Abstract
Time to flowering and maturity in soybean is controlled by loci E1 to E5, and E7 to E9. These loci were assigned to molecular linkage groups (MLGs) except for E5. This study was conducted to map the E5 locus using F2 populations expected to segregate for E5. F2 populations were subjected to quantitative trait locus (QTL) analysis for days to flowering (DF) and maturity (DM). In Harosoy-E5 × Clark-e2 population, QTLs for DF and DM were found at a similar position with E2. In Harosoy × Clark-e2E5 population, QTLs for DF and DM were found in MLG D1a and B1, respectively. In Harosoy-E5Dt2 × Clark-e2 population, a QTL for DF was found in MLG B1. Thus, results from these populations were not fully consistent, and no candidate QTL for E5 was found. In Harosoy × PI 80837 population, from which E5 was originally identified, QTLs corresponding to E1 and E3 were found, but none for E5 existed. Harosoy and PI 80837 had the e2-ns allele whereas Harosoy-E5 had the E2-dl allele. The E2-dl allele of Harosoy-E5 may have been generated by outcrossing and may be responsible for the lateness of Harosoy-E5. We conclude that a unique E5 gene may not exist.
Keywords: E5 gene, maturity gene, QTL mapping, soybean
Introduction
Eight loci have been reported to control the time to flowering and maturity in soybean in temperate zones: E1 and E2 (Bernard 1971); E3 (Buzzell 1971); E4 (Buzzell and Voldeng 1980); E5 (McBlain and Bernard 1987); E7 (Cober and Voldeng 2001), E8 (Cober et al. 2010); E9 (Kong et al. 2014). Segregation for late maturity was reported in a backcross, Harosoy (6) × PI 80837 made to transfer Pd1 (dense pubescence) to Harosoy (McBlain and Bernard 1987). A late BC5F4 progeny plant was designated as L64-4830. The F2 population of L64-4830 × Harosoy segregated for a single locus for lateness. Test crosses of L64-4830 with Harosoy maturity-allele NILs that have different alleles of loci E1, E2 and e3 indicated that the maturity locus was not any of these loci. The new locus was designated as E5, and the genetic effect of E5 on time to flowering and maturity was similar to that of E2 (McBlain and Bernard 1987).
Maturity loci, by controlling time to flowering and maturity, enable soybean cultivars to adapt to various regions with different daylengths. In addition, based on studies using NILs for maturity loci, maturity genes are involved in chilling tolerance in terms of quality and yield of seed. Chilling temperatures (about 15°C) during flowering period induce browning and cracking of the seed coats (Takahashi and Asanuma 1996). Genetic analysis suggest that a few major genes were involved in the tolerance to chilling temperatures and one of these genes was closely associated with maturity (Takahashi and Abe 1994). A QTL having a large effect on browning of the seed coat was identified at a similar position with E1 (Githiri et al. 2007). To evaluate the separate effect of E1 to E5 on the intensity of seed coat browning and cracking, Harosoy (e1 e2 E3 E4 e5 E7) and its NILs for E1 to E5 were subjected to chilling treatments (Takahashi and Abe 1999). The intensity of browning was slightly reduced by E2 and e4, and profoundly reduced by E1 and E5, but it was not affected by e3. The degree of cracking was slightly increased by e3 and drastically reduced by e4, E1, and E5. The E7 allele had inhibitory effects for both browning and cracking (Benitez et al. 2004). Thus, E5 had a large impact on the deterioration of seed coats induced by low temperatures. Further, Harosoy NILs with an allelic combination of E1e3e4 and NILs with e1E3E4 had similar time to maturity and similar number of pods under control conditions (Takahashi et al. 2005). However, the NILs with E1e3e4 had higher pod numbers than the NILs with e1E3E4 under chilling treatments, resulting in a higher seed yields.
The linked loci E1 and E7, E2, E3, E4, E8 and E9 loci were assigned to MLGs C2 (chromosome (chr) 6), O (chr 10), L (chr 19), I (chr 20), C1 (chr 4) and J (chr 16), respectively (Abe et al. 2003, Cober and Voldeng 2001, Cober et al. 2010, Cregan et al. 1999, Kong et al. 2014, Xia et al. 2012). Three cDNA copies for phytochrome A exist and two of them correspond to E3 and E4 (Liu et al. 2008, Watanabe et al. 2009). E2 encodes a GIGANTEA-like protein (Watanabe et al. 2011). GIGANTEA is a nuclear-localized protein that functions upstream of CONSTANS and FLOWERING LOCUS T encoding a florigen (Lin et al. 2007, Mizoguchi et al. 2005). E1 encodes a protein containing a bipartite nuclear localization signal and a region distantly related to the B3 domain, but its functional mechanism is uncertain (Xia et al. 2012). Based on nucleotide polymorphisms among E1 to E4 genes, amplicon length polymorphism (ALP) and derived cleaved amplified polymorphic sequence (dCAPS) markers were developed to classify alleles of these loci (Tsubokura et al. 2014). However, there is no map information for E5 locus. This study was conducted to determine the location of E5 using DNA markers.
