Abstract
Magnetic resonance images of the spinal cord play an important role in studying neurological diseases, particularly multiple sclerosis, where spinal cord atrophy can provide a measure of disease progression and disability. Current practices involve segmenting the spinal cord manually, which can be an inconsistent and time-consuming process. We present an automatic segmentation method for the spinal cord using a combination of deformable atlas based registration and topology preserving classification. Using real MR data, our method is shown to be highly accurate when compared to segmentations by manual raters. In addition, our results always maintain the correct topology of the spinal cord, therefore providing segmentations more consistent with the known anatomy.
Index Terms: Topology-preserving segmentation, digital homeomorphism, spinal cord segmentation, Magnetic resonance imaging, Magnetization transfer images
1. INTRODUCTION
The analysis of spinal cord magnetic resonance (MR) images is used for studying a number of neurological diseases [1], particularly multiple sclerosis, where spinal cord atrophy has been shown to correlate with disability [2] and as a measure for assessing the effects of potential neuroprotective therapies [3]. Recent work has shown correlations between sensorimotor dysfunction and magnetization transfer (MT) signals in dorsal and lateral white matter columns in spinal cord MT-weighted images [4]. Current analysis, however, is generally performed using manual segmentations of the images. The relatively small size of the spinal cord causes manual segmentations to be time-consuming, making it difficult and costly to analyze large quantities of data. In addition, manual raters are often inconsistent, may carry bias, and require sufficient training.
Quite a number of methods have been proposed for automatic segmentation of the spine in CT images [5–7], and semi-automatic segmentation of the spinal cord in MR images [8–11]. However, only a few attempts have been made to design a fully automatic segmentation method. Nieniewski [12] proposed a morphological watershed approach, and more recently, Koh [13] presented a method specifically for T2-weighted images of the lumbar region using gradient vector flow fields. Both of these methods, however, requires a manually or heuristically chosen starting point to initialize the algorithm. In addition, the methods were only tested on T2-weighted images, and may not necessarily be adaptable for use with other modalities with lower contrast, such as MT-weight images.
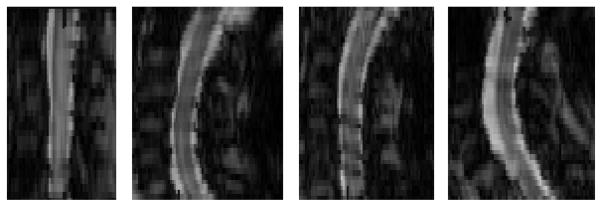
There are several difficulties associated with automatic segmentation of the spinal cord in MR images. Its inherent non-rigidness causes the shape and curvature of the spinal cord in the image to be highly dependent on the subjects and their position within the scanner. This makes atlas-based registration approaches for segmentation difficult, since the accuracy of the registration is diminished if the image being segmented differs too greatly from the atlas. This is particularly true for methods that use rigid or affine registration to construct statistical atlases from manual segmentation, since the probabilities cannot be calculated properly with the various possible curvatures of the spinal cord. Fig. 1 shows several examples of this variability.
Fig. 1.
Sagittal views of axially acquired magnetization transfer MR images of the spinal cord from four different subjects at approximately the same field of view. This demonstrates the wide variability in the shape and curvature of the spinal cord in the images.
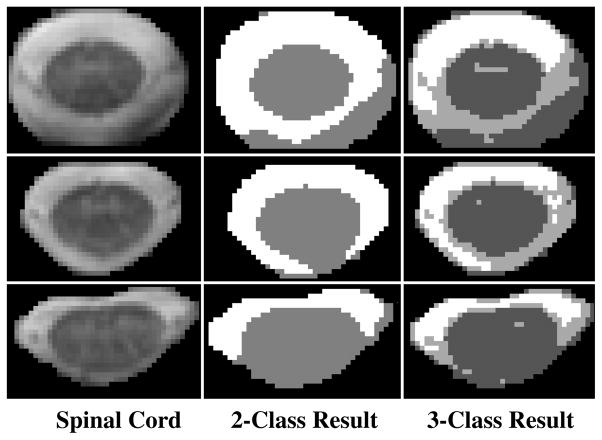
Additional problems for segmentation include the presence of nerve roots, partial voluming with the dura mater, limited resolution and possible influences on the image from the vertebral bodies. These issues all contribute to difficulties when using existing segmentation tools that rely on correctly classifying the tissue intensities. Fig. 2 shows an example of results from a standard tissue classification method [14] that is capable of accounting for gain field inhomogeneity. To demonstrate the difficulty, the intensity classification was performed only on the spinal cord and cerebrospinal fluid (CSF) with all other surrounding tissues and structures in the image removed. We can see that even without the influence from the surrounding tissues, the results were still poor in many areas.
