Abstract
Efforts are underway to develop staple crops with improved levels of provitamin A carotenoids to help combat dietary vitamin A deficiency (VAD), which has afflicted the health of resource-poor people in the developing world. As a staple crop for more than 500 million people in sub-Saharan Africa, cassava enriched with provitamin A carotenoids could have a widespread nutritional impact. To this effect, 13 provitamin A clones were evaluated in a randomized complete block design in six environments to assess genotype by environment interaction (GEI) effects for total carotenoid (TCC) and dry matter content (DMC) in roots. Additive main effect and multiplicative interaction analysis showed significant variation among genotypes for TCC, DMC, fresh root weight and harvest index. Environmental effects were non-significant for TCC, but GEI effects were significantly large for all traits measured. There were significant temporal increments for all traits measured within 12 months after planting. TCC correlated negatively with DMC, illustrating an important challenge to overcome when developing provitamin A cassava varieties without compromising DMC, which is a major farmer-preference trait. Nonetheless, best performing genotypes were identified for TCC, DMC and FRW, and these could constitute genetic resources for advancement or developing breeding populations through hybridization.
Keywords: cassava, provitamin A carotenoid content, genotype and environment effects, breeding
Introduction
Cassava (Manihot esculenta Crantz) is the second most widely produced and consumed crop in Uganda. The crop is grown by most smallholder farmers throughout the country due to its ability to yield better than other staple food crops under conditions of extended drought and poor soils (Ceballos et al. 2011, El-Sharkawy 2007). Despite this important feature, it is common for cassava cultivars to display high sensitivity to differences in environmental conditions (Akinwale et al. 2011). The phenomenon of differential genotypic responses under varying environments is referred to as genotype by environment interaction (GEI). Abiotic and biotic stresses influence expression of genes that control key agronomic traits, which gives rise to GEI (Kang 2002). For example, during a typical 12-month growing period, cassava can experience overlapping and/or contrasting environmental stresses, thus exacerbating the extent of GEI. As such, GEI remains of interest in most plant breeding programmes. Subsequently, systematic evaluation of GEI effects for a given trait is useful for understanding varietal stability and hence strategic deployment of varieties (Acquaah 2012).
It is for these reasons that several univariate and multivariate statistical models have been developed for stability analyses and/or understanding GEI (Eberhart and Russell 1966, Gauch et al. 2008, Gauch 2013). Over the past years, excellent reviews highlighting weaknesses, strengths and best practices of these stability and/or GEI models have been undertaken (Crossa 1990, Piepho 1994, Ye et al. 2013). It suffices to note that a number of studies on cassava have opted for additive main effect and multiplicative interaction (AMMI) for assessment of GEI effects on, among other traits, carotenoid content (Maroya et al. 2012), early bulking of storage roots (Agyeman et al. 2015) and resistance to cassava brown streak disease (CBSD) (Pariyo et al. 2015). AMMI allows exhaustive data analysis by performing regular analysis of variance (ANOVA) and estimating interaction effects through principal component analysis (PCA), which somewhat increases precision in trait estimates and enables reliable selections (Gauch et al. 2008, Hongyu et al. 2014). A complementary analytical tool to visualise GEI is the genotype plus genotype by environment (GGE) biplot (Yan and Tinker 2006). The polygon view of a GGE biplot is the best way to assess the interaction patterns between genotypes and environments and to effectively interpret a biplot (Yan and Kang 2002). Genotypes that occupy vertices of the polygon are the best performers for a given trait in a specific environment. The GGE biplot allows identification of stable and best performing genotypes in test environments, which is an important decision-making tool for identifying crop varieties for subsequent release (Farshadfar et al. 2013, Rao et al. 2011).
It is commonplace for cassava breeders to evaluate advanced breeding lines (as many as 30) in several environments (as many as 10) to account for GEI when identifying genotypes with high and stable performance (Akinwale et al. 2011, Maroya et al. 2012). Studies by Akinwale et al. (2011), Tumuhimbise et al. (2014) and Agyeman et al. (2015) have indicated considerable variation in fresh root yield across varying environmental conditions. Ssemakula and Dixon (2007) noted low influence of GEI on carotenoid content in cassava roots at harvest, based on analysis of 28 genotypes in five environments evaluated over two growing cycles. A much later study on performance of 18 provitamin A clones across five environments in Nigeria indicated significant interaction between genotypes and test environments for carotenoid content (Maroya et al. 2012). Advancement of improved cassava clones for on-farm production would require subjecting such clones to systematic evaluation under diverse environments to identify better adapted genotypes (Fukuda et al. 2002, Nassar and Ortiz 2006).
Recently, the national cassava breeding programme in Uganda initiated a breeding objective tailored towards developing provitamin A cassava that expresses high levels of other farmer preference traits, especially dry matter content (DMC) (Esuma et al. 2012). It is envisioned that this initiative will culminate into deployment of provitamin A cassava varieties for purposes of improving nutrition among populations vulnerable to vitamin A deficiency. This food-based intervention to alleviate micronutrient deficiency could have a sustainable impact in developing countries, including Uganda, where food fortification and supplementation have been less impacting due to poor social infrastructures and high poverty levels (Boy et al. 2009, Mayer et al. 2008, Thompson and Amoroso 2011). Therefore, this study was undertaken to (1) assess the GEI for DMC and carotenoid content, (2) evaluate the effect of crop age on DMC and carotenoid content in cassava roots and (3) identify stable genotypes for high carotenoid and DMC levels.
Materials and Methods
Study genotypes
Thirteen genotypes of diverse genetic background were evaluated in this study (Table 1). Ten of the genotypes were selected from sets of germplasm previously acquired from the International Center for Tropical Agriculture (CIAT) and the International Institute for Tropical Agriculture (IITA). The CIAT and IITA materials were clones at advanced stages of selection for fresh root yield and carotenoid content. The other three genotypes were yellow-flesh landraces obtained from farmer fields in Uganda.
Table 1.
Provitamin A cassava genotypes used to study genotype by environment interaction for carotenoid and dry matter content
| Genotype | Code | Status | Source |
|---|---|---|---|
| 91-01730 | G1 | Improved | IITAa |
| ANDIFEKU | G2 | Landrace | Uganda |
| CPCR15B-26 | G3 | Improved | CIATb |
| MAYAYA | G4 | Landrace | Uganda |
| MH02-073HS | G5 | Improved | IITA |
| MH04-2757 | G6 | Improved | IITA |
| MH05-0452 | G7 | Improved | IITA |
| MH07-0529 | G8 | Improved | IITA |
| MM01-0014 | G9 | Improved | IITA |
| MM01-1003 | G10 | Improved | IITA |
| MM06-0466 | G11 | Improved | IITA |
| MM06-2862 | G12 | Improved | IITA |
| BUSIA | G13 | Landrace | Uganda |
International Institute of Tropical Agriculture;
International Center for Tropical Agriculture.
Experimental sites and design
Trials were conducted over two seasons between May 2012 and December 2014, each lasting 15 months, in three locations situated in different agroecological zones: Abi (north-western savannah grassland), Bulindi (in western savannah grasslands) and Namulonge (Lake Victoria crescent). Season 2012/2013 trials were planted in May 2012 and season 2013/2014 trials were planted in September 2013. Each cropping season was considered an environment, giving a total of six environments (Table 2). Temperature and rainfall data were recorded during the experimentation period as well as soil nutrient profile of the fields prior to planting the trials; these varied within a range reported to be ideal for cassava production (Cadavid 2012, Hauser et al. 2014) (Table 3). Trials were laid out in randomized complete block design with three replications, each consisting of seven rows of seven plants, giving a plot size of 49 plants. Planting was done at a spacing of 1 × 1 m, giving a density of 10,000 plants ha−1. To increase chances of sprouting and uniform plant establishment, all stakes used for planting were generated from the middle portions of mature stems. Adjacent plots were separated by a gap of 2 m alleys. Weeding was done as necessary and experiments entirely rain fed.