Materials and Methods
Plant materials
Canadian cultivar Harosoy, NILs with genetic background of Harosoy or US cultivar Clark, and PI 80837 were used (Table 1). PI 80837 is a plant introduction collected from Akita Prefecture, Japan (landrace Mejiro). Germplasm information is detailed in the Germplasm Resources Information Network (GRIN) website at https://npgsweb.ars-grin.gov/gringlobal/accessiondetail.aspx?id=1118475. Harosoy has gray normal pubescence (t pd1), Clark has tawny normal pubescence (T pd1) and PI 80837 has gray dense pubescence (t Pd1). Harosoy and Clark (e1 E2 E3 E4 e5 E7) differ only at the E2 locus at the known maturity loci. Three cross combinations were made by pollinating Harosoy (or Harosoy NIL) with pollen from Clark NIL to develop F2 populations that, among maturity genes, were expected to differ in only E5 (Table 2). Another F2 population Harosoy × PI 80837 that is identical with the original cross from which E5 was identified (McBlain and Bernard 1987), was included in the test. Seeds of the NILs and PI 80837 were obtained from the USDA Soybean Germplasm Collections. The NILs were produced by crossing donor cultivars with lines having the respective alleles and backcrossing the progeny up to BC5 (Bernard et al. 1991). Flowers of Harosoy or Harosoy NILs were emasculated one day before opening and were pollinated with Clark NILs or PI 80837. Hybridity of the F1 plants was ascertained by either color or density of pubescence.
Table 1.
Soybean materials used in this study
| Line name | Line designation | Genotype | Pedigree |
|---|---|---|---|
| Harosoy | – | e1 e2 E3 E4 e5 E7 dt2 t pd1 | – |
| Harosoy-E5 | L64-4830 | e1 e2 E3 E4 E5 E7 dt2 t pd1 | Harosoy (6) × PI 80837 |
| Harosoy-E5Dt2 | L62-812 | e1 e2 E3 E4 E5 E7 Dt2 t pd1 | Harosoy (6) × PI 80837 |
| Clark | – | e1 E2 E3 E4 e5 E7 dt2 T pd1 | – |
| Clark-e2 | L62-1392 | e1 e2 E3 E4 e5 E7 dt2 T pd1 | Clark (6) × PI 86024 (e2) |
| Clark-e2E5 | L94-1110 | e1 e2 E3 E4 E5 E7 dt2 T pd1 | L63-3117 (Clark-e2) (6) × L64-4830 |
| PI 80837 | – | E5 Dt2 t Pd1 | – |
Table 2.
F2 populations used in this study
| Cross combination | Year of crossing | Date of planting | Cultivation method | Location | Number of | |
|---|---|---|---|---|---|---|
|
| ||||||
| Parents | F2 plants | |||||
| Harosoy-E5 × Clark-e2 | 2007 | June 20, 2008 | Pot | Tsukuba | 4 | 98 |
| Harosoy × Clark-e2E5 | 2010 | June 16, 2011 | Pot | Tsukuba | 8 | 110 |
| Harosoy-E5Dt2 × Clark-e2 | 2010 | May 25, 2011 | Field | Sapporo | 4 | 119 |
| Harosoy × PI 80837 | 2012 | June 27, 2013 | Pot | Tsukuba | 8 | 104 |
Plant cultivation
Pot experiments were conducted from June to October at the National Institute of Crop Science, Tsukuba, Japan (36°06′N, 140°05′E). Five seeds from each of the parents and three F2 seeds were planted in pots (12.5 cm diameter) filled with 2.5 kg soil (low-humic andosols) supplemented with ammonium sulfate (0.8 g), monocalcium phosphate (1.6 g), fused magnesium phosphate (3.2 g) and potassium sulfate (0.8 g). One week after emergence, seedlings were thinned to one per pot and grown in an unheated vinyl plastic greenhouse. Pots were distributed at random in the greenhouse and repositioned twice a week. Field experiments were carried out from May to October at Hokkaido University, Sapporo, Japan (43°25′N, 143°32′E). Seeds were sown in paper pots on May 25 and the seedlings were transplanted to the field on June 5 in 2011. N, P, and K were applied at 3.0, 4.4, and 8.3 g m−2, respectively. Plants were individually planted 25 cm apart within rows that were spaced 60 cm apart. The number of parents and F2 plants used in each experiment are listed in Table 2. Number of days from planting to opening of the first flower (R1) (Fehr et al. 1971) was recorded for individual plants. In pot experiments, days from planting to maturity (R8), when 95 percent of pods had mature color, were also recorded.
Seeds of the parental lines were planted on June 7, 2012 in the field at the National Institute of Crop Science. N, P and K were applied at levels similar to those in the field of Hokkaido University. Eleven to 14 plants for each line were individually grown at a spacing of 70 cm between rows and 10 cm between plants. Days to flowering (R1) were individually recorded and subjected to analysis of variance. Means of days to flowering among lines were compared with Tukey’s HSD test using the Statistica software 03J (StatSoft).
