Summary
SnoN regulates multiple signaling pathways including TGFβ/Smad and p53 and displays both pro-oncogenic and anti-oncogenic activities in human cancer. We have observed previously that both its intracellular localization and expression levels are sensitive to cell density, suggesting that it may crosstalk to Hippo signaling. Here we report that indeed SnoN interacts with multiple components of the Hippo pathway to inhibit the binding of Lats2 to TAZ and the subsequent phosphorylation of TAZ, leading to TAZ stabilization. Consistently, SnoN enhances the transcriptional and oncogenic activities of TAZ, and reducing SnoN decreases TAZ expression as well as malignant progression of breast cancer cells. Interestingly, SnoN itself is downregulated by Lats2 that is activated by the Scribble basolateral polarity protein. Thus, SnoN is a critical component of the Hippo regulatory network that receives signals from the tissue architecture and polarity to coordinate the activity of intracellular signaling pathways.
Keywords: SnoN, Hippo, TAZ, Scribble, cell polarity, breast cancer, EMT
Introduction
SnoN was discovered as a proto-oncoprotein based on its close homology to v-Ski, the transforming protein of the Sloan–Kettering virus and its cellular homolog c-Ski (Givol et al., 1995, Nomura et al., 1989, Boyer et al., 1993, Pearson-White, 1993, Pearson-White and Crittenden, 1997). In mammals, SnoN is expressed ubiquitously in embryos and adult tissues (Deheuninck and Luo, 2009, Jahchan and Luo, 2010, Zhu and Luo, 2012). It is essential for embryonic angiogenesis and regulates mammary gland alveologenesis and onset of lactation in adult animals (Zhu et al., 2013, Jahchan et al., 2012). Deregulation of SnoN activity results in premature ageing and cellular transformation (Pan et al., 2012, Pan et al., 2009). SnoN possesses both pro-oncogenic and anti-oncogenic activities in mammalian tumorigenesis. Supporting its role as an oncogene, the human snoN gene is located within the 3q26 amplicon that is frequently amplified in many cancer types, and its expression is elevated in many human cancer cell lines and some tissues (Jahchan and Luo, 2010, Deheuninck and Luo, 2009, Zhu and Luo, 2012). In agreement with this, overexpression of SnoN in the mouse mammary gland accelerates the formation of aggressive multi-focal adenocarcinomas and pulmonary metastasis induced by the Polyoma middle T antigen (Jahchan et al., 2010). Interestingly, while reducing SnoN expression by shRNA in human lung or breast cancer cells inhibits tumor growth both in vitro and in vivo, it enhances epithelial to mesenchymal transition (EMT) of lung and breast cancer cells in vitro and tumor metastasis in vivo, indicating that SnoN may also possess an anti-oncogenic activity (Zhu et al., 2007). Indeed, in some human cancer tissues, SnoN expression is downregulated as cancer becomes more malignant (Jahchan et al., 2013). Consistent with these, overexpression of SnoN inhibits oncogene-induced transformation of primary mouse embryonic fibroblasts (MEF) by activating a p53- and PML-dependent premature senescence pathway, and mice expressing a high level of SnoN are resistant to papilloma development in a two-step skin carcinogenesis model and display senescence in vivo (Pan et al., 2009).
SnoN has been shown to regulate the activity of several important signaling pathways. SnoN interacts with Smad2, Smad3 and Smad4 and represses their ability to activate TGFβ responsive genes through disrupting the functional heteromeric Smad complexes, recruiting a transcription co-repressor complex and blocking the binding of transcriptional co-activators to the Smads (Stroschein et al., 1999, Sun, 1999, Wu et al., 2002, He et al., 2003). SnoN has also been reported to interact with and activate p53 in the PML nuclear bodies in response to various cellular stress signals, leading to premature senescence and inhibition of tumorigenesis (Pan et al., 2009, Pan et al., 2012). In addition, SnoN stabilizes STAT5 in the mammary gland to enhance prolactin signaling, enabling onset of lactation (Jahchan et al., 2012) and promotes BMP9/10-induced activation of Smad1/5 through facilitating ALK1 activation in endothelial cells (Zhu et al., 2013). Of these activities, the ability of SnoN to antagonize the growth inhibitory pathway of TGFβ/Smad signaling may account for its pro-oncogenic activity and its ability to promote p53 activation may contribute to its anti-oncogenic activity (Zhu and Luo, 2012, Jahchan and Luo, 2010).
The expression and localization of SnoN in mammalian cells are tightly controlled and can be altered during malignant progression (Krakowski et al., 2005, Jahchan et al., 2013, Villanacci et al., 2008, Chia et al., 2006). SnoN expression is upregulated at the level of transcriptional activation in response to a variety of stimuli including TGFβ and cellular stress signals, and downregulated via the ubiquitin- and proteasome-dependent degradation pathway through the action of several E3 ligases including anaphase promoting complex, Smurf2 and Arkadia (Jahchan et al., 2012, Deheuninck and Luo, 2009, Luo, 2004, Zhu and Luo, 2012)Although originally identified as a nuclear protein, SnoN is later found to localize predominantly in the cytoplasm in non-malignant epithelial tissues and cells, but become nuclear in highly malignant tissues (Krakowski et al., 2005). During the investigation of mechanisms underlying the alteration of SnoN localization, we discovered that cell density played a critical role in this regulation, suggesting the possible involvement of the Hippo pathway.
The Hippo pathway is an evolutionarily conserved signaling pathway that senses cell density, polarity and adhesion information as well as a variety of signals from the extracellular environment to regulate cell proliferation and growth, survival, differentiation and stem cell self-renewal (Yu and Guan, 2013, Pan, 2010, Schroeder and Halder, 2012). In mammals, the Hippo core kinase cassette consists of two serine/threonine kinases Mst1 or Mst2 and Lats1 or Lats2 and two adapter proteins Sav and Mob. In response to upstream signals, Mst complexes with Sav to phosphorylate and activate Lats, which in association with Mob, phosphorylates the key transcription factors TAZ and YAP (Zhao et al., 2007, Ota and Sasaki, 2008, Hao et al., 2008, Dong et al., 2007, Lei et al., 2008). TAZ and YAP are transcription factors that when recruited to target promoters through binding to the DNA-binding cofactor TEAD (Vassilev et al., 2001), induce transcription programs that regulate cell proliferation, survival, differentiation, EMT and cancer stem cell expansion (Ota and Sasaki, 2008, Zhang et al., 2009, Zhang et al., 2008, Zhao et al., 2008, Cordenonsi et al., 2011, Lian et al., 2010). When phosphorylated by Lats, TAZ and YAP are sequestered in the cytoplasm and/or degraded, resulting in the inhibition of their target gene expression (Yu and Guan, 2013). TAZ and YAP are well-known oncogenes whose expression is elevated in many types of human cancer (Harvey et al., 2013). In particular, TAZ is highly upregulated in invasive human breast cancer cell lines and in over 20% of breast cancer tissues (Chan et al., 2008). High levels of TAZ correlate with breast tumors of higher histological grade and increased invasiveness as well as expanded cancer stem cell compartment (Cordenonsi et al., 2011). Furthermore, overexpression of TAZ, especially the constitutively active TAZS89A, in breast cancer cells promotes EMT, cancer stem cell self-renewal and tumor invasion (Cordenonsi et al., 2011).
In this study, we uncovered SnoN as a regulator of the Hippo pathway. We showed that SnoN enhanced the stability of TAZ in a Lats2-dependent manner to promote oncogenic transformation and EMT in mammary epithelial cells and tumor growth in vivo. The expression levels and subcellular localization of SnoN in mammary epithelial cells are also tightly regulated by the Hippo kinases that are activated by the Scribble polarity protein. Our study thus has uncovered a regulatory loop for SnoN and Hippo signaling that may play important roles in breast cancer development.