Table 2.
Geographical characteristics of environments for the GEI study on accumulation of carotenoids and dry matter content in cassava
| Location | Latitude | Longitude | Altitude | Cropping season | Ea |
|---|---|---|---|---|---|
| Abib | 2°36′33.3″N | 31°1′28.4″E | 1,060 m | May 2012–Aug 2013 | E1 |
| Bulindic | 1°27′58.9″N | 31°26′39.1″E | 1,157 m | May 2012–Aug 2013 | E2 |
| Namulonged | 0°31′13.7″N | 32°37′36.0″E | 1,164 m | May 2012–Aug 2013 | E3 |
| Abi | Sep 2013–Dec 2014 | E4 | |||
| Bulindi | Sep 2013–Dec 2014 | E5 | |||
| Namulonge | Sep 2013–Dec 2014 | E6 |
Environments in which GEI trials were conducted;
Abi Zonal Agricultural Research and Development Institute;
Bulindi Zonal Agricultural Research and Development Institute;
National Crops Resources Research Institute (NaCRRI).
Table 3.
Soil and weather characteristics of the six environments of the genotype by environment interaction trials
| Parameter | Criticald | E1e | E2 | E3 | E4 | E5 | E6 |
|---|---|---|---|---|---|---|---|
| pH | 4.00–8.00* | 6.10 | 6.20 | 5.70 | 5.60 | 6.0 | 6.10 |
| OMa (%) | 3.00 | 3.12 | 2.12 | 5.60 | 5.10 | 3.60 | 3.90 |
| N (%) | 0.20 | 0.31 | 0.36 | 0.41 | 0.24 | 0.29 | 0.19 |
| P (ppm) | 10.00* | 1.90 | 2.21 | 2.10 | 0.90 | 4.80 | 4.80 |
| Ca (ppm) | 50.00* | 3,724 | 3,689 | 4,833 | 4,933 | 4,224 | 3,724 |
| Mg (ppm) | 14.30* | 742 | 644 | 1,215 | 1,221 | 681 | 581 |
| K (ppm) | 58.50* | 356 | 389 | 255 | 164 | 630 | 639 |
| Zn (ppm) | 1.00 | 2.22 | 2.01 | 1.70 | 1.30 | 4.10 | 3.41 |
| B (ppm) | 0.20* | 0.04 | 0.04 | 0.02 | 0.01 | 0.06 | 0.08 |
| Cu (ppm) | 5.00 | 3.13 | 3.00 | 2.00 | 2.01 | 3.10 | 3.10 |
| Fe (ppm) | 50.00* | 189 | 172 | 182 | 191 | 172 | 189 |
| Mn (ppm) | 20.00 | 144 | 138 | 189 | 180 | 156 | 165 |
|
| |||||||
| Rainfall mm | 1,278 | 1,300 | 1,306 | 1,343 | 1,370 | 1,396 | |
| Min Tb (°C) | 17.6 | 18.2 | 18.4 | 18.7 | 18.0 | 19.0 | |
| Max Tc (°C) | 30.4 | 31.1 | 29.2 | 29.5 | 28.3 | 29.1 | |
Organic matter content;
Minimum temperature;
Maximum temperature;
Critical values for levels of nutrients required for crop growth: values with asterisks are critical for cassava (Cadavid 2012);
Six environments (E1–E6) as defined in Table 2.
Data collection
For each trial, total carotenoid content (TCC), DMC, fresh root weight (FRW) and harvest index (HI) were measured at 6, 9, 12, and 15 months after planting (MAP). Inner five rows of each experimental plot constituted a net plot of 25 plants so that six random plants were uprooted during every sampling time to take measurement on the traits. Biomass from harvested plants was bulked to estimate yield components by separately weighing the fresh roots and foliage (FSW). HI was computed from the measure of FRW and FSW as:
Three roots, each randomly picked from three of the six harvested plants, were used for preparing homogenous samples for measurement of DMC and TCC. These roots were washed to remove soil particles, peeled, washed under running water and dried with a paper towel. The dried roots were cut longitudinally into quarters. The opposite quarters from all three roots were pooled, chopped into small pieces and mixed manually to obtain a homogeneous sample. Homogenous samples (200 g) were taken for measurement of DMC by drying the sample in an oven at a temperature of 105°C for 24 hours. The dried samples were reweighed to obtain their DMC, calculated as:
where DSW = dry sample weight and FSW = fresh sample weight. TCC was measured using the UV/visible spectrophotometry method described by Rodriguez-Amaya and Kimura (2004). Briefly, approximately 10 g of the homogenous root sample was weighed and transferred into a mortar. To aid grinding, 3 g Hyflosupercel (celite) was added to the sample and the mixture ground in 50 ml cold acetone, using a pestle. The resultant solution was filtered into a conical flask through glass wool in a thistle funnel. This procedure was repeated 2–3 times until the residue was free of any colour. The extract was transferred into a 500 ml separating funnel with a Teflon stopcock, containing about 40 ml of petroleum ether. To remove acetone from the extract, double distilled water was added gently along the slanting surface of the funnel so that emulsion formation was avoided. The aqueous phase was discarded and the procedure repeated 3–4 times to get rid of acetone residues. The petroleum ether phase was transferred to a 50 ml volumetric flask through a funnel containing glass wool and 15 g of anhydrous sodium sulphate to remove the residual water and the extract made up to 50 ml with petroleum ether. Absorbance of this extract was measured at 450 nm using a spectrophotometer (Specord 210, Analytikjena model Torre Boldone BG, Italy) and TCC calculated as:
where: A = absorbance, V = total extract volume (ml), W = sample weight (g), 2,592 = β-carotene absorption coefficient in petroleum ether. All procedures for carotenoid extraction and measurement were performed in a dark room and samples were analyzed within 24 hours of harvesting.
Data analysis
Datasets for each environment and sampling point were initially analyzed independently and error variances tested for homogeneity using Hartley’s Fmax test (Hartley 1950), but differences were non-significant (P < 0.05). Therefore, un-weighted combined AMMI analysis was performed across environments by exploiting the features of interaction principal component axes (IPCA) in version 36.5.1 of the Agrobase software (Agronomix Software 2013), using the model:
where: Yge = trait value of genotype g in environment e, μ = grand mean, αg = genotype deviation from the grand mean, βe = environment deviation from the grand mean, N = number of interaction principal components (IPC) considered, λn = singular value for the IPC n, γgn = the element of eigenvector for genotype g and IPC n, δen = the element of eigenvector for environment e and IPC n and ɛge = random error (Gauch 2006, 2013, Gauch et al. 2008). The IPCs were extracted from AMMI model. The AMMI analysis showed that mean squares for interaction principal component axis two (IPCA2) were non-significant for all traits evaluated at 12 MAP, which is the optimal crop age for phenotypic evaluation of cassava. Thus, the AMMI1 model was adopted and biplots of the IPCA1 scores versus genotype and environment means were presented for measured traits. The AMMI analyses were complemented with GGE biplot analysis. The first two principal components were used to obtain GGE biplots using the PBTools software (PBTools 2014). To generate a biplot for visual analysis of multi-environment data, the singular values were partitioned into genotype and environment eigenvectors so that the GGE biplot model was rewritten as:
where: Yge = trait value of genotype g in environment e, μ = grand mean, βe = environment deviation from the grand mean, N = number of interaction principal components (IPC) considered, λn = singular value for the IPC n, γgn = the element of eigenvector for genotype g and IPC n, δen = the element of eigenvector for environment e and IPC n and ɛge = random error (Gauch et al. 2008, Yan and Kang 2002). Collectively, AMMI and GGE biplots were used to assess the performance and interaction patterns of genotypes and environments. Based on AMMI, a genotype with absolute IPCA1 value close to zero indicated low interaction and was considered to be stable while genotypes with greater absolute IPCA1 values were considered to have high sensitivity to environmental changes.