DNA extraction and SSR analysis
Total DNA was extracted from trifoliolate leaves of the parents and the F2 population by the CTAB method (Murray and Thompson 1980). A total of 94 F2 plants were randomly selected and used for analysis, because PCR reaction plates and the electrophoresis apparatus were designed for multiples of 96 samples (94 F2 plants and two parents). SSR markers developed by USDA (Song et al. 2004) or by the Kazusa DNA Research Institute (Hisano et al. 2007) were used for screening of polymorphisms between the parents. The PCR mixture contained 20 ng of genomic DNA, 2.25 pmol of primer, 625 pmol of nucleotides, and 0.125 unit of ExTaq in 1 × ExTaq Buffer supplied by the manufacturer (Takara Bio, Ohtsu, Japan) in a total volume of 5 μL. An initial 4 min denaturation at 95°C was followed by 35 cycles of 1 min denaturation at 95°C, 1 min annealing at 49°C, and 1 min extension at 68°C. PCR was performed in an Applied Biosystems 9700 thermal cycler (Applied Biosystems, Foster City, CA). PCR products were separated in 8% nondenaturing acrylamide gels, and the fragments were visualized by staining with ethidium bromide. To investigate the possibility of outcrossing, SSR marker genotypes of Harosoy-E5 (5 markers for each MLG) were compared with Harosoy (recurrent parent) and PI 80837 (donor parent).
Linkage mapping and QTL analysis
Markers were tested by Chi-square analyses for segregation in 1:2:1 or 3:1 ratio. A linkage map was constructed using MAPMAKER/EXP. ver. 3.0 (Lander et al. 1987) with a threshold LOD score of 3.0. Designation of MLGs followed Cregan et al. (1999). QTL analysis was performed by composite interval mapping (Zeng 1993) using the QTL Cartographer version 2.5 (Wang et al. 2007). The threshold LOD score was determined by permutation test with 1000 repetitions corresponding to genome-wide 5% level of significance.
Genotyping of E1 to E4 genes
Total DNA was extracted from trifoliolate leaves of genotypes listed in Table 1 along with reference cultivars having specific maturity alleles (Bay for E1 and E2-dl alleles, Tachinagaha for e3-tr allele, Moshidou Gong 503 for e3-Mo allele and Tokei 780 for e4-SORE-1 allele) (Liu et al. 2008, Tsubokura et al. 2014). Alleles of E1 to E4 were determined by ALP or dCAPS analyses following previous reports (Liu et al. 2008, Tsubokura et al. 2014). Alleles of E1 gene (E1 or e1-as) were determined by PCR (primers: G33snpTaqcutF and G33snpTaqcutR1) and TaqαI digestion (marker designation: E1_TaqαI). Allele of E2 gene (e2-ns or other alleles) was determined by PCR (primers: SoyGI_dCAPaMs19300FW and SoyGI_dCAPa19440RV) and DraI digestion (marker designation: E2_DraI). Allele of E2 gene (E2-in or E2-dl) was determined by PCR amplicon size (primers: E2_15345FW and E2_15856RV) (marker designation: E2_InDel). Allele of E3 gene (e3-tr or other alleles) was determined by PCR amplicon size (primers: E3_08557FW, E3_09908RV, E3Ha_1000RV, and e3tr_0716RV) (marker designation: E3_Mix). Allele of E3 gene (E3-Ha or e3-Mo) was determined by PCR (primers: E3_08094FW and E3_08417RV) and MseI digestion (marker designation: E3_MseI). Allele of E4 gene (E4 or e4-SORE-1) was determined by PCR amplicon size (primers: PhyA2-for, PhyA2-Rev/E4, PhyA2-Rev/e4) (marker designation: E4_Mix). PCR products for E2_InDel and E4_Mix were separated in agarose gels of 2 and 1%, respectively. The other products were separated in 8% nondenaturing acrylamide gels. A diagnostic marker for the E2 gene (E2_DraI) was taken for the Harosoy-E5 × Clark-e2 population. Diagnostic markers for E1 (E1_TaqαI) and E3 genes (E3_Mix) were taken for the Harosoy × PI 80837 population.
Results
Distribution of days to flowering and maturity
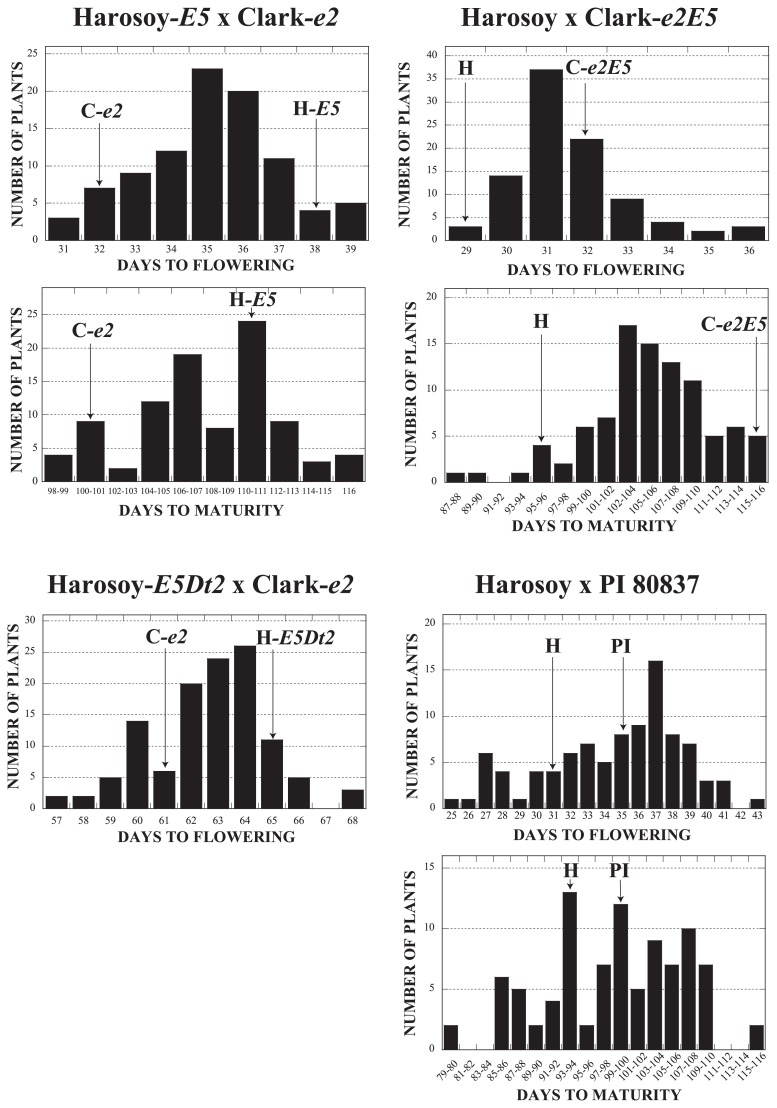
