, MDA-MB-468 cells, which confirmed that the mTOR mutations are sufficient to promote dominant resistance (Extended Data Figures 2a-d).
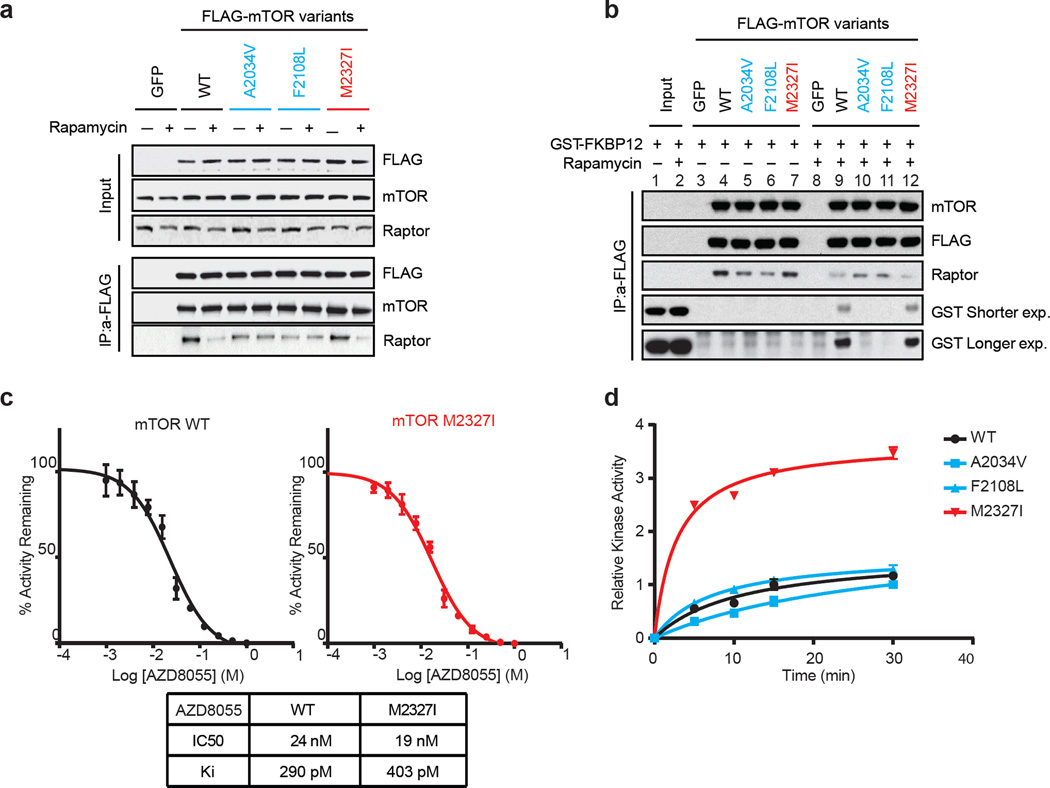
FRB domain mutations have been found in untreated patients (Extended Data Table 2) and previous random mutagenesis screens in yeast have shown that single amino acid changes in the mTOR FRB domain confer rapamycin resistance9–12. The RR mutants identified in this screen exhibit a similar mechanism of resistance by disrupting interaction of mTOR with FKBP12-rapamycin complex in cells and in vitro (Figures 2a, b).
Figure 2. Non-overlapping mechanisms of resistance mediated by mTOR mutations.
a, mTOR-FLAG Wild-Type (WT) and variants were transfected into 293H cells. Cells were treated with rapamycin and lysates were immunoprecipitated (IP) with an anti-FLAG antibody. mTORC1 complex formation was assessed by immunoblotting. b, 293H cells were transfected and complex isolated as described in a, and an in vitro competition assay was performed followed by immunoblotting. For gel source data, see Supplemental Figure 2. c, Varying concentrations of AZD8055 were tested in vitro on WT and M2327I mTOR followed by a kinase reaction (see Methods). The IC50s were determined by fitting to a standard 4-parameter logistic using GraphPad Prism V.5. The diagram shows the mean of n=3 data. The error bars represent the standard deviation between experiments. d, 293H cells were transfected and the complex was isolated as described in a. An in vitro kinase assay was performed and the level of P-AKT (S473) was determined by immunoblotting. Dots represent on each curve the relative P-AKT at different time points. The kinase activity curves were generated using Pad Prism v.6 after densitometry analysis was performed. All experiments were repeated at least three times.
In contrast to the FRB domain mutations found in RR cells which line the rapalog/FKBP binding pocket, analysis of the recently solved structure of the mTOR kinase domain in complex with the TORKi, PP242, (PDB:4JT5)13 revealed that M2327 is >15Å away from the inhibitor, suggesting either an allosteric mechanism of reduced TORKi affinity or that this mutation causes resistance through a mechanism that does not involve reduced drug binding. Indeed, both WT and M2327I mTOR bind AZD8055 with similar affinities (Figure 2c). We asked whether the M2327I mutation in the mTOR kinase domain altered the kinetic properties of the kinase. As shown in Figure 2d, the M2327I mutant has a 3-fold increase in mTOR kinase activity compared to the WT and RR mutants. This is consistent with the higher P-S6K (T389), P-AKT (S473) and P-4EBP1 Ser 65 basal levels observed in these cells (Extended Data Figure 1d).
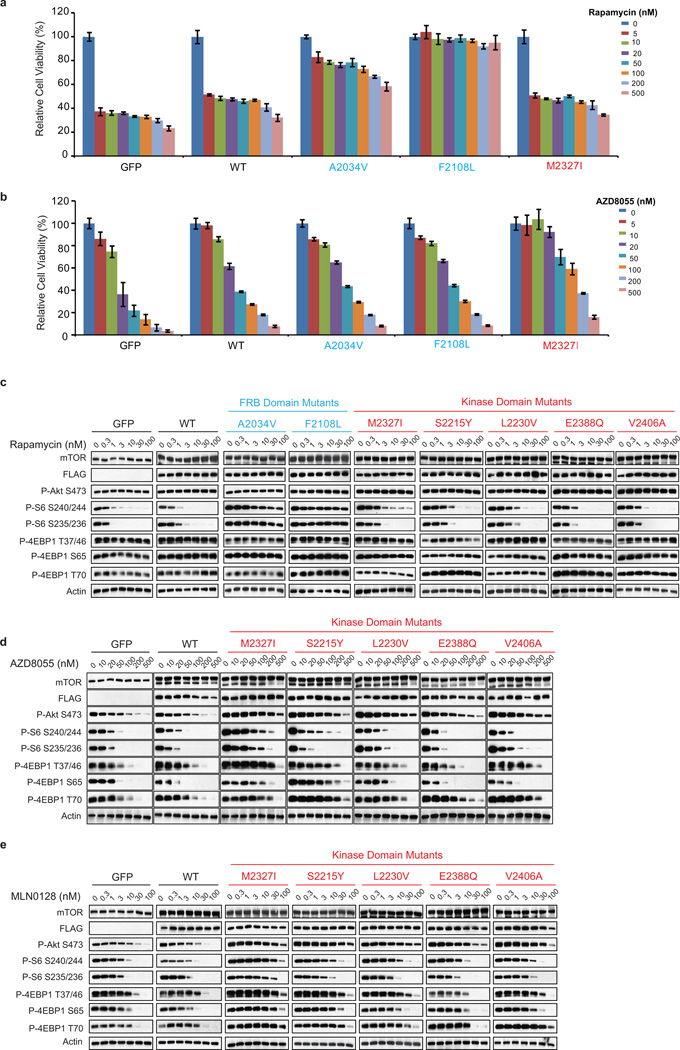
The emergence of a hyperactive mTOR kinase domain mutation (M2327I) that could theoretically confer a growth advantage led us to wonder if similar mutations might pre-exist in drug-naïve patient tumors. Indeed, the precise M2327I mutation as well as other mTOR kinase domain mutations have been identified in 5 untreated patients (Extended Data Tables 1, 3) 14, 15. To determine if additional mTOR kinase domain mutants were also hyperactive and insensitive to TORKi, various mTOR kinase domain mutations that occur in patients were inducibly expressed in MDA-MB-468 cells and tested for sensitivity to the TORKis AZD8055 and MLN0128 (Extended Data Figures 2d, 2e). The concentrations of drug required to inhibit mTORC1 and mTORC2 substrates in these cells were 3 to 30 fold higher than those required in WT cells, although not all substrates show precisely the same dose response.