In stability analysis, it is possible to find a highly stable genotype that is not necessarily the best performer for traits of interest. To overcome this challenge, the genotype selection index (GSI) was adopted, which simultaneously selects for performance and stability (Farshadfar et al. 2013). For a given genotype, GSI is the sum of the corresponding rankings for mean performance and the AMMI stability value (ASV). The ASV is a measure of the stability of a genotype based on weighted IPCA1 and IPCA2 scores. Lower values of ASV indicate greater stability of the genotype (Purchase et al. 2000). For this study, IPCA2 axes were non-significant for all traits at 12 MAP, the optimum age of physiological maturity of cassava; therefore GSI was modified such that ranking was only based on IPCA1 as indicated below:
where: GSIg = genotype stability index for genotype g across locations for each trait, RIPCA1g = rank of genotype g across environments based on IPCA1 and RYg = rank of genotype g based on mean performance across locations. Genotypes with the lowest GSI for a given trait were considered to have the highest combined performance and stability (Farshadfar et al. 2013). Estimates of the variance components were used to calculate heritability of traits, such that:
where: H2 = broad sense heritability, = variance component for genotype effects, = variance component for interaction between genotype and environment and = variance component for residual effects.
Results
Analysis of variance
Combined AMMI analysis showed varying levels of significance for the mean squares of traits measured across the crop age (Table 4). The genotype mean squares were highly significant (P < 0.001) for all traits evaluated at different crop ages, indicating wide phenotypic variability in genotypes used in this study. There were significant (P < 0.05) GEI mean squares for all traits measured at every sampling age of the crop, except for DMC and FRW at 15 MAP. However, environmental differences were nonsignificant for TCC at 12 and 15 MAP. The IPCA1 mean squares were significant, with varying levels of significance, for all traits at all crop ages. IPCA2 mean squares were highly significant (P < 0.001) for TCC at 6 MAP and DMC at 9 MAP, very significant (P < 0.01) for DMC at 9 MAP, significant (P < 0.05) for FRW at 9 MAP and TCC at 15 MAP (P < 0.01). IPCA2 mean squares were non-significant for HI at all crop ages. For all traits evaluated in this study, both IPCA1 and IPCA2 accounted for more than 80% of the total variation observed in GEI, which was confirmed by the significant GEI effects for traits (with exception of HI). Overall, H2 was high, with the smallest value of 0.64 observed for HI. Coefficient of variation (CV) ranged from 2.98–20.37%, which was within the acceptable range.
Table 4.
AMMI analysis of 13 cassava genotypes phenotyped in six environments in Uganda
| Source of variation | DFa | TCC6b | DMC6c | FRW6d | HI6e |
|---|---|---|---|---|---|
| Environment (E) | 5 | 2.82*** | 4.53* | 3.81*** | 0.02** |
| Genotype (G) | 12 | 8.40*** | 26.72*** | 3.16*** | 0.03*** |
| GEIf | 60 | 0.41*** | 0.77** | 0.04*** | 0.04* |
| IPCA1g | 16 | 1.37*** | 1.56** | 0.10*** | 0.02** |
| IPCA2h | 14 | 0.12*** | 1.27 | 0.02 | 0.00 |
| Residual | 144 | 0.03 | 0.75 | 0.01 | 0.00 |
| CVi (%) | 4.46 | 4.68 | 9.16 | 5.34 | |
| H2j | 0.71 | 0.89 | 0.68 | 0.64 | |
| %GEI due to IPCA1 | 88.46 | 54.43 | 78.47 | 58.10 | |
| %GEI due to IPCA2 | 6.87 | 38.76 | 12.14 | 30.99 | |
|
| |||||
| TCC9 | DMC9 | FRW9 | HI9 | ||
| E | 5 | 0.39*** | 4.99** | 7.55*** | 0.08** |
| G | 12 | 21.03*** | 49.47*** | 36.23*** | 0.13*** |
| GEI | 60 | 0.23*** | 2.36*** | 0.21*** | 0.11** |
| IPCA1 | 16 | 0.75*** | 6.40*** | 0.63*** | 0.06** |
| IPCA2 | 14 | 0.07 | 2.21*** | 0.13* | 0.00 |
| Residual | 144 | 0.05 | 0.68 | 0.07 | 0.01 |
| CV (%) | 3.97 | 2.99 | 6.76 | 20.24 | |
| H2 | 0.81 | 0.89 | 0.74 | 0.71 | |
| %GEI due to IPCA1 | 85.20 | 72.32 | 78.98 | 78.26 | |
| %GEI due to IPCA2 | 6.91 | 21.89 | 14.16 | 9.32 | |
|
| |||||
| TCC12 | DMC12 | FRW12 | HI12 | ||
| E | 5 | 1.15 | 11.09** | 2.93*** | 0.15*** |
| G | 12 | 29.37*** | 33.53*** | 51.06*** | 0.26*** |
| GEI | 60 | 0.32*** | 1.90*** | 0.37*** | 0.13** |
| IPCA1 | 16 | 0.75*** | 4.19*** | 0.98*** | 0.08** |
| IPCA2 | 14 | 0.27 | 1.81 | 0.29 | 0.18 |
| Residual | 144 | 0.13 | 0.83 | 0.08 | 0.01 |
| CV (%) | 4.84 | 2.98 | 5.64 | 20.37 | |
| H2 | 0.78 | 0.94 | 0.71 | 0.68 | |
| %GEI due to IPCA1 | 61.87 | 58.89 | 71.77 | 89.74 | |
| %GEI due to IPCA2 | 19.51 | 22.28 | 18.41 | 7.94 | |
|
| |||||
| TCC15 | DMC15 | FRW15 | HI15 | ||
| E | 5 | 0.29 | 40.59*** | 2.91 | 0.18*** |
| G | 12 | 28.46*** | 57.57*** | 51.46*** | 0.32*** |
| GEI | 60 | 0.39*** | 1.11 | 0.37 | 0.11*** |
| IPCA1 | 16 | 0.82*** | 2.44** | 0.98** | 0.03* |
| IPCA2 | 14 | 0.48** | 1.38 | 0.29 | 0.00 |
| Residual | 144 | 0.17 | 1.06 | 0.08 | 0.01 |
| CV (%) | 5.48 | 3.34 | 5.63 | 19.99 | |
| H2 | 0.86 | 0.91 | 0.78 | 0.71 | |
| %GEI due to IPCA1 | 55.37 | 58.77 | 71.37 | 85.43 | |
| %GEI due to IPCA2 | 28.71 | 29.21 | 18.56 | 8.32 | |
Degrees of freedom;
Total carotenoid content;
Dry matter content;
Fresh root weight;
Harvest index;
Genotype by environment interaction;
Interaction principal component axis 1;
Interaction principal component axis 2;
Coefficient of variation;
Broad sense heritability; Numbers after each trait acronym refer to the crop age (MAP) of data collection.
significant at P < 0.05, P < 0.01 and P < 0.001, respectively.
A comparison of mean values of traits across crop ages indicated significant increments in trait values at different cassava growth ages, but differences between mean values at 12 and 15 MAP were non-significant (Table 5), except for HI. Nonetheless, there were some genotypes that showed significant increase in levels of TCC (G1, G7, G8 and G12) and DMC (G2, G4, G6 and G10) from 6–15 MAP. There was no increment in FRW from 12–15 MAP.
Table 5.