The frequency distribution of DM and DF of F2 populations as well as their mean parental values are presented in Fig. 1. The Harosoy-E5Dt2 × Clark-e2 population was grown in field at Sapporo. The other populations were grown in pots at Tsukuba. In the Harosoy-E5 × Clark-e2 population, Clark-e2 flowered 6 days earlier and matured 10 days earlier than Harosoy-E5. DF and DM of F2 plants were distributed without apparent transgression. DM is generally subject to environmental factors, and the appearance of late-maturing plants may not be attributable to transgressive effects. In the Harosoy × Clark-e2E5 population, Harosoy flowered 3 days earlier and matured 20 days earlier than Clark-e2E5. Transgressive segregation was observed in F2 plants; 3 plants flowered 4 days later than Clark-e2E5 and one plant matured 8 days earlier than Harosoy. In the Harosoy-E5Dt2 × Clark-e2 population, Clark-e2 flowered 4 days earlier than Harosoy-E5Dt2. Transgressive segregation was observed in F2 plants; two plants flowered 4 days earlier than Clark-e2 and three plants flowered 3 days later than Harosoy-E5Dt2. In the Harosoy × PI 80837 population, Harosoy flowered 4 days earlier and matured 5 days earlier than PI 80837. Transgressive segregation of F2 plants was most evident in this population; one plant flowered 6 days earlier than Harosoy and one plant flowered 8 days later than PI 80837. Further, two plants matured 15 days earlier than Harosoy and two plants matured 17 days later than PI 80837. In every population, it was difficult to classify the F2 plants into early and late categories for either DF or DM. Therefore, QTL analysis was performed to map the maturity genes. Days to flowering of the parental lines are shown in Table 3. Harosoy, Harosoy-E5Dt2 and Clark-e2 composed an early-flowering group. Harosoy-E5Dt2, Clark-e2 and Clark-e2E5 composed an intermediate group that partially overlapped with the early-flowering group. Harosoy-E5 and PI 80837 were classified into the late-flowering group. Days to flowering of Harosoy-E5 was 10 d later than Harosoy. In contrast, days to flowering of Clark-e2E5 and Harosoy-E5Dt2 were not significantly different from Clark-e2 and Harosoy, respectively.
Fig. 1.
Frequency distribution of days to flowering and maturity in four soybean F2 populations. Mean values of days to flowering and maturity of the parents are shown by arrows. The Harosoy-E5Dt2 × Clark-e2 population was grown in field at Sapporo. The other populations were grown in pots at Tsukuba.
Table 3.
Days to flowering of soybean lines at Tsukuba, Japan in 2012
| Line | Days to flowering |
|---|---|
| Harosoy | 35.3aa |
| Harosoy-E5 | 45.2c |
| Harosoy-E5Dt2 | 36.0ab |
| Clark-e2 | 37.1ab |
| Clark-e2E5 | 37.8b |
| PI 80837 | 47.2c |
Means within a column followed by the same letter are not significantly different (P = 0.05) according to Tukey’s HSD test.
Linkage groups
In the four F2 populations, 155 to 209 markers that were polymorphic between the parents and distinctly segregated in the F2 population were used for linkage mapping (Table 4). A total of 142 to 199 markers were linked to comprise 27 to 36 linkage groups spanning 1524 to 2555 cM (Supplemental Fig. 1).
Table 4.
Linkage groups obtained from four soybean F2 populations
| Cross combination | Number of polymorphic marker | Number of linked marker | Number of linkage group | Total map length (cM) |
|---|---|---|---|---|
| Harosoy-E5 × Clark-e2 | 162 | 142 | 3 | 1524 |
| Harosoy × Clark-e2E5 | 155 | 154 | 31 | 1741 |
| Harosoy-E5Dt2 × Clark-e2 | 185 | 177 | 27 | 1982 |
| Harosoy × PI 80837 | 209 | 199 | 35 | 2555 |
QTLs for days to flowering and maturity
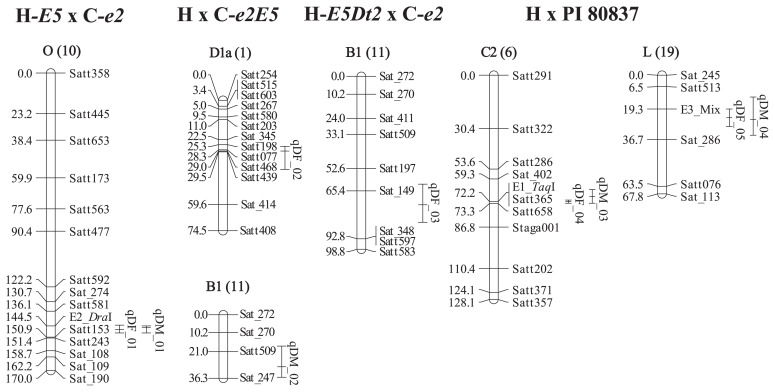
In the Harosoy-E5 × Clark-e2 population, large QTLs for DF (qDF_01) and DM (qDM_01) were found in the vicinity of E2_DraI around the end of MLG O (chr 10) (Table 5, Fig. 2). The qDF_01 locus had a LOD score of 26.44, accounting for 51.2% of phenotypic variation. The qDM_01 locus had a LOD score of 19.51, accounting for 29.5% of phenotypic variation. The alleles from Harosoy-E5 increased DF and DM at the QTLs.