These data suggest that the hyperactivation of mTOR kinase by single amino acid mutations found in drug-naïve patients can reduce the sensitivity to ATP-competitive mTOR inhibitors in cells. These findings highlight the need for a new class of mTOR inhibitor capable of targeting both drug-naïve (pre-existing) mTOR mutant-driven cancers, as well as emergent resistant mutations.
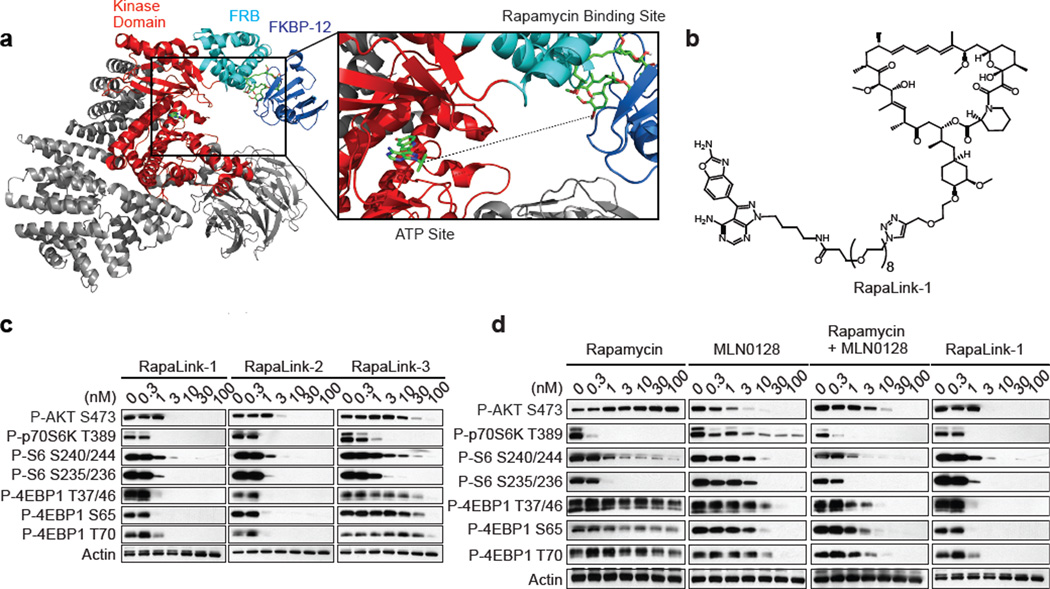
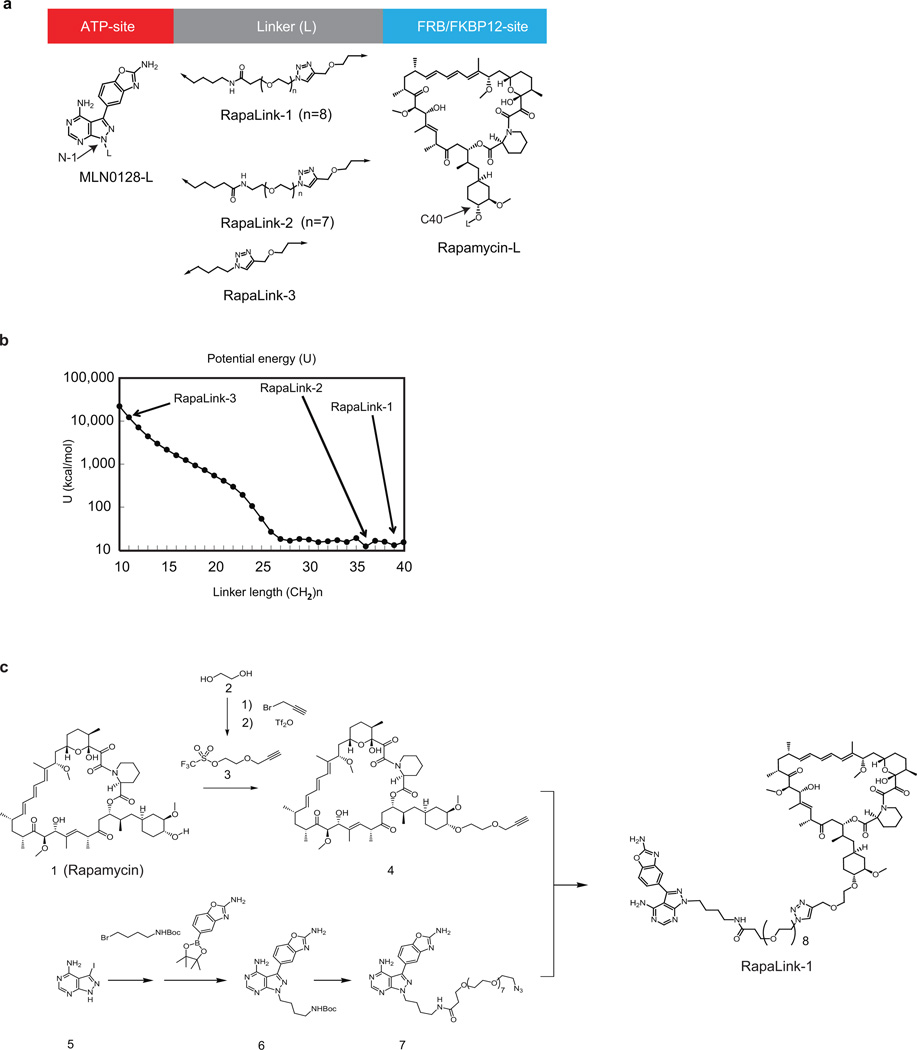
We developed a molecular model of mTOR in complex with rapamycin-FKBP12 using the FRB domain as the common domain in two available mTOR crystal structures (1FAP & 4JT5) (Figure 3a). This model revealed the juxtaposition of the rapamycin and TORKi binding sites and suggested an avidity-based approach to overcome drug-resistant mutations in either the FRB or the kinase domain. A bivalent mTOR inhibitor consisting of a rapamycin-FRB binding element appropriately linked to a TORKi would be expected to inhibit the RR class of FRB-domain mutants because the TORKi binding site would provide high affinity recognition. For the TKi-R class of kinase domain mutations, a bivalent inhibitor would be predicted to be similarly potent by virtue of an intact rapamycin binding site. We reasoned that binding at one site would position the second half of the ligand in close proximity for binding to the second site, thus overcoming point mutations that diminish drug binding (as found in RR cells) or which hyperactivate the kinase (as found in TKi-R cells)16. To develop a new bivalent class of mTOR inhibitors, we required a non-perturbing, strain-free linker between rapamycin and a TORKi, such that the resulting inhibitor can simultaneously bind to both sites. Analysis of our mTOR-rapamycin-FKBP12 model revealed that the hydroxyl group at the C40 position of rapamycin is exposed to solvent and is oriented toward the ATP binding site of mTOR (Figure 3a). Analysis of the TORKi (PP242) bound structure (PDB:4JT5) revealed the N-1 position of the pyrazole ring is oriented toward rapamycin and exposed to solvent (Extended Data Figure 3a). We selected MLN0128 as the TORKi as it is a highly selective17 structural analog of PP242, and is currently in clinical trials.
Figure 3. RapaLink-1 is a potent mTOR inhibitor.
a, Molecular model constructed by two available co-crystal structures, mTOR catalytic domain bearing TORKi PP242 (4JT5) and mTOR FRB domain/rapamycin/FKBP12 (1FAP). Dotted line represents a guide line for the linker design of bivalent mTOR inhibitors. b, RapaLink-1 structure is displayed. c, MCF-7 cells were treated with RapaLink-1, -2, and -3 or (d) with rapamycin, MLN0128, combination of rapamycin and MLN0128 or RapaLink-1 for 4 hours followed by immunoblotting. The rapamycin panel is the same shown in Figure 1b and the RapaLink-1 panel is the same shown in Figure 3c. For gel source data, see Supplemental Figure 3.
To determine the optimum linker length between the chosen sites we used the molecular modeling program MOE18 to evaluate the potential energy of a methylene-based cross-linker with lengths from 10 to 40 heavy atoms. This analysis revealed that 27 atoms would be the minimal length required to span the two ligand-binding sites (Extended Data Figure 3b). We incorporated a polyethylene glycol unit of varying lengths and used the azide-alkyne cycloaddition reaction to synthesize RapaLink-1, -2, and -3 (Figure 3b, Extended Data Figure 3c and Methods in SI). Our modeling suggested that RapaLink-3, with an 11-heavy atom linker would be too short to allow optimal binding to both sites simultaneously while RapaLink-1 and -2, which contain 39 and 36 heavy atom linkers, respectively, would allow simultaneous bivalent binding to the mTOR-FKBP12 complex.