Mean of four traits measured at different crop ages in 13 genotypes across six environments in Uganda
| Trait | Agea | G1 | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 | G10 | G11 | G12 | G13 | Mean |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCCb | 6 | 3.5d | 2.6c | 3.5c | 3.3c | 5.1c | 4.1c | 4.1d | 4.0d | 4.9c | 4.0c | 3.9c | 3.9d | 3.0c | 3.8c |
| 9 | 5.0c | 3.5b | 5.1b | 4.7b | 7.6b | 6.4b | 5.6c | 5.5c | 6.7b | 6.1b | 5.8b | 5.4c | 4.2b | 5.5b | |
| 12 | 6.8b | 5.0a | 7.5a | 6.5a | 9.8a | 8.5a | 7.3b | 7.3b | 8.7a | 8.4a | 7.8a | 6.7a | 5.9a | 7.4a | |
| 15 | 7.2a | 5.2a | 7.5a | 6.7a | 9.9a | 8.5a | 7.6a | 7.7a | 8.9a | 8.5a | 7.8a | 7.3b | 5.8a | 7.6a | |
|
| |||||||||||||||
| DMCc | 6 | 17.5c | 20.8d | 19.1c | 20.1d | 16.4c | 18.8d | 18.7c | 18.7c | 18.2c | 19.9d | 17.7c | 18.3c | 17.3c | 18.6c |
| 9 | 26.0b | 29.8c | 28.0b | 29.1c | 24.5b | 28.0c | 28.9b | 26.9b | 25.9b | 30.1c | 26.6b | 26.5b | 27.7b | 27.5b | |
| 12 | 29.6a | 31.6b | 32.3a | 32.0b | 27.7a | 29.7b | 30.1a | 31.0a | 30.4a | 32.5b | 29.4a | 29.7a | 31.0a | 30.5a | |
| 15 | 29.8a | 33.6a | 31.8a | 32.9a | 27.7a | 31.6a | 30.1a | 30.4a | 30.7a | 33.8a | 28.8a | 29.9a | 30.9a | 30.9a | |
|
| |||||||||||||||
| FRWd | 6 | 1.7c | 0.9c | 0.8c | 0.4c | 1.3c | 1.2c | 1.5c | 1.8c | 1.2c | 1.8c | 1.2c | 1.8c | 1.0c | 1.3c |
| 9 | 5.7b | 1.8b | 2.4b | 1.2b | 4.6b | 3.2b | 4.1b | 4.9b | 2.7b | 5.0b | 4.2b | 5.6b | 2.8b | 3.8b | |
| 12 | 6.9a | 2.9a | 3.0a | 1.6a | 6.1a | 5.4a | 5.1a | 6.0a | 4.7a | 6.3a | 5.8a | 7.1a | 3.7a | 5.0a | |
| 15 | 7.0a | 2.9a | 3.0a | 1.7a | 6.1a | 5.5a | 5.1a | 6.0a | 4.8a | 6.3a | 5.9a | 7.1a | 3.8a | 5.0a | |
|
| |||||||||||||||
| HIe | 6 | 0.3c | 0.2b | 0.2b | 0.1b | 0.2c | 0.2c | 0.2c | 0.2d | 0.2c | 0.3c | 0.2b | 0.2c | 0.2c | 0.2d |
| 9 | 0.4b | 0.4a | 0.2b | 0.2a | 0.4b | 0.3b | 0.3b | 0.4c | 0.4b | 0.4b | 0.4a | 0.4b | 0.4b | 0.3c | |
| 12 | 0.5a | 0.4a | 0.3a | 0.2a | 0.5a | 0.3b | 0.3b | 0.5b | 0.5a | 0.5a | 0.4a | 0.4b | 0.4b | 0.4b | |
| 15 | 0.5a | 0.4a | 0.3a | 0.2a | 0.6a | 0.4a | 0.4a | 0.7a | 0.5a | 0.5a | 0.4a | 0.5a | 0.6a | 0.5a | |
Crop age (MAP) at which traits were measured;
Total carotenoid content (μg g−1);
Dry matter content (%);
Fresh root weight (kg plant−1);
Harvest index. For each genotype, means with the same letter are not statistically different; G1–G13 are genotypes defined in Table 1.
Mean performance of genotypes
Total carotenoid content
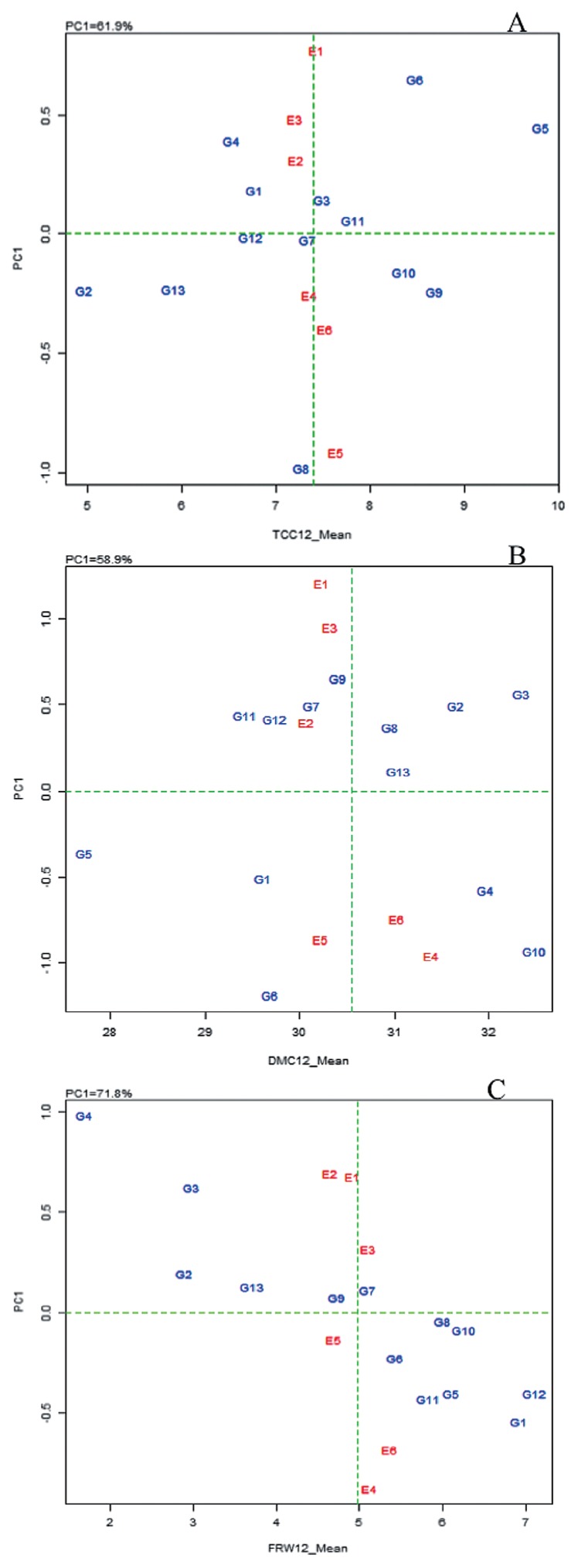
Based on GSI for TCC, genotype MH02-073HS ranked highest, while ANDIFEKU ranked lowest for the trait (Table 6). The improved genotypes introduced from CIAT and IITA had higher TCC values compared to Ugandan landraces (ANDIFEKU, MAYAYA and BUSIA). MM06-2862 was the most stable genotype across all test environments, based on its low value of 0.015 for absolute IPCA1 score. Meanwhile, MH07-0529 ranked lowest for stability based on the high value (0.980) for IPCA1 score. However, all the absolute values of IPCA1 scores were close to zero, with a small range (0.015–0.890), which indicated a generally stable performance across the six environments. Based on GSI, genotype MM06-0466 ranked as the best performer for TCC while genotypes ANDIFEKU, MAYAYA and MH07-0529 performed poorest. The test environments showed low absolute IPCA1 scores for TCC: E1 = 0.774, E2 = 0.309, E3 = 0.486, E4 = −0.255, E5 = −0.916 and E6 = −0.397. The AMMI1 biplot scattered these environments close to the axis for mean TCC, suggesting low interaction effects for environments (Fig. 1A).
Table 6.