Table 5.
QTLs responsible for days to flowering and maturity observed in four soybean F2 populations
| Cross combination | Phenotype (days to) | QTL name | Linkage groupa | Proximal marker | Position (cM)b | LOD score | Additive effectc | Dominance effect | Variance explained (%) |
|---|---|---|---|---|---|---|---|---|---|
| Harosoy-E5 × Clark-e2 | Flowering | qDF_01 | O (10) | E2_DraI | 146.5 | 26.44 | 2.38 | 0.43 | 51.2 |
| Maturity | qDM_01 | O (10) | E2_DraI | 145.5 | 19.51 | 4.74 | 1.64 | 29.5 | |
| Harosoy × Clark-e2E5 | Flowering | qDF_02 | D1a (1) | Satt468 | 29.0 | 5.71 | −0.98 | −0.20 | 14.6 |
| Maturity | qDM_02 | B1 (11) | Satt509 | 30.0 | 8.48 | −4.26 | 0.35 | 32.3 | |
| Harosoy-E5Dt2 × Clark-e2 | Flowering | qDF_03 | B1 (11) | Sat_149 | 73.4 | 6.46 | 1.59 | −0.18 | 25.5 |
| Harosoy × PI 80837 | Flowering | qDF_04 | C2 (6) | E1_TaqI | 72.2 | 37.15 | −4.48 | 2.07 | 69.5 |
| qDF_05 | L (19) | E3_Mix | 24.3 | 15.87 | 2.90 | 0.27 | 19.5 | ||
| Maturity | qDM_03 | C2 (6) | E1_TaqI | 69.3 | 6.76 | −5.71 | 4.45 | 56.2 | |
| qDM_04 | L (19) | E3_Mix | 25.3 | 4.47 | 5.14 | 1.14 | 14.3 |
Linkage group name is followed by chromosome number in parenthesis.
Distance from top of linkage group.
Additive effects of each QTL are those of Harosoy or Harosoy-NIL allele in contrast to pollen-parent allele.
Fig. 2.
Linkage groups containing QTLs for days to flowering and maturity found in four soybean F2 populations. The name of the linkage group followed by the chromosome number in parenthesis is indicated at the top. Distances of markers (cM) from the top of each linkage group are shown on the left. Length of vertical bars is equal to the one-LOD likelihood confidence interval. Horizontal lines in the middle of the bars indicate the positions of the QTL peak.
In the Harosoy × Clark-e2E5 population, a QTL for DF (qDF_02) was found between Satt198 and Satt077 in MLG D1a (chr 1). The locus had a LOD score of 5.71, accounting for 14.6% of phenotypic variation. The allele from Clark-e2E5 increased DF at the QTL. Further, a QTL for DM (qDM_02) was found between Satt509 and Sat_247 in MLG B1 (chr 11). The locus had a LOD score of 8.48, accounting for 32.3% of phenotypic variation. The allele from Clark-e2E5 increased DM at the QTL. In the Harosoy-E5Dt2 × Clark-e2 population, a QTL for DF (qDF_03) was found between Sat_149 and Sat_348 in MLG B1. The locus had a LOD score of 6.46, accounting for 25.5% of phenotypic variation. The allele from Harosoy-E5Dt2 increased DF at the QTL.
In the Harosoy × PI 80837 population, large QTLs for DF (qDF_04) and DM (qDM_03) were found in the vicinity of E1_TaqαI in MLGs C2 (chr 6). The qDF_04 locus had a LOD score of 37.15, accounting for 69.5% of phenotypic variation. The qDM_03 had a LOD score of 6.76 accounting for 56.2% of phenotypic variation. The alleles from PI 80837 increased DF and DM at the QTLs. In addition, QTLs for DF (qDF_05) and DM (qDM_04) were found in the vicinity of E3_Mix in MLG L (chr 19). The qDF05 locus had a LOD score of 15.87, accounting for 19.5% of phenotypic variation. The allele from PI 80837 decreased DF. The qDM_04 locus had a LOD score of 4.47, accounting for 14.3% of phenotypic variation. The allele from PI 80837 increased DM.