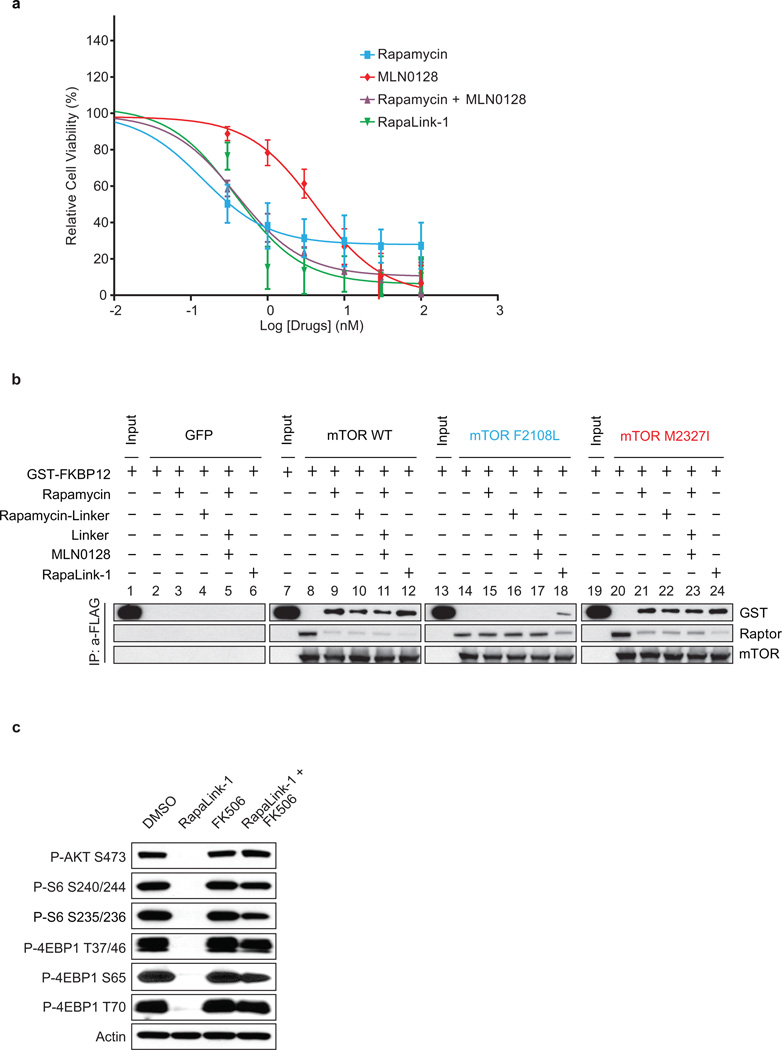
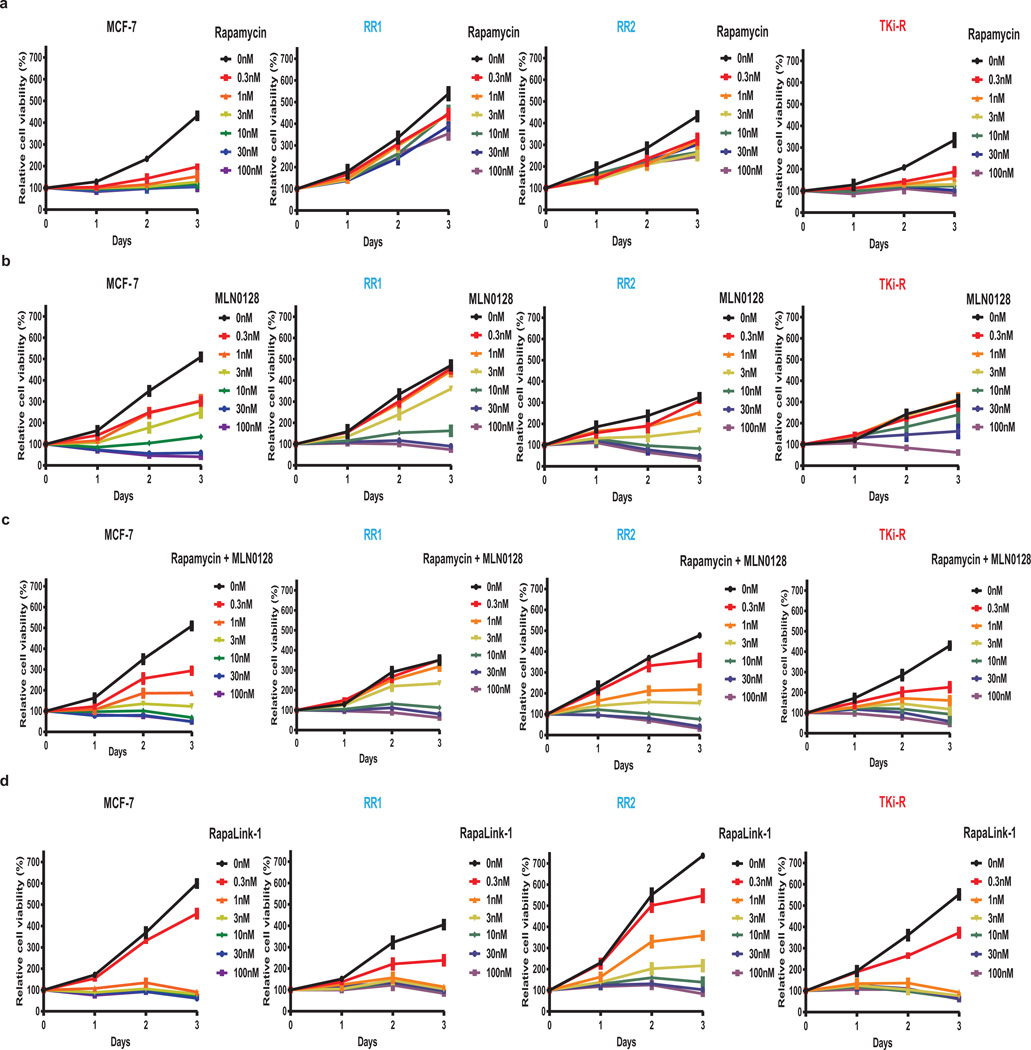
Cells were treated with increasing concentrations of either RapaLink-1, 2, or 3 and the effects on mTOR signaling were assessed by Western blotting. We observed that both RapaLink-1 and -2 inhibited the phosphorylation of both mTORC1 and mTORC2 targets at doses between 1 and 3 nM (Figure 3c). However, RapaLink-3, which contains the shortest linker showed diminished potency against the phosphorylation of 4EBP1 (T37/46/70 and S65) and AKT (S473) while still inhibiting P-S6 (S240/244 and S235/236). This is consistent with the prediction that a longer linker is necessary to allow simultaneous binding to both drug sites and indicates that rapamycin binding is dominant over MLN0128 binding due to the preferential inhibition of P-S6 over P-4EBP1. Consistent with its strong signaling inhibition (Figure 3d), RapaLink-1 potently inhibited the growth of MCF-7 cells at levels comparable to rapamycin or a combination of rapamycin with MLN0128 (Extended Data Figures 4a, 5).