Ranking of 13 cassava genotypes based on the genotype selection index for total carotenoid content
| Genotype | TCCa | RTCCb | IPCA1c | RIPCA1d | GSIe | RGSIf |
|---|---|---|---|---|---|---|
| 91-01730 | 6.76 | 9 | 0.185 | 6 | 15 | 8 |
| ANDIFEKU | 4.95 | 13 | −0.237 | 8 | 21 | 11 |
| CPCR15B-26 | 7.49 | 6 | 0.146 | 4 | 10 | 4 |
| MAYAYA | 6.52 | 11 | 0.393 | 10 | 21 | 11 |
| MH02-073HS | 9.81 | 1 | 0.450 | 11 | 12 | 7 |
| MH04-2757 | 8.48 | 3 | 0.651 | 12 | 15 | 8 |
| MH05-0452 | 7.32 | 7 | −0.023 | 2 | 9 | 2 |
| MH07-0529 | 7.26 | 8 | −0.980 | 13 | 21 | 11 |
| MM01-0014 | 8.68 | 2 | −0.238 | 9 | 11 | 5 |
| MM01-1003 | 8.36 | 4 | −0.162 | 5 | 9 | 2 |
| MM06-0466 | 7.82 | 5 | 0.060 | 3 | 8 | 1 |
| MM06-2862 | 6.73 | 10 | −0.015 | 1 | 11 | 5 |
| BUSIA | 5.91 | 12 | −0.231 | 7 | 19 | 10 |
| LSD0.05g | 0.35 | 0.08 |
Total carotenoid content (μg g−1);
Rank of genotypes solely based on the mean TCC;
Interaction principal component axis 1;
Rank of genotypes based on absolute value of IPCA1;
Genotype selection index;
Rank of genotypes based on GSI;
Least significant difference at 5% confidence level.
Fig. 1.
AMMI1 biplot for mean total carotenoid content (TCC), mean dry matter content (DMC) and mean fresh root weight (FRW) (A, B, C, respectively) against PC1 scores for 13 cassava genotypes evaluated in six environments in Uganda. TCC12_Mean = mean of TCC (μg g−1) at 12 MAP; DMC_Mean = mean of DMC (%) at 12 MAP; FRW_Mean = mean of FRW (ky plat−1) at 12 MAP; genotype (G1–G13) and environment (E1–E6) names are as defined in Tables 1, 2, respectively.
Dry matter content
DMC was highest in genotype MM01-1003 and lowest in MH02-073HS (Table 7). Overall, landraces ANDIFEKU, MAYAYA and BUSIA had higher DMC compared to the carotene-rich genotypes introduced from CIAT and IITA. BUSIA had the lowest absolute value for IPCA1 (−0.115), ranking as the most stable genotype across the test environments. MH04-2757 ranked lowest for stability, based on the absolute IPCA1 score of 1.183. With the exception of MH04-2757, all other genotypes had absolute values for IPCA1 scores close to zero, which indicated that the performance of genotypes was generally stable for DMC across the six test environments. Interestingly, BUSIA ranked highest based on GSI for DMC, which matched its rank based on stability. Therefore, BUSIA was the best performer for DMC, whereas MH04-2757 ranked as the poorest performer for the trait. Environments also showed low absolute IPCA1 scores for DMC: E1 = −1.211, E2 = −0.397, E3 = −0.949, E4 = 0.956, E5 = 0.856 and E6 = 0.744. The absolute values were generally close to zero, except for E1, indicating low interaction between genotypes and test environments, which was consistent with the corresponding AMMI biplot display pattern (Fig. 1B).
Table 7.
Ranking of 13 cassava genotypes based on the genotype selection index for dry matter content
| Genotype | DMCa | RDMCb | IPCA1c | RIPCA1d | GSIe | RGSIf |
|---|---|---|---|---|---|---|
| 91-01730 | 29.60 | 11 | 0.503 | 8 | 19 | 12 |
| ANDIFEKU | 31.64 | 4 | −0.493 | 6 | 10 | 3 |
| CPCR15B-26 | 32.25 | 2 | −0.562 | 9 | 11 | 4 |
| MAYAYA | 31.97 | 3 | 0.578 | 10 | 13 | 5 |
| MH02-073HS | 27.72 | 13 | 0.361 | 2 | 15 | 8 |
| MH04-2757 | 29.69 | 10 | 1.183 | 13 | 23 | 13 |
| MH05-0452 | 30.13 | 8 | −0.494 | 7 | 15 | 8 |
| MH07-0529 | 30.95 | 6 | −0.370 | 3 | 9 | 2 |
| MM01-0014 | 30.41 | 7 | −0.654 | 11 | 18 | 11 |
| MM01-1003 | 32.48 | 1 | 0.925 | 12 | 13 | 5 |
| MM06-0466 | 29.42 | 12 | −0.443 | 5 | 17 | 10 |
| MM06-2862 | 29.74 | 9 | −0.420 | 4 | 13 | 5 |
| BUSIA | 31.04 | 5 | −0.115 | 1 | 6 | 1 |
| LSD0.05g | 0.50 | 0.13 |
Dry matter content (%);
Rank of genotypes based on the mean DMC;
Interaction principal component axis 1 for DMC;
Rank of genotypes based on absolute IPCA1 for DMC;
Genotype selection index;
Rank of genotypes based on GSI,
Least significant difference at 5% confidence level.
Fresh root weight
MM06-2862 ranked highest for mean FRW, while MAYAYA ranked lowest for the trait (Table 8). Overall, landraces ANDIFEKU, MAYAYA and BUSIA had low ranks for FRW compared to introductions from CIAT and IITA. MH07-0529 had the lowest absolute value for IPCA1 score (0.042), making it the most stable genotype for FRW; but MAYAYA had the highest absolute value of IPCA1 score (0.985), implying it was the most unstable and specifically adapted genotype. On the other hand, genotype MH07-0529 and MM01-1003 were the best overall performers for FRW, based on the GSI. MAYAYA, which is a landrace, had the lowest rank for all parameters used for assessing the genotypes’ performance. The IPCA1 scores for environments were also relatively low for FRW: E1 = 0.675, E2 = 0.696, E3 = 0.320, E4 = −0.873, E5 = −0.136 and E6 = −0.682, indicating low environmental interaction effects for this trait (Fig. 1C).
Table 8.
Ranking of 13 cassava genotypes based on the genotype selection index for fresh root weight
| Genotype | FRWa | RFRWb | IPCA1c | RIPCA1d | GSIe | RGSIf |
|---|---|---|---|---|---|---|
| 91-01730 | 6.91 | 2 | −0.538 | 11 | 13 | 6 |
| ANDIFEKU | 2.88 | 12 | 0.193 | 6 | 18 | 11 |
| CPCR15B-26 | 2.98 | 11 | 0.624 | 12 | 23 | 12 |
| MAYAYA | 1.60 | 13 | 0.985 | 13 | 26 | 13 |
| MH02-073HS | 6.10 | 4 | −0.404 | 9 | 13 | 6 |
| MH04-2757 | 5.42 | 7 | −0.223 | 7 | 14 | 8 |
| MH05-0452 | 5.09 | 8 | 0.112 | 4 | 12 | 5 |
| MH07-0529 | 6.00 | 5 | −0.042 | 1 | 6 | 1 |
| MM01-0014 | 4.73 | 9 | 0.074 | 2 | 11 | 4 |
| MM01-1003 | 6.25 | 3 | −0.084 | 3 | 6 | 1 |
| MM06-0466 | 5.83 | 6 | −0.428 | 10 | 16 | 10 |
| MM06-2862 | 7.10 | 1 | −0.402 | 8 | 9 | 3 |
| BUSIA | 3.71 | 10 | 0.131 | 5 | 15 | 9 |
| LSD0.05g | 0.76 | 0.16 |
Fresh root weight (kg plant−1) based on average of 10 plants plot−1;
Rank of genotypes based on the mean FRW;
Interaction principal component axis 1 for FRW;
Rank of genotypes based on IPCA1 for FRW;
Genotype selection index;
Rank of genotypes based on GSI,
Least significant difference at 5% confidence level.