Genotype of maturity genes
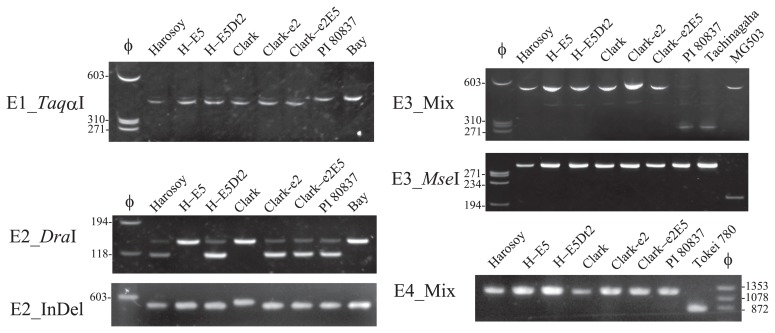
PCR with E1_TaqαI primers produced a 444 bp fragment in all materials including the reference cultivar Bay. TaqαI digestion generated a 412 bp fragment in Harosoy, Harosoy-E5, Harosoy-E5Dt2, Clark, Clark-e2, Clark-e2E5 whereas products of PI 80837 and Bay were undigested (Fig. 3). The results suggest that PI 80837 and Bay have the dominant allele (E1) whereas the other materials have the recessive allele (e1-as).
Fig. 3.
Results of ALP and dCAPS analyses to determine genotype of soybean maturity genes E1 to E4. Marker designations are indicated to the left of each panel. φ, molecular marker φx174/HaeIII; H-E5, Harosoy-E5; H-E5Dt2, Harosoy-E5Dt2. The migration of size markers (bp) is shown to the left or right of the gel.
PCR with E2_DraI primers produced a 142 bp fragment in all materials. DraI digestion generated a 115 bp fragment in Harosoy, Harosoy-E5Dt2, Clark-e2, Clark-e2E5 and PI80837 whereas products of Harosoy-E5, Clark and Bay were undigested. These results suggest that Harosoy-E5 had the dominant allele (E2) similar to Clark and Bay, whereas Harosoy, Harosoy-E5Dt2, Clark-e2, Clark-e2E5 and PI 80837 had the e2-ns allele. PCR with E2_InDel primers generated a 544 bp fragment only in Clark whereas the other materials had a 512 bp fragment. The results suggest that Clark had the E2-in allele whereas Harosoy-E5 and Bay had the E2-dl allele.
PCR with E3_Mix primers produced a 274 bp fragment in PI 80837 and Tachinagaha whereas the primers generated a 558 bp fragment in other materials. The results suggest that PI 80837 and Tachinagaha have the e3-tr allele. PCR with E3_MseI primer generated a 324 bp fragment in all materials. MseI digestion generated a 223 bp fragment only in Moshidou Gong 503 whereas products of the other materials were unaffected. The results suggest that Moshidou Gong 503 had the e3-Mo allele and the other materials had the E3-ha allele. PCR with E4_Mix primers produced a 837 bp fragment in Tokei 780 whereas a 1,229 bp fragment was generated from the other materials. The results suggest that Tokei 780 had the e4-SORE1 allele whereas the other materials had the E4 allele. Alleles of the maturity genes in the soybean lines are summarized in Table 6.
Table 6.
Alleles of maturity genes in soybean lines
| Line | E1 | E2 | E3 | E4 |
|---|---|---|---|---|
| Harosoy | e1-as | e2-ns | E3-Ha | E4 |
| Harosoy-E5 | e1-as | E2-dl | E3-Ha | E4 |
| Harosoy-E5Dt2 | e1-as | e2-ns | E3-Ha | E4 |
| Clark | e1-as | E2-in | E3-Ha | E4 |
| Clark-e2 | e1-as | e2-ns | E3-Ha | E4 |
| Clark-e2E5 | e1-as | e2-ns | E3-Ha | E4 |
| PI 80837 | E1 | e2-ns | e3-tr | E4 |
Genotype of SSR markers
Out of 100 SSR markers polymorphic between Harosoy and PI 80837, 95 markers of Harosoy-E5 had a genotype of Harosoy and one marker (Satt148 in MLG I) had a genotype of PI 80837. The results indicate that Harosoy-E5 is a NIL of Harosoy. However, four markers of Harosoy-E5 (Sat_183 in MLG D1b, Satt049 in MLG I, Satt406 in MLG J and Satt592 in MLG O) had genotypes different from Harosoy or PI 80837 (Supplemental Table 1), suggesting that an outcrossing event may have occurred during the development of this NIL.
Discussion
In the Harosoy-E5 × Clark-e2 population, QTLs having high LOD scores, qDF_01 and qDM_01, were found in the vicinity of the E2 gene (Watanabe et al. 2011). The results suggest a possibility that E5 is located in the close vicinity of E2, or E5 is an allele of the E2 locus. However, ALP and dCAPS analyses revealed that Harosoy and PI 80837 had the e2-ns allele whereas Harosoy-E5 had the E2-dl allele. Harosoy-E5 was originated from a cross between PI 80837 and Harosoy so it must have the e2-ns allele. We suspected seed contamination during propagation and obtained another batch of seed samples from the USDA germplasm collection. However, the analytical results were identical to the previous batch.