Result
The expression and localization of SnoN are regulated by cell density
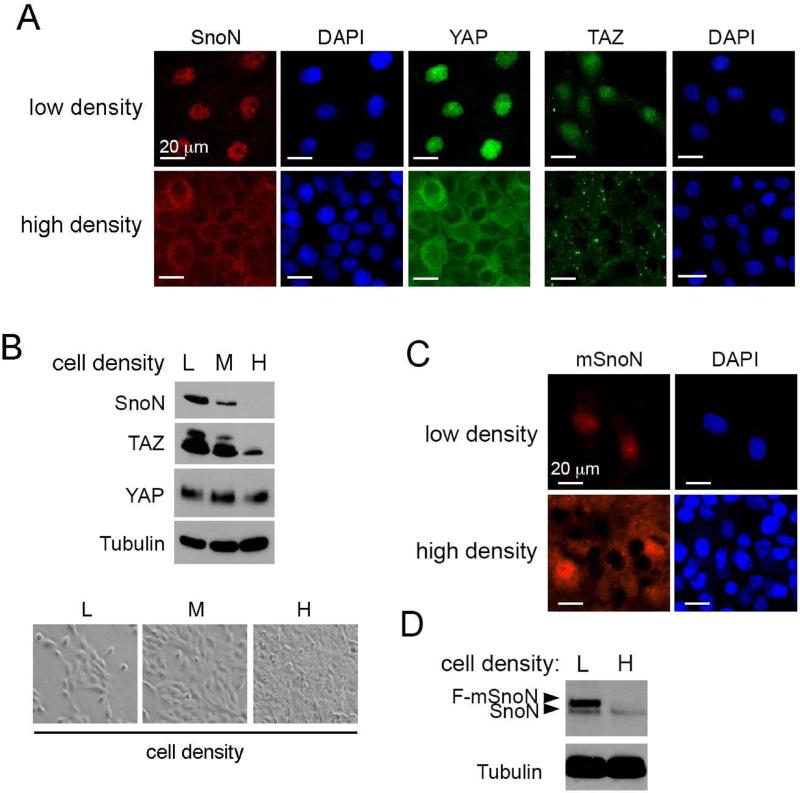
We have reported previously that SnoN exhibits predominantly cytoplasmic localization in nonmalignant mammary tissues but is nuclear in some malignant tissues and cell lines (Krakowski et al., 2005). In the process of investigating the differential intracellular localization of SnoN, we noticed that SnoN localization was highly sensitive to cell density: it exhibited predominantly nuclear localization at the low density but became largely cytoplasmic at a high cell density (Figure 1A). More importantly, the overall intensity of SnoN stain at the high density was much weaker than that at the low density, suggesting that the expression level of SnoN may also be subjected to regulation by cell density. Indeed, Western blotting analysis confirmed that SnoN abundance decreased dramatically with the increase in culture density (Figure 1B). This change in localization in response to cell density is reminiscent of the behavior of TAZ and YAP, that also display nuclear localization at the low density but become largely cytoplasmic at the high cell density [(Zhao et al., 2007) and Figure 1A]. In addition, the expression level of TAZ, but not YAP, was also reduced with the increase in cell density, showing an interesting correlation with SnoN expression (Figure 1B). In contrast, the abundance of Ski, a SnoN homolog, did not change with increase in cell density (Figure S1). Interestingly, the localization and expression level of a mutant SnoN defective in Smad binding (mSnoN) (Zhu et al., 2007, He et al., 2003) was also regulated by cell density (Figure 1C&D), similar to WT SnoN, suggesting that this regulation is likely to be independent of the ability of SnoN to regulate the Smad proteins.
Figure 1. SnoN localization and expression are sensitive to cell density.
(A) Immunofluorescence staining of SnoN (red) and YAP or TAZ (green) in MCF10A cells at low or high cell density. Nuclei were stained with DAPI. Bar, 20 μm. (B) Western blotting analysis of the levels of SnoN, TAZ and YAP (top) in MCF10A cells cultured at low (L), medium (M) or high (H) cell density (bottom). Tubulin was used as a loading control. (C&D) Flag-mSnoN was stably expressed in MCF10A cells, and its localization examined by immunofluorescence staining with anti-Flag (C) and levels detected by Western blotting with anti-SnoN (D).
See also Figure S1.
SnoN regulates TAZ levels in epithelial cell lines and in mammary tissue in vivo
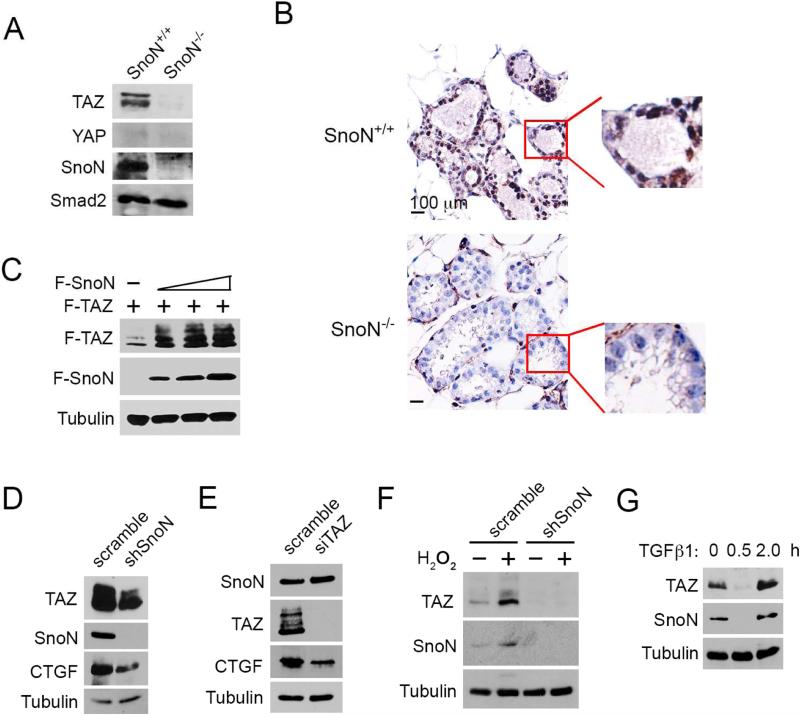
The correlation between SnoN and TAZ expression has also been found in vivo. In the mouse mammary gland, SnoN expression is sharply elevated at late pregnancy, and this high level of SnoN is required for alveologenesis (Jahchan et al., 2012). We found that in the WT mammary gland during late pregnancy (day 18.5), TAZ was expressed at a much higher level than YAP, which was barely detectable (Figure 2A, left). Interestingly, in SnoN−/− glands, TAZ expression was markedly diminished (Figure 2A&B). Thus, SnoN appears to regulate TAZ expression in the mammary epithelial cells in vivo.
Figure 2. SnoN regulates TAZ levels in both mammary tissues and epithelial cell lines.
(A) Total proteins from pregnancy day 18.5 SnoN+/+ and SnoN−/− mammary glands were subjected to Western blotting analyses. Smad2 was used as a loading control. (B) Immunohistochemistry staining of TAZ in pregnancy day 18.5 SnoN+/+ and SnoN−/− mammary sections. Boxed area highlighted the TAZ staining in ductal epithelial cells. Bar, 100 μm. (C) Lysates from cells transfected with a fixed amount of Flag-TAZ (F-TAZ) and increasing amounts (0, 0.5, 1, 2 μg) of Flag-SnoN (F-SnoN) were assessed by Western blotting with anti-Flag. Tubulin was used as a loading control. (D) TAZ and CTGF expression in parental and shSnoN MCF10A cells was analyzed by Western blotting. (E) Reducing TAZ by siTAZ did not affect SnoN levels as assessed by Western blotting. (F) MCF10A cells stably expressing shSnoN or scramble control were treated with 70 μM H2O2 for 24 h, and levels of SnoN and TAZ analyzed by Western blotting. (G) Cells were stimulated with 100 pM TGFβ for the indicated time, and levels of SnoN and TAZ analyzed by Western blotting.
To confirm directly that SnoN could regulate TAZ expression, we transfected 293T cells with a fixed amount of TAZ and increasing amounts of SnoN. As shown in Figure 2C, elevated SnoN expression caused a corresponding increase in TAZ levels. Furthermore, knocking down endogenous SnoN expression in MCF10A cells by shRNA reduced endogenous levels of TAZ as well as that of its target, CTGF (Figure 2D). Finally, knocking down TAZ by siTAZ did not alter SnoN levels (Figure 2E). Thus, SnoN is likely to act upstream of TAZ to regulate its expression.
SnoN expression can be induced by various cellular stress signals, such as oxidative stress and oncogene overexpression (Pan et al., 2009, Pan et al., 2012). Since SnoN regulates TAZ expression, we speculated that these signals might also upregulate TAZ expression. Indeed, when MCF10A cells were treated with H2O2 (oxidative stress), TAZ expression was markedly elevated, in good correlation with the pattern of SnoN expression (Figure 2F). Moreover, when SnoN was knocked down, both the basal level of TAZ and the induction of TAZ expression were attenuated significantly (Figure 2F), confirming that SnoN acts upstream of TAZ. Similarly, TAZ abundance was dynamically regulated by TGFβ in a manner mirroring that of SnoN levels (Figure 2G). As reported previously, TGFβ treatment resulted in a rapid degradation of SnoN within 30 min followed by a potent induction due to increased transcription 2 h later (Figure 2G) (Stroschein et al., 1999, Zhu et al., 2005). In parallel to this, the TAZ level was also reduced immediately after TGFβ treatment followed by an increase 2 h later (Figure 2G). Taken together, these data support the notion that SnoN regulates TAZ expression.