Identifying superior genotypes and mega environments
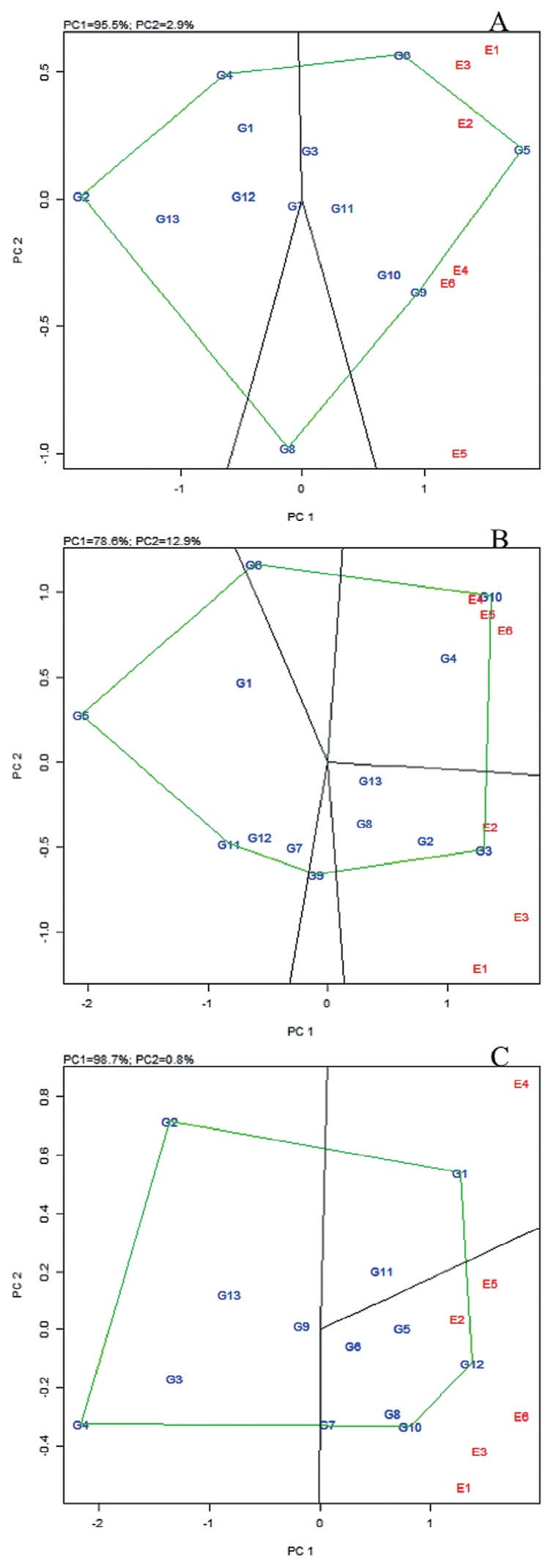
For this section, results for TCC, DMC and FRW at 12 MAP are presented, as these were the focus traits. The following were the vertex genotypes: G2 (ANDIFEKU), G4 (MAYAYA), G5 (MH02-073HS), G6 (MH04-2757) and G9 (MM01-0014) for TCC; G3 (CPCR15B-26), G5 (MH02-073HS), G6 (MH04-2757), G9 (MM01-0014), G10 (MM01-1003) and G11 (MM06-0466) for DMC; G1 (91-01730), G2 (ANDIFEKU), G4 (MAYAYA), G10 (MM01-1003) and G12 (MM06-2862) for FRW (Fig. 2). Another important feature of GGE biplots is that they indicate environmental groupings, suggesting possible existence of different mega environments. For example, two mega environments are suggested for each trait evaluated in this study (Fig. 2). For TCC, the first mega environment had environments E1, E2 and E3, with genotype G5 (MH02-073HS) and G6 (MH04-2757) as the best performer and the second mega environment had environments E4 and E6, with genotype G9 (MM01-0014) performing best. For DMC, the first mega environment was constituted by E2 in which G3 (CPCR15B-26) was the most superior genotype and the second mega environment contained E4, E5 and E6, with genotype G10 (MM01-1003) as the best performer. The first mega environment for FRW was the group of E1, E3 and E6 in which G10 (MM01-1003) and G12 (MM06-2862) performed best, while the second mega environment had E4 with G1 as the best yielder. In practice, it is difficult to have a mega environment per trait; data generated in this study could suggest (within limits) E4 as a candidate mega environment for provitamin A cassava trials.
Fig. 2.
Polygon views of the GGE biplots based on symmetrical scaling for the which-won-where pattern of genotypes and environments for total carotenoid content (A), dry matter content (B) and fresh root weight (C). Genotype (G1–G13) and environment (E1–E6) names are as defined in Tables 1, 2, respectively.
Phenotypic correlations among traits studied
Phenotypic values at 12 and 15 MAP were used for the correlation analysis because they reflected optimal levels of the traits. For all traits, there were positively significant correlations between mean values at different crop ages, suggesting temporal accumulation of phenotypes studied. DMC had a significant negative correlation with TCC (Table 9).
Table 9.
Spearman correlation coefficients among three traits phenotyped for 13 cassava genotypes in six environments in Uganda
| Trait | TCC12a | TCC15 | DMC12b | DMC15 | FRW12c |
|---|---|---|---|---|---|
| TCC15 | 0.91*** | ||||
| DMC12 | −0.35** | −0.36** | |||
| DMC15 | −0.39** | −0.41** | 0.68*** | ||
| FRW12 | 0.14 | 0.15 | −0.11 | −0.14 | |
| FRW15 | 0.13 | 0.18 | −0.04 | −0.08 | 0.89*** |
Total carotenoid content;
Dry matter content;
Fresh root weight; Numbers after the trait acronyms indicate crop age at harvest.
significant at P < 0.05, P < 0.01 and P < 0.001, respectively.
Discussion
The overall objective of this study was to assess the stability of 13 provitamin A clones by examining their performance for TCC, DMC, FRW and HI in six environments. The significant variation for TCC, DMC and FRW in these genotypes presents an important opportunity to exploit in cassava breeding in Uganda. This variability could form the basis for making good progress in genetic improvement of cassava for these traits through hybridization and selection. Environmental effects were non-significant for TCC at 12 and 15 MAP but highly significant for both DMC and FRW, indicating significant variation in mean performance of genotypes for the latter traits in different environments. This could suggest that carotenoids content in roots reaches a plateau when a cassava plant attains physiological maturity.
An obvious deduction from the low environmental effect on TCC is that selection for the trait can effectively be achieved by evaluating target genotypes in one location. However, a typical cassava variety selection scheme involves screening no less than five candidate genotypes for fresh root yield and DMC that are key drivers of variety adoption (Abdoulaye et al. 2014, Fukuda et al. 2002, Owusu and Donkor 2012). Therefore, breeding programmes targeting development of provitamin A rich cassava varieties could use initial on-station trials for identifying carotene-rich genotypes that can later be subjected to multi-locational evaluations where focus shifts to other traits highly influenced by environmental effects. This strategy would save costs while increasing precision to identify best performers for root yield and DMC.
The AMMI analysis for all traits showed that more than 50% of the variation in GEI sum of squares (SS) was accounted for by IPCA1. Subsequently fitted IPCAs, notably IPCA2, were non-significant, indicating that they largely captured random error. These results compare well with those of Gauch (2006), which showed that significant IPCA1 and subsequent axes in AMMI capture interaction exclusively in a monotonic sequence that decreases from the largest component in the first axis to the smallest component in the last axis. Therefore the significance of IPCA1 scores provided the necessary confidence for considering use of AMMI biplots for visual assessment of the genotype and location performances and their interactions (Gauch et al. 2008).