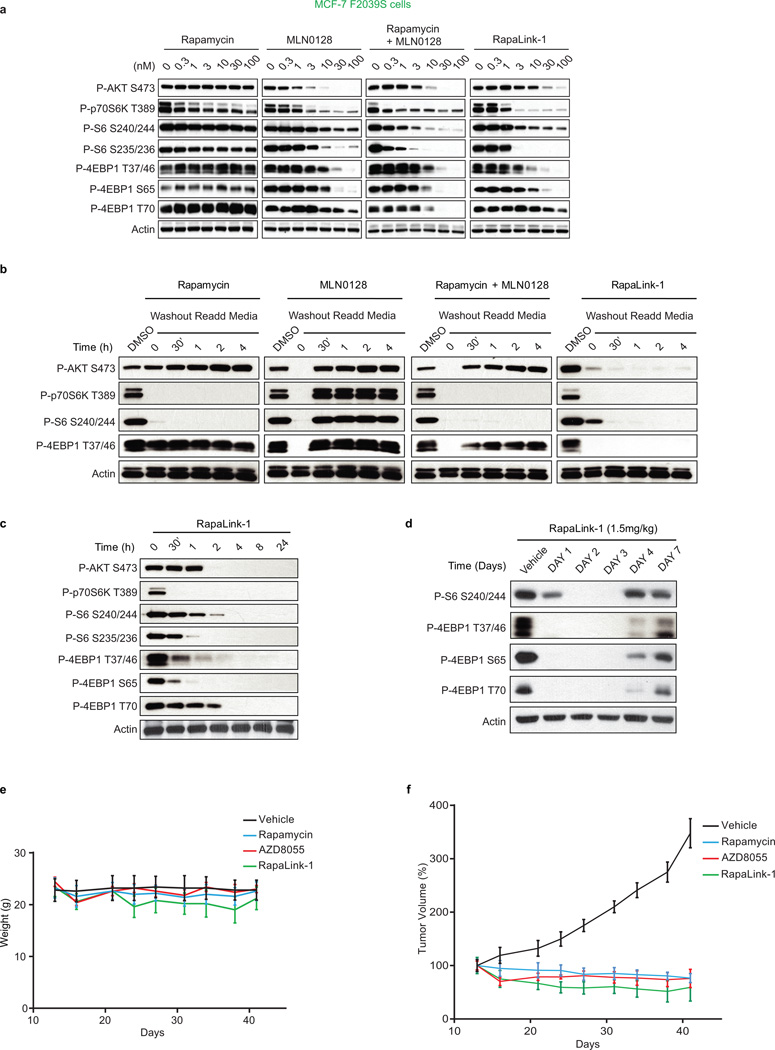
We further tested the requirement of both halves of RapaLink-1 to simultaneously bind mTOR. First, we measured the ability of RapaLink-1 to recruit FKBP12 to mTOR by performing an in vitro FKBP12 binding assay; we show that RapaLink-1 is indeed able to recruit GST-FKBP12 to mTOR WT (Extended Data Figure 4b, lane 12). Moreover, we used the FKBP12 competitive ligand, FK506, to pharmacologically block RapaLink-1 from interacting with FKBP12 and thus mTOR. We observed that FK506 completely rescued the phosphorylation of mTORC1 and C2 substrates upon RapaLink-1 treatment (Extended Data Figure 4c). Last, we isolated MCF-7 RapaLink-1-resistant cells; these cells harbor a mutation located in the mTOR FRB domain at position F2039S. As shown in Extended Data Figure 6a, rapamycin treatment did not inhibit P-S6K (T389) and P-S6 (S240/244 and S235/236) in the mTOR F2039S cells as observed in the MCF-7 cells. Moreover, these cells displayed a decreased sensitivity to MLN0128, combination of rapamycin and MLN0128 as well as RapaLink-1. Taken together, these data demonstrate that RapaLink-1:FKBP12’s binding to the FRB domain is necessary for simultaneous binding to the ATP-site of mTOR and therefore for RapaLink-1-dependent inhibition of mTOR signaling.
While the design of bivalent inhibitors for therapeutic use has had mixed success due to the poor pharmaceutical properties of the hybrid molecules19, FKBP12-binding hybrids have actually been used to improve the pharmaceutical properties of small-molecule inhibitors unrelated to TORKi. These FK506-based hybrids exploit the high intracellular concentration of FKBP12, specifically in blood cells, and the high affinity of FK506 for FKBP12 to create a reservoir of drug that prolongs serum half-life20. In agreement with the improved pharmaceutical properties of previous FKBP12-binding hybrids, RapaLink-1 showed prolonged inhibition of mTOR signaling in vitro (Extended Data Figures 6b, c) as well as in vivo after a tolerable dose of 1.5 mg/kg, which lasted for over 4 days (Extended Data Figures 6d, e) and was able to inhibit the growth of WT mTOR MCF-7 xenografts as well as the current clinical mTOR inhibitors (Extended Data Figure 6f).
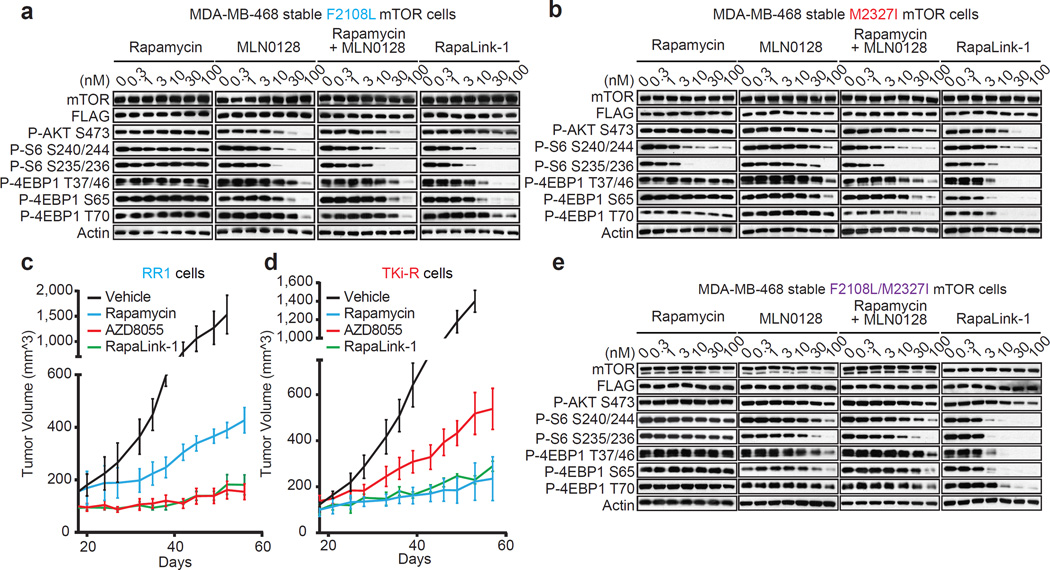
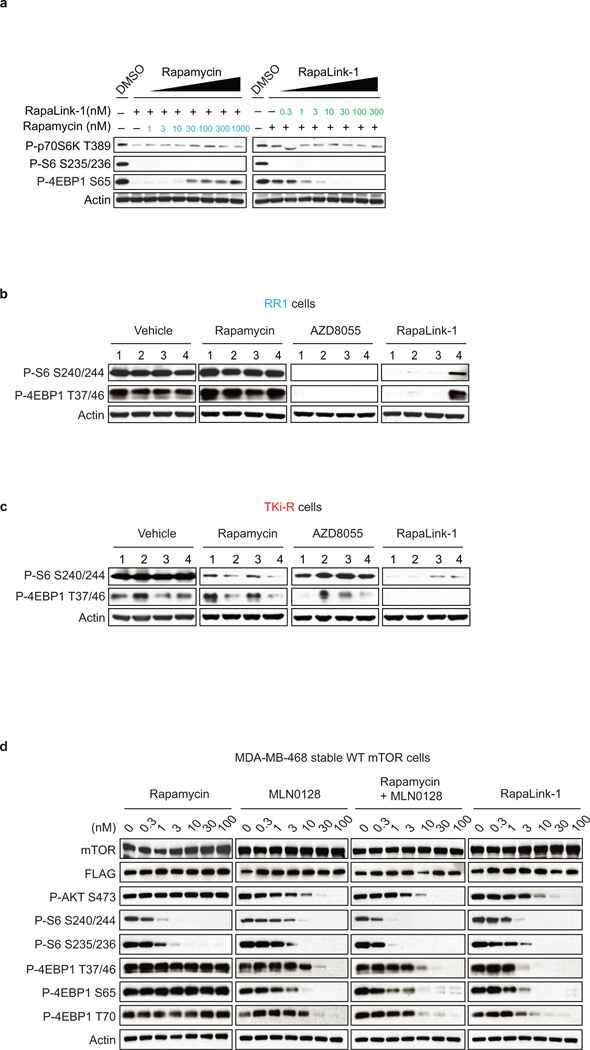
To assess whether RapaLink-1 could block mTOR signaling of the F2108L mTOR and M2327I mTOR drug resistant mutants, MDA-MB-468 cells expressing the alleles were treated with either rapamycin, MLN0128, a combination of both drugs or RapaLink-1. Consistent with RapaLink-1:FKBP12 complex’s ability to bind mTOR FRB and Kinase Domain mutants (Extended Data Figure 4b: lanes 18 and 24), and increased avidity (Extended Data Figure 7a); RapaLink-1 at low doses (3–10 nM) was the only drug regimen capable of inhibiting mTOR signaling in both F2108L mTOR and M2327I mTOR expressing cells (Figures 4a, b). Mouse xenografts of MCF-7 cells expressing the RR1 mutant A2034V mTOR showed significantly less sensitivity to rapamycin yet maintained full sensitivity to AZD8055 and RapaLink-1 treatment (Extended Data Figure 7b, Figure 4c). Similarly, xenografts with MCF-7 cells expressing the TKi-R mutant, M2327I mTOR, showed significantly less sensitivity to AZD8055 treatment, yet retained full sensitivity to rapamycin and RapaLink-1 (Extended Data Figure 7c, Figure 4d). The dosing of RapaLink-1 may be limited by toxicity, which can only be tested in the clinic. However, our preclinical data and that of others21 suggests that mTOR kinase inhibitors can be given safely when administered intermittently and are more effective than daily dosing schedules.