SnoN promotes TAZ protein stability
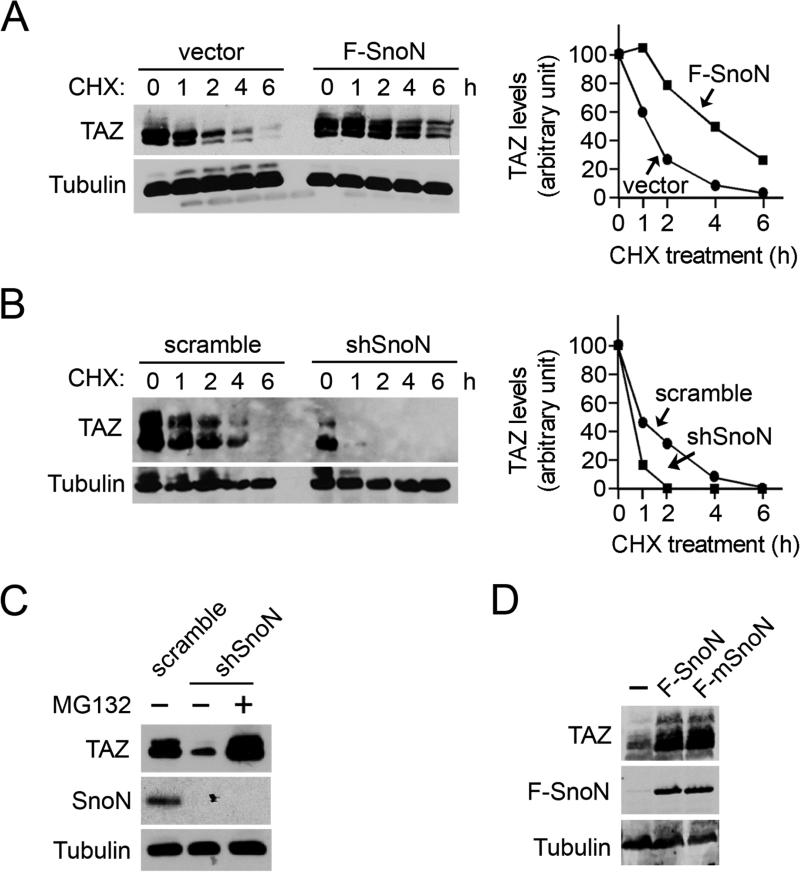
Semi-quantitative RT-PCR analysis indicated that the mRNA levels of TAZ were indistinguishable between WT and SnoN−/− mammary glands (Figure S2A). Consistent with this, knocking down SnoN in MCF10A cells had no effect on the level of TAZ mRNA (Figure S2B), suggesting that the attenuated TAZ expression in tissues and cells lacking SnoN was likely due to a decrease in protein stability. In support of this, the half-life of TAZ was significantly extended upon SnoN overexpression (Figure 3A) and decreased upon SnoN knockdown (Figure 3B). In addition, treatment of shSnoN-expressing cells with the proteasome inhibitor MG132 restored TAZ levels (Figure 3C), confirming that SnoN stabilizes TAZ by blocking proteasome-dependent degradation of TAZ. This stabilization of TAZ by SnoN is independent of the interaction of SnoN with the Smad proteins since mSnoN also upregulated TAZ levels as effectively as WT SnoN (Figure 3D).
Figure 3. SnoN promotes TAZ stability.
(A) Parental MCF10A cells or those stably expressing F-SnoN were treated with 100 μg/ml cycloheximide (CHX) for the indicated times. TAZ levels were measured by Western blotting with anti-TAZ and quantified in the graph shown to the right. (B) MEFs expressing scramble control or shSnoN were subjected to CHX treatment as described above in A. (C) TAZ level was measured in lysates prepared from MCF10A cells expressing shSnoN or scramble control after treatment with or without 20 μM MG132 for 6 h. (D) mSnoN enhanced TAZ expression as efficiently as WT SnoN as shown by Western blotting with anti-TAZ and anti-Flag, respectively.
See also Figure S2A&B.
SnoN enhances the transcription and oncogenic activities of TAZ
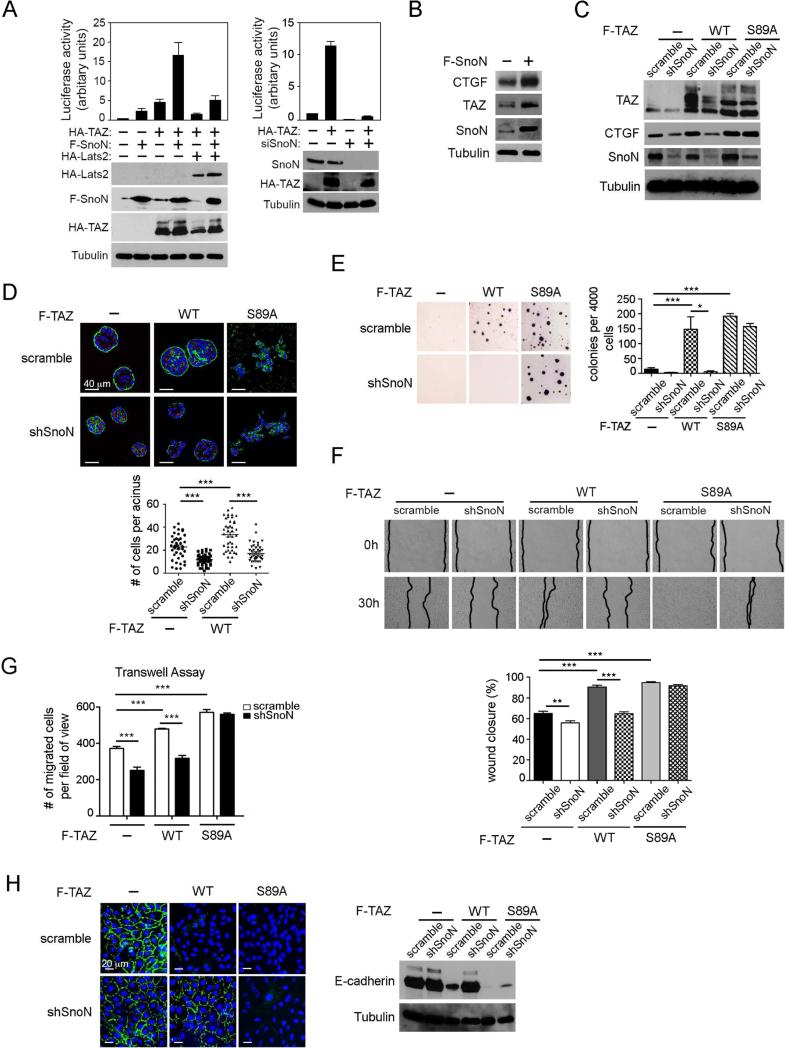
We next examined whether SnoN enhanced the ability of TAZ to activate transcription and promote oncogenic transformation of mammary epithelial cells. In a TAZ-responsive luciferase reporter assay, SnoN stimulated transcriptional activation by either endogenous or ectopically expressed TAZ (Figure 4A, left), while knocking down SnoN expression by either siRNA or shRNA reduced both basal as well as TAZ-induced transcriptional activation (Fig. 4A, right and Figure S2C). Moreover, SnoN antagonized the ability of Lats2 to repress transcriptional activation by TAZ (Figure 4A, left). Consistent with this, we also observed that expression of endogenous CTGF, a direct TAZ target gene, increased significantly in MCF10A cells overexpressing SnoN (Figure 4B). Thus, SnoN can enhance the transcription activity of TAZ.
Figure 4. SnoN enhances the transcription and transforming activities of TAZ.
(A) The TAZ-dependent luciferase activity was measured in 293 cells expressing F-SnoN or HA-TAZ either alone or together in the presence or absence of HA-Lats2, or siSnoN in the presence or absence of HA-TAZ. Each data point represents mean ±SEM from two independent wells. (B) Expression levels of CTGF, TAZ and SnoN in MCF10A cells stably expressing F-SnoN or control vector were examined by Western blotting. Tubulin was used as a loading control. (C) Western blotting analysis of TAZ abundance in MCF10A cells expressing WT or S89A TAZ together with or without shSnoN. (D) MCF10A cells stably expressing WT TAZ or TAZS89A together with or without shSnoN were cultured in 3D IrECM for six days before staining for α-integrin (green) as a basolateral marker and GM130 (red) as an apical marker. Nuclei are stained with DAPI (blue). Top: Confocal images are representative of three independent experiments. Scale bar, 40 μm. Bottom: The average size of acini was determined by the number of nuclei in each acinus. Forty acini from three independent experiments were quantified for each cell line and presented in the graph. (E-H). MCF10A cells stably expressing WT TAZ or TAZS89A together with or without shSnoN were subjected to: (E) Soft agar assay; colonies were stained with MTT, quantified and shown in the graph to the right; (F) Wound healing assay: wound closure was monitored at 0 h and 30 h and quantified in the graph (bottom); (G) Cell migration assay: A Transwell assay was performed for 8 h, and the migrated cells were stained and counted. (H) E-cadherin localization was determined by immunofluorescence staining (green, left), and expression level by Western blotting (right). Bar, 20 μm. Data in the graphs in D-G were derived from at least three independent experiments and are presented as means ± SEM [analysis of variance (ANOVA), Newman-Keuls multiple comparison test; *P < 0.05, ** P < 0.01, ***P < 0.001].
See also Figure S2C and Figure S3.
TAZ is a known oncogene whose expression is upregulated in breast cancer tissues (Chan et al., 2008), and high levels of TAZ correlate with breast tumors of higher histological grade and increased invasiveness as well as increased cancer stem cells content (Cordenonsi et al., 2011). When overexpressed in the untransformed MCF10A cells (Figure 4C), TAZ induced the formation of larger acini with increased number of cells per acinus, but preserving their polarity in the 3D laminin-rich extracellular matrix (lrECM), while control MCF10A cells underwent morphological differentiation to form polarized and organized multi-cellular acinar-like structures (Figure 4D). Overexpression of TAZ also induced anchorage-independent growth (Figure 4E) and promoted various aspects of EMT including increased cell motility and migration as well as reduced and mislocalized E-cadherin expression (Figure 4F-H). Reducing SnoN expression in these TAZ-expressing cells by either stably expressed shSnoN (Figure 4C) or transiently expressed siSnoN (Figure S3A) consistently returned the acini to the similar size as control MCF10A acini (Figure 4D and Figure S3B), abolished anchorage-independent growth (Figure 4E and Figure S3C) and reduced cell motility and migration (Figure 4F&G and Figure S3D). In addition, shSnoN in TAZ-overexpressing cells restored E-cadherin expression and partially corrected the localization of E-cadherin back to cell-cell junction (Figure 4H and Figure S3E). Taken together, these data indicate that SnoN promotes TAZ-induced cell growth, EMT and transformation.