It was evident in this study that ranking of genotypes based on stability alone was not consistent with their mean performance for traits. In this regard, GSI proved to be a more reliable selection criterion for identifying best performers when conducting multi-location evaluation trials. Using this selection criterion, MM06-0466 and BUSIA were identified as the best yielders for TCC and DMC, respectively. Landraces used in this study were generally the poorest performers for TCC and FRW, but best performers for DMC. Landraces offer the advantage of having alleles that enhance adaptation to local environments. Therefore, landraces studied here constitute invaluable cassava genetic backgrounds for introgressing TCC and FRW from the introductions.
In this study, genotypes were evaluated for 15 months, which means natural growth conditions in the first three environments differed from those in the last three. The first three environments (E1, E2 and E3) were the 2012–2013 trials evaluated in the three experimental sites whereas the last three environments (E4, E5 and E6) were a repeat of the trials evaluated during the 2013–2014 season. Actually, 2012–2013 trials were planted in May while 2013–2014 trials were planted in September, which suggests differences environmental conditions experienced by the plants during the entire growth cycle. This observation appeared to account for the characteristic clustering of environments in AMMI1 biplots for all traits studied, in which E1, E2 and E3 tended to group together and distant from E4, E5 and E6 that also grouped together.
The pattern of temporal variation in levels of TCC, DMC and FRW are important to note. Mean values for these traits progressively increased up to 12 MAP, suggesting that cassava harvested at this age would give optimal levels for these traits. The quest for early maturing cassava is gathering rapid pace as the crop’s relevance for food security becomes more prominent (Bassey and Harry 2013, Tumuhimbise et al. 2012), but this is likely to be met with costs associated with less than optimal production for most of the traits before 12 MAP. Besides, other biotic constraints such as CMD and CBSD impact negatively on cassava’s potential for early bulking (Tumuhimbise et al. 2014). In an earlier study, Ngeve (2003) noted continuous increase in root yield between 6–16 MAP, with the fresh root yield increasing by up to 9.3 t ha−1 from 8 MAP to 12 MAP. As breeders pursue the development of early bulking cassava varieties, it is imperative that selection be made to exploit the crop’s potential to accumulate other important traits, including TCC and DMC at an equally early age.
However, synthesis and accumulation of TCC and DMC in cassava roots appear to be driven by genetic factors expressed along the growth stages of cassava, which are significantly influenced by prevailing environmental conditions (Asafu-Agyei and Osafo 2000). Sagrilo et al. (2008) associated the steady increase of DMC in cassava roots with temporal partitioning and accumulation of assimilates into the storage parts, with the peak influenced by the amount of vegetative growth. They indicated that the highest carbohydrate proportions were allocated to the storage roots during periods of low vegetative growth. The current study suggested 12 MAP as the age at which optimal levels of TCC, DMC and FRY can be realised in cassava roots. In comparison to findings of the current study, Tumuhimbise et al. (2014) reported the possibility of achieving economically meaningful fresh root yield (up to 25 t ha−1) at 9 MAP, but some of the genotypes evaluated in that study had a genetic background of early bulking. Nonetheless, such genotypes could provide the genetic resource for combining TCC and DMC with early bulking, through hybridization. Based on genotypes evaluated in the current study, it would make economic sense to harvest cassava at 12 MAP, which eliminates maintenance costs required for further weeding, releases the land for production of other crops and guarantees good quality of planting material for the coming season as their storage period would be short (Ngeve 2003).
The strong negative correlation between DMC and TCC in the current study is undesirable and appears to be contrary to similar results with Latin American germplasm evaluated at CIAT. It is worth noting that combined selection for both DMC and carotenoid content in Latin America has been underway much longer than in Africa. Thus, such negative correlations could have been lost during the several cycles of recombination (Ceballos et al. 2013). Indeed, high DMC would be an important feature for cassava breeding efforts targeting generation of provitamin A varieties that are acceptable to farmers. DMC in the 13 genotypes studied here was less than that in varieties commonly grown by farmers in Uganda (Kawuki et al. 2011). To translate investments in cassava biofortification research into impact on human nutrition, breeding efforts will need to focus on developing varieties that combine high levels of both DMC and TCC in high-yielding genetic backgrounds. Nonetheless, the best performing genotypes identified in this study could form the material for such genetic improvement through hybridization.
On a positive note, best performing genotypes were identified for each of the traits studied. For example, overall performance of the genotypes based on combined ranking indicated that genotypes MM01-1003, MM06-2862, MH07- 0529 and MH05-0452 were the top four performers for all the traits studied. Such genotypes could be of immediate importance for further evaluation and/or use in breeding. It suffices to note that the improved genotypes evaluated in this study were a set drawn from an advanced breeding population, which means they could have attained stability for important agronomic traits including FRW and DMC. Therefore, it is recommended that such genotypes be further screened for resistance to CBSD and other biotic stresses, for the possibility of advancing best clones for on-farm production.
Acknowledgement
The research was supported by funds from East African Agricultural Productivity Project, a grant from the World Bank to the Government of Uganda. The authors are grateful to Dr. Edward Kanju of IITA, Tanzania, for providing some of the provitamin A clones used in this study.
Literature Cited
- Abdoulaye, T., Abass, A., Maziya-Dixon, B., Tarawali, G., Okechukwu, R., Rusike, J., Alene, A., Manyong, V. and Ayedun, B. (2014) Awareness and adoption of improved cassava varieties and processing technologies in Nigeria. J. Dev. Agric. Econ. 6: 67–75. [Google Scholar]
- Acquaah, G. (2012) Principles of Plant Genetics and Breeding. Second ed. John Willey and Sons Ltd, UK. [Google Scholar]
- Agronomix Software (2013) AGROBASE Generation II User’s Guide. Release version 36.5.1 in MS-SQL. Agronomix Software Inc, Winnipeg, MB, Canada. [Google Scholar]
- Agyeman, A., Parkes, E.Y. and Peprah, B.B. (2015) AMMI and GGE biplot analyses of root yield performance of cassava genotypes in forest and coastal ecologies. Int. J. Agric. Pol. Res. 3: 122–132. [Google Scholar]
- Akinwale, M.G., Akinyele, B.O., Odiyi, A.C. and Dixon, A.G.O. (2011) Genotype × environment interaction and yield performance of 43 improved cassava (Manihot esculenta Crantz) genotypes at three agro-climatic zones in Nigeria. Br. Biotechnol. J. 1: 68–84. [Google Scholar]
- Asafu-Agyei, J.N. and Osafo, D.M. (2000) Dry matter production and distribution model in intercropped cassava. Ghana J. Agric. Sci. 34: 27–37. [Google Scholar]