Figure 4. RapaLink-1 reverses resistance due to mTOR FRB and kinase domain mutations.
a, MDA-MB-468 cells inducibly expressing mTOR F2108L or M2327I (b) or F2108L/M2327I mTOR double mutant (e) were treated as in Figure 3d and followed by immunoblotting. For gel source data, see Supplemental Figures 4, 5 and 6. All experiments were repeated at least three times. c, Mice bearing RR1 or TKi-R (d) xenograft tumors (n=5 for each group) were randomized to 4 different groups; (1) Vehicle (M, W, F); (2) rapamycin (10 mg/kg; M, W, F); (3) AZD8055 (75 mg/kg; M, W, F); (4) RapaLink-1 (1.5 mg/kg; weekly). Tumor size was measured by caliper two times per week. The results were reported as tumor volume (mm^3) ± standard deviation.
It is reasonable to anticipate that patients bearing hyperactive mTOR kinase domain mutations, who originally respond to rapalogs, may eventually relapse due to the emergence of a second FRB mutation, as previously observed5. To test if RapaLink-1 would be an effective mTOR inhibitor in this case, MDA-MB-468 cells expressing F2108L/M2327I mTOR mutations were generated. As expected, mTOR substrates were resistant to rapamycin, MLN0128 and to a combination of both treatments in the F2108L/M2327I double-mutant cells. Yet, the signaling of these double mutant cells remained as sensitive as the mTOR WT cells to RapaLink-1 treatment (Figure 4e, Extended Data Figure 7d).
Through exploitation of both the ATP and the FRB binding sites of mTOR, we have developed a new class of mTOR inhibitor which potently inhibits tumor growth and signaling in WT mTOR expressing cells as well as in cells that have acquired resistance to either rapalogs or ATP-competitive inhibitors or both. Such inhibitors have been developed for GPCRs22 (termed bitopic ligands) but have not been exploited in protein kinase inhibitor design. Interestingly, the only other bitopic kinase inhibitor we are aware of is the natural CDK2/cyclin-A inhibitor p27. The peptidic inhibitor spans the cyclin box and extends into the ATP site of CDK2, creating high affinity highly specific inhibitor23. Perhaps other allosteric sites near the ATP pocket on kinases could be similarly exploited such as the PIF pocket24 or even by bridging two adjacent ATP pockets in kinase complexes such as KSR-MEK25.
Methods
Cell culture and reagents
All cell lines were obtained from the American Type Culture Collection (ATCC). MCF-7 and MDA-MB-468 (ATCC catalog number: HTB-22 and HTB-132 respectively) breast cancer cell lines were maintained in a 1:1 mixture of DME: F12 medium supplemented with 4 mM glutamine, 100 units/mL each of penicillin and streptomycin, and 10% heat-inactivated fetal bovine serum (FBS) and incubated at 37°C in 5% CO2. The MDA-MB-468 inducible expression cells were maintained in the same medium with addition of 50 µg/ml hygromycin and 0.2 µg/mL puromycin. The HEK-293 cells (ATCC catalog number: CRL-1573) were maintained in DMEM medium with glutamine, antibiotics and 10% FBS. The cell lines resulted negative for mycoplasma contamination. AZD8055 was obtained from AstraZeneca Pharmaceuticals, rapamycin was purchased from EMD Bioscience. RAD001, KU006, WY354, PP242, MLN0128 were purchased from Tocris. Doxycycline was purchased from Sigma Aldrich. Puromycin and hygromycin stock solution were purchased from Invitrogen. Drugs were dissolved in DMSO to yield 10 mM stock and stored at −20°C.
Selection of drug resistant clones
Cell lines resistant to rapamycin (RR1 and RR2) and AZD8055 (TKi-R) were generated by exposing the parental breast cancer cell line MCF-7 to a high dose of drug (500 nM of either rapamycin or AZD8055) for 3 months of continuous drug exposure (change of media every 3 days); the cells were then sent to sequencing. MCF-7-RapaLink-1-resistant cells were generated by exposing MCF-7 cells to RapaLink-1 (10 nM) for 9 months of continuous drug exposure (change of media once per week); the cells were then sent to sequencing.
gDNA Sequencing
We profiled genomic alterations in 279 key cancer-associated genes using our IMPACT assay (Integrated Mutation Profiling of Actionable Cancer Targets), which utilizes solution phase hybridization-based exon capture and massively parallel DNA sequencing. Custom oligonucleotides were designed to capture all protein-coding exons and select introns of 279 commonly implicated oncogenes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies. We prepared barcoded sequence libraries (New England Biolabs, Kapa Biosystems) for DNA from the MCF-7 parental cell line and drug-resistant subclones, and we performed exon capture on barcoded pools by hybridization (Nimblegen SeqCap). 250 ng of genomic DNA was input for library construction. Libraries were pooled at equimolar concentrations (100 ng per library), combined with barcoded libraries from a separate project, and input to a single exon capture reaction as previously described26. DNA was subsequently sequenced on an Illumina HiSeq 2000 to generate paired-end 75-bp reads. We achieved a mean unique sequence coverage of 487X per sample.
Method for Construction of Docking Modeling
The coordinate of the crystal structure of rapamycin with FKBP12 and FRB domain was retrieved from the Protein Data Bank (accession code 1FAP). The coordinate of the crystal structure of ATP-competitive mTOR inhibitor (PP242) with mTORΔN and mLST8 was retrieved from the Protein Data Bank (accession code 4JT5). Two co-crystal structures were aligned and amino acids and water molecules in FRB domain of 1FAP were deleted. The coordinate of co-crystal structure unavailable ATP-competitive mTOR inhibitor (MLN0128) was manually constructed by modifying the coordinate of the co-crystal structure of its analog (PP242). The obtained modeling containing rapamycin and MLN0128 was energy-minimized using the MMFF94x force field in Molecular Operating Environment (MOE, described below)21 to provide a template structure. During the minimization procedure, the following conditions were adopted. The dielectric constant was set to 4 × r, where r is the interatomic distance. The residues, which are 9 Å away from compound, were fixed. And atomic charges for the protein and the compounds were set according to the AMBER99 and the AM1-BCC method, respectively. A crosslinker tethering rapamycin with an ATP-competitive mTOR inhibitor was manually constructed and energy-minimized using the MMFF94x force field in MOE to provide the initial conformation. The obtained initial conformation was subjected to conformation search using LowModeMD in MOE (Iterative limitation was set as 30). Values of potential energies (kcal/mol) of automatically created conformation(s) were averaged.
Code availability
MOE (version 2013.0801, Chemical Computing Group, Montreal, Canada), Scientific Vector Language (SVL) source.
Cell proliferation assay
The effect of the drug on cell proliferation was determined using a CellTiter-Glo Luminescent Cell Viability Assay kit (Promega, Madison, WI), which is based on quantification of the cellular ATP level. Cells were plated in 96-well plates at a density of 2,000–5,000 cells (8 replicates per condition). The following day, cells were treated with a range of drug concentrations prepared by serial dilution. After 3–5 days of treatment, 100 ml of prepared reagent was added to each well. The contents of the wells were mixed on a plate shaker for 1 hour, and then luminescence was measured by an Analyst AD (Molecular Devices, Sunnyvale, CA). The relative growth was normalized to the untreated samples in each group. The growth or inhibition curves and IC50 values were calculated with Graph Pad Prism v.6.