Previous studies indicate that TAZ stability is regulated by ubiquitin-mediated proteasomal degradation and requires phosphorylation on Ser89 by Lats1 or Lats2 (Liu et al., 2010). To determine whether SnoN regulates TAZ stability in a Lats2-dependent manner, we next tested whether SnoN also affected the expression of TAZS89A mutant that is refractory to Lats2 phosphorylation and inhibition (Lei et al., 2008). Interestingly, the expression levels (Figure 4C and Figure S3A) and oncogenic activities of TAZS89A were not affected by SnoN knockdown. While MCF10A cells expressing TAZS89A formed disorganized and irregular colonies with fully disrupted apical-basal polarity when cultured in 3D IrECM (Figure 4D and Figure S3B) and underwent robust growth in soft-agar (Figure 4E and Figure S3C), knocking down SnoN had no effect on the transforming phenotypes of TAZS89A cells. Similarly, while TAZS89A strongly promoted cell motility, migration and downregulation of E-cadherin, reducing SnoN expression did not reverse any of these processes (Figure 4F-H and Figure S3D&E), unlike its effect on WT TAZ-expressing cells. Thus, SnoN enhances the transcription and oncogenic activities of TAZ likely through a Lats2-dependent mechanism.
SnoN binds to Hippo signaling molecules and inhibits TAZ phosphorylation
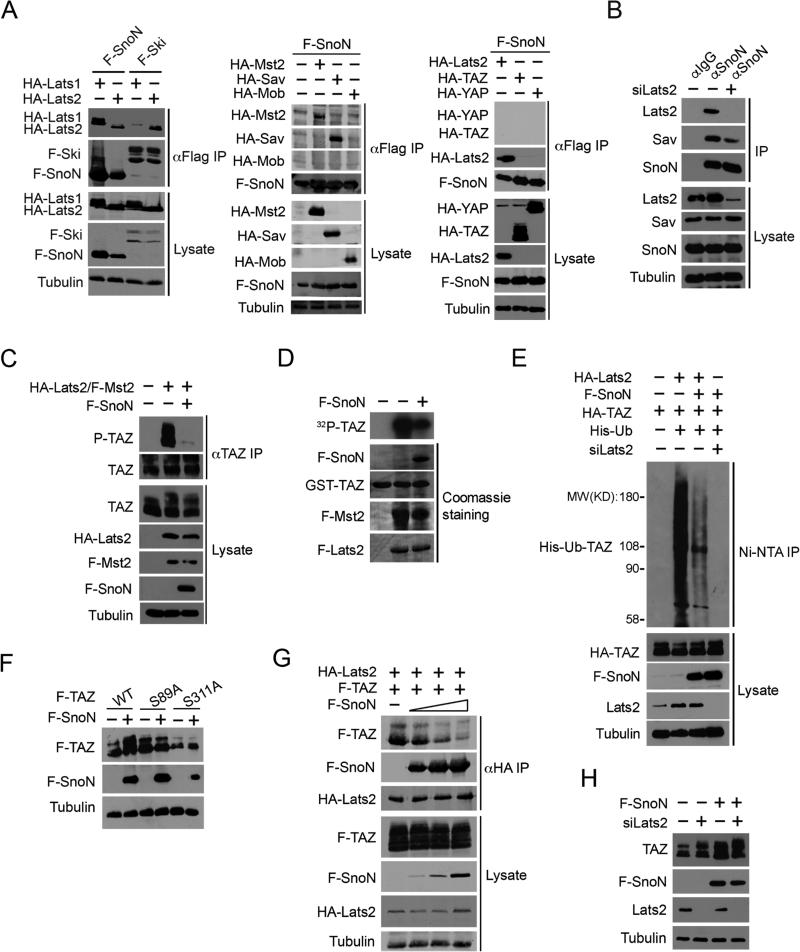
We next explored the mechanisms by which SnoN enhances TAZ expression. We found that SnoN did not bind to TAZ or YAP (Figure 5A), suggesting the possibility that SnoN may affect TAZ stabilization through the Hippo kinase complex. In an independent line of research, we identified Ski as a regulator of the Hippo signaling by binding to multiple members of the Hippo core kinase complex (Rashidian et al., 2015). Since SnoN is highly homologous to Ski, we examined whether SnoN could interact with key components of Hippo complex by a co-immunoprecipitation assay in both transfected cells and under the endogenous condition. In transfected cells, SnoN bound to multiple Hippo signaling proteins including Lats1, Lats2, Mst2, and Sav, but not to TAZ, YAP or Mob (Figure 5A). The endogenous SnoN also co-immunoprecipitated with Lats2 and Sav specifically and efficiently in MCF10A cells (Figure 5B), but not with Mst2 (data not shown), suggesting that the interaction between SnoN and Mst2 might be indirect. This binding preference of SnoN differs from that of Ski. Ski interacts with Lats2, but not Lats1, while SnoN bound to both Lats1 and Lats2 (Figure 5A). Unlike Ski, SnoN did not bind to endogenous Mst2, Mob and TEAD (Figure 5A, Figure S4 (Rashidian et al., 2015)). Thus, SnoN interacts with selective members of the Hippo core kinase complex.
Figure 5. SnoN interacts with the Hippo kinase complex and inhibits the ability of Lats2 to phosphorylate TAZ.
(A) F-SnoN or F-Ski was isolated by immunoprecipitation (IP) with anti-Flag from 293T cells transfected with F-SnoN or F-Ski together with Hippo signaling molecules as indicated, and proteins associated with F-SnoN or F-Ski were detected by Western blotting with anti-HA antibodies (upper panels). The abundance of these proteins in the lysates was shown in the lower panels. Tubulin was used as a loading control. (B) MCF10A cells were transfected with either scramble control or siLats2 and treated with 20 μM MG132 for an additional 6 h. Interactions between endogenous SnoN and Hippo signaling proteins were examined by IP with anti-SnoN or anti-IgG followed by Western blotting with antibodies specific for individual Hippo components (upper). The abundance of these proteins in the cell lysates was assessed by Western blotting (lower). (C) HA-Lats2 and F-Mst2 were transfected into MCF10A cells in the absence or presence of F-SnoN and treated with 20 μM MG132 for 6 h. Phosphorylated TAZ was isolated by IP with anti-TAZ followed by Western blotting with anti-Phospho-S89 TAZ. (D) In vitro kinase assay. Lats2 and Mst2 were IP from cells transfected with F-Lats2 and F-Mst2 either alone or together with F-SnoN and subjected to in vitro kinase assay using GST-TAZ as a substrate. 32P-TAZ was detected by auto-radiography (top panel). The abundance of GST-TAZ, F-Lats2, F-Mst2 and F-SnoN was measured by Coomassie blue staining (lower panels). (E) 293T cells transfected with various constructs indicated were treated with 20 μM MG132 for 6 h, and His-Ub-TAZ was pulled down by the Ni-NTA column and detected by Western blotting with anti-HA (top panel). The abundance of proteins in the cell lysates was assessed by Western blotting (lower panels). (F) WT, S89A or S311A TAZ were transfected into 293T cells alone or together with F-SnoN, and the expression levels of these proteins analyzed by Western blotting with anti-Flag. (G) 293T cells were transfected with a fixed amount of F-TAZ and HA-Lats2 and increasing amounts (0, 0.5, 1, 2 μg) of F-SnoN and then treated with MG132 for 6 h. SnoN and TAZ that associated with Lats2 were isolated by IP with anti-HA and detected by Western blotting with anti-Flag. (H) Lats2 siRNAs were transfected into control or F-SnoN-overexpressing MCF10A cells. Endogenous TAZ and Lats2 levels were detected by Western blotting.
See also Figure S4.
We next asked whether SnoN affected the Lats2-dependent phosphorylation of TAZ using an antibody specific for phospho-S89 TAZ. In 293T cells transfected with or without Lats2 and Mst2, SnoN readily inhibited phosphorylation of endogenous TAZ on Ser89 by Lats2 and Mst2 (Figure 5C). Similarly, in an immune complex in vitro kinase assay to measure TAZ phosphorylation, SnoN markedly inhibited TAZ phosphorylation by Lats2 and Mst2 (Figure 5D). This reduced TAZ phosphorylation in the presence of SnoN was accompanied by a significant decrease in the ubiquitination of TAZ, which was completely abolished in the absence of Lats2, as expected (Figure 5E), indicating that SnoN most likely promotes stabilization of TAZ by blocking Lats2-dependent phosphorylation and ubiquitination of TAZ. Consistent with this, SnoN failed to stabilize two TAZ mutants, S89A and S311A that are refractory to phosphorylation by Lats2 (Figure 5F).
SnoN could inhibit TAZ phosphorylation by decreasing the expression and/or kinase activity of Lats2 and Mst2 or by disrupting the interaction between Lats2 and TAZ. We found that SnoN did not affect the expression or kinase activity of Lats2 and Mst2 (Figure 5G and data not shown). Rather, it appeared to disrupt the Lats2-TAZ interaction. In cells transfected with fixed amounts of Lats2 and TAZ and varying amounts of SnoN, increasing levels of SnoN caused a corresponding decrease in the binding of Lats2 to TAZ (Figure 5G). Consistent with this, while knocking down Lats2 by siRNA increased the basal TAZ level, it impaired further stabilization of TAZ by SnoN (Figure 5H). Thus, SnoN enhances TAZ stability by disrupting the interaction between TAZ and Lats2 and preventing TAZ phosphorylation by Lats2, leading to decreased ubiquitination and degradation of TAZ. Although Lats2 was used in the experiments described in this study, the results are also true for Lats1.