In the Harosoy × Clark-e2E5 population, DF differed by only 3 days but DM differed by 12 days among the parents. In this population, QTLs for DF (qDF_02) and DM (qDM_02) were found. Consistent with the parental differences, LOD score of qDM_02 was larger than qDF_02. In the Harosoy-E5Dt2 × Clark-e2 population, a QTL for DF (qDF_03) was observed in MLG B1. A gene for type of stem termination (Dt2) that has a potential effect on maturity (Bernard 1972) is expected to segregate in the population. However, qDF_03 was mapped to a different MLG from Dt2 located in MLG G (chr 18) (Ping et al. 2014). Though the three populations are expected to segregate for the same gene, results of QTL analysis were not consistent. Harosoy-E5 flowered 11 days later and matured 13 days later compared with Harosoy at Tsukuba, suggesting that the effects of E5 were comparable to those of E1 and E2 at this location (Takahashi and Abe 1999). In these populations, however, QTLs with large effects (qDF_01 and qDM_01) corresponded to E2. The effects of the other QTLs were too small to consider them as E5.
The Harosoy × PI 80837 population must segregate for E5 if a unique E5 gene exists. Two QTLs for DF (qDF4 and qDF5) and two QTLs for DM (qDM_03 and qDM_04) were found in this population. The QTLs qDF_04 and qDM_03 were found in the vicinity of the E1 gene (Xia et al. 2012). dCAPS analysis revealed that PI 80837 has the E1 allele whereas Harosoy had the e1-as allele. Further, a large LOD score of qDF_04 was consistent with a large effect of the E1 gene (Takahashi and Abe 1999). The results suggest that qDF_04 and qDM_03 may correspond to E1. The qDF_05 and qDM_04 loci were found in the vicinity of E3 gene (Watanabe et al. 2009). ALP and dCAPS analyses revealed that Harosoy has the E3-ha allele and PI 80837 has the e3-tr allele. The results suggest that qDF_05 and qDM_04 may correspond to E3. The effect of E3 was not evident compared to E1 and E2, probably because of the short daylength at the location of experiments (Takahashi and Abe 1999). Harosoy and PI 80837 probably have allelic combinations of e1 e2 E3 E4 and E1 e2 e3 E4, respectively. Transgressive segregation observed in their F2 population may have been caused by appearance of plants with double-recessive and double-dominant alleles of E1 and E3 loci. Thus, no candidate QTL corresponding to the E5 gene was found in this population. Furthermore, all of the QTLs found in the other populations (Harosoy-E5 × Clark-e2, Harosoy × Clark-e2E5 and Harosoy-E5Dt2 × Clark-e2) may not correspond to E5, because the E5 gene was expected to segregate in the Harosoy × PI 80837 population.
Harosoy-E5 has an allelic combination of e1-as E2-dl E3-Ha E4. Harosoy-E5 may have been generated by outcrossing with pollen having the E2-dl allele. The finding that four SSR markers of Harosoy-E5 had genotypes different from Harosoy or PI 80837 strongly suggests outcrossing. Lateness of Harosoy-E5 may be attributable to the E2-dl allele. Test crosses for E2 locus were made by crosses between Harosoy-E5 and Harosoy-E2 (L74-21 or L74-27) (McBlain and Bernard 1987). L74-21 and L74-27 were developed by crossing Harosoy six times with Clark (Bernard et al. 1991) which has the E2-in allele. The test crosses may have generated plants having a heterozygous E2-in E2-dl genotype. Crossing experiments between plants with E2-dl and E2-in allele should be performed to check if transgressive late segregants would appear as reported previously (McBlain and Bernard 1987). The two dominant alleles E2-in and E2-dl have many nucleotide polymorphisms in the 5′ upstream region, exons, introns and 3′ downstream region (Tsubokura et al. 2014). Harosoy-E5 flowered 3d later and matured 9 d later than Harosoy-E2 in Tsukuba (Takahashi and Abe 1999). The frequency of seeds with low-temperature induced seed coat deterioration of Harosoy-E5 (browning: 29.4%, cracking: 38.9%) was lower than Harosoy-E2 (browning: 55.9%, cracking: 69.2%). The different effects of Harosoy-E2 (L64-4584) and Harosoy-E5 are possibly caused by allelic differences between E2-in and E2-dl, or differences in unidentified genetic backgrounds. Under the same environments, days to flowering of Harosoy-E5 were substantially later than Harosoy, whereas days to flowering of Clark-e2E5 and Harosoy-E5Dt2 were similar to Clark-e2 and Harosoy, respectively. Clark-e2E5 and Harosoy-E5Dt2 may have been developed by introducing other genomic regions affecting time to flowering and maturity, partially because backcrossing was done without using genetic markers.
In conclusion, Harosoy-E5 may have been generated by unexpected outcrossing with pollen having an E2-dl allele. The allele may be responsible for lateness of Harosoy-E5. Thus, we conclude that a unique E5 gene does not exist, though the existence of a maturity locus in the gap of the linkage groups cannot be excluded. Some of the QTLs found in this study have not yet been reported. Analysis of the pertinent genes may be useful to further understand the control mechanism for time to flowering and maturity in soybean.
Supplementary Information
Acknowledgements
We thank Dr. R.L. Nelson at USDA/ARS University of Illinois for supplying the seeds of the NILs and PI 80837. We are grateful to Dr. Joseph G. Dubouzet (New Zealand) for critical reading of the manuscript. This study was partially supported by the Japanese Government (MEXT) Scholarship to A. Dissanayaka, T.O. Rodriguez and F. Rojas Rodas, the scholarship from the China Scholarship Council to S. Di and F. Yan, and the Invitation Fellowship Programs for Research in Japan from the Japan Society for the Promotion of Science (JSPS) to S.M. Githiri.