Immunoblot analysis
Cells were washed with PBS once, disrupted on ice for thirty minutes in NP-40 (50 mM Tris [pH 7.4], 1% NP-40, 150 mmol/L NaCl, 40 mmol/L NaF) or RIPA lysis buffer (Thermo Scientific) supplemented with protease and phosphatase inhibitors (Pierce Chemical) and cleared by centrifugation. Protein concentration was determined with BCA reagent from Pierce. Equal amounts of protein (10 to 50 µg) in cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes (GE healthcare), immunoblotted with specific primary and secondary antibodies and detected by chemiluminescence with the ECL detection reagents from Amersham Biosciences. Antibodies for P-AKT (S473) (#4060L), P-p70S6K (T389) (#9234L), P-S6 (S240/244) (#5364L) and P-S6 (S235/236) (#4858L), P-4EBP1 (T37/46) (#9459L), P-4EBP1 (S65) (#9451L), P-4EBP1 (T70) (#9455L), β-actin (#4970S), mTOR (#2972S) and Raptor (#2280S) were purchased from Cell Signaling Technology. The FLAG (#F1804) antibody was purchased from Sigma. The GST antibody (#sc-138) was from Santa Cruz.
Retrovirus based gene inducible expression cell system
The mTOR genes were sub-cloned into TTIGFP-MLUEX vector harboring tet-regulated promoter. Mutations were introduced by using the site-directed Mutagenesis Kit (Stratagene) as previously described27. The retrovirus encoded the rtTA3 or mTOR genes were packaged in Phoenix-AMPHO cells. The medium containing virus was filtered with 0.45 PVDF filters followed by incubation with the target cells for 6 hours. The cells were then cultured in virus-free medium for 2 days. The cells were selected with puromycin (2 µg/ml) or hygromycin (500 µg/ml) for 3 days. The positive infected cell populations were further sorted with transiently expressed GFP marker after being exposed to 1 µg/ml doxycycline and the sorted positive cells were cultured and expanded in medium without doxycycline but with antibiotics at a maintaining dose until the following assays.
Transient Transfections
Cells were seeded at 60 mm or 100 mm plates and transfected the following day using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The ratio between DNA and lipofectamine was 1 µg DNA to 3 µL lipofectamine.
In vitro FKBP12 binding assay
Cells expressing FLAG tagged WT or mutant mTOR were collected and lysed with 0.3% CHAPS buffer. The mTOR complexes were pulled down with anti-FLAG antibody conjugated agarose. Then, the beads bound complexes were incubated with recombinant FKBP12 (Fisher Scientific) (250 nM) or FKBP12 (250 nM) and rapamycin (250 nM) at 4°C for 30 minutes. After incubation, the beads were washed 5 times with CHAPS buffer. The protein complexes were eluted with 1X Laemmli Buffer and assayed by Western blotting.
Sequencing Sanger
The cDNA were generated by mRNA isolated from cell pellets with SV total RNA isolation kit, SV minipreps DNA purification kit and ImProm-II™ Reverse Transcription System kit from Promega Inc. The mTOR cDNA was amplified with the oligos listed in the table below. The PCR products were subjected to gel purification and sequenced by Genewiz, Inc.
Mutagenesis
All the mTOR mutants were generated by QuikChange II Site-Directed Mutagenesis Kit obtained from Agilent and confirmed by Sanger sequencing.
mTOR in vitro kinase assay
Active mTOR kinases were expressed in 293 cells and isolated by immunoprecipitation with anti-FLAG beads in 0.3% CHAPS buffer. The AKT recombinant protein was acquired from AstraZeneca Pharmaceuticals. The in vitro kinase assays was performed with 250 µM ATP at 37°C for 20 minutes.
In vitro kinase inhibition assay
Concentration-response curves with a concentration range of 1,000 to 0.97 nM and 2-fold serial dilution were constructed by dispensing a 100 µM DMSO solubilised stock of AZD8055 into white 384-well medium-binding microplates (Greiner Bio-One, UK) using an HP D3000 Digital Dispenser (HP, Palo Alto, CA, USA). The kinase reaction was performed as described above using 200 µM ATP, 1.5 µM peptide substrate and either 5 nM wild-type mTOR or 2 nM M2327I mutant mTOR. The IC50 values were calculated from initial rate data before being corrected for competition with ATP using the Cheng-Prussoff equation and assuming the compound is fully ATP competitive28. The IC50s were determined by fitting to a standard 4-parameter logistic using GraphPad Prism V.5 (GraphPad Software Inc., San Diego, CA).
Animal studies
All in vivo studies were conducted in accordance with guidelines approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee (IACUC). The maximal tumor volume permitted by MSKCC IACUC is 2,000 mm3; this limit was not exceeded in any of the experiments. Eight-week-old athymic nu/nu female mice (Harlan Laboratories) were injected subcutaneously with 10 million cells together with matrigel (BD Biosciences). 17β-estradiol pellets (0.72 mg/90 days release) (Innovative Research of America) were implanted subcutaneously three days before tumor cell inoculation. Once tumors reached an average volume of 100 mm3, mice were randomized (n= 5 mice per group) to receive rapamycin (10 mg/kg), AZD8055 (75 mg/kg), RapaLink-1 (1.5 mg/kg) or vehicle only as control. Sample size was chosen based on previous experiments. Rapamycin was formulated in DMSO and delivered i.p., AZD8055 was formulated in 30% captisol, and administered orally, RapaLink-1 was formulated in 6% DMSO and 30% captisol and delivered by i.p. Mice treated with RapaLink-1 were also given saline s.c. injections twice/day along with water supplemented with 5% glucose. Tumors were measured twice weekly using calipers, and tumor volume was calculated using the formula: length × width2 × 0.52. Samples were lysed and processed as previously published29.
Statistical Analysis
Results are mean values ± standard deviation. Investigators were not blinded when assessing the outcome of the in vivo experiments. All cellular experiments were repeated at least three times.
Extended Data
Extended Data Figure 1. Acquired-mTOR mutations promote resistance to mTOR inhibitors in MCF-7 cells.
a, The RNA from MCF-7 parental, RR1, RR2 and TKi-R cells was isolated and the RT-PCR products were submitted to Sanger sequencing at Genewiz, Inc. b, MCF-7 parental, RR1, RR2 and TKi-R cells were treated with either DMSO or 50 nM of RAD001 for 4 hours. Immunoblot analyses were performed on mTOR effectors. c, MCF-7 parental, RR1, RR2 and TKi-R cells were treated with either DMSO as a control or 500 nM of either KU006, WY354, PP242 mTOR inhibitors or with different doses of MLN0128 (d) for 4 hours. Immunoblot analyses were performed on mTOR effectors. All cellular experiments were repeated at least three times.
Extended Data Figure 2. Acquired-mTOR mutations promote resistance to mTOR inhibitors in MDA-MB-468 cells.
a, Dose-dependent cell growth inhibition of the MDA-MB-468 cells expressing GFP, WT mTOR or different mTOR variants (A2034V, F2108L and M2327I) upon rapamycin or AZD8055 treatment (b). Cells were pretreated for 24 hours with doxycycline (1 µg/mL) to induce the expression of exogenous mTOR. The cell growth was determined as described in Figure 1d. c, MDA-MB-468 cells expressing GFP, WT mTOR or different mTOR variants were treated with different concentration of rapamycin, AZD8055 (d) or with MLN0128 (e) for 4 hours. Immunoblot analyses were performed on mTOR effectors. All cellular experiments were repeated at least three times.
Extended Data Figure 3. Synthesis of the mTOR bivalent inhibitor RapaLink-1.
a, Compound design of RapaLink-1, -2, and -3 possessing a polyethylene glycol unit of varying lengths. b, Calculated potential energies U (kcal/mol) of modeled compounds of varying methylene (CH2)n linker lengths for bivalent interactions with the catalytic site and the FKBP12 site. c, A convergent synthetic route for a bivalent mTOR inhibitor RapaLink-1.
Extended Data Figure 4. RapaLink-1 requires FKBP12 for binding to mTOR FRB domain.
a, Dose-dependent cell growth inhibition curves of the MCF-7 parental cell line treated with rapamycin, MLN0128, combination of rapamycin and MLN0128 or RapaLink-1. The cell growth was determined as described in Figure 1d. b, mTOR-FLAG WT and variants were transfected into 293H cells. The mTORC1 complex was isolated, and an in vitro competition assay in the presence of FKBP12 was performed as described in Figure 2b. c, MCF-7 cells were treated with either DMSO, RapaLink-1 (10 nM), FK506 (10 µM) or combination of both for 24 hours at which time the cells were collected. Immunoblot analyses were performed on mTOR signaling. All experiments were repeated at least three times.