SnoN functions upstream of TAZ in breast cancer cells
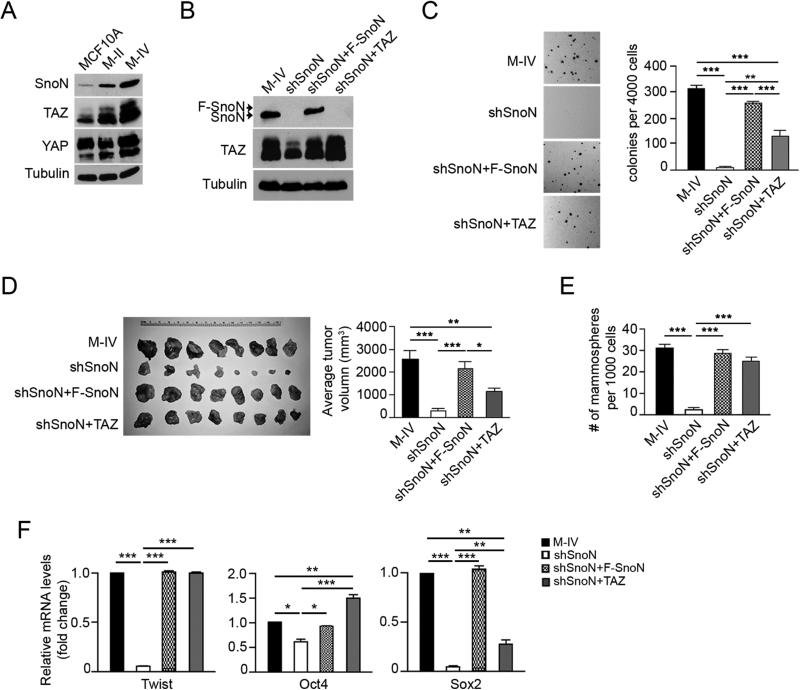
TAZ is upregulated as breast cancer cells progress to more malignant stages (Cordenonsi et al., 2011). Because high levels of SnoN have also been reported in malignant human breast cancer cell lines (Zhu et al., 2007), we asked whether SnoN contributed to the elevated TAZ levels in these breast cancer cells. In the MCF10A breast cancer progression model containing the non-transformed MCF10A cells, Ras-transformed MCF10A-T1k cells (M-II) and their metastatic and malignant derivative, MCF10A-CA1a cells (also known as M-IV), TAZ is expressed at a low level in the untransformed MCF10A, gradually increased in M-II cells and significantly upregulated in the metastatic M-IV cells (Figure 6A), as reported previously (Cordenonsi et al., 2011). Interestingly, SnoN level was also upregulated in M-IV cells, mirroring that of TAZ, while YAP or Ski expression showed no correlation with that of TAZ or SnoN (Figure 6A and Figure S5). Reducing SnoN levels in M-IV cells by either shSnoN or siSnoN decreased TAZ expression (Figure 6B and Figure S6A). Importantly, this downregulation of endogenous TAZ could be rescued efficiently by a shRNA-resistant Flag-tagged SnoN cDNA (F-SnoN) (Figure 6B).
Figure 6. SnoN functions upstream of TAZ in breast cancer cells.
(A) The abundance of SnoN, TAZ and YAP in MCF10A series as shown by Western blotting. (B) Western blotting analysis of endogenous TAZ and SnoN in M-IV cells stably expressing various constructs. (C) Soft agar colonies were stained and quantified in the graph to the right. (D) M-IV cells stably expressing various constructs were injected subcutaneously into the nude mice. Tumors from each mouse were isolated (left), and their sizes measured and quantified (right). (E) Two rounds of mammosphere formation assay were carried out in M-IV cells expressing various constructs. The numbers of mammospheres formed in the second round culture were counted, quantified and shown in the graph. (F) qRT-PCR analysis of breast cancer stem cell markers after normalization to β-actin. Data in C-F were derived from at least three independent experiments and are presented as means ± SEM [analysis of variance (ANOVA), Newman-Keuls multiple comparison test; *P < 0.05, ** P < 0.01, ***P < 0.001].
See also Figure S5 and Figure S6.
M-IV cells are highly transformed and readily form soft-agar colonies in vitro and grow tumors in vivo in a xenograft mouse model (Figure 6C-D). They also contain a CSC population that readily forms mammospheres and expresses various CSC markers including Oct4, Sox2 and Twist (Figure 6E-F) (Cordenonsi et al., 2011). Knocking down SnoN in these cells by shRNA or siRNA significantly impaired the anchorage-independent growth (Figure 6C and Figure S6B) and xenograft tumor growth (Figure 6D), similar to what has been reported for MDA-MB-231 cells previously (Zhu et al., 2007), and markedly diminished CSC self-renewal, as shown by the mammosphere formation assay (Figure 6E and Figure S6C), and expression of CSC markers (Figure 6F and Figure S6D). These impaired transforming activities were again effectively rescued by F-SnoN (Figure 6C-F).
These data support a pro-oncogenic role of SnoN in malignant human breast cancer cell lines. To determine whether these activities of SnoN were mediated by TAZ, we restored TAZ levels in M-IV/shSnoN cells (Figure 6B) and asked whether this could rescue some of the transforming phenotypes. Although at a level higher than that of endogenous TAZ in M-IV cells, ectopically expressed WT TAZ only partially restored anchorage-independent growth and xenograft tumor growth in vivo (Figure 6C-D), suggesting that TAZ is likely only one of the downstream effectors of SnoN in promoting tumor growth. This is consistent with our previously published study suggesting that the ability of SnoN to antagonize the cytostatic response of the TGFβ/Smads is also partially responsible for its mitogenic activity (Zhu et al., 2007). Interestingly, the restored TAZ rescued nearly 80% of the mammoshpere forming ability and markedly increased expression of several CSC markers, including Twist1, Oct4 and Sox2 in TAZ restored cells that was lost in shSnoN cells (Figure 6E&F). Thus, the ability of SnoN to promote CSC renewal is mainly mediated by TAZ.
Taken together, these data support the model that SnoN acts upstream of TAZ to enhance its ability to promote malignant progression of breast cancer cells.
SnoN is regulated by the Scribble polarity protein via Lats2
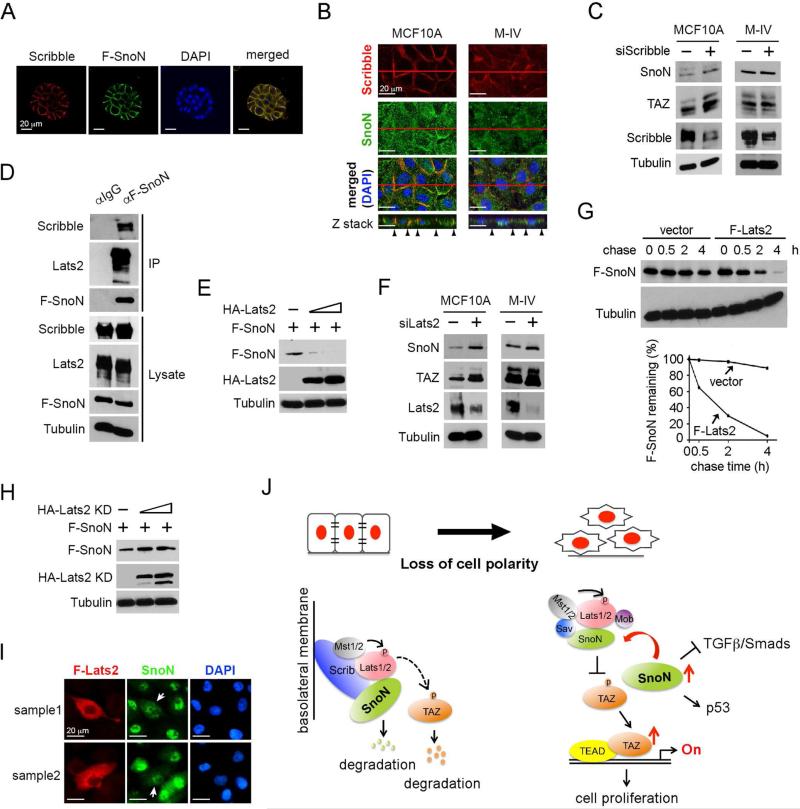
Our earlier observation that SnoN expression and localization are sensitive to cell density suggests that SnoN itself may be regulated by the Hippo pathway in response to signals from the polarity and junction complexes. Previous studies have shown that the Lats kinases can be activated by multiple polarity and cell adhesion complexes including the apically-localized Crumb complex, Par3/par6/aPKC complex, the Cadherin-catenin complex and the basal laterally-localized Scribble complex (Schroeder and Halder, 2012). To assess which of these complexes is likely involved in regulation of SnoN, we first determined the localization of SnoN in polarized mammary epithelial cells. In the 3D MCF10A acini that showed well-established basolateral and apical polarity, SnoN was found at the basolateral domain, similar to the localization of Scribble (Figure 7A). In the conventional 2D culture, a fraction of SnoN was also found to co-localize with Scribble at the cell-cell junction of untransformed MCF10A cells, but not in malignant M-IV cells (Figure 7B), hinting the potential involvement of the Scribble complex. Indeed, knocking down Scribble by siRNA upregulated SnoN and TAZ expression in MCF10A cells, but had no effect on SnoN and TAZ expression in M-IV cells (Figure 7C), supporting the model that Scribble regulates SnoN expression in untransformed cells.