Literature Cited
- Abe, J., Xu, D.H., Miyano, A., Komatsu, K., Kanazawa, A. and Shimamoto, Y. (2003) Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci. 43: 1300–1304. [Google Scholar]
- Benitez, E.R., Funatsuki, H., Kaneko, Y., Matsuzawa, Y., Bang, S.W. and Takahashi, R. (2004) Soybean maturity gene effects on seed coat pigmentation and cracking in response to low temperatures. Crop Sci. 44: 2038–2042. [Google Scholar]
- Bernard, R., Nelson, R. and Cremeens, C. (1991) USDA soybean genetic collections: isoline collection. Soyb. Genet. Newsl. 18: 27–57. [Google Scholar]
- Bernard, R.L. (1971) Two major genes for time of flowering and maturity in soybeans. Crop Sci. 11: 242–244. [Google Scholar]
- Bernard, R.L. (1972) Two genes affecting stem termination in soybeans. Crop Sci. 12: 235–239. [Google Scholar]
- Buzzell, R.I. (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can. J. Genet. Cytol. 13: 703–707. [Google Scholar]
- Buzzell, R.I. and Voldeng, H.G. (1980) Inheritance of insensitivity to long daylength. Soyb. Genet. Newsl. 7: 26–29. [Google Scholar]
- Cober, E.R. and Voldeng, H.D. (2001) A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41: 698–701. [Google Scholar]
- Cober, E.R., Molnar, S.J., Charette, M. and Voldeng, H.D. (2010) A new locus for early maturity in soybean. Crop Sci. 50: 524–527. [Google Scholar]
- Cregan, P.B., Jarvik, T., Bush, A.L., Shoemaker, R.C., Lark, K.G., Kahler, A.L., Kaya, N., VanToai, T.T., Lohnes, D.G., Chung, L.et al. (1999) An integrated genetic linkage map of the soybean genome. Crop Sci. 39: 1464–1490. [Google Scholar]
- Fehr, W.R., Caviness, C.E., Burmood, D.T. and Pennington, J.S. (1971) Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 11: 929–931. [Google Scholar]
- Githiri, S.M., Yang, D., Khan, N.A., Xu, D., Komatsuda, T. and Takahashi, R. (2007) QTL analysis of low temperature-induced browning in soybean seed coats. J. Hered. 98: 360–366. [DOI] [PubMed] [Google Scholar]
- Hisano, H., Sato, S., Isobe, S., Sasamoto, S., Wada, T., Matsuno, A., Fujishiro, T., Yamada, M., Nakayama, S., Nakamura, Y.et al. (2007) Characterization of the soybean genome using EST-derived microsatellite markers. DNA Res. 14: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F.J., Nan, H.Y., Cao, D., Li, Y., Wu, F.F., Wang, J.L., Lu, S.J., Yuan, X.H., Cober, E.R., Abe, J.et al. (2014) A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 54: 2529–2535. [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E. and Newberg, L.A. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lin, M.K., Belanger, H., Lee, Y.J., Varkonyi-Gasic, E., Taoka, K.I., Miura, E., Xoconostle-Cazares, B., Gendler, K., Jorgensen, R.A., Phinney, B.et al. (2007) FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19: 1488–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K. and Abe, J. (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180: 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlain, B.A. and Bernard, R.L. (1987) A new gene affecting the time of flowering and maturity in soybeans. J. Hered. 78: 160–162. [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J.et al. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping, J.Q., Liu, Y.F., Sun, L.J., Zhao, M.X., Li, Y.H., She, M.Y., Sui, Y., Lin, F., Liu, X.D., Tang, Z.X.et al. (2014) Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26: 2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q.J., Marek, L.F., Shoemaker, R.C., Lark, K.G., Concibido, V.C., Delannay, X., Specht, J.E. and Cregan, P.B. (2004) A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109: 122–128. [DOI] [PubMed] [Google Scholar]
- Takahashi, R. and Abe, J. (1994) Genetic and linkage analysis of low temperature-induced browning in soybean seed coats. J. Hered. 85: 447–450. [DOI] [PubMed] [Google Scholar]
- Takahashi, R. and Asanuma, S. (1996) Association of T gene with chilling tolerance in soybean. Crop Sci. 36: 559–562. [Google Scholar]
- Takahashi, R. and Abe, J. (1999) Soybean maturity genes associated with seed coat pigmentation and cracking in response to low temperatures. Crop Sci. 39: 1657–1662. [Google Scholar]
- Takahashi, R., Benitez, E.R., Funatsuki, H. and Ohnishi, S. (2005) Soybean maturity and pubescence color genes improve chilling tolerance. Crop Sci. 45: 1387–1393. [Google Scholar]
- Tsubokura, Y., Watanabe, S., Xia, Z.J., Kanamori, H., Yamagata, H., Kaga, A., Katayose, Y., Abe, J., Ishimoto, M. and Harada, K. (2014) Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 113: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.C., Basten, J. and Zeng, Z.B. (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC: (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). [Google Scholar]
- Watanabe, S., Hideshima, R., Xia, Z.J., Tsubokura, Y., Sato, S., Nakamoto, Y., Yamanaka, N., Takahashi, R., Ishimoto, M., Anai, T.et al. (2009) Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Xia, Z.J., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., Takahashi, R., Anai, T., Tabata, S., Kitamura, K.et al. (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188: 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., Anai, T., Sato, S., Yamazaki, T., Lu, S.et al. (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 109: E2155–E2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z.B. (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.