Extended Data Figure 5. RapaLink-1 is a potent mTOR inhibitor in WT and mutant mTOR cells.
a, MCF-7, (b) RR1, (c) RR2, (d) TKi-R cells were treated with different concentrations of rapamycin, MLN0128, combination treatment or RapaLink-1 over 3 days. The cell growth was determined as described in Figure 1d. Each dot and error bar on the curves represents mean ± standard deviation (n=8).
Extended Data Figure 6. RapaLink-1 has a prolonged intracellular half-life in WT mTOR cells.
a, MCF-7 F2039S cells were treated with different concentrations of rapamycin, MLN0128, combination treatment or RapaLink-1 for 4 hours at which time the cells were collected. Immunoblot analyses were performed on mTOR signaling. b, MCF-7 cells were treated for 4 hours with either DMSO control, 30 nM of rapamycin, 30 nM of MLN0128, combination of 30 nM of both or 30 nM of RapaLink-1 for 4 hours at which time the treatments were washed out 3 times with PBS and fresh media was readded for the indicated times. Immunoblot analyses were performed on mTOR effectors. c, MCF-7 cells were treated with 10 nM of RapaLink-1 and collected at the indicated times. Immunoblot analyses were performed as described above. All experiments were repeated at least three times. d, Mice bearing MCF-7 xenograft tumors were treated with one single dose of Vehicle or RapaLink-1 (1.5 mg/kg), tumors were collected at different days after treatment as indicated. Immunoblot analyses were performed on mTOR effectors. e, The weight of the mice treated in the efficacy study f is reported here. f, Mice bearing MCF-7 xenograft tumors were treated as described in Figure 4 c (n=5 for each group). The results were reported as % tumor volume ± standard deviation.
Extended Data Figure 7. RapaLink-1 is a more potent mTOR inhibitor than rapamycin.
a, MCF-7 cells were treated for 4 hours with either RapaLink-1 (10 nM) or rapamycin (10 nM) with simultaneous addition of increasing doses of either rapamycin or RapaLink-1 (left and right panels respectively). Immunoblot analyses were performed on mTOR effectors. b, Mice bearing RR1 or TKi-R (c) xenograft tumors were treated for 24 hours with a single dose of either vehicle, rapamycin (10 mg/kg), AZD8055 (75 mg/kg) or RapaLink-1 (1.5 mg/kg) (n=4 for each group). Immunoblot analyses were performed on mTOR effectors. d, MDA-MB-468 cells inducibly expressing mTOR WT were treated with either rapamycin, MLN0128, combination of rapamycin and MLN0128 or RapaLink-1 for 4 hours. Immunoblot analyses were performed on mTOR effectors with the indicated antibodies. Rapamycin and MLN0128 panels are the same shown for WT in Extended Data Figure 2c and e respectively.
Extended Data Table 1. mTOR mutations found in human patient samples.
List of some FRB and Kinase Domain mTOR mutations found in human patient samples. Data was collected from the cBio-portal, MSKCC.
| References | Histology | mTOR mutations | Domains | Treatments |
|---|---|---|---|---|
| TCGA-B0-4852-01 | ccRCC (TCGA) | E2033V | FRB | Baseline |
| Wagle et al. 2014 | Thyroid | F2108L | FRB | Everolimus |
| TCGA-DU-6393-01 | Glioma (TCGA) | M2327I | Kinase | Baseline |
| TCGA-A3-3347-01 | ccRCC (TCGA) | M2327I | Kinase | Baseline |
| P-00006559-T01-IM5 | Colorectal Cancer (MSKCC-IMPACT) |
M2327I | Kinase | Baseline |
| P-0000645-T01-IM3 | Bladder Urothelial Carcinoma (MSKCC-IMPACT) |
M2327I | Kinase | Baseline |
| ccRCC_28 | ccRCC (U Tokyo) | M2327I | Kinase | Baseline |
| P-0000614-T01-IM3 | Endometrial Cancer (MSKCC-IMPACT) |
M2327I | Kinase | Baseline |
Extended Data Table 2. List of FRB domain mutations found in human patient samples.
Data was collected from the cBioPortal, MSKCC.
| References | Cancer Study | Mutations |
|---|---|---|
| P-0004920-T01-IM5 | MSK-IMPACT | E2014K |
| SNU738_CENTRAL | CCLE | L2016R |
| BL41_HAEATOPOIE… | CCLE | I2017T |
| TCGA-KM-8441-01 | chRCC (TCGA) | I2017T |
| P-0003073-T01-IM5 | MSK-IMPACT | I2017S |
| TCGA-66-2787-01 | Lung squ (TCGA) | W2023G |
| BCP1_HAEMATOPOIE… | CCLE | G2030V |
| TCGA-B0-4852-01 | ccRCC (TCGA) | E2033V |
| TCGA-B5-A11E-01 | Uterine (TCGA) | A2056V |
| TCGA-G3-A25U-01 | Liver (TCGA) | M2057I |
| TCGA-AZ-6598-01 | Colorectal (TCGA) | Q2063fs |
| H072999 | Liver (AMC) | K2066R |
| NCIH 1792_LUNG | CCLE | Q2072 |
| P-0000112-T01-IM3 | MSK-IMPACT | Q2072R |
| S05-31806-TP-NT | CSCC (Dana-Farber) | X2073_splice |
| P-0005266-T01-IM5 | MSK-IMPACT | R2076L |
| TCGA-EE-A2GC-06 | Melanoma (TCGA) | D2077E |
| P-0002050-T01-IM3 | MSK-IMPACT | W2084C |
| TCGA-AX-A0J0 | Uterine (TCGA) | Y2088D |
| TCGA-CQ-5327 | Head & neck (TCGA) | M2089I |
| P-0005159-T01-IM5 | MSK-IMPACT | L2097F |
| TCGA-D8-A1Y0-01 | Breast (TCGA) | W2101L |
Extended Data Table 3. List of mTOR kinase domain mutations found in human patient samples.
Data was collected from the cBioPortal, MSKCC.
| References | Cancer Study | Mutations |
|---|---|---|
| TCGA-C5-A1BM-01 | Cervical (TCGA) | D2191H |
| TCGA-BR-4370-01 | Stomach (TCGA) | R2193C |
| P-0004376-T01-IM5 | MSK-IMPACT | F2202L |
| TCGA-BT-A0YX-01 | Bladder (TCGA) | L2209V |
| TCGA-B0-4810-01 | ccRCC (TCGA) | A2210P |
| JHUEM7_ENDOMETRIUM | CCLE | S2215Y |
| TCGA-JW-A5VL-01 | Cervical (TCGA) | S2215Y |
| TCGA-FU-A3HZ-01 | Cervical (TCGA) | S2215Y |
| TCGA-AA-A00K-01 | Colorectal (TCGA) | S2215Y |
| TCGA-A6-6141-01 | Colorectal (TCGA) | S2215Y |
| TCGA-F4-6806-01 | Colorectal (TCGA) | S2215F |
| TCGA-DM-A1D4 | Colorectal (TCGA) | S2215F |
| S12-23181-TP-NT | CSCC (Dana-Farber) | S2215F |
| TCGA-CJ-5679-01 | ccRCC (TCGA) | S2215Y |
| TCGA-A4-7828-01 | pRCC (TCGA) | S2215Y |
| P-0000208-T01-IM3 | MSK-IMPACT | S2215Y |
| P-0000208-T02-IM5 | MSK-IMPACT | S2215Y |
| P-0005214-T01-IM5 | MSK-IMPACT | S2215Y |
| MEL-UKRV-Mel-20 | Melanoma (Broad) | S2215Y |
| TCGA-BS-A0UF-01 | Uterine (TCGA) | S2215Y |
| TCGA-BS-A0UV-01 | Uterine (TCGA) | S2215Y |
| TCGA-BG-A0VX | Uterine (TCGA) | S2215Y |
| TCGA-CS-5396-01 | Glioma (TCGA) | L2216P |
| HEC251_ENDOMETRIUM | CCLE | R2217W |
| TCGA-A6-4105-01 | Colorectal (TCGA) | Q2223K |
| TCGA-CJ-4887-01 | ccRCC (TCGA) | L2230V |
| SNU1196_BILIARY | CCLE | S2231W |
| KMH2_HAEMATOPOIE | CCLE | T2232I |
| CW2_LARGE_INTEST | CCLE | G2238D |
| TCGA-JW-A5VL-01 | Cervical (TCGA) | W2239C |
| P-0000614-T01-IM3 | MSK-IMPACT | W2239Ffs*39 |
| DS-bla-084 | Bladder (MSKCC 2014) | P2241S |
| MSKCC-0296_R | Bladder (DFARBER_MSKCC 2014) | P2241S |
| TCGA-DK-A6B6-01 | Bladder (TCGA) | P2241S |
| TCGA-JW-A5VL-01 | Cervical (TCGA) | P2241S |
| TCGA-97-7554-01 | Lung adeno (TCGA) | R2251Q |
| HEC1A_ENDOMETRIUM | CCLE | R2254M |
| SNU520_STOMACH | CCLE | H2265P |
| TCGA-75-5126-01 | Lung adeno (TCGA) | R2266P |
| TCGA-34-5239-01 | Lung squ (TCGA) | A2272S |
| TCGA-EB-A3Y7-01 | Melanoma (TCGA) | A2272V |
| P-0005309-T01-IM5 | MSK-IMPACT | Q2282P |
| TCGA-BR-8363-01 | Stomach (TCGA) | T2294A |
| P-0001171-T01-IM3 | MSK-IMPACT | D2298H |
| DS-bla-037 | Bladder (MSKCC 2014) | E2311K |
| P-0001042-T01-IM3 | MSK IMPACT | A2325V |
| P-0003529-T01-IM5 | MSK-IMPACT | V2330I |
| TCGA-AA-3666-01 | Colorectal (TCGA) | I2333M |
| TCGA-B0-5691-01 | Renal clear cell (TCGA) | L2334V |
| TCGA-HU-A4GU-01 | Stomach (TCGA) | R2339_splice |
| L540_HAEMATOPOIE | CCLE | H2340R |
| TCGA-HB-A43Z-01 | Sarcoma (TCGA) | S2342Y |
| NCIH446_LUNG | CCLE | M2345V |
| ESCC-148T | Esophagus sq (ICGC) | G2351E |
| LUAD-FH5PJ | Lung adeno (BROAD) | H2355R |
| S12-23181-TP-NT | CSCC (Dana-Farber) | G2359R |
| TCGA-E9-A54Y-01 | Breast (TCGA) | R2368Q |
| HEC108-ENDOMETRIUM | CCLE (Novartis/Broad 2012) | R2368Q |
| P-0002079-T01-IM3 | MSK-IMPACT | L2383F |
| P-0001042-T01-IM3 | MSK-IMPACT | T2384I |
| TCGA-BH-A0HP-01 | Breast (TCGA) | E2388Q |
| SW48_LARGE_INTES | CCLE | V2389_splice |
| COSM51966 | Kidney | V2406A |
| TCGA-A7-A5ZV-01 | Breast (TCGA) | D2412H |
| H061142 | Liver (AMC) | D2412V |
| TCGA-EK-A3GK-01 | Cervical (TCGA) | E2419K |
| CLL147 | CLL (BROAD) | A2420P |
| P-0003254-T01-IM5 | MSK-IMPACT | A2420V |
| P-0004798-T01-IM5 | MSK-IMPACT | A2420V |
| TCGA-FS-A4FC-06 | Melanoma (TCGA) | A2420V |
| TCGA-ER-A42L-06 | Melanoma (TCGA) | D2424N |
| ST486-HAEMATOPOI | CCLE | L2427R |
| TCGA-06-0122-01 | GBM (TCGA) | L2427Q |
| TCGA-KN-8437-01 | chRCC (TCGA) | L2427R |
| P-0005355-T01-IM5 | MSK-IMPACT | L2427R |
| P-0005044-T01-IM5 | MSK-IMPACT | L2427Q |
| P-0000839-T01-IM3 | MSK-IMPACT | L2427Q |
| MOLTI6_HAEMATOP | CCLE | R2430M |
| TCGA-G4-6322-01 | Colorectal (TCGA) | R2441Q |
Supplementary Material
Acknowledgments
N.R. would like to thank the National Institutes of Health (NIH) (P01 CA094060) for funding as well as the Breast Cancer Research Foundation Grant and the National Cancer Institute Cancer Center Support Grant P30 CA008748, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center, and the Team up for a Cure Fund. K.M.S would like to thank the NIH P50 AA017072 and the Stand Up 2 Cancer Lung Cancer Dream Team and the Howard Hughes Medical Institute for funding. We would like to thank Radha Mukherjee, Sarit Schwartz, Jack Taunton and Bryan Roth for helpful comments.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
V.R.O, M.O., Y.Z., C.N., N.R, and K.M.S. conceived the project, designed and analyzed the experiments, and wrote the manuscript. V.R.O, M.O., Y.Z., C.N., C.M., A.B., W.W. D.B., S.C. T.K. performed and supervised the laboratory experiments. H.W. and M.B. performed and supervised the IMPACT sequencing and analysis. E.D. designed and supervised the in vivo experiments.
Author Information
Reprints and permissions information is available at www.nature.com/reprints.
K.M.S. is an inventor on patents related to MLN0128 held by the University of California, San Francisco (UCSF), and sublicensed to Takeda Pharmaceuticals. NR and K.M.S. are consultants and M.O. is an employee at Takeda Pharmaceuticals Company Limited, which is conducting MLN0128 clinical trials. C.M.W., D.G.B, S.C., T.K. are employees at AstraZeneca which are conducting AZD2014 (mTOR kinase inhibitor) trials. K.M.S. and M.O. are inventors on a patent application held by (UCSF) and licensed to Kura Oncology. K.M.S. is a shareholder in Kura Oncology, K.M.S. and N.R. are consultants to Kura Oncology.
References
- 1.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 2.Basu B, et al. First-in-Human Pharmacokinetic and Pharmacodynamic Study of the Dual m-TORC 1/2 Inhibitor AZD2014. Clin Cancer Res. 2015;21:3412–3419. doi: 10.1158/1078-0432.CCR-14-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer G, et al. Genome Sequencing Identifies a Basis for Everolimus Sensitivity. Science. 2012;338:221–221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagle N, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagle N, et al. Response and Acquired Resistance to Everolimus in Anaplastic Thyroid Cancer. N Engl J Med. 2014;371:1426–1433. doi: 10.1056/NEJMoa1403352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowling RJO, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown EJ, et al. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara K, et al. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz MC, Heitman J. Tor Mutations Confer Rapamycin Resistance by Preventing Interaction with Fkbp12-Rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, et al. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grabiner BC, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molecular Operating Environment (MOE) [Google Scholar]
- 19.Szczepankiewicz BG, et al. Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Using a Linked-Fragment Strategy. J Am Chem Soc. 2003;125:4087–4096. doi: 10.1021/ja0296733. [DOI] [PubMed] [Google Scholar]
- 20.Marinec PS, Chen L, Barr KJ. FK506-binding protein (FKBP) partitions a modified HIV protease inhibitor into blood cells and prolongs its lifetime in vivo. Proc Natl Acad Sci USA. 2009;106:1336–1341. doi: 10.1073/pnas.0805375106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel MR, Hamilton E, LoRusso PM, Gluck WL, Jones SF, Kittaneh M, et al. A phase I study evaluating continuous and intermittent AZD2014 in combination with fulvestrant in patients with ER+ advanced metastatic breast cancer (abstract). Proceedings of the AACR 106th Annual Meeting; 2015 April 18–22; Philadelphia, PA: AACR; 2015. Abstract nr CT233.25. [Google Scholar]
- 22.Valant C, Robert Lane J, Sexton PM, Christopoulos A. The Best of Both Worlds? Bitopic Orthosteric/Allosteric Ligands of G Protein–Coupled Receptors. Annu Rev Pharmacol Toxicol. 2012;52:153–178. doi: 10.1146/annurev-pharmtox-010611-134514. [DOI] [PubMed] [Google Scholar]
- 23.Russo AA, Jeffrey PD, Patten AK, Massagué J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 24.Wei L, et al. Design and synthesis of benzoazepin-2-one analogs as allosteric binders targeting the PIF pocket of PDK1. Bioorg Med Chem Lett. 2010;20:3897–3902. doi: 10.1016/j.bmcl.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Brennan DF, et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 26.Juric D, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Z, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell. 2015;28:370–383. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng AC, Eksterowicz J, Geuns-Meyer S, Sun Y. Analysis of Kinase Inhibitor Selectivity using a Thermodynamics-Based Partition Index. J Med Chem. 2010;53:4502–4510. doi: 10.1021/jm100301x. [DOI] [PubMed] [Google Scholar]
- 29.Rodrik-Outmezguine VS, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.