Figure 7. SnoN is regulated by Scribble in a Lats2-dependent manner.
(A) SnoN localized at the basolateral membrane in 3D culture. MCF10A/F-SnoN cells were cultured in 3D IrECM for 6 days, fixed and stained with anti-Scrib (red) or anti-Flag (green). Nuclei were stained with DAPI (blue). Bar, 20 μm. (B) Confocal images showed that SnoN (green) co-localized with Scribble (red) at the basolateral junction in MCF10A cells in 2D, but not in M-IV cells where both proteins have lost the membrane localization. The top panels showed the XY images and the bottom panels were the Z stacks. Black arrowheads indicated the lateral membrane domains. Bar, 20 μm. (C) Reducing Scribble expression led to upregulation of SnoN and TAZ expression in MCF10A cells, but not in M-IV cells. (D) MCF10A cells stably expressing F-SnoN were cultured until reached high cell density and treated with 20 μM MG132 for 6 h. F-SnoN and the associated Scribble and Lats2 were analyzed by co-IP assay. (E) Protein levels were determined by Western blotting in cells transfected with fixed amount of F-SnoN and increasing amounts of HA-Lats2. (F) Reducing Lats2 expression led to increased SnoN and TAZ expression in both MCF10A and M-IV cells. (G) Pulse-chase assay. 35S-labeled F-SnoN was immunoprecipitated from 293T cells transfected with F-SnoN together with or without F-Lats2 and detected by auto-radiography. Quantification of the autoradiograph was shown at the bottom. (H) Cells were transfected with F-SnoN and increasing amounts of HA-Lats2 (KD) and subjected to Western blotting analysis. (I) Immunofluorescence staining of SnoN (green) and F-Lats2 (red) in MCF10A cells showed that overexpression of F-Lats2 (white arrow) resulted in nuclear reduction of SnoN. Images from two cell fields were shown (sample 1 and 2). Bar, 20 μm. (J) A model of mutual regulatory interaction between SnoN and Hippo pathway.
Scribble has been shown to activate the Hippo core kinase complex at the basolateral membrane by directly binding to Lats2 as well as Mst and Mob in MCF10A cells (Cordenonsi et al., 2011). Since SnoN also binds to Lats2, we next examined whether SnoN could interact with Scribble and be subjected to downregulation by the basolaterally activated Hippo kinases. Indeed, at the high cell density, stably expressed SnoN interacted with both endogenous Scribble and Lats2 in MCF10A cells (Figure 7D). In addition, Lats2 affected both SnoN protein levels and localization. As shown in Figure 7E-7F, increasing amount of Lats2 caused a corresponding decrease in the protein levels of SnoN, while knocking down Lats2 resulted in an increase in SnoN and TAZ levels, suggesting that Lats2 downregulates SnoN expression. Lats2 did not affect the mRNA level of SnoN (data not shown), but reduced the half-life of SnoN in a pulse-chase assay (Figure 7G). This activity required the kinase activity of Lats2, as introduction of the kinase inactive mutant of Lats2 (KD) did not reduce SnoN expression (Figure 7H). Lats2 also altered the localization of SnoN. In MCF10A cells cultured at a low density when Hippo signaling is not activated, SnoN was predominantly nuclear. When Lats2 was overexpressed, SnoN was found in both cytoplasm and nucleus, and the intensity of SnoN stain was also decreased (Figure 7I), consistent with its downregulation. Thus, activation of Hippo signaling and Lats2 by the Scribble polarity protein downregulates SnoN expression and prevents its nuclear localization.
Based on the data shown here, we propose a model of mutual regulatory interaction between SnoN and the Hippo kinases (Figure 7J). In a normal tissue or cultured under the high cell density condition, epithelial cells establish proper cell polarity and adhesion, and Hippo signaling is activated by various polarity complexes including the Scribble complex. Through its interaction with Scribble, majority of SnoN are recruited to the basolateral domain where it is downregulated by the active Lats2, while a low level of SnoN may exist in the ctyoplasm to regulate TAZ stability. This downregulation of SnoN as well as TAZ by junction-activated Hippo kinases eventually results in inhibition of cell proliferation. In this way, epithelial tissue architecture and cell polarity can restrict the activity of proliferative signals. When cell polarity is disrupted, as often occurs during malignant transformation and progression or during tissue injury and damage, SnoN is no longer repressed by the Scribble-associated Hippo complex and accumulates rapidly in the cells. This elevated SnoN in cells can orchestrate a number of signaling activities including further enhancing TAZ stabilization by inhibiting the Hippo complex that may be activated by other pathways (such as growth factor signaling), repressing TGFβ/Smad activity or promoting p53 activation. In breast cancer cells, this results in upregulation of TAZ, which in turn promotes cell proliferation and EMT, while repressing the growth inhibitory signals of TGF β/Smads, ultimately leading to malignant progression. In this capacity, SnoN functions as an important mediator that transmits signals from the tissue architecture and cell polarity to regulate the activity of various intracellular signaling pathways.
Discussion
In this study, we have revealed a mechanism for mutual regulation between SnoN and Hippo signaling. Our data showed that SnoN directly bound to the Hippo core kinase complex and blocked the binding of Lats2 to TAZ, resulting in inhibition of TAZ phosphorylation and its subsequent degradation. Through this mechanism, SnoN enhances TAZ expression and its transcriptional activity. In the mammary gland in vivo, SnoN controls the TAZ level at late stage of pregnancy, and this regulation is necessary for mammary epithelial cell proliferation and differentiation. In breast cancer cells, the upregulation of SnoN may contribute to elevated TAZ expression and enhanced TAZ activities to promote breast cancer transformation and CSC expansion. Depletion of SnoN expression significantly reduced TAZ protein levels as well as TAZ-induced oncogenic transformation and EMT in vitro and tumor growth in xenograft models in vivo. Our study thus has identified SnoN as a positive regulator of TAZ in normal mammary gland function and breast cancer development. Given the ubiquitous expression of SnoN and TAZ in most tissue and cell types, it is likely that this regulation also occurs in other tissue and cell types beyond the mammary gland.
Previously Varelas et al has reported that TAZ controls nucleocytoplasmic shuttling of Smad2/3-4 and also forms a transcription complex with Smad2/3-4 on TGFβ target promoters to stimulate transcription (Varelas et al., 2008). Given the ability of SnoN to regulate TGFβ/Smad signaling, an obvious question is whether the SnoN-TAZ regulation is mediated by the Smad proteins. We found that a mutant SnoN defective in binding to the Smads was equally capable of upregulating TAZ expression as the WT SnoN, indicating that the upregulation of TAZ by SnoN is independent of SnoN-Smad regulation. Additionally, the model of TAZ-Smad regulation was recently challenged by Nallet-Staub F. et al to be cell-line specific; it only occurred in the EpH4 murine mammary epithelial cell line but not true in almost all other human cell lines tested (Nallet-Staub et al., 2015). Nallet-Staub further argued that Hippo signaling and TAZ do not directly regulate TGFβ signaling. Although TAZ and Smad2/3-4 may co-exist in the same transcription complex on specific promoters, whether the interaction is direct or not awaits further study. Thus, the SnoN regulation of Hippo/TAZ signaling we revealed in this study is independent of its antagonism of the Smad proteins.
In addition to enhancing TAZ expression, SnoN itself, in particular its expression level and intracellular localization, is sensitive to cell density and is regulated by cell polarity through the Hippo pathway. This regulation appears to be mediated by the basolateral polarity protein Scribble and requires its activation of the Lats2 kinase. SnoN is recruited to the basolateral membrane through its interaction with Scribble, where it is downregulated by Lats2 through a yet-to-be-identified mechanism. Interestingly, TAZ has been proposed to be downregulated by the same Scribble/Mst2/Lats2 complex in untransformed epithelial cells (Cordenonsi et al., 2011). Through these events, proper cell polarity can control the activity of important signaling molecules. When cell polarity is disrupted during pathological processes such as malignant progression or tissue injury, SnoN is no longer repressed by the Scribble-associated Hippo complex and accumulates in the cells. Since SnoN is a potent regulator of several signaling pathways including TAZ/YAP, TGFβ/Smads and p53, this elevated SnoN in cells can orchestrate a number of signaling activities that ultimately promote cell proliferation, EMT and CSC expansion. Thus, SnoN may function as a key controlling node through which proper tissue organization and architecture can dictate the activity and outcomes of a number of important intracellular signaling pathways.