- Bassey, E.E. and Harry, G.I. (2013) Screening cassava (Manihot esculenta Crantz) genotypes for tuber bulking, early maturity and optimum harvesting time in Uyo, south eastern Nigeria. Peak J. Agric. Sci. 1: 83–88. [Google Scholar]
- Boy, E., Mannar, V., Pandav, C., de Benoist, B., Viteri, F., Fontaine, O. and Hotz, C. (2009) Achievements, challenges, and promising new approaches in vitamin and mineral deficiency control. Nutr. Rev. 67: 24–30. [DOI] [PubMed] [Google Scholar]
- Cadavid, L.F. (2012) Soils and fertilizers for the cassava crop. In: Ospina, B. and Ceballos H. (eds.) Cassava in the Third Millennium: Modern Production, Processing, Use, and Marketing Systems. CIAT, Cali, Colombia, pp. 113–137. [Google Scholar]
- Ceballos, H., Ramirez, J., Bellotti, A.C., Jarvis, A. and Alvarez, A. (2011) Adaptation of cassava to changing climates. In: Yadav, S.S., Redden R., Hatfield J.L., Lotze-Campen H. and Hall A.J.W. (eds.) Crop Adaptation to Climate Change. Blackwell Publishing, Hoboken, NJ, pp. 411–425. [Google Scholar]
- Ceballos, H., Morante, N., Sánchez, T., Ortiz, D., Aragón, I., Chávez, A.L., Pizarro, M., Calle, F. and Dufour, D. (2013) Rapid cycling recurrent selection for increased carotenoids content in cassava roots. Crop Sci. 53: 2342–2351. [Google Scholar]
- Crossa, J. (1990) Statistical analyses of multilocation trials. Adv. Agron. 44: 55–85. [Google Scholar]
- Eberhart, S.A. and Russell, W.A. (1966) Stability parameters for comparing varieties. Crop Sci. 6: 36–40. [Google Scholar]
- El-Sharkawy, M.A. (2007) Physiological characteristics of cassava tolerance to prolonged drought in the tropics: implications for breeding cultivars adapted to seasonally dry and semiarid environments. Braz. J. Plant Physiol. 19: 257–286. [Google Scholar]
- Esuma, W., Rubaihayo, P., Pariyo, A., Kawuki, R., Wanjala, B., Nzuki, I., Harvey, J.J.W. and Baguma, Y. (2012) Genetic diversity of provitamin A cassava in Uganda. J. Plant Stud. 1: 60–71. [Google Scholar]
- Farshadfar, E., Rashidi, M., Mahdi, M.J. and Zali, H. (2013) GGE biplot analysis of genotype × environment interaction in chickpea genotypes. Eur. J. Exp. Biol. 3: 417–423. [Google Scholar]
- Fukuda, W.M.G., Silva, S.O. and Iglesias, C.A. (2002) Cassava breeding. Crop Breed. Appl. Biot. 2: 617–638. [Google Scholar]
- Gauch, H.G. (2006) Statistical analysis of yield trials by AMMI and GGE. Crop Sci. 46: 1488–1500. [Google Scholar]
- Gauch, H.G., Piepho, H. and Annicchiarico, P. (2008) Statistical analysis of yield trials by AMMI and GGE: further considerations. Crop Sci. 48: 866–889. [Google Scholar]
- Gauch, H.G. (2013) A simple protocol for AMMI analysis of yield trials. Crop Sci. 53: 1860–1869. [Google Scholar]
- Hartley, H.O. (1950) The use of range in analysis of variance. Biometrika 37: 271–280. [PubMed] [Google Scholar]
- Hauser, S., Wairegi, L., Asadu, C.L.A., Asawalam, D.O., Jokthan, G. and Ugbe, U. (2014) Cassava System Cropping Guide: Africa Soil Health Consortium. CAB International, Nairobi. [Google Scholar]
- Hongyu, K., García-Peña, M., Araújo, L.B. and Dias, C.T.S. (2014) Statistical analysis of yield trials by AMMI analysis of genotype × environment interaction. Biom. Lett. 51: 89–102. [Google Scholar]
- Kang, M.S. (2002) Quantitative Genetics, Genomics, and Plant Breeding. CABI Publishing, New York, USA. [Google Scholar]
- Kawuki, R.S., Pariyo, A., Amuge, T., Nuwamanya, E., Ssemakula, G., Tumwesigye, S., Bua, A., Baguma, Y., Omongo, C., Alicai, T.et al. (2011) A breeding scheme for local adoption of cassava (Manihot esculenta Crantz). J. Plant Breed. Crop Sci. 3: 120–130. [Google Scholar]
- Maroya, N.G., Kulakow, P., Dixon, A.G.O., Maziya-Dixon, B. and Bakare, M.A. (2012) Genotype × environment interaction of carotene content of yellow-fleshed cassava genotypes in Nigeria. J. Life Sci. 6: 595–601. [Google Scholar]
- Mayer, J.E., Pfeiffer, W.H. and Beyer, P. (2008) Biofortified crops to alleviate micronutrient malnutrition. Curr. Opin. Plant Biol. 11: 166–170. [DOI] [PubMed] [Google Scholar]
- Nassar, N.M.A. and Ortiz, R. (2006) Cassava improvement: challenges and impacts. J. Agric. Sci. 145: 163–171. [Google Scholar]
- Ngeve, J.M. (2003) Cassava root yields and culinary qualities as affected by harvest age and test environment. J. Sci. Food Agric. 83: 249–257. [Google Scholar]
- Owusu, V. and Donkor, E. (2012) Adoption of improved cassava varieties in Ghana. Agric. J. 7: 146–151. [Google Scholar]
- Pariyo, A., Baguma, Y., Alicai, T., Kawuki, R., Kanju, E., Bua, A., Omongo, C.A., Gibson, P., Osiru, D.S., Mpairwe, D.et al. (2015) Stability of resistance to cassava brown streak disease in major agro-ecologies of Uganda. J. Plant Breed. Crop Sci. 7: 67–78. [Google Scholar]
- PBTools (2014) Biometrics and Breeding Informatics, Version 1.4. PBGB Division. International Rice Research Institute, Los Banos, Laguna. [Google Scholar]
- Piepho, H.P. (1994) Best linear unbiased prediction (BLUP) for regional yield trials: a comparison to additive main effects and multiplicative interaction (AMMI) analysis. Theor. Appl. Genet. 89: 647–654. [DOI] [PubMed] [Google Scholar]
- Purchase, J.L., Hatting, H. and Van Deventer, S.C. (2000) Genotype × environment interaction of winter wheat (Triticum aestivum L.) in South Africa: II. Stability analysis of yield performance. S. Afr. J. Plant Soil 17: 101–107. [Google Scholar]
- Rao, P.S., Reddy, P.S., Rathore, A., Reddy, B.V.S. and Panwar, S. (2011) Application GGE biplot and AMMI model to evaluate sweet sorghum (Sorghum bicolor) hybrids for genotype × environment interaction and seasonal adaptation. Indian J. Agric. Sci. 81: 438–444. [Google Scholar]
- Rodriguez-Amaya, D.B. and Kimura, M. (2004) HarvestPlus Handbook for Carotenoid Analysis. International Food Policy Research Institute (IFPRI) and International Center for Tropical Agriculture (CIAT), Washington, DC and Cali. [Google Scholar]
- Sagrilo, E., Filho, P.S.V., Pequeno, M.G., Gonçalves-Vidigal, M.C. and Kvitschal, M.V. (2008) Dry matter production and distribution in three cassava (Manihot esculenta Crantz) cultivars during the second vegetative plant cycle. Braz. Arch. Biol. Technol. 51: 1079–1087. [Google Scholar]
- Ssemakula, G. and Dixon, A. (2007) Genotype × environment interaction, stability and agronomic performance of carotenoid-rich cassava clones. Scientific Sci. Res. Essays 2: 390–399. [Google Scholar]
- Thompson, B. and Amoroso, L. (2011) Combating Micronutrient Deficiencies: Food-Based Approaches. CAB International and FAO, Oxfordshire, UK. [Google Scholar]
- Tumuhimbise, R., Melis, R., Shanahan, P. and Kawuki, R. (2012) Farmers’ perceptions on early storage root bulking in cassava (Manihot esculenta Crantz) in east and central Uganda and their implication for cassava breeding. World J. Agric. Sci. 8: 403–408. [Google Scholar]
- Tumuhimbise, R., Melis, R., Shanahan, P. and Kawuki, R. (2014) Genotype × environment interaction effects on early fresh storage root yield and related traits in cassava. Crop J. 2: 329–337. [Google Scholar]
- Yan, W. and Kang, M.S. (2002) GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists and Agronomists. CRC Press, New York. [Google Scholar]
- Yan, W. and Tinker, N. (2006) Biplot analysis of multi-environment trial data: principles and applications. Can. J. Plant Sci. 86: 623–645. [Google Scholar]
- Ye, G., McNeil, D.L. and Hill, G.D. (2013) Methods for analysing multi-site plant variety trials II. Selection for yield and stability. Agron. Soc. N.Z. 31: 25–33. [Google Scholar]