Fig. 2.
Two and three-class segmentations using a standard tissue classification tool [14] on several axial slices of magnetization transfer weighted MR images. The spinal cord is the dark structure at the center of the image and the CSF is the light structure surrounding the spinal cord.
To address these problems, we present a fully automatic intensity based method for segmentation of the spinal cord (and surrounding CSF) that uses a combination of an atlas-based deformable registration approach with a topology preserving classification technique.
2. METHODS
2.1. Overview
Our method for spinal cord segmentation involves two primary steps. First we use a deformable registration technique to align an intensity atlas to the target image to be segmented. The mapping found from this alignment is then applied to a topology template and statistical atlas associated with the intensity atlas. This provides the initialization for the second step, which uses a topology preserving classification approach, similar to that presented in [15] for brain segmentation. This technique iteratively estimates the tissue intensity classification and uses it to evolve the topology template using homeomorphic thinning and growing. With each iteration, the topology template is modified to be closer (and eventually converging) to the segmentation of the spinal cord.
The primary advantage of using a topology constraint derives from the well defined topology of the spinal cord and CSF. From the anatomy, we know that the spinal cord and CSF together have a cylindrical topology, where the CSF completely surrounds the spinal cord. By maintaining this constraint, the algorithm prevents misclassification that might break this topology, such as the examples shown in Fig. 2, where areas exterior to the CSF were misclassified as spinal cord.
2.2. Image Acquisition
The MR data used in our experiments were of the cervical spinal cord, acquired from the C2–C6 vertebral body levels. The scans were performed with the same 3-Tesla Intera scanner (Philips Medical Systems, Best, The Netherlands) using body coil excitation and two-element phased array surface coil reception. Each image was composed of 2.25mm slices with nominal in-plane resolution of 0.6 x 0.6mm. The images were MT-weighted, using a MT prepulse applied 1.5 kHz off resonance (24 ms, five-lobed sincgauss pulse with maximum amplitude 9.5 mT), as described in [16]. Other parameters: repetition time 110 ms, echo time 13 ms, flip angle 9, echo planar imaging factor 3 and SENSE acceleration factor 2.
2.3. Atlas Alignment using Deformable Registration
Deformable registration is used to initially align an intensity atlas to the target image. This is performed using an adaptation [17] of the ABA registration algorithm presented in [18], which models a deformation field, v(x), using a summation of radial basis functions (RBFs), Φ(x):
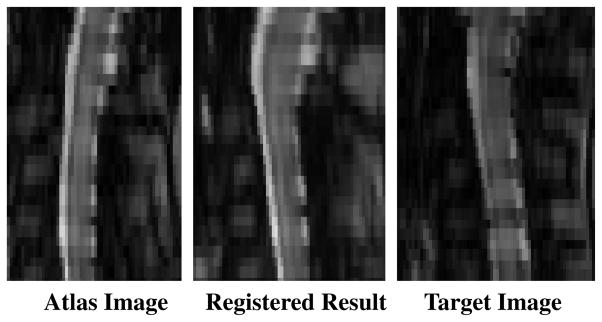
where ci are the center locations of the RBFs and wi are the coefficients being optimized. The algorithm attempts to maximize the normalized mutual information (NMI) [19] between two images, which has been shown to be a robust variant of the mutual information similarity metric commonly used for registration of MR images [20]. Fig. 3 shows an example of the result from this initial atlas registration.
Fig. 3.
Sagittal slices showing an example of the results achieved from the initial registration of the intensity atlas to the target image.
Once the deformation field is found between the intensity atlas and the target image, the mapping is directly applied to the statistical atlas. However, the deformation may not necessarily maintain the topology of the template. To address this, a homeomorphic approximation of the deformation field is constructed using the iterative method presented in [21]. This allows the template to be transformed without changing its initial topology.
2.4. Atlases Construction
For the purpose of our experiments, a subject was randomly chosen from the data to serve as the intensity atlas and its manual segmentation was used as the topology template. Four other subjects were then deformably registered to the intensity atlas using the same method described in Section 2.3. The corresponding mappings were applied to the manual segmentation for these subjects. The four transformed manual segmentations along with the original segmentation for the intensity atlas were then used to create the statistical atlas by calculating the probability of the correct segmentation at each voxel in the image. The resulting probabilities were Gaussian-smoothed to reduce discrete drop-offs in the atlas.
2.5. Homeomorphic Thinning and Growing
The final step in our method follows very closely the homeomorphic thinning and growing technique presented in [15] for brain segmentation, which describes the following in much greater detail. The approach first uses the initialization of the topology template to estimate the membership of each voxel, which represents the likelihood that the intensity at the voxel belongs to a particular tissue class. Then each structures in the template is thinned using a fast marching algorithm that removes the voxels with the lowest membership from the structure. After the structures are thinned, voxels are grown back by expanding the structure towards the voxels with the highest membership and prior probabilities from the statistical atlas.
Both the thinning and growing process are done while maintaining a digital homeomorphism criterion [15], which checks that the changes to the template do not affect its topology. This guarantees that the resulting segmentation always has the same topology as the starting template. Following the thinning and growing step, the memberships are recalculated since the template has changed to better represent the segmentation. This process is repeated until convergence, at which point the template represents a good approximation of the true segmentation of the spinal cord.
3. RESULTS
Twenty MT-weighted MR images of the cervical spinal cord were used to evaluate our method. Each image had a corresponding segmentation performed by a manual rater, and four of the images had repeated segmentations by two different raters. The automatic results from our method were compared with the manual segmentations by calculating their Dice overlap: , which is the volume of the overlap divided by the average of the volume of each segmentation, with a value of one indicating perfect overlap.
Table 1 shows the average and standard deviation of the Dice overlap of the spinal cord and CSF when using just the atlas registration, when using our method, and when comparing between two manual raters. We can see that for both structures, our method dramatically improved the results from using just the atlas registration, and is overall very comparable to the overlap achieved between two manual raters.
Table 1.
Mean and standard deviation(in parentheses) of Dice overlap compared to manual segmentations.
| Method | CSF | Spinal Cord | Spinal Cord + CSF |
|---|---|---|---|
|
| |||
| Atlas Registration | 0.711 (0.070) | 0.821 (0.065) | 0.732 (0.064) |
| Topology Preserving Seg. | 0.843 (0.034) | 0.904 (0.035) | 0.866 (0.029) |
| 2nd Manual Rater | 0.884 (0.008) | 0.937 (0.011) | 0.903 (0.003) |
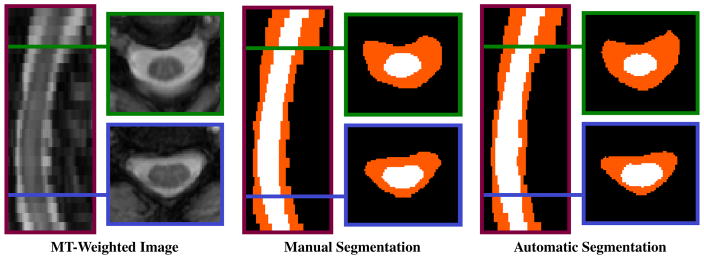
Qualitative assessment was also performed by visually inspecting the automatic results in regions that were identified as difficult to segment during the manual segmentation. The results for these regions (examples shown in Fig. 4) were found to be of roughly the same quality as the manual segmentations.
Fig. 4.
Axial and sagittal views of an MT-weighted image and its segmentation produced manually and automatically.
4. DISCUSSION AND CONCLUSION
We have introduced a fully automatic algorithm for segmentation of the spinal cord in magnetic resonance images. Our results showed a high level of accuracy compared to segmentations performed by manual raters.
Our method applies the concepts of homeomorphic segmentation presented in [15] to the segmentation of the spinal cord by using deformable registration to accommodate for the shape and curvature variabilities in the spinal cord when aligning and constructing the necessary atlases. This assures that the results from our method will always have the correct topology of the spinal cord, therefore providing segmentations more consistent with the known anatomy.
Although only MT images were used in our experiments, our method is not dependent on any specific modality. By simply switching the intensity atlas, the method can be easily applied to any MR imaging modality where there is sufficient contrast in the spinal cord.
Future work will focus on segmentation of finer structures in the spinal cord, such as the gray matter and the individual columns of the white matter.
Acknowledgments
This work was supported by the Intramural Research Program of NINDS. Data acquisition was supported by the National Multiple Sclerosis Society under a Tissue Repair grant to Peter Calabresi (Johns Hopkins University).
Contributor Information
Min Chen, Email: mchen55@jhu.edu.
Aaron Carass, Email: aaron_carass@jhu.edu.
Pierre-Louis Bazin, Email: pbazin1@jhmi.edu.
Daniel S. Reich, Email: daniel.reich@nih.gov.
Jerry L. Prince, Email: prince@jhu.edu.
References
- 1.Doi K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Computerized Medical Imaging and Graphics. 2007;31:198–211. doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin X, Tench CR, Evangelou N, Jaspan T, Constantinesc CS. Measurement of spinal cord atrophy in multiple sclerosis. Journal of NeuroImaging. 2004;14(3 Supple):20S–26S. doi: 10.1177/1051228404266265. [DOI] [PubMed] [Google Scholar]
- 3.Kalkers NF, Barkhof F, Bergers E, van Schijndel R, Polman CH. The effect of the neuroprotective agent riluzole on mri parameters in primary progressive multiple sclerosis: a pilot study. Multiple Sclerosis. 2002;8(6):532–533. doi: 10.1191/1352458502ms849xx. [DOI] [PubMed] [Google Scholar]
- 4.Zackowski KM, Smith SA, Reich DS, et al. Sensorimotor dysfunction in multiple sclerosis and column-specific magnetization transferimaging abnormalities in the spinal cord. Brain. 2009;132:1200–1298. doi: 10.1093/brain/awp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archip NA, Erard PJ, Egmont-Petersen M, Haefliger JM, Germond JF. A knowledge-based approach to automatic detection of the spinal cord in ct images. IEEE Trans Med Imag. 2002;21:1504–1516. doi: 10.1109/TMI.2002.806578. [DOI] [PubMed] [Google Scholar]
- 6.Karangelis G, Zimeras S. An accurate 3d segmentation method of the spinal canal applied to ct data. Proc Bildverarbeitung fr die Medizin. 2002 [Google Scholar]
- 7.Burnett SSC, Starkschalla G, Stevens CW, Liao Z. A deformable-model approach to semi-automatic segmentation of ct images demonstrated by application to the spinal canal. Medical Physics. 2004;31:251–263. doi: 10.1118/1.1634483. [DOI] [PubMed] [Google Scholar]
- 8.Coulon O, Hickman SJ, Parker GJ, Barker GJ, Miller DH, Arridge SR. Quantification of spinal cord atrophy from magnetic resonance images via a b-spline active surface model. Magnetic Resonance in Medicine. 2002;47:1176–1185. doi: 10.1002/mrm.10162. [DOI] [PubMed] [Google Scholar]
- 9.Uitert RV, Bitter I, Butma JA. Semi-automatic spinal cord segmentation and quantification. CARS 2005: Computer Assisted Radiology and Surgery. 2005:224–229. [Google Scholar]
- 10.McIntosh C, Hamarneh G. Spinal crawlers: Deformable organisms for spinal cord segmentation and analysis. Medical Image Computering and ComputerAssisted Intervention (MICCAI ‘06) 2006:808–815. doi: 10.1007/11866565_99. [DOI] [PubMed] [Google Scholar]
- 11.Horsfield MA, Salab S, Neemac M, Absintab M, Bakshic A, Sormanid MP, Roccab MA, Bakshic R, Filippib M. Rapid semi-automatic segmentation of the spinal cord from magnetic resonance images: Application in multiple sclerosis. NeuroImage. 2010;50:446–455. doi: 10.1016/j.neuroimage.2009.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieniewski M, Serneels R. Segmentation of spinal cord images by means of watershed and region merging together with inhomogeneity correction. Machine Graphics and Vision International Journal. 2002;11:101–121. [Google Scholar]
- 13.Koh T, Kim T, Chaudhary V, Dhillon G. Automatic segmentation of the spinal cord and the dural sac in lumbar mr images using gradient vector flow field. International Conference of the IEEE Engineering in Medicine and Biology Society; 2010. [DOI] [PubMed] [Google Scholar]
- 14.Pham DL, Prince JL. Adaptive fuzzy segmentation of magnetic resonance images. IEEE Trans Med Imag. 1999;18(9):737–752. doi: 10.1109/42.802752. [DOI] [PubMed] [Google Scholar]
- 15.Bazin PL, Pham DL. Homeomorphic brain image segmentation with topological and statistical atlases. Medical Image Analysis. 2008;12:616–625. doi: 10.1016/j.media.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SA, Jones CK, Gifford A, et al. Reproducibility of tract-specific magnetization transfer and diffusion tensor imaging in the cervical spinal cord at 3 tesla. NMR in Biomedicine. 2009;23(2):207–217. doi: 10.1002/nbm.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, Carass A, Wheeler B, Liu X, Prince JL. Multi-channel enhancement of the adaptive bases algorithm,” in. 16th Annual Meeting of the Organization for Human Brain Mapping; 2010. [Google Scholar]
- 18.Rohde GK, Aldroubi A, Dawant BM. The Adaptive Bases Algorithm for intensity based nonrigid Image Registration. IEEE Trans Med Imag. 2003;22(11):1470–1479. doi: 10.1109/TMI.2003.819299. [DOI] [PubMed] [Google Scholar]
- 19.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32(1):71–86. [Google Scholar]
- 20.Wells WM, III, Viola P, Atsumi H, Nakajima S, Kikinis R. Multimodal volume registration by maximation of mutual information. Medical Image Analysis. 1996;1(1):35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 21.Bazin PL, Ellingsen LM, Pham DL. Digital homeomorphisms in deformable registration. Proceedings of the International Conference on Information Processing in Medical Imaging (IPMI07); 2007. [DOI] [PubMed] [Google Scholar]