One surprising observation is that SnoN and Ski exert opposite effects on TAZ and Hippo signaling, although the two proteins share considerable sequence homology and some common functional features. In chicken and quail embryo fibroblasts, both SnoN and Ski promote transformation and terminal differentiation (Boyer et al., 1993, Givol et al., 1995). Both proteins bind to the Smad proteins and are potent negative regulators of TGFβ signaling (Stroschein et al., 1999, Sun, 1999). However, more recent studies reveal that the two proteins also display important functional and regulatory differences. Ski and SnoN modulate different aspects of embryonic development and regulate distinct tissue-specific functions in adult animals, due to the distinct expression pattern. For example, SnoN regulates embryonic angiogenesis (Zhu et al., 2013) while Ski is essential for development of the neurocrest lineage, bone and skeletal muscle (Berk et al., 1997). In the adult animal, SnoN, but not Ski, is critical for alveologenesis and onset of lactation through its ability to promote STAT5 signaling and TAZ [(Jahchan et al., 2012) & Fig. 2]. We have recently reported that Ski inhibits TAZ function and expression in breast cancer cells through both Lats2-dependent and Lats2-indepenent mechanisms (Rashidian et al., 2015). Our current finding that SnoN is a positive regulator of TAZ by blocking its phosphorylation by Lats2 adds another support to the notion that SnoN and Ski play non-redundant roles in many biological processes. Although both SnoN and Ski bind to multiple components of the Hippo core kinase complex, the functional outcomes of these bindings are different. Ski mainly functions as a scaffold protein to facilitate the interaction between Mst2 and Lats2 through Sav to enhance Lats2 activation, while SnoN prevents TAZ from binding to Lats2. Finally, Pot et al recently reported that SnoN interacted with NDR1 (also known as STK38), a member of the NDR/Lats family of Ser/Thr kinases that also includes Lats2 (Hergovich and Hemmings, 2009), to antagonize TGFβ-dependent transcription and inhibits growth arrest (Pot et al., 2013). We also noticed that SnoN interacted strongly with all four members of the NDR/Lats family, including NDR1 and NDR2 in addition to Lats1and Lats2 (unpublished data). However, NDR1/2 is not required for the regulation of TAZ protein stability by SnoN. Different from SnoN, Ski binds to NDR1 and Lats2 strongly, to NDR2 weakly, but does not bind to Lats1 (Rashidian et al., 2015). The structural basis for the differential regulation of the Hippo and NDR/Lats kinases by Ski and SnoN is not clear. It's known that the WW domain and PPxY motif are critical for mediating interactions between many Hippo pathway components. However, Ski and SnoN do not contain the WW domains or a typical PPxY motif that is recognized by the Lats2 WW domain (data not shown). It's possible that the less conserved C-terminal domain of SnoN and Ski may be responsible for their functional differences.
TAZ functions as an oncogene in human breast cancer to promote cancer cell proliferation, survival, EMT and CSC expansion (Barron and Kagey, 2014, Harvey et al., 2013, Zhao et al., 2011). The ability of SnoN to enhance TAZ expression and activity provides yet another mechanism for the pro-oncogenic activity of SnoN, in addition to its ability to antagonize the cytostatic activity of TGFβ/Smad pathway(Deheuninck and Luo, 2009). The observation that TAZ was downregulated in breast cancer cells expressing shSnoN and that overexpression of WT TAZ partially restored transformation in vitro and tumor growth in vivo supports the idea that the regulation of TAZ by SnoN partially contributes to the pro-mitogenic activity of SnoN. Interestingly, in contrast to the partial rescue of the transforming phenotypes of shSnoN cells, TAZ fully restored the mammosphere forming ability of shSnoN cells, indicating that the ability of SnoN to regulate breast CSC expansion is mainly mediated by the TAZ pathway. Finally, TAZ and SnoN seem to exert different effects on EMT in cancer cell lines. SnoN suppresses EMT in malignant breast cancer or lung cancer cells (Zhu et al., 2007), while TAZ promotes it (Lei et al., 2008). This differential effects of SnoN and TAZ on EMT are likely due to the involvement of other signaling pathways regulated by SnoN and by TAZ in cancer cells, which depending on the cancer or tissue context, may produce different and complex biological outcomes.
In summary, our study has revealed a role of SnoN in regulating the activity of the Hippo/TAZ pathway in breast cancer cells. The ability of SnoN to regulate multiple signaling pathways suggests that it may function to coordinate or balance the activities of various signaling pathways to enable alveologenesis in the mammary gland and regulate breast cancer development.
Experimental Procedures
Transfection, infection, immunoprecipitation and Luciferase assay
Stable MCF10A cells lines were generated by retroviral infection as described previously (Rashidian et al., 2015). Immunoprecipitation and western blotting were performed as previously described (Zhu et al., 2007).
To measure TAZ/YAP transcriptional activity, a total of 2.5 μg of DNA (including 0.05 μg 8xGT-IIC-δ51LucII Luciferase reporter construct (Ota and Sasaki, 2008) and/or the indicated plasmids or 70 pmol siRNAs) was transiently transfected into 293T cells by Lipofectamine 2000. The luciferase activity was measured at 36 h after transfection.
Immunofluorescence, Immunohistochemistry
Cells cultured in 2D or 3D IrECM or tissue samples were fixed, permeabilized and processed as previously described (Jahchan et al., 2012).
Pulse chase assay
Cells grown in a 60 mm dish were incubated in methionine-free DMEM/10% and dialyzed FBS for 20 min and pulse-labeled with 300 μCi [35S] methionine in the same medium for 30 min. The cells were then chased with the complete medium and lysed as described before (Stroschein et al., 1999).
In vivo ubiquitination assay
293T cells were transfected with HA-TAZ, His-ubiquitin and various constructs and treated with 20 μM MG132 for 6 h. Cells were lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole) and sonicated. The lysates were incubated with 50 μl His-select® HF Nickel Affinity beads (Sigma) at room temperature for 3 h, and the beads were washed with buffer A, buffer B (1.5 M guanidine-HCl, 25 mM Na2PO4/NaH2PO4, 20 mM Tris-Cl [pH 6.8], 17.5 mM imidazole) and buffer TI (25 mM Tris-Cl [pH 6.8], 20 mM imidazole) and boiled in 2× SDS-PAGE loading buffer containing 200 mM imidazole. His-tagged proteins were analyzed by western blotting with specific antibodies.
Wound healing assay
A plastic tip was used to generate a wound across the confluent cell monolayer. The wound closure was measured after 30 h as previously described ((Rashidian et al., 2015).
Soft agar assay
4000 cells were suspended in 0.2 ml medium containing 0.375% Bacto Agar (BD) and overlaid on the hardened 0.66% agar bottom layer in a well of a 24-well cluster. Fresh medium (0.2 ml) containing 0.375% agar was added to each well once a week for 4 weeks. The colonies were visualized by staining with 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Sigma) for 4 h at 37°C.
Three dimensional (3D) Morphogenesis assay
Differentiation of MCF10A cells in the 3D IrECM was performed as described before (Jahchan et al., 2012). After 6 days in the 3D culture, morphological differentiation was examined by phase contrast microscopy and by immunofluorescence staining for α6-integrin (MAB1378, Chemicon) and Golgi matrix protein of 130 (GM130, BD Pharmingen) as markers of basal lateral and apical polarity, respectively. Microscopy was performed on a Zeiss LSM710 confocal microscope at the Berkeley Biological Imaging Facility. The images were generated by serial confocal cross sections.
Two-round mammosphere formation assay
1×103 M-IV cells was plated as single-cell suspensions in ultra-low attachment culture plates in serum-free DMEM-F12 medium supplemented with B27, 10 ng/ml bFGF, 20 ng/ml EGF, 5 μg/ml insulin (Santa Cruz Biotechnology) and 0.4% bovine serum albumin (Sigma) as described previously (Cordenonsi et al., 2011). After 5 days, mammospheres were collected by gentle centrifugation (800 rpm) for 5 min, dissociated in 0.05% trypsin and 0.53 mM EDTA-Na (Invitrogen) at 37°C for 10 min and re-plated at a density of 500 cells/ml for an additional 5 days.
In vivo xenograft assay
For tumor injection into the 6-week old female athymic BALB/c mice (Charles River Laboratories), 2×106 M-IV cells in a 200 μl suspension of 1:1 Matrigel:buffer were injected subcutaneously into each site on the left and right flanks of nude mice, using a 26-gauge needle fitted with a 1-ml sterile syringe. Tumor growth was monitored once a week. Mice were sacrificed 5 weeks after injection, and tumors were surgically isolated. Tumor volume (V) was calculated by using the formula: V = length × width × thickness.
Statistical Analysis
All data were derived from at least three independent experiments and are presented as means ± SEM. Comparisons among groups were performed by one-way ANOVA (Newman-Keuls multiple comparison test) with Prism 5 software. p values are shown when relevant (*p < 0.05; **p < 0.01, ***p < 0.001).
Further experimental details for the above procedures and additional assays are provided in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
SnoN promotes TAZ stability by blocking its phosphorylation by Lats2
SnoN enhances the transcriptional and oncogenic activities of TAZ
SnoN is downregulated by the Scribble polarity complex through the Hippo kinases
Acknowledgments
We thank Dr. Kun-Liang Guan and Dr. Alain Mauviel for providing cDNAs of components of Hippo pathway and Dr. Hiroshi Sasaki for the 8xGT-IIC-δ51LucII construct. We also thank Steve Ruzin and Denise Schichnes at CNR Biological imaging facility at UC-Berkeley for assistance with microscopy. This study is supported by National Institutes of Health grants R21 CA187632 and DOD BRCP grant BC140444P1 to K. Luo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Q. Z. and K. L. designed the research and wrote the paper. K. L. supervised data analysis; Q. Z. performed research and analyzed data; E.L.S. generated most of the MCF10A cell lines and performed luciferase assays and pulse chase assays; N. J. performed part of the 3D Morphogenesis assays; X. J. and A. X. performed in vivo xenograft assays.
The authors declare no conflicts of interest and no competing financial interests.
References
- BARRON DA, KAGEY JD. The role of the Hippo pathway in human disease and tumorigenesis. Clin Transl Med. 2014;3:25. doi: 10.1186/2001-1326-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERK M, DESAI SY, HEYMAN HC, COLMENARES C. Mice lacking the ski proto oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–39. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOYER PL, COLMENARES C, STAVNEZER E, HUGHES SH. Sequence and biological activity of chicken snoN cDNA clones. Oncogene. 1993;8:457–66. [PubMed] [Google Scholar]
- CHAN SW, LIM CJ, GUO K, NG CP, LEE I, HUNZIKER W, ZENG Q, HONG W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–8. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- CHIA JA, SIMMS LA, COZZI SJ, YOUNG J, JASS JR, WALSH MD, SPRING KJ, LEGGETT BA, WHITEHALL VL. SnoN expression is differently regulated in microsatellite unstable compared with microsatellite stable colorectal cancers. BMC Cancer. 2006;6:252. doi: 10.1186/1471-2407-6-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORDENONSI M, ZANCONATO F, AZZOLIN L, FORCATO M, ROSATO A, FRASSON C, INUI M, MONTAGNER M, PARENTI AR, POLETTI A, DAIDONE MG, DUPONT S, BASSO G, BICCIATO S, PICCOLO S. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–72. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- DEHEUNINCK J, LUO K. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 2009;19:47–57. doi: 10.1038/cr.2008.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONG J, FELDMANN G, HUANG J, WU S, ZHANG N, COMERFORD SA, GAYYED MF, ANDERS RA, MAITRA A, PAN D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIVOL I, BOYER PL, HUGHES SH. Isolation and characterization of the chicken c-sno gene. Gene. 1995;156:271–6. doi: 10.1016/0378-1119(95)00066-f. [DOI] [PubMed] [Google Scholar]
- HAO Y, CHUN A, CHEUNG K, RASHIDI B, YANG X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- HARVEY KF, ZHANG X, THOMAS DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- HE J, TEGEN SB, KRAWITZ AR, MARTIN GS, LUO K. The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J Biol Chem. 2003;278:30540–7. doi: 10.1074/jbc.M304016200. [DOI] [PubMed] [Google Scholar]
- HERGOVICH A, HEMMINGS BA. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors. 2009;35:338–45. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- JAHCHAN NS, LUO K. SnoN in mammalian development, function and diseases. Curr Opin Pharmacol. 2010;10:670–5. doi: 10.1016/j.coph.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAHCHAN NS, OUYANG G, LUO K. Expression profiles of SnoN in normal and cancerous human tissues support its tumor suppressor role in human cancer. PLoS One. 2013;8:e55794. doi: 10.1371/journal.pone.0055794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAHCHAN NS, WANG D, BISSELL MJ, LUO K. SnoN regulates mammary gland alveologenesis and onset of lactation by promoting prolactin/ STAT5 signaling. Development. 2012;139:3147–3156. doi: 10.1242/dev.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAHCHAN NS, YOU YH, MULLER WJ, LUO K. Transforming growth factor-beta regulator SnoN modulates mammary gland branching morphogenesis, postlactational involution, and mammary tumorigenesis. Cancer Res. 2010;70:4204–13. doi: 10.1158/0008-5472.CAN-10-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRAKOWSKI AR, LABOUREAU J, MAUVIEL A, BISSELL MJ, LUO K. Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc Natl Acad Sci U S A. 2005;102:12437–42. doi: 10.1073/pnas.0504107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEI QY, ZHANG H, ZHAO B, ZHA ZY, BAI F, PEI XH, ZHAO S, XIONG Y, GUAN KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–36. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAN I, KIM J, OKAZAWA H, ZHAO J, ZHAO B, YU J, CHINNAIYAN A, ISRAEL MA, GOLDSTEIN LS, ABUJAROUR R, DING S, GUAN KL. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–18. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU CY, ZHA ZY, ZHOU X, ZHANG H, HUANG W, ZHAO D, LI T, CHAN SW, LIM CJ, HONG W, ZHAO S, XIONG Y, LEI QY, GUAN KL. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285:37159–69. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- NALLET-STAUB F, YIN X, GILBERT C, MARSAUD V, BEN MIMOUN S, JAVELAUD D, LEOF EB, MAUVIEL A. Cell density sensing alters TGF-beta signaling in a cell-type-specific manner, independent from Hippo pathway activation. Dev Cell. 2015;32:640–51. doi: 10.1016/j.devcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Sasamoto S, Ishii S, Date T, Matsui M, Ishizaki R. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res. 1989;17:5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OTA M, SASAKI H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–69. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- PAN D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN D, ZHU Q, CONBOY MJ, CONBOY IM, LUO K. SnoN activates p53 directly to regulate aging and tumorigenesis. Aging Cell. 2012;11:902–11. doi: 10.1111/j.1474-9726.2012.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN D, ZHU Q, LUO K. SnoN functions as a tumour suppressor by inducing premature senescence. EMBO J. 2009;28:3500–13. doi: 10.1038/emboj.2009.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEARSON-WHITE S. SnoI, a novel alternatively spliced isoform of the ski protooncogene homolog, sno. Nucleic Acids Res. 1993;21:4632–8. doi: 10.1093/nar/21.19.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEARSON-WHITE S, CRITTENDEN R. Proto-oncogene Sno expression, alternative isoforms and immediate early serum response. Nucleic Acids Res. 1997;25:2930–7. doi: 10.1093/nar/25.14.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POT I, PATEL S, DENG L, CHANDHOKE AS, ZHANG C, BONNI A, BONNI S. Identification of a Novel Link between the Protein Kinase NDR1 and TGFbeta Signaling in Epithelial Cells. PLoS One. 2013;8:e67178. doi: 10.1371/journal.pone.0067178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASHIDIAN J, LE SCOLAN E, JI X, ZHU Q, MULVIHILL MM, NOMURA D, LUO K. Ski regulates Hippo and TAZ signaling to suppress breast cancer progression. Sci Signal. 2015;8:ra14. doi: 10.1126/scisignal.2005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHROEDER MC, HALDER G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol. 2012;23:803–11. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- STROSCHEIN SL, WANG W, ZHOU S, ZHOU Q, LUO K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286:771–4. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- SUN Y, LIU X, NG-EATON E, LODISH HF, WEINBERG RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc Natl Acad Sci U S A. 1999;96:12442–7. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARELAS X, SAKUMA R, SAMAVARCHI-TEHRANI P, PEERANI R, RAO BM, DEMBOWY J, YAFFE MB, ZANDSTRA PW, WRANA JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–48. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- VASSILEV A, KANEKO KJ, SHU H, ZHAO Y, DEPAMPHILIS ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–41. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VILLANACCI V, BELLONE G, BATTAGLIA E, ROSSI E, CARBONE A, PRATI A, VERNA C, NIOLA P, MORELLI A, GRASSINI M, BASSOTTI G. Ski/SnoN expression in the sequence metaplasia-dysplasia-adenocarcinoma of Barrett's esophagus. Hum Pathol. 2008;39:403–9. doi: 10.1016/j.humpath.2007.07.009. [DOI] [PubMed] [Google Scholar]
- WU JW, KRAWITZ AR, CHAI J, LI W, ZHANG F, LUO K, SHI Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell. 2002;111:357–67. doi: 10.1016/s0092-8674(02)01006-1. [DOI] [PubMed] [Google Scholar]
- YU FX, GUAN KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–71. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG H, LIU CY, ZHA ZY, ZHAO B, YAO J, ZHAO S, XIONG Y, LEI QY, GUAN KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–62. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG L, REN F, ZHANG Q, CHEN Y, WANG B, JIANG J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–87. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO B, TUMANENG K, GUAN KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO B, WEI X, LI W, UDAN RS, YANG Q, KIM J, XIE J, IKENOUE T, YU J, LI L, ZHENG P, YE K, CHINNAIYAN A, HALDER G, LAI ZC, GUAN KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO B, YE X, YU J, LI L, LI W, LI S, YU J, LIN JD, WANG CY, CHINNAIYAN AM, LAI ZC, GUAN KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Q, KIM YH, WANG D, OH SP, LUO K. SnoN facilitates ALK1-Smad1/5 signaling during embryonic angiogenesis. J Cell Biol. 2013;202:937–50. doi: 10.1083/jcb.201208113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Q, KRAKOWSKI AR, DUNHAM EE, WANG L, BANDYOPADHYAY A, BERDEAUX R, MARTIN GS, SUN L, LUO K. Dual role of SnoN in mammalian tumorigenesis. Mol Cell Biol. 2007;27:324–39. doi: 10.1128/MCB.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Q, LUO K. SnoN in regulation of embryonic development and tissue morphogenesis. FEBS Lett. 2012 doi: 10.1016/j.febslet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Q, PEARSON-WHITE S, LUO K. Requirement for the SnoN oncoprotein in transforming growth factor beta-induced oncogenic transformation of fibroblast cells. Mol Cell Biol. 2005;25:10731–44. doi: 10.1128/MCB.25.24.10731-